Abstract
Summary
Recurrent fusions of ETS genes are considered driving mutations in a diverse array of cancers, including Ewing’s sarcoma, acute myeloid leukemia, and prostate cancer. We investigate the mechanisms by which ETS fusions mediate their effects, and find that the product of the predominant ETS gene fusion, TMPRSS2:ERG, interacts in a DNA-independent manner with the enzyme poly(ADP-ribose) polymerase 1 (PARP1) and the catalytic subunit of DNA protein kinase (DNA-PKcs). ETS gene-mediated transcription and cell invasion require PARP1 and DNA-PKcs expression and activity. Importantly, pharmacological inhibition of PARP1 inhibits ETS positive, but not ETS negative, prostate cancer xenograft growth. Finally, overexpression of the TMPRSS2:ERG fusion induces DNA damage, which is potentiated by PARP1 inhibition in a manner similar to that of BRCA1/2-deficiency.
Keywords: Prostate, Rearrangement, Gene Fusion, TMPRSS2, ERG, DNA-PKcs, PARP1
INTRODUCTION
ETS transcription factors are aberrantly expressed in a diverse array of cancers including prostate, breast, melanoma and Ewing’s sarcoma (Jane-Valbuena et al., 2010; Jeon et al., 1995; Shurtleff et al., 1995; Sorensen et al., 1994; Tognon et al., 2002; Tomlins et al., 2005). In prostate cancer, recurrent gene fusions of the androgen-regulated gene, TMPRSS2, to the oncogenic ETS transcription factor ERG are present in approximately 50% of prostate cancers (Brenner and Chinnaiyan, 2009; Kumar-Sinha et al., 2008; Tomlins et al., 2005). Although ERG is the predominant ETS gene rearrangement observed, other ETS transcription factors are found at a much lower frequency in prostate cancer, including ETV1 (Tomlins et al., 2005), ETV4 (Tomlins et al., 2006) and ETV5 (Helgeson et al., 2008). ETS gene fusions appear early in prostatic disease during the transition from high-grade prostatic intraepithelial neoplasia (PIN) lesions to invasive carcinoma (Helgeson et al., 2008; Hermans et al., 2008; Klezovitch et al., 2008; Tomlins et al., 2007a; Wang et al., 2008) and are formed by several mechanisms including interstitial deletion and genomic insertion (Perner et al., 2007). In prostate cell lines devoid of the TMPRSS2:ERG gene fusion, androgen receptor-induced proximity can trigger topoisomerase-2β-mediated TMPRSS2:ERG gene fusion formation (Haffner et al., 2010), which is significantly enhanced by genotoxic stresses such as ionizing radiation (Lin et al., 2009; Mani et al., 2009).
Once an ETS gene fusion is formed through genomic rearrangement, the subsequent overexpression of an ETS gene fusion protein can contribute to cancer progression through several different mechanisms. For example, TMPRSS2-ERG gene fusion expression is required for cell growth in cell line models that harbor an endogenous gene fusion both in vitro and in vivo (Helgeson et al., 2008; Sun et al., 2008; Tomlins et al., 2007a; Wang et al., 2008). Likewise, ETS proteins are active transcription factors that drive cellular invasion through the induction of a transcriptional program highly enriched for invasion-associated genes (Helgeson et al., 2008; Hermans et al., 2008; Klezovitch et al., 2008; Tomlins et al., 2007a; Wang et al., 2008). Genetically-engineered mice expressing ERG or ETV1 under androgen regulation exhibit PIN-like lesions, but do not develop frank carcinoma, suggesting that additional collaborating mutations may be required for de novo carcinogenesis (Carver et al., 2009; King et al., 2009; Klezovitch et al., 2008; Kumar-Sinha et al., 2008; Tomlins et al., 2007a; Zong et al., 2009). Importantly, overexpression of ERG leads to accelerated carcinogenesis in mouse prostates with deletion of the tumor suppressor PTEN (Carver et al., 2009; King et al., 2009). Additionally, in a transplant model, mouse prostate epithelial cells that are forced to overexpress both ERG and the androgen receptor gene form invasive prostate cancer (Zong et al., 2009). This suggests that ERG rearrangements can function to accelerate prostate carcinogenesis.
Given the functional consequences of ETS gene rearrangements in prostate cancer progression, a critical question remains unanswered: are ETS gene fusions therapeutic targets, either directly or indirectly? Given the difficulties in targeting nuclear transcription factors using conventional therapeutic strategies (Darnell, 2002), we hypothesized that associated enzymes critical for ERG function may instead serve as viable therapeutic targets to inhibit ETS-positive prostate cancer cell growth.
RESULTS
Identification of ERG interacting proteins by mass spectrometry
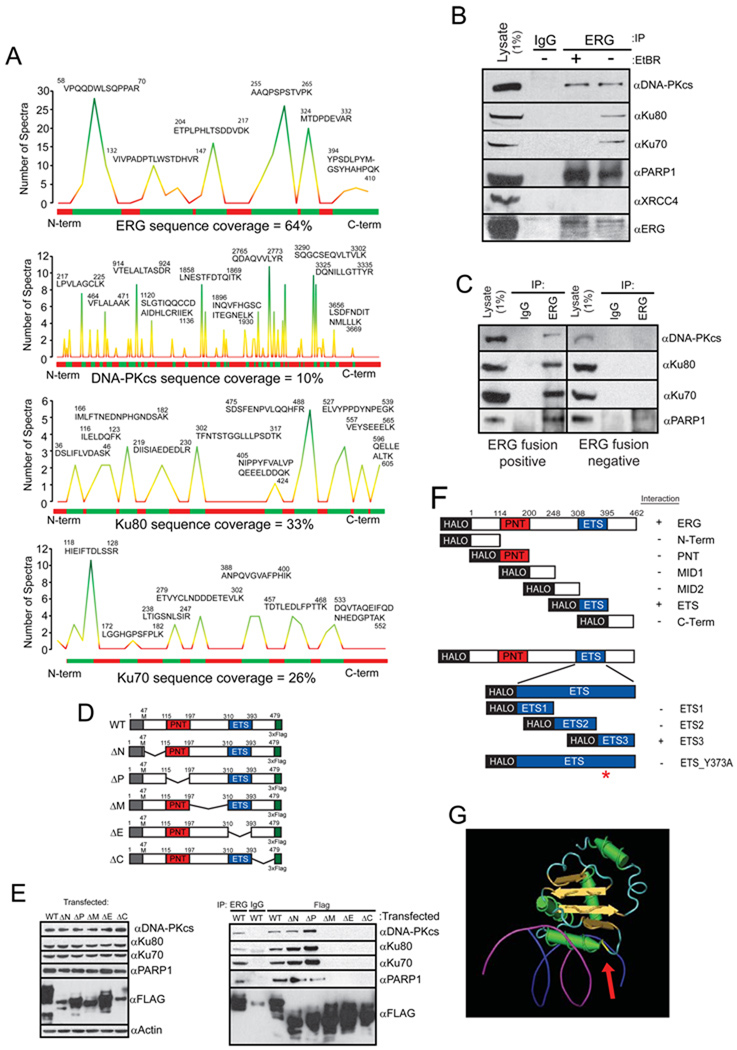
To identify ERG-interacting proteins that may serve as rational therapeutic targets and explore the mechanism by which ETS gene fusions mediate their effects, we performed mass spectrometric (MS) analysis of proteins that interact with the most prevalent ETS gene fusion product, ERG (encoded from TMPRSS2 exon 1 fused to ERG exon 2 (Tomlins et al., 2005)). VCaP prostate cancer cells (which harbor a TMPRSS2:ERG rearrangement) or human embryonic kidney cells (HEK) 293 cells were infected with either adenoviral V5- or FLAG-epitope-tagged ERG expression vectors, respectively. Immunoprecipitation (IP) was completed in 8 biological replicates to isolate protein-protein interactions as described by schematic (Figure S1A). As expected, the interaction bait, ERG, was the top scoring protein identified in the pull-down with 64.4% coverage with 17 tryptic peptides scanned over 500 times (Figure 1A, Table S1). Interestingly, three of the next four interacting proteins of high confidence and high sequence coverage identified were components of the DNA-dependent protein kinase complex and included the large catalytic subunit of a phosphatidylinositol 3/4 (PI3/4)-kinase called DNA-dependent protein kinase (DNA-PKcs) (10% coverage) and its known interacting subunits Ku70 (26% coverage) and Ku80 (34% coverage) (Figure 1A, Table S1). Interactions were confirmed with an independent antibody (Figure S1B) and IPs performed from VCaP cells demonstrated an endogenous association that occurs in the absence of ectopic overexpression (Figure 1B and Figure S1C).
Figure 1. The TMPRSS-ERG gene fusion product interacts with PARP1 and the DNA-PK complex.
(A) MS analysis of proteins interacting with ERG. Histograms show peptide coverage of ERG, DNA-PKcs, Ku70 and Ku80.
(B) ERG, DNA-PKcs, PARP1 but not Ku70 or Ku80, interact independent of DNA. IP performed from VCaP cells which naturally harbor the ERG translocation.
(C) ERG, DNA-PKcs, PARP1, Ku70 and Ku80 associate in ERG gene fusion positive human prostate cancer tissues. Representative ERG positive and negative prostate cancers shown of three pairs of tissues.
(D) Schematic of TMPRSS2-ERG gene fusion tiling deletion expression vectors.
(E) IP of DNA-PKcs, Ku70, Ku80 and PARP1 from HEK293 cells transfected with ERG expression vectors depicted in (D). Input Western is shown on the left, IP-Western shown on the right. All IPs were performed with FLAG antibody unless otherwise indicated.
(F) Schematic representation of halo-tagged ERG fragment vectors. The construct were transcribed using wheat germ extracts and halo-tagged protein was purified. Proteins were then incubated with purified DNA-PKcs and IP-westerns performed. Fragments able to IP DNA-PKcs are indicated with a ‘+’.
(G) ETS1:DNA crystal from (Garvie et al., 2001) used to demonstrate physical location of Tyrosine373 (from ERG) relative to the DNA binding residues.
1% of the total cell lysate used for IP was added to the input lane. Representative experiments are shown.
To identify additional proteins participating in the ERG:DNAPK complex, we assessed our list of ERG interactors for other proteins known to interact with DNA-PKcs, Ku70, or Ku80 and identified two peptides for poly-(ADP-ribose) polymerase (PARP1): VVSEDFLQDVSASTK and QQVPSGESAILDR. Importantly, we demonstrated that PARP1 endogenously associated with ERG in VCaP cells (Figure 1B). We then performed reverse IPs using antibodies against DNA-PKcs, PARP1 and Ku80 and showed that each antibody was able to detect ERG-V5 protein (Figure S1D). To detect the PARP1:ERG interaction with the endogenous TMPRSS2:ERG gene fusion product, we used agarose-coupled PARP1 antibody to perform the IP-Western, which confirmed that PARP1 interacts with the gene fusion product in an endogenous setting (Figure S1E).
As DNA-PKcs only binds with Ku70 and Ku80 in the presence of DNA (Spagnolo et al., 2006), we tested the dependence of the ERG:PARP1 and ERG:DNA-PKcs interactions on intact DNA by performing the IP in the presence of 100µM ethidium bromide. This treatment disrupted the interaction between ERG, Ku70 and Ku80, but not the interaction between ERG and either PARP1 or DNA-PKcs, demonstrating that the ERG:PARP1 and ERG:DNA-PKcs interactions are DNA-independent (Figure 1B). As a control, we tested whether ERG would bind another PI3/4 kinase family member, ATR, or another protein known to interact with the DNAPK complex, XRCC4. Consistent with our IP-MS data, we were unable to detect an interaction between ATR or XRCC4 and ERG by IP-Western blot analysis (Figure S1C and Figure 1B, respectively).
We next assessed whether the ERG:PARP1 and ERG:DNA-PKcs interactions occur in human prostate cancer tissues. ERG-IP showed enrichment for DNA-PKcs, Ku70, Ku80 and PARP1 in ERG gene fusion-positive, but not in ETS gene fusion-negative prostate cancer tissues (Figure 1C, Figure S1F). Interestingly, the lack of detectable ERG:PARP1 interaction in tissue without ETS gene rearrangement is likely due to the near absent ERG expression in rearrangement-negative prostate cancer (Park et al., 2010), as when overexpressed, wild type (WT) ERG interacts with PARP1 in cell lines that do not harbor the translocation (Figure S1G). Additional IP-Westerns were performed to test the dependence of the ERG:PARP1 interaction on DNA in human prostate cancer tissues. Importantly, the interaction occurred independent of DNA in all three independent human tissues (Figure S1H).
Next, we sought to map the interactions and created a series of flag tagged ERG expression vectors with tiling deletions including: the N-terminus (deletion of AA: 47–115, predicted molecular weight 44.6 kDa), pointed domain (AA: 115–197, 43.4 kDa), the middle amino acids (197–310, 41.6 kDa), the ETS domain (AA: 310–393, 43.7 kDa) or the C-terminus (AA: 393–479, 43.6 kDa) and labeled the constructs ΔN, ΔP, ΔM, ΔE, ΔC, respectively as depicted in Figure 1D. IP following transient transfection demonstrated that the interactions between ERG, DNA-PKcs, Ku70, Ku80 and PARP1 occurred in the C-terminal half of the ERG protein (Figure 1E). To further map the ERG:PARP1 and ERG:DNA-PKcs interactions and to confirm that both PARP1 and DNA-PKcs interact with other ETS family member proteins, we performed IP-Western blot analysis in HEK293 cells transfected with either ERG-FLAG, ETS1-FLAG, SPI1-FLAG or ETV1-FLAG expression vectors, which were selected for their sequence relationship to ERG (Figure S1I). In all four experiments, pull downs confirmed the interactions (Figure S1G, S1J, S1K and S1L). We then created N-terminal halo-tagged expression vectors for in vitro purification of these ETS genes. Subsequent IP-westerns demonstrated that all 4 of these proteins bind directly to DNA-PKcs (Figure S1M). Given the sequence alignment of these 4 ETS proteins and the large tiling deletion data, our data suggested that the interactions occur through the ETS DNA binding domain.
To definitively map the ERG:DNA-PKcs and ERG:PARP1 interactions, we utilized HALO-tagged WT ERG and six individual HALO-tagged fragments spanning the entire ERG protein (Yu et al., 2010). As expected, IP-Western blot demonstrated that the ERG:DNA-PKcs interaction occurred through the ETS DNA binding domain. To further map the interaction between ERG and DNA-PKcs, we utilized a series of three HALO-tagged fragments that tiled the ETS domain, which localized the interaction to the final 28 amino acids of the ETS domain (Figure 1F and S1N). Importantly, although the crystal structure of the ETS domain from ERG has not yet been reported, the crystal structure of another ETS factor that we demonstrated interacts with DNA-PKcs, ETS1, has been published (Garvie et al., 2001). Based on homology with other interacting ETS proteins and structural information, we predicted that the interaction was dependent on the amino acids, YYDKN. By site directed mutagenesis of each residue to alanine, we demonstrated that the Y373A mutant was unable to precipitate DNA-PKcs suggesting that this interaction is mediated by Tyrosine 373 (Figure 1F and S1P). Analysis of the ETS1 structure shows that Y373 is adjacent to the arginine residues that fit into the DNA groove and that Y373 is accessible to potential interacting proteins (Figure 1G).
After demonstrating that ERG interacts with DNA-PKcs directly through amino acid Y373, we sought to map the ERG:PARP1 interaction. However, purified ERG was only able to interact with purified PARP1 in the absence of ethidium bromide (Figure S1O). Because the interaction occurred in cells independent of ethidium bromide, this suggests that the ERG:PARP1 interaction is mediated by other proteins. This is consistent with the results from our IP-MS experiment in which few PARP1 peptides were identified suggesting that the ERG:PARP1 interaction is mediated by an intermediate protein such as DNA-PKcs.
PARP1 and DNA-PKcs are required for ERG-mediated transcription
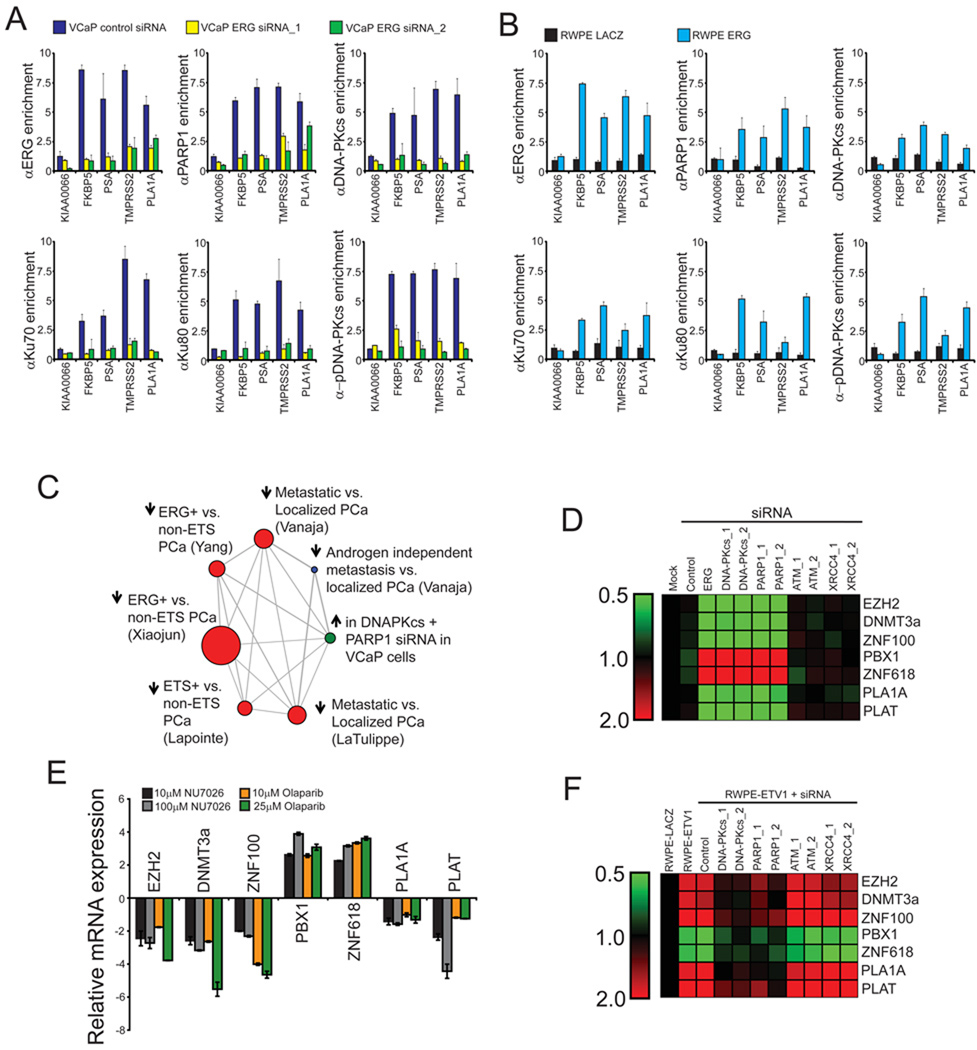
Given that the interaction of DNA-PKcs and PARP1 with ERG occurs through the ETS domain, we hypothesized that both PARP1 and DNA-PKcs function as co-regulators of ERG transcriptional activity. Thus, we performed chromatin immunoprecipitation (ChIP) assays in VCaP cells and assessed enrichment of known ERG-targets including the PLA1A promoter and the FKBP5, PSA and TMPRSS2 enhancers. These experiments demonstrated that ERG, DNA-PKcs, activated DNA-PKcs (assessed by T2609 phosphorylation), Ku70, Ku80 and PARP1 bind to these sites, but not to the negative control gene KIAA0066 (Tomlins et al., 2008) (Figure S2A). Interestingly, this enrichment was disrupted by ERG siRNA (Figure 2A and Figure S2B) and ChIP performed in ERG-overexpressing RWPE cells supporting a model in which ERG recruits PARP1 and DNA-PKcs to specific genomic loci during transcription (Figure 2B). Consistent with this hypothesis, serial ChIP reactions (ERG, then PARP1 or DNA-PKcs) demonstrated that an ERG:PARP1 complex and an ERG:DNA-PKcs complex are both present at ERG-regulated loci (Figure S2C). Although it was not possible to perform re-ChIP experiments with the PARP1 and DNA-PKcs antibodies, IP-Western blot analysis confirmed that PARP1 and DNA-PKcs interact in a DNA-independent manner in VCaP cells (Figure S2D). Likewise, this experiment suggests that DNA-PKcs binding to ERG does not disrupt the ERG:DNA interaction.
Figure 2. PARP1 and DNA-PKcs are required for ERG-regulated transcription.
(A) ChIP of PARP1 and the DNAPK complex shows an association with ERG-regulated targets including the PLA1A promoter as well as FKBP5 PSA and TMPRSS2 enhancers, but not the negative control gene KIAA0066. ChIPs were performed in VCaP cells treated with control or one of two independent ERG siRNAs for 48 hours prior to cross-linking.
(B) ChIP performed as in (A), but with stable RWPE-ERG or -LACZ cells.
(C) Data from gene expression arrays was analyzed by molecular concept mapping. The gene set analyzed is the set of genes that were greater than 2-fold differential in all three siRNA treatments relative to control. This gene set was used to determine the correlation of genes regulated by ERG, DNA-PKcs and PARP1 in VCaP cells with published microarray data. Node size is proportional to the number of genes in the set and edges represent statistically significant associations (p < 0.01). Arrow directionality represents gene sets either being induced or repressed.
(D) VCaP cells were treated with siRNA as indicated 48 hours prior to RNA isolation. qPCR was then run to confirm gene expression changes identified in the microarray experiment. Data is shown as a heat map with siRNA treatments along the x-axis and genes whose expression was analyzed by qPCR along the y-axis. Shades of green represent down-regulation of gene expression while shades of red represent up-regulation.
(E) VCaP cells were treated with either NU7026 or Olaparib for 48 hours as indicated and qPCR analysis of ERG-target genes identified from gene expression microarray experiment was performed.
(F) As in (D), except stable RWPE-ETV1 cells were used. All qPCR experiments were run three times in quadruplicate.
All bar graphs are shown with +/− SEM unless otherwise indicated. See also Figure S2 and Table S2.
To test whether DNA-PKcs and PARP1 are required for ERG-mediated transcriptional activation, we constructed a PLA1A promoter reporter. Transfection of the reporter into RWPE cells treated with either LACZ or ERG adenovirus and siRNA (Figure S2E) indicated that both DNA-PKcs (p = 1.99 × 10−3) and PARP1 (p = 2.37 × 10−3) are required for ERG-induced activation of PLA1A (Figure S2E) in RWPE cells. In contrast, inhibition of the related PI3/4-like kinase, ATM, had no significant effect on ERG activity.
While ATM and ATR repair DNA strand breaks through different pathways, DNA-PKcs is specifically required for NHEJ (Weterings and Chen, 2007). In this process, DNA-PKcs, Ku70 and Ku80 form a complex on the broken DNA end which facilitates DNA end processing and rejoining in a multi-step procedure that requires the XRCC4/DNA Ligase IV complex. In fact, XRCC4 and DNA Ligase IV are both independently required for execution of NHEJ in mammalian cells, as targeted inactivation of either gene leads to NHEJ defects in mouse cells (Barnes et al., 1998; Frank et al., 1998). Subsequently, we used siRNA to knockdown XRCC4 (Figure S2E) to evaluate the necessity of effective execution of NHEJ for ERG-induced transcriptional activation of the PLA1A promoter. Because knockdown of XRCC4 had no effect on ERG activity, the experiment further suggests a NHEJ-independent role for DNA-PKcs in ERG-mediated transcription (Figure S2E).
Given the importance of PARP1 and DNA-PKcs for ERG-mediated transcription, we sought to explore the global effects of inhibiting PARP1 and DNA-PKcs on the ERG-transcriptome. To do this, we used Agilent Whole Genome Oligo Expression Arrays to profile RNA from VCaP cells treated with either DNA-PKcs or PARP1 siRNA (knockdown confirmed in Figure S2I). Our analysis revealed 50 and 252 unique features that were greater than two-fold down- and upregulated, respectively, in both the PARP1 and DNA-PKcs siRNA-treated samples (Table S2). Venn diagram analysis was used to show the overlap of differential gene sets to genes regulated by ERG in VCaP cells (Tomlins et al., 2008) (p < 0.0001 for all interactions unless indicated, hypergeometric test) (Figure S2F, S2G). To then understand how this gene signature is related to existing signatures, we uploaded our expression signature into Oncomine Concepts Map (OCM) (Rhodes et al., 2007; Tomlins et al., 2007b) to identify human tissue gene signatures that are enriched for genes upregulated by DNA-PKcs and PARP1 siRNA in VCaP cells (genes repressed by PARP1 and DNA-PKcs). This provided unbiased validation that the tissue-based gene signatures most highly enriched with our gene set were the genes repressed in ETS-positive as compared to ETS-negative prostate cancer; Xiaojun et. al. (Tomlins et al., 2007b) [OR = 3.08, p =1.40 × 10−15], Yang et. al. (Tomlins et al., 2007b) [OR = 2.91, p =3.30 × 10−10] and Lapointe et. al. (Lapointe et al., 2004) [OR = 3.33, p =2.30 × 10−6] (Figure 2C). Interestingly, the gene signature also showed significant overlap with the set of genes repressed in metastatic as compared to localized prostate cancer suggesting that repression of these genes is important for prostate cancer progression; (Vanaja et al., 2003) [OR = 2.99, p =1.5 × 10−10], (LaTulippe et al., 2002) [OR = 3.31, p =1.50 × 10−6] (Figure 2C). Treatment of VCaP cells with siRNA confirmed gene expression changes as predicted by the gene expression arrays (Figure 2D) as did treatment with either the small molecule DNA-PKcs kinase inhibitor, NU7026, or the small molecule PARP1 inhibitor, Olaparib (Figure 2E). Analysis of siRNA treated RWPE-ETV1 cells (Figure S2J) confirmed that DNA-PKcs and PARP1 regulated ETV1 transcriptional activity as well (Figure 2F). Taken together these data suggest that PARP1 and DNA-PKcs play a role in modulating transcriptional activity of a number of ETS target genes, some of which are differentially expressed between localized and metastatic disease.
PARP1 and DNA-PKcs are required for ERG-mediated cell invasion, intravasation and metastasis
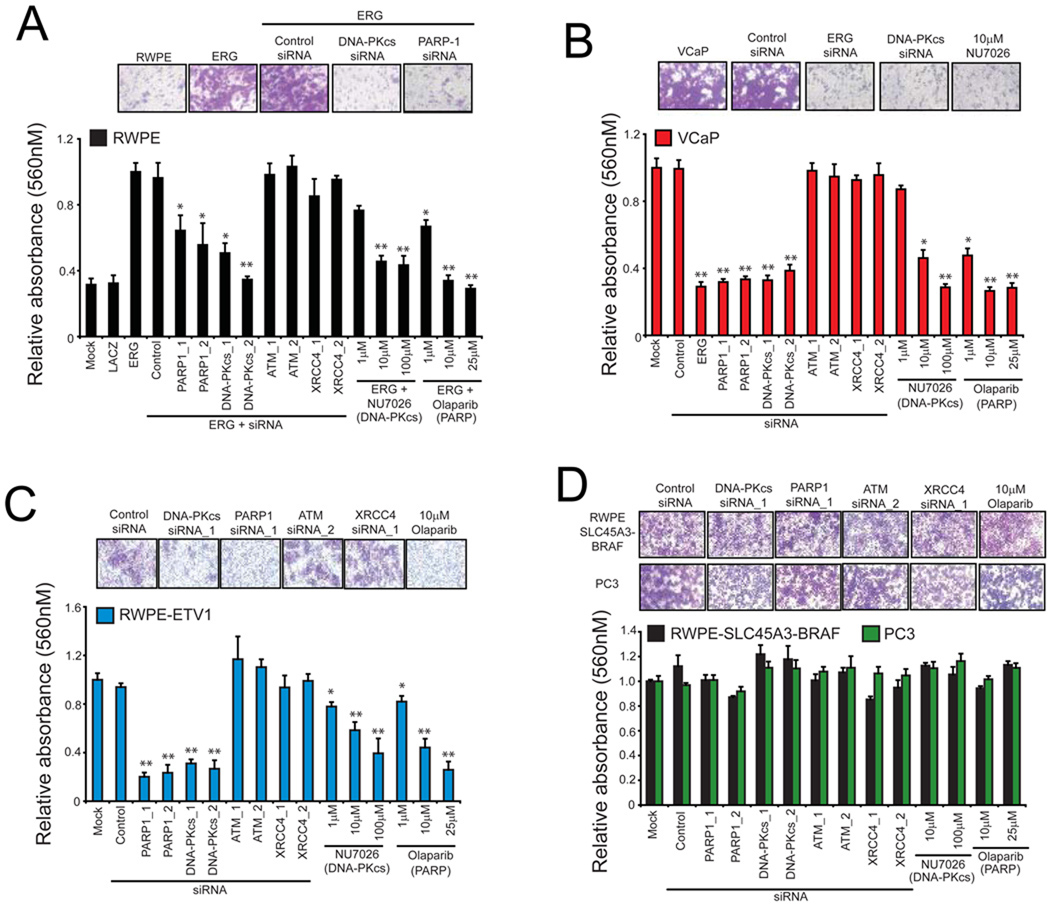
Inhibition of PARP1 and DNA-PKcs altered ERG-transcriptional activity of several progression-associated genes such as EZH2, here we tested the role of these enzymes in ERG-induced cell invasion. Both DNA-PKcs siRNA (Figure S2H and S2I) and NU7026 attenuated invasion in RWPE cells transduced with ERG (Figure 3A) and VCaP cells (Figure 3B) [p < 0.01 for DNA-PKcs siRNA or NU7026 > 10µM]. Likewise, we found that treatment with either PARP1 siRNA (Figure S2H and S2I) or Olaparib led to a significant reduction in ERG-driven RWPE and VCaP cells invasion (Figