Abstract
The human skeleton is affected by mutations in Low-density lipoprotein Receptor-related Protein 5 (LRP5). To understand how LRP5 influences bone properties, we generated mice with inducible Lrp5 mutations that cause high bone mass and low bone mass phenotypes in humans. We conditionally-induced Lrp5 mutations in osteocytes and found that bone properties in these mice were comparable to bone properties in mice with inherited mutations. We also conditionally-induced an Lrp5 mutation in cells that contribute to the appendicular skeleton, and not to the axial skeleton, and we observed bone properties were altered in the limbs, and not in the spine. These data indicate that Lrp5 signaling functions locally and suggest increasing LRP5 signaling in mature bone cells as a strategy to treat human low bone mass disorders, such as osteoporosis.
The skeleton is influenced by environmental, genetic, neurologic, endocrine, paracrine, and autocrine factors. Efforts to identify pathways that affect bone have been facilitated by genetic studies in individuals with abnormally low or abnormally high bone mass1–6. An important role for LRP5 was identified using this approach. Individuals with the Osteoporosis-Pseudoglioma syndrome, a low bone mass disorder, have loss-of-function mutations in LRP51,7, while heterozygous missense mutations in LRP5 have been observed in individuals with dominantly inherited high bone mass (HBM)2,3,8.
The mechanism by which LRP5 regulates bone mass has not been fully delineated. Studies in several laboratories indicate that LRP5 can function as a co-receptor in the canonical Wnt signaling cascade in vivo and ex vivo1,3,9–12. Furthermore, mice with genetic alterations in other components of the canonical Wnt signaling pathway have alterations in bone mass, consistent with this pathway’s importance in bone mass accrual 10,13–16. A direct role for LRP5 in osteoblast-lineage cells has been proposed based on studies in mice that over-express LRP5 cDNAs driven by a rat type I collagen promoter17. However, phenotypes resulting from transgene-driven over-expression of a protein may not accurately reflect the endogenous protein’s function.
We generated two lines of Lrp5 knockin mice, each containing a missense mutation associated with human HBM. Both missense mutants had been shown to be comparable to wild-type LRP5 in their ability to transduce canonical Wnt signaling in transfected cells9. However, the mutants differ in their efficiency of trafficking to the cell surface, and in their interactions with the chaperone protein MESD and the extracellular inhibitors DKK1 and SOST9,11,18. We designed the Lrp5 knockin alleles to function as HBM alleles following Cre-mediated recombination. This enabled us to compare the effect of inheriting an HBM Lrp5 allele with the effect of conditionally-activating an HBM Lrp5 allele in a cell-type-specific or tissue-specific manner. We also generated mice with floxed wild-type (WT) Lrp5 alleles that could be converted to knockout alleles following Cre-mediated recombination. This enabled us to compare the effect of inheriting inactive Lrp5 with the effect of conditionally inactivating Lrp5 in a cell-type-specific or tissue-specific manner.
RESULTS
HBM Lrp5 alleles increase bone mass
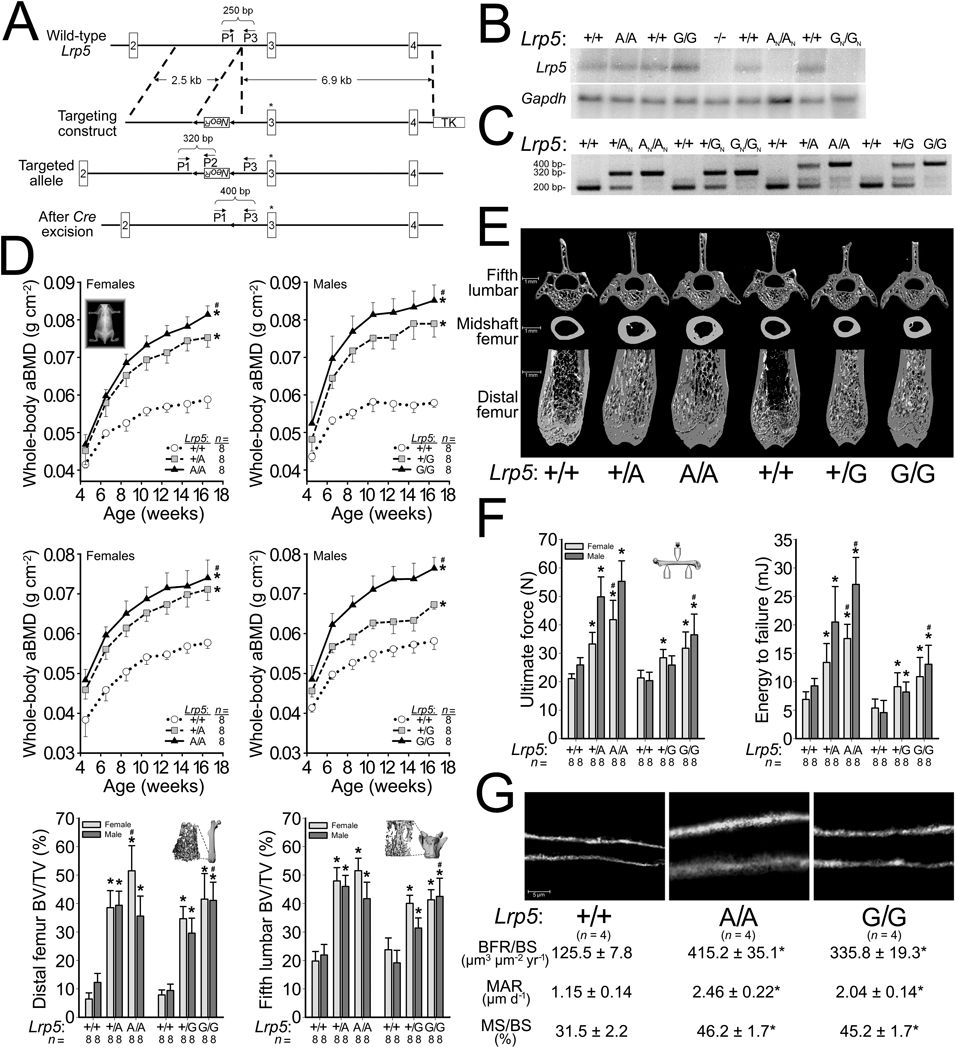
Exon 3 of mouse Lrp5 encodes the residues we mutated to HBM alleles (p.G171V and p.A214V). Our targeting vector incorporated a neomycin-resistance-cassette (NeoR) flanked by LoxP sites (Fig. 1a). Since NeoR is driven by a strong promoter and transcribed in the opposite direction of Lrp5, we anticipated that NeoR would interfere with Lrp5 transcription and thereby cause NeoR-containing Lrp5 alleles (i.e., GN and AN) to be poorly expressed. Mice with the Lrp5 genotypes GN/GN or AN/AN had reduced Lrp5 expression compared to WT mice (Fig. 1b). Cre-excision of NeoR (i.e., G and A) enhanced expression, so that mice with genotypes G/G or A/A had Lrp5 expression that was comparable to WT mice (Fig. 1b). We designed a PCR assay to distinguish Lrp5 WT (+), NeoR-containing (GN or AN), and HBM (G or A) alleles (Fig. 1c).
Figure 1.
Generation and characterization of HBM Lrp5 knockin mice. (a) Schematic depicting the targeting and genotyping strategies for HBM Lrp5 alleles. The 5’ targeting arm contains a neomycin-resistance cassette (NeoR) flanked by LoxP sites (arrowheads). The 3’ targeting arm begins in intron 2 and extends into intron 4, and is followed by a thymidine kinase (TK) cassette. Site-directed mutagenesis altered specific amino acid residues encoded by Lrp5 exon 3 (asterisk). The relative locations and orientation of primers (P1, P2, and P3) used for PCR genotyping and their expected amplimer sizes are noted. (b) Autoradiographs of a northern blot containing whole bone total RNA from mice with different Lrp5 genotypes initially hybridized with a radioactive Lrp5 cDNA probe (upper) and subsequently with a Gapdh cDNA probe which serves as a loading control (lower). (c) Photograph of an agarose gel containing PCR amplimers derived from genomic DNA of mice with different Lrp5 genotypes. (d) Graphs depicting the areal bone mineral density (aBMD) measured by DEXA in mice with different Lrp5 genotypes followed to 16.5-weeks-old (top and middle rows). Graphs depicting the percent trabecular bone volume in the total volume (BV/TV) of the distal femurs and 5th lumbar vertebrae of 16.5-wk-old male and female mice with different Lrp5 genotypes (bottom row). (e) Representative µCT scan images obtained from 16.5-wk-old mice with different Lrp5 genotypes. (f) Graphs depicting biomechanical properties of whole femurs in a 3-point bending assay from 16.5-wk-old mice with different Lrp5 genotypes. (g) Representative images of new bone formation assessed by double calcein labeling of mice with different Lrp5 genotypes. Bone formation rates/bone surface area (BFR/BS), mineral apposition rates (MAR), and mineralizing surface/bone surface (MS/BS). The numbers of mice studied (n =) are indicated, as are error bars equal to 1 s.d. Asterisks (*) indicate a significant difference (p < 0.05) when compared to WT mice, whereas # indicates a significant difference (p < 0.05) when compared to heterozygous HBM Lrp5 mice.
Having created conditional alleles that can be converted to HBM alleles by Cre, we investigated whether mice with inherited HBM alleles recapitulated the human HBM phenotype. Mice with HBM alleles had increased bone mass (Fig. 1d,e and Supplementary Fig. S1 and Table S1), bone strength (Fig. 1f), and bone formation rates (Fig. 1g), compared to WT mice. When we intercrossed mice with HBM Lrp5 to transgenic βatGal mice, a Wnt reporter mouse strain that expresses LacZ in response to canonical Wnt signaling19, we observed increased LacZ expression in cortical bone homogenates from βatGal mice with HBM Lrp5 compared to βatGal mice with WT Lrp5; we also observed increased Axin2 expression, a known Wnt target gene20, in the HBM Lrp5 mice (Supplementary Fig. S1).
Activating HBM Lrp5 in bone cells increase bone mass
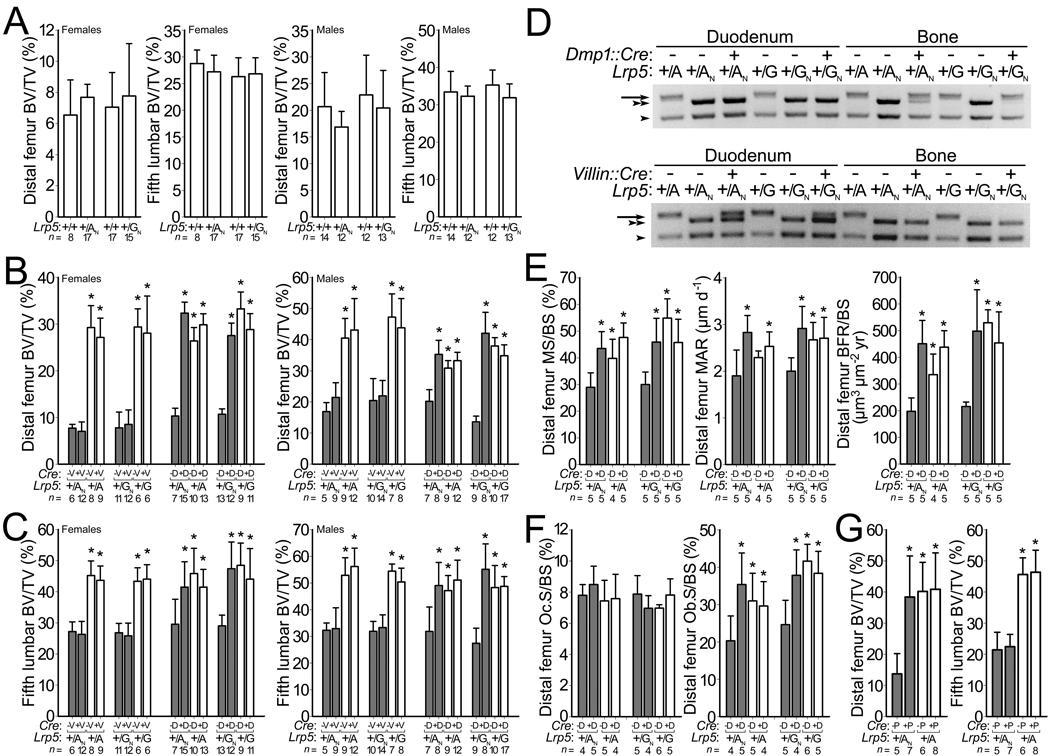
Mice heterozygous for alleles GN or AN had bone mass comparable to WT mice (Fig. 2a and Supplementary Figs. S2, S3, and Table S2), indicating that decreased expression of the GN or AN allele was partially compensated by its increased function. Since Cre converts alleles GN and AN to alleles G and A, respectively, we tested whether increasing HBM Lrp5 expression in osteocytes would affect bone mass. We crossed mice with G/GN or A/AN genotypes to mice hemizygous for a transgene, Dmp1::Cre, that expresses Cre in osteocytes21. Offspring that inherited the HBM allele (G or A) had high bone mass, independent of whether they inherited Dmp1::Cre (Fig. 2b,c). Offspring that inherited the NeoR-containing allele (GN or AN), and not Dmp1::Cre, had normal bone mass (Fig. 2b,c and Supplementary Figs. S3 and S4). Offspring that inherited the NeoR-containing allele (GN or AN) and Dmp1::Cre had increased bone mass (Fig. 2b,c and Supplementary Figs. S3 and S4), supporting a local role for Lrp5 signaling in bone. We confirmed that the Dmp1::Cre transgene was active in bone (Figs. 2d and 3). Furthermore, we noted that conversion in osteocytes of alleles GN and AN to alleles G and A, respectively, had the same effect on bone as did inheriting G and A alleles (Fig. 2e,f and Supplementary Fig. S4).
Figure 2.
Effect of conditionally activating NeoR-containing HBM Lrp5 alleles. Graphs depicting (a) femoral and vertebral trabecular BV/TV in WT mice and in mice with NeoR-containing HBM Lrp5 alleles. (b) femoral trabecular BV/TV and (c) vertebral trabecular BV/TV in mice with (shaded bars) and without (unshaded bars) inherited NeoR-containing HBM Lrp5 alleles (+/AN or +/GN, and +/A or +G, respectively), and with and without Villin::Cre (+V and −V, respectively) or Dmp1::Cre (+D and −D, respectively) transgenes. (d) Photographs of agarose gels containing PCR amplimers derived from mouse genomic DNA extracted from either duodenum or femur cortex of mice with different Lrp5 and Cre-transgene genotypes. PCR amplimers correspond to the sizes depicted in Fig. 1a. (top) amplimers from the Dmp1::Cre cross. (bottom) amplimers from the Villin::Cre cross. WT allele (arrowhead), AN or GN allele (double arrowhead), A or G allele (arrow). (e) Graphs depicting fluorochrome-derived bone formation parameters in the distal femur from 9-wk-old female mice administered double calcein labeling. X-axis group notations follow those described for panel b. (f) Graphs depicting the proportion of distal femur trabecular bone surface covered by osteoclasts (left) and osteoblasts (right). (g) Graphs depicting femur and vertebra trabecular BV/TV in 12-wk-old HBM Lrp5 mice with (shaded bars) and without (unshaded bars) inherited NeoR-containing alleles (+/AN and +/A), respectively, and with and without the Prx1::Cre transgene (+P and −P, respectively). MS/BS, MAR, BFR/BS are as defined in Fig. 1. The numbers of mice studied (n =) are indicated, as are error bars equal to 1 s.d. Asterisks (*) indicate a significant difference (p < 0.05) when compared to NeoR-containing littermates that did not inherit a Cre-transgene.
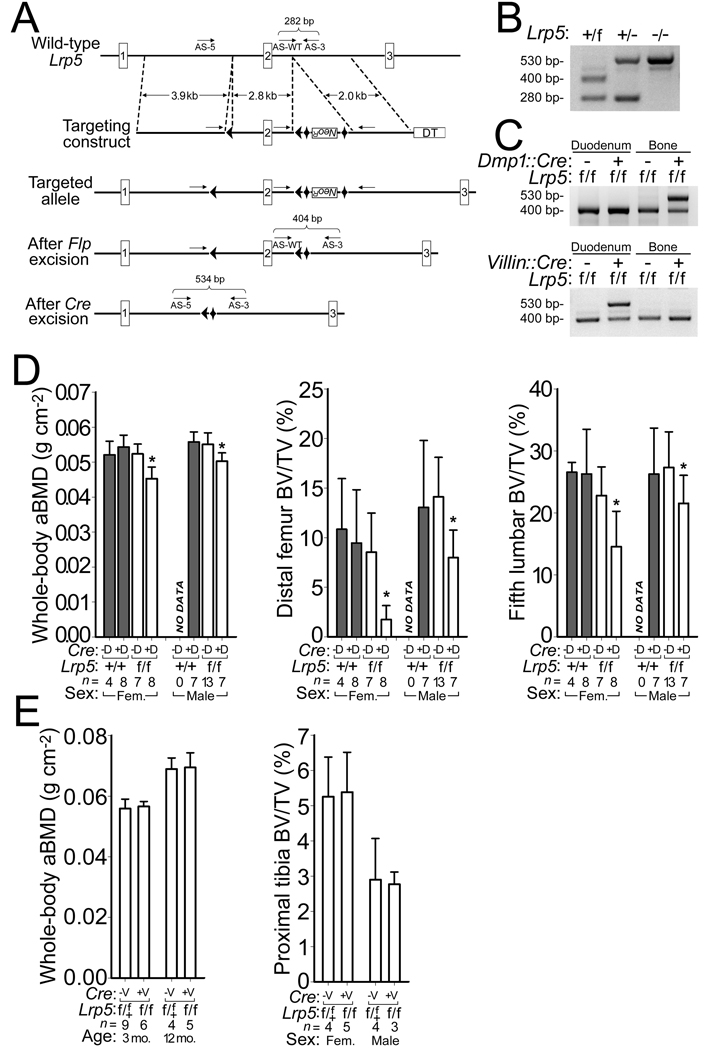
Figure 3.
Generation and characterization of mice with a conditional knockout allele of Lrp5. (a) Schematic depicting the creation of the Lrp5 floxed allele. LoxP sites (arrowheads), the neomycin-resistance cassette (NeoR), FRT sites (diamonds), and the diptheria toxin (DT) cassette are shown. The relative locations and orientation of the 3 primers (arrows) used for PCR genotyping and their expected amplimer sizes are noted. (b) Photograph of agarose gel depicting PCR amplimers for WT (+), floxed (f), and knockout (−) Lrp5 alleles from genomic DNA of mice with different Lrp5 genotypes. (c) Photographs of agarose gels containing PCR amplimers derived from mouse genomic DNA extracted from either duodenum or femur cortex of floxed Lrp5 mice, with or without the Dmp1::Cre transgene (top) and with or without the Villin::Cre transgene (bottom). (d) Graphs depicting whole-body aBMD (left), femoral trabecular BV/TV (middle), and vertebral trabecular BV/TV (right) in 16-wk-old mice homozygous for WT or floxed Lrp5 alleles, with or without the Dmp1::Cre transgene. (e) Graphs depicting whole-body aBMD and tibial trabecular BV/TV in 3-mo-old and 12-mo-old mice heterozygous or homozygous for floxed Lrp5 alleles with or without the Villin::Cre transgene. The numbers of mice studied (n =) are indicated, as are error bars equal to 1 s.d. Asterisks (*) indicate a significant difference (p < 0.05) when compared to floxed Lrp5 littermates that did not inherit the Cre transgene.
Dmp1::Cre was active in some non-bone tissues (Supplementary Fig. S5). This observation led us to test whether Lrp5 signaling outside of bone was responsible for the increased bone mass. We began by evaluating the role of Lrp5 in the intestine, since a model in which Lrp5 affects bone mass by regulating serotonin production in the duodenum has been proposed22. In this model, Lrp5 has no direct role in bone, but rather affects bone mass accrual by regulating the expression of the enzyme tryptophan hydroxylase 1 (Tph1) in the duodenum22. Tph1 is the rate-limiting enzyme for peripheral serotonin (5HT) synthesis and the intestine is the principal source of 5HT found in blood23,24; another tryptophan hydroxylase (Tph2) produces 5HT in the central nervous system24–26. We crossed mice with G/GN or A/AN genotypes to mice hemizygous for a transgene, Villin::Cre, that drives Cre expression in intestinal stem cells, from which 5HT producing enterochromaffin cells are derived27. Conversion of allele GN or AN to alleles G or A, respectively, in the intestine had no effect on bone mass (Fig. 2b,c and Supplementary Fig. S3). Villin:Cre drove Cre expression in the intestine and not in the bone (Figs. 2d and 3, and Supplementary Fig. S5).
While the Villin::Cre experiments excluded intestinal Lrp5 activity in the hormonal regulation of bone mass, they did not exclude Lrp5 activity at other non-bone sites in the hormonal regulation of bone mass. Therefore, we tested whether Lrp5 acted locally or systemically to affect bone mass by crossing mice with A/AN genotypes to mice hemizygous for a transgene, Prx1::Cre, that expresses Cre in cells that give rise to the appendicular skeleton, but not to the axial skeleton28 (Supplementary Fig. S6). Conversion of AN to A in cells that contribute to the appendicular skeleton increased bone mass in the limb, but not in the spine (Fig. 2g and Supplementary Fig. S6).
Loss of Lrp5 in bone cells decreases bone mass
We created a conditional knockout allele of Lrp5 by flanking exon 2 with LoxP sites (Fig. 3a). Deletion of this exon by Cre-mediated recombination resulted in production of an Lrp5 transcript that was frame-shifted and had a premature termination codon shortly after the signal peptide (data not shown). We developed a PCR assay for Lrp5 WT (+), floxed (f), and Cre-excised (−) Lrp5 alleles (Fig. 3b). Bone mass in homozygous floxed mice (f/f) was indistinguishable from WT (Fig 3). Bone mass in mice homozygous for Cre-excised alleles (−/−) was low, similar to other lines of Lrp5 knockout mice10,29 (data not shown). We conditionally inactivated Lrp5 by crossing homozygous floxed mice (f/f) to homozygous floxed mice that were also hemizygous for Dmp1::Cre. We confirmed that Dmp1::Cre inactivated Lrp5 in bone and not in duodenum (Fig. 3c). Compared to their littermates, mice lacking Lrp5 activity in osteocytes had reduced bone mass (Fig. 3d and supplemental Fig. S5). We tested whether inactivation of Lrp5 in the intestine would decrease bone mass by performing a similar cross with Villin::Cre and observed no significant effect on bone mass (Fig. 3e and Fig. S5d).
Lrp5 genotype does not affect peripheral serotonin levels
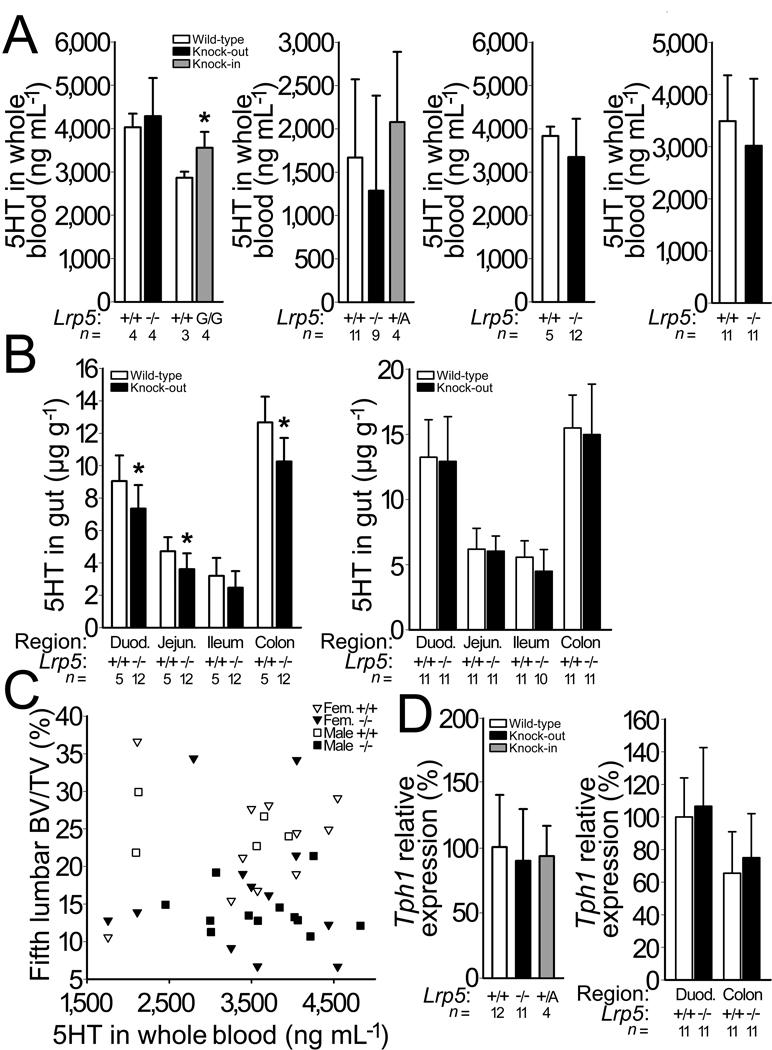
Our data support a mechanism in which Lrp5 functions via the canonical Wnt pathway in osteocytes to regulate bone mass, instead of regulating bone mass indirectly via other tissues. However, Lrp5 could also regulate serotonin production in the gut. We measured blood 5HT in mice with different Lrp5 genotypes, but saw no association between genotype and 5HT levels (Fig. 4a and Supplementary Fig. S7). We also measured 5HT levels of different regions of the intestine in Lrp5 WT and knockout mice and found only a small decrease in 5HT in knockout mice (Fig. 4b), which is opposite to that predicted by the model in which lack of Lrp5 increases Tph1 expression22. We also did not observe a correlation between whole blood 5HT levels and bone mass in individual mice (Fig. 4c). Lastly, using real-time PCR we quantified the level of Tph1 transcript in RNA extracted from the duodenums of mice with different Lrp5 genotypes, but detected no differences (Fig. 4d).
Figure 4.
Effect of Lrp5 genotype on serotonin (5HT) levels and on Tph1 expression. (a) Graphs depicting whole blood 5HT measured by HPLC in 6-mo-old Lrp5 WT and knockout mice that had been backcrossed to C57BL6/J, and in Lrp5 WT and HBM knockin (G/G) mice on a mixed 129Sv/C57BL/6J background (far left), in 3-mo-old Lrp5 WT, knockout and HBM knockin (+/A) mice on a mixed 129Sv/C57BL/6J genetic background (middle left), in 3-mo-old male (middle right) and 13-mo-old female (far right) WT and knockout littermates on a mixed 129SvEvBrd/ C57BL/6J-Tyrc-Brd background. (b) Graphs depicting the quantity of 5HT extracted from several regions of the intestine, beginning in the duodenum and proceeding through the jejunum, ileum, and proximal colon in 3-mo-old male (left) and in 13-mo-old female (right) Lrp5 WT and knockout littermates on a mixed 129SvEvBrd/ C57BL/6J-Tyrc-Brd background. (c) Scattergram depicting vertebral trabecular BV/TV and whole blood 5HT measurements in individual Lrp5 WT (open symbols) and knockout (filled symbols) littermates. Correlations between BV/TV and whole blood serotonin were r2=0.13 (p = 0.16) for male mice, and r2=0.02 (p = 0.53) for female mice. (d) Graphs depicting normalized Tph1 transcript levels in duodenum RNA extracts from Lrp5 WT, knockout, and HBM knockin (+/A) mice on a mixed 129Sv/C57BL/6J genetic background (left) and duodenum and colon RNA extracts from Lrp5 WT and knockout mice on a 129SvEvBrd/C57BL/6J-Tyrc-Brd background (right) with Gapdh serving as the internal control. The mean Tph1 expression level for Lrp5 WT duodenum is set as 100%. The numbers of mice studied (n =) are indicated, as are error bars equal to 1 s.d. Asterisks (*) indicate a significant difference (p < 0.05) compared to WT mice.
Peripheral serotonin synthesis does not affect bone mass
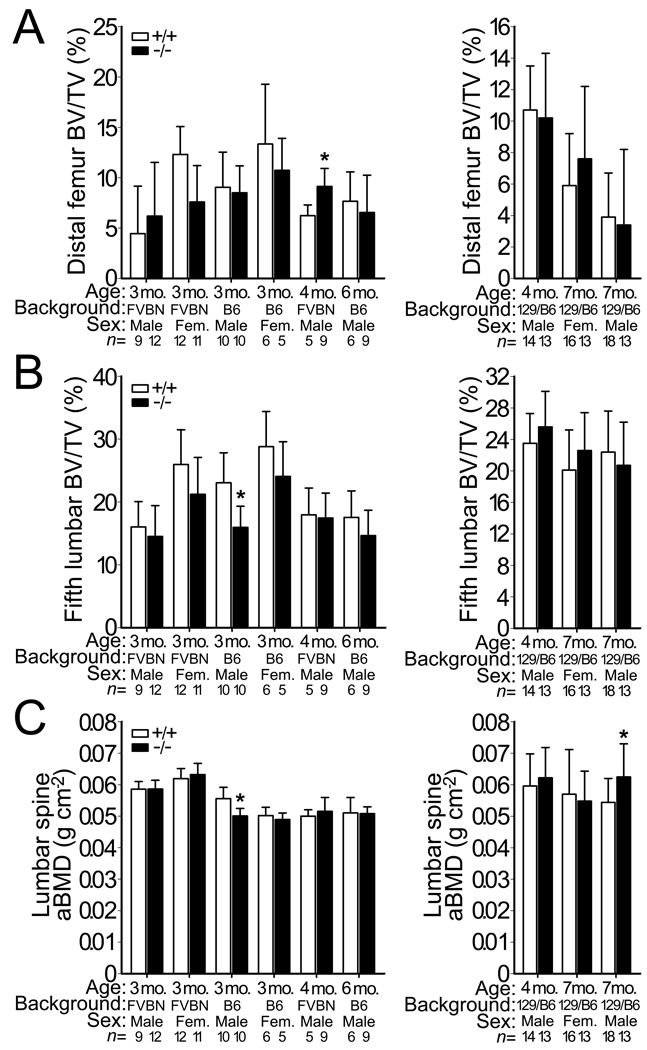
The model22 by which Lrp5 controls serotonin synthesis and regulates bone mass via an endocrine, rather than a Wnt-based local mechanism was unexpected30 and suggested bone health could be improved by pharmacologically inhibiting intestinal serotonin synthesis or the hormone’s action on osteoblasts31. In fact, pharmacologic inhibition of Tph1 in the intestine was reported to increase bone mass in ovariectomized (OVX) mice and rats to the same degree as an FDA approved anabolic bone therapy, intermittent parathyroid hormone (teriparatide) treatment32. Because 5HT in whole blood is nearly absent in Tph1−/− mice compared with WT mice23,24, we measured bone mass in Tph1−/− mice. After correcting for multiple testing, there were no significant differences in bone mass between Tph1−/− and WT mice (Fig. 5 and Supplementary Table S3).
Figure 5.
Bone mass in WT and Tph1−/− mice. (a) Graphs depicting femoral trabecular BV/TV in WT and Tph1−/− mice on either FVB/N or C57BL/6 backgrounds (left) or on a mixed 129SvEvBrd/C57BL/6J-Tyrc-Brd background (right). (b) Graphs depicting the vertebral trabecular BV/TV of the 5th lumbar vertebra in the same mice described in panel a. (c) Graphs depicting lumbar spine aBMD, as measured by DEXA, in the same mice described in panel a. The numbers of mice studied (n =) are indicated, as are 1 s.d. error bars. Asterisks (*) indicate a significant difference (p < 0.05) compared to WT mice using an unpaired t-test; none of these differences remain significant after correcting for multiple testing.
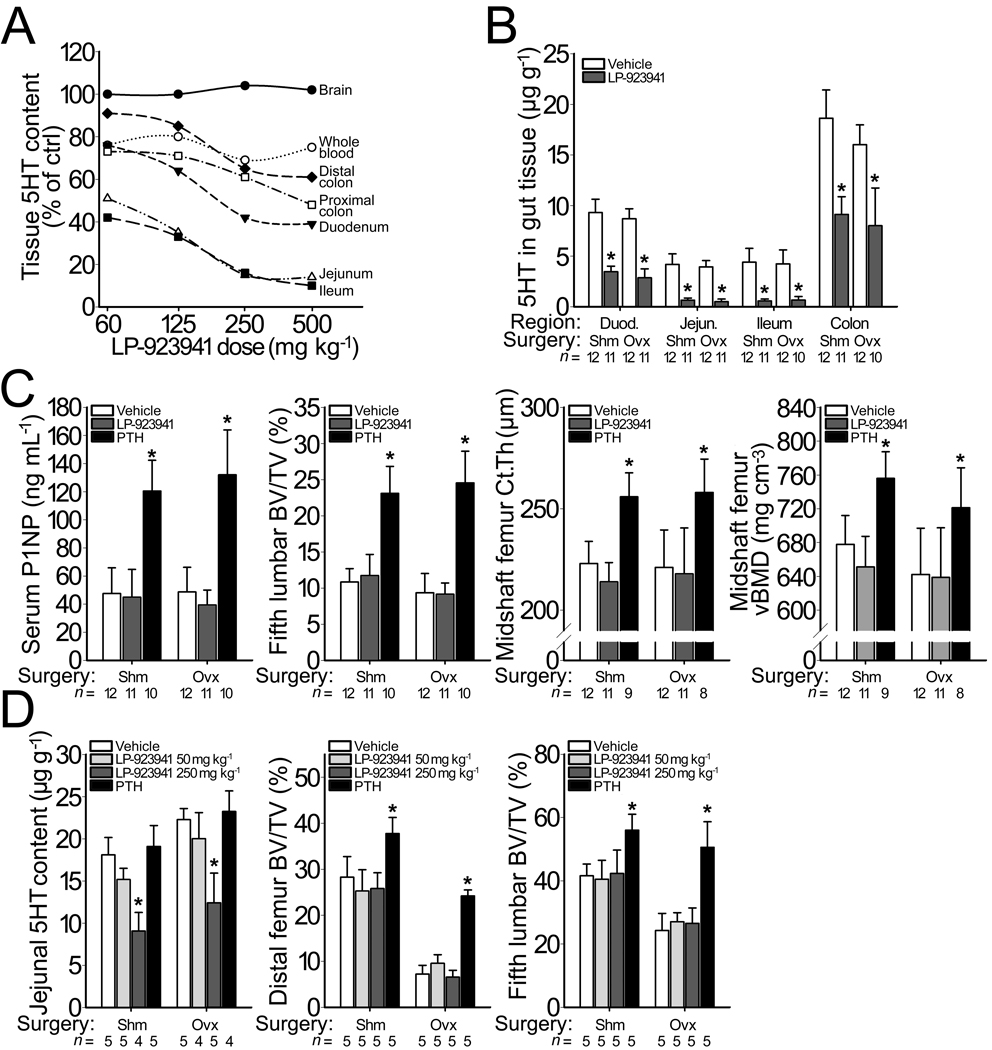
It is possible that Tph1−/− mice activate compensatory pathways that keep bone mass at WT levels. This could explain why an increase in bone mass was reported when Tph1 was conditionally inactivated only in the intestine22 or when a small molecule was used to inhibit Tph1 in the intestine32. Therefore, we treated intact and OVX mice and rats for 6 weeks with a small molecule Tph1 inhibitor, LP-923941, which is the active enantiomer of the compound LP-533401 (Supplementary Table S4) previously reported to increase bone mass32. The LP-923941 dose of 250 mg kg−1 day−1 employed in our pharmacology studies was selected from a preliminary 7-day dose-response study in mice (Fig. 6a).
Figure 6.
Bone mass following pharmacologic inhibition of Tph1 activity. (a) Graph depicting dose dependent changes in 5HT content, compared to vehicle-treated controls, in 9-wk-old WT female C57BL/6 mice after receiving daily doses of LP-923941 for 7 days. A daily dose of 250 mg/kg lowered 5HT levels in whole blood and in intestine, but not in brain. (b) Graph depicting changes in the intestinal 5HT content of sham-operated (SHM) and ovariectomized (OVX) mice that received vehicle or LP-923941 (250 mg/kg/day) for 6 wks. OVX alone reduced 5HT content in the duodenum and colon by ~12% (p < 0.05) compared to SHM mice. Treatment with LP-923941 significantly reduced serotonin content equally in all regions of the intestine in SHM and in OVX mice. (c) Effect of treatment with LP-923941 (250 mg/kg/day) or teriparatide (80 µg/kg/day), the 1–34 residue amino-terminal fragment of human parathyroid hormone (PTH), on serum PINP levels, a marker of bone formation (left), vertebral trabecular BV/TV (middle left), midshaft femur cortical thickness (middle right), and midshaft femoral volumetric BMD (right) in SHM and in OVX mice. (d) Effect of treating SHM and OVX rats with LP-923941 (50 or 250 mg/kg/day) or teriparatide (80 µg/kg/day) for 6 weeks on jejunal 5HT content (left), femoral trabecular BV/TV (middle), and vertebral trabecular BV/TV (right). The numbers of mice studied (n =) are indicated, as are error bars equal to 1 s.d. Asterisks (*) indicate a significant difference (p < 0.05) compared to vehicle treated mice.
Treating intact and OVX mice with LP-923941 for 6 weeks reduced 5HT content in whole blood by 42% at 2 weeks (2715 versus 4813 ng ml−1, p < 0.001) and 44% at 6 weeks (2543 versus 4359 ng ml−1, p < 0.001), without influencing brain 5HT content (0.49 versus 0.50 µg gm−1) or turnover as indicated by 5-hydroxyindoleacetic acid levels (0.231 versus 0.229 µg gm−1). The ability of LP-923941 to reduce 5HT synthesis in all segments of the intestine was not influenced by OVX (p < 0.001, Fig. 6b) and was similar in the 7-day dose-response and 6-week pharmacology studies. Reductions in whole blood serotonin levels and intestinal serotonin content did not influence serum P1NP levels (a marker of bone formation), trabecular bone mass, or cortical bone mass in either sham operated (SHM) or OVX mice (Fig. 6c and Supplementary Fig. S8). In contrast, teriparatide increased bone formation and bone mass in SHM and OVX mice (Fig. 6c and Supplementary Fig. S8).
We also treated SHM and OVX rats for 6 weeks with LP-923941, which resulted in a reduction of intestinal 5HT content by 51% and 46%, respectively, compared to vehicle treated controls (Fig. 6d). The reduction of intestinal 5HT was not accompanied by any change in trabecular or cortical bone mass of the femur nor in trabecular bone mass of the 5th lumbar vertebra (Fig. 6d), whereas daily injections with teriparatide significantly increased bone mass in the femur and vertebral body of SHM and OVX rats (Fig. 6d).
DISCUSSION
Our in vivo data indicate that Lrp5 acts in osteocytes and perhaps some late-stage osteoblasts to effect changes in bone mass via canonical Wnt signaling. Axin2, a downstream target of Wnt signaling, is more highly expressed in bone from HBM mice, as is a LacZ reporter of canonical Wnt signaling (Supplemental Fig. S1). These results are consistent with in vitro studies of LRP5-mediated Wnt signaling, which reported that HBM alleles are less inhibited by the endogenous inhibitors DKK1 and SOST than are WT alleles9,11,33,34 (Supplemental Fig. S10). DKK1 is expressed by many cells including osteocytes and a complete lack of Dkk1 is incompatible with life; however, mice with partial loss-of-function mutations in DKK1 have increased bone mass consistent with this protein acting as a negative regulator of bone formation16,35. SOST is principally expressed by mature osteocytes, and when genetically absent in humans and in mice causes a phenotype that is similar to the phenotype caused by HBM mutations in LRP54,5,15. The strong expression of SOST by mature bone cells suggests it functions as a tonic negative regulator of new bone formation. Mechanical loading of bone, a potent inducer of new bone formation in vivo, reduces SOST expression by osteocytes36 and increases the expression of Wnt target genes37,38. Therefore, by being less sensitive to endogenous inhibitors, HBM Lrp5 mutations likely induce new bone formation in the absence of, or at lower, mechanical load. Conversely, mice with Lrp5 loss-of-function mutations exhibit a blunted anabolic response to mechanical load39, which is consistent with insufficient Wnt signaling in Lrp5−/− osteocytes despite the reduction in SOST expression that follows mechanical loading (Supplemental Fig. S10).
LRP5 is also expressed during osteoblast commitment and differentiation1,40. Therefore, similar to other signaling pathways that influence cellular differentiation at several stages41,42, LRP5 signaling may influence other aspects of osteoblast differentiation. This speculation is compatible with studies which have noted, in the absence of canonical Wnt signaling, that differentiating mesenchymal stem cells adopt chondrogenic and adipogenic fates instead of osteoblastic fates43–46. However, these studies blocked canonical Wnt signaling by conditionally inactivating β-catenin. LRP5 and its most closely related mammalian paralog, LRP6, are both able to transduce Wnt signaling in vitro, and have overlapping and non-redundant roles in vivo during gastrulation and skeletal patterning10,47. Therefore, LRP6 may be able to compensate for LRP5 during some stages of osteoblast differentiation.
We found that activation of HBM alleles in osteocytes caused the same increase in bone mass as having the HBM alleles active in all cells (Fig. 2). Conversely, Dmp1::Cre-mediated inactivation of Lrp5 caused a decrease in bone mass compared to non-inactivated littermates (Fig. 3). Taken together, these data indicate that LRP5 signaling by mature bone cells regulates bone mass and, in the case of the HBM mutation, is sufficient to recapitulate the inherited HBM phenotype. Further support for this conclusion derives from studies in which we conditionally activated HBM Lrp5 in cells that contribute to the appendicular skeleton, but not to the axial skeleton. Bone mass increased only at skeletal sites where HBM Lrp5 was activated (Fig. 2).
Our results do not support a model in which Lrp5 regulates bone mass via the regulation of peripheral 5HT synthesis in the duodenum22. When we conditionally activated HBM Lrp5 or inactivated WT Lrp5 in 5HT producing enterochromaffin cells, we found no effect on bone mass (Figs. 2 and 3). In addition, we found no Lrp5 genotype-specific differences in whole blood 5HT levels (Fig. 4) and we did not observe an effect of genetically or pharmacologically lowering peripheral 5HT levels on bone mass (Figs. 5 and 6).
Currently, we do not know the origin of the difference in results between our study and prior studies22,32. Perhaps differences in the design of the Lrp5 conditional alleles, the transgenic mice that were used to produce cell-type-specific Cre-recombination within bone and intestine, the assays employed to measure 5HT levels, the use of conditional versus global Tph1 knockout alleles, the vivariums in which the animals were raised, and the trial design for pharmacologic inhibition of Tph1 in OVX mice and rats account for the differing results (also see Supplementary Material).
We created mice with knockin HBM alleles in which the gene’s intron-exon structure remained intact. The earlier study inserted an HBM Lrp5 cDNA with a carboxyl-terminal FLAG-tag into the 1st exon of Lrp522. We used a Dmp1::Cre transgene that is expressed in osteocytes and some late-stage osteoblasts, whereas the other study22 employed a 2.3Col1a1::Cre transgene that is expressed earlier during osteoblastic differentiation48. We used a different Villin::Cre transgenic line than the earlier study22, although both lines expressed Cre in intestinal stem cells. Nevertheless, it remains possible that off-target sites of Cre expression account for some of the divergent results between our study and the other study22.
We found no association between Lrp5 genotype and serum 5HT level. However, serum 5HT levels depend upon the efficiency of 5HT release from platelets during clot formation, which is affected by the collection and clotting method, and the clotting time49. As such, serum measures can be unreliable within and across studies. In contrast, as long as the capacity of platelets to store serotonin is not exceeded, whole blood 5HT levels correlate well with free circulating 5HT levels50. Therefore, we also measured whole blood 5HT by HPLC23. Whole blood 5HT levels did not correlate with Lrp5 genotype or with bone mass. Because whole blood 5HT levels can be influenced by other factors, such as inflammation and infection, it is possible that environmental differences between the present study and the earlier study22 account for the divergent 5HT results.
We studied bone mass in Tph1−/− mice, whereas Tph1 was conditionally inactivated using Villin::Cre in the previous study22. Tph1−/− mice may have activated compensatory pathways to normalize bone mass, whereas mice with conditional Tph1 inactivation may not. We tested this possibility by pharmacologically inhibiting 5HT synthesis in the intestine. We used the active enantiomer of the small molecule inhibitor previously reported to affect bone mass32. We studied mice that had undergone OVX a year before receiving the pharmacologic agent and found no effect on bone mass compared to vehicle treated controls (Fig. 6); in contrast, teriparatide had a strong anabolic effect. We examined aged OVX mice since women 1 decade after menopause are a major target population for anabolic osteoporosis therapy. However, our OVX model differed from that used in the earlier study in which mice had undergone OVX less than 2 months before receiving the pharmacologic agent32. Therefore, we utilized an OVX rat model in which we began pharmacological Tph1 inhibition 5 weeks after OVX surgery; this is within the time frame employed in the previous study32. Again, we observed no effect of Tph1 inhibition on bone mass compared to vehicle treated controls, whereas teriparatide had a robust bone anabolic effect (Fig. 6). However, because we only studied the active enantiomer, we cannot exclude the possibility that the inactive enantiomer increases bone mass via a mechanism that is independent of Tph1 inhibition.
Although Lrp5 genotype may affect peripheral serotonin synthesis in some contexts22, in the present study we did not observe Lrp5-mediated effects on peripheral 5HT levels or Tph1-mediated effects on bone mass. Therefore we think it unlikely that the mechanism by which Lrp5 normally affects bone mass involves intestinal 5HT synthesis. Instead, our data indicate that LRP5 functions in bone to control bone mass and are consistent with the receptor participating in the adaptive response of bone to mechanical load (Wolff’s Law)51. Therapies aimed at enhancing these functions of LRP5 in humans should benefit individuals who have skeletal fragility due to low bone mass.
METHODS
Generation of new genetically modified mouse strains
We created Lrp5 conditional HBM and knockout alleles by homologous recombination in ES cells. We used targeting vectors containing different HBM-associated mutations (p.G171V or p.A214V) for the HBM knockin mice. We included a floxed neomycin-resistance cassette (NeoR) for selection (Fig. 1a). We bred knockin mice that retained NeoR and mice in which NeoR had been Cre-excised. We created a floxed knockout allele, by placing flanking LoxP sites in the same orientation around exon 2. We included an FRT-flanked neomycin selection cassette that we subsequently excised (Fig. 3a). We bred mice that retained the floxed exon 2 and mice in which exon 2 had been Cre-excised. Genotyping was performed by PCR. Other mice employed in these studies have been described previously: EIIa::Cre52, FLPer53, Cmv::Cre54, Dmp1::Cre21, Villin::Cre27, Prx1::Cre28, βatGal19, Lrp5−/−10,29,55, Tph1−/−23,24, and Rosa26mTmG56. Details are included in the Supplementary Material.
Assessment of bone properties
We performed quantitative histomorphometry in mice as previously described39. We analyzed one 6 µm coronal section from the distal femur of each mouse for mineralizing surface (MS/BS; %), mineral apposition rate (MAR; µm day−1), and bone formation rate (BFR; µm3 µm−2 year−1). We measured 3-dimensional morphometric properties in the cortical and trabecular bone of mouse distal femoral metaphyses and 5th lumbar vertebrae by microcomputed tomography (µCT). We performed whole body dual energy X-ray absorptiometry (DEXA) to measure areal bone mineral density (aBMD; gm cm−2) and bone mineral content (BMC; gm) for the post-cranial skeleton, for the lumbar spine (L3–L5, inclusive), and for individual bones. We performed biomechanical testing on the left femurs by loading them to failure in monotonic compression using a crosshead speed of 0.2 mm s−1, during which we collected force and displacement measurements every 0.005 s. From the force versus displacement curves, were calculated ultimate force (in N), yield force (in N), stiffness (in N mm−1), and energy to failure (in mJ). Details are included in the Supplementary Material.
Measurement of serotonin levels, and Tph1, LacZ, and Axin2 mRNA expression
We measured whole blood serotonin using two methods23,57. For serum serotonin measures, we used blood that was collected from the retro-orbital sinus and clotted at room temperature. We recovered serum by centrifugation at 4°C, 17,000 × g for 25 minutes and then assayed serotonin by competitive ELISA. We extracted and measured serotonin from different segments of intestine as previously described23. We performed quantitative RT-PCR for intestinal Tph1 expression using total RNA recovered from the duodenum. We performed quantitative RT-PCR for LacZ and Axin2 expression using RNA recovered from cortical bone tissue of 8-wk-old male and female βatGal transgenic mice with Lrp5 WT and A/A genotypes. Details are included in the Supplementary Material.
Pharmacological inhibition of gut Tph1 activity
We treated SHM and OVX mice and rats with the Tph1 inhibitor LP923941. This compound is the active enantiomer of the Tph1 inhibitor LP-533401 examined previously23,32. Compared to LP-533401, LP-923941 has approximately twice the potency in enzymatic and cell based assays, with the inactive enantiomer having greatly reduced potency (Supplementary Table S4). We treated animals by oral gavage for 6 weeks with 250 mg kg−1 day−1 of LP923941 or with vehicle-alone. To serve as a positive control for bone formation, we subcutaneously injected SHM and OVX mice and rats for 6 weeks with teriparatide (hPTH 1–34) at a dose of 80 µg kg−1 day−1 or with vehicle-alone. For the P1NP assay, we collected blood in heparinized capillary tubes by retro-orbital bleeding. We measured P1NP using a Rat/Mouse P1NP EIA. Details are included in the Supplementary Material.
Statistical analyses
We tested longitudinal data for differences among genotypes using single classification repeated measures ANOVA. We tested cross-sectional data (e.g. µCT, biomechanical properties, serotonin measurements) across genotypes for significant differences using single classification ANOVA, followed by the Sheffe post-hoc comparison to probe pairwise comparisons in the event that the omnibus ANOVA was significant. We performed statistical calculations in StatView 5.0. We performed two- or three-factor ANOVAs for the pharmacology studies using SPSS, version 11.5.0. We set the experiment-wise error rate at α=0.05 for all tests. Details are included in the Supplementary Material.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the other members of their laboratories for technical support: B. Newby, K. Kurek and E. Boyden for assistance with the HBM mouse studies, G. Zhou for assistance with histomorphometry, M. Niedecker and E. Kleinschmidt for assistance with the radiography, the Case Transgenic and Targeting Facility, K. Sisson and P. Swiatek of the VAI Mouse Germline Modification Core, B. Eagleson and the staff of the VAI Vivarium, and J. Bardenhagen, J. Greer, S. Jeter-Jones, J. Liu, M.K. Shadoan, D.D. Smith, W. Xiong and A. Yu of Lexicon Pharmaceuticals, Inc. and F. Bourgondien, R. Zhang, and S. Yeh of Merck Sharp & Dohme Research Laboratories. This work was supported by the following grants: NIH grant AR53237 (to AGR), PHS Career Development Award UL 1RR025761-02 (to PJN), NIH grant GM74241 and a Leukemia and Lymphoma Society Scholarship (both to XH), NIH grant AR053293 and the Van Andel Research Institute (to BOW). MLW is an investigator with the Howard Hughes Medical Institute. Employees of Lexicon Pharmaceuticals, Inc. (RB, QL, FM, DRP, QY, and BZ) and Merck Sharp & Dohme Research Laboratories (HG and JAG) have received compensation in the form of salary and stock options.
REFERENCES
- 1.Gong Y, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 2.Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyden LM, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 4.Balemans W, et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum Mol Genet. 2001;10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- 5.Brunkow ME, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morello R, et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 7.Ai M, Heeger S, Bartels CF, Schelling DK. Clinical and molecular findings in osteoporosis-pseudoglioma syndrome. Am J Hum Genet. 2005;77:741–753. doi: 10.1086/497706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Wesenbeeck L, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003;72:763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmen SL, et al. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res. 2004;19:2033–2040. doi: 10.1359/JBMR.040907. [DOI] [PubMed] [Google Scholar]
- 11.Semenov MV, He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006;281:38276–38284. doi: 10.1074/jbc.M609509200. [DOI] [PubMed] [Google Scholar]
- 12.Kato M, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokubu C, et al. Skeletal defects in ringelschwanz mutant mice reveal that Lrp6 is required for proper somitogenesis and osteogenesis. Development. 2004;131:5469–5480. doi: 10.1242/dev.01405. [DOI] [PubMed] [Google Scholar]
- 14.Nakanishi R, et al. Secreted frizzled-related protein 4 is a negative regulator of peak BMD in SAMP6 mice. J Bone Miner Res. 2006;21:1713–1721. doi: 10.1359/jbmr.060719. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res. 2008;23:860–869. doi: 10.1359/jbmr.080216. [DOI] [PubMed] [Google Scholar]
- 16.Morvan F, et al. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J Bone Miner Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- 17.Babij P, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18:960–974. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. 2004;24:4677–4684. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maretto S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jho EH, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, et al. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86:320–325. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 22.Yadav VK, et al. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell. 2008;135:825–837. doi: 10.1016/j.cell.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, et al. Discovery and characterization of novel tryptophan hydroxylase inhibitors that selectively inhibit serotonin synthesis in the gastrointestinal tract. J Pharmacol Exp Ther. 2008;325:47–55. doi: 10.1124/jpet.107.132670. [DOI] [PubMed] [Google Scholar]
- 24.Walther DJ, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- 25.Alenina N, et al. Growth retardation and altered autonomic control in mice lacking brain serotonin. Proc Natl Acad Sci U S A. 2009;106:10332–10337. doi: 10.1073/pnas.0810793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savelieva KV, et al. Genetic disruption of both tryptophan hydroxylase genes dramatically reduces serotonin and affects behavior in models sensitive to antidepressants. PLoS One. 2008;3:e3301. doi: 10.1371/journal.pone.0003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 28.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 29.Iwaniec UT, et al. PTH stimulates bone formation in mice deficient in Lrp5. J Bone Miner Res. 2007;22:394–402. doi: 10.1359/jbmr.061118. [DOI] [PubMed] [Google Scholar]
- 30.Long F. When the gut talks to bone. Cell. 2008;135:795–796. doi: 10.1016/j.cell.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Rosen CJ. Serotonin rising--the bone, brain, bowel connection. N Engl J Med. 2009;360:957–959. doi: 10.1056/NEJMp0810058. [DOI] [PubMed] [Google Scholar]
- 32.Yadav VK, et al. Pharmacological inhibition of gut-derived serotonin synthesis is a potential bone anabolic treatment for osteoporosis. Nat Med. 2010;16:308–312. doi: 10.1038/nm.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balemans W, et al. The binding between sclerostin and LRP5 is altered by DKK1 and by high-bone mass LRP5 mutations. Calcif Tissue Int. 2008;82:445–453. doi: 10.1007/s00223-008-9130-9. [DOI] [PubMed] [Google Scholar]
- 34.Ellies DL, et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21:1738–1749. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald BT, et al. Bone mass is inversely proportional to Dkk1 levels in mice. Bone. 2007;41:331–339. doi: 10.1016/j.bone.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robling AG, et al. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem. 2008;283:5866–5875. doi: 10.1074/jbc.M705092200. [DOI] [PubMed] [Google Scholar]
- 37.Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone. 2008;42:606–615. doi: 10.1016/j.bone.2007.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson JA, et al. Wnt/beta-catenin signaling is a normal physiological response to mechanical loading in bone. J Biol Chem. 2006;281:31720–31728. doi: 10.1074/jbc.M602308200. [DOI] [PubMed] [Google Scholar]
- 39.Sawakami K, et al. The Wnt co-receptor LRP5 is essential for skeletal mechanotransduction but not for the anabolic bone response to parathyroid hormone treatment. J Biol Chem. 2006;281:23698–23711. doi: 10.1074/jbc.M601000200. [DOI] [PubMed] [Google Scholar]
- 40.Dong Y, et al. Molecular cloning and characterization of LR3, a novel LDL receptor family protein with mitogenic activity. Biochem Biophys Res Commun. 1998;251:784–790. doi: 10.1006/bbrc.1998.9545. [DOI] [PubMed] [Google Scholar]
- 41.Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 42.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 44.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Hu H, et al. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 46.Kang S, et al. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 47.Kelly OG, Pinson KI, Skarnes WC. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. 2004;131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- 48.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 49.Ingebretsen OC, Bakken AM, Farstad M. Liquid chromatography of serotonin and adenine nucleotides in blood platelets, illustrated by evaluation of functional integrity of platelet preparations. Clin Chem. 1985;31:695–698. [PubMed] [Google Scholar]
- 50.Anderson GM, Stevenson JM, Cohen DJ. Steady-state model for plasma free and platelet serotonin in man. Life Sci. 1987;41:1777–1785. doi: 10.1016/0024-3205(87)90696-5. [DOI] [PubMed] [Google Scholar]
- 51.Wolff J. The law of bone remodelling. Berlin, New York: Springer-Verlag; 1892. [Google Scholar]
- 52.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- 54.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clement-Lacroix P, et al. Lrp5-independent activation of Wnt signaling by lithium chloride increases bone formation and bone mass in mice. Proc Natl Acad Sci U S A. 2005;102:17406–17411. doi: 10.1073/pnas.0505259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 57.Tenner K, Qadri F, Bert B, Voigt JP, Bader M. The mTPH2 C1473G single nucleotide polymorphism is not responsible for behavioural differences between mouse strains. Neurosci Lett. 2008;431:21–25. doi: 10.1016/j.neulet.2007.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.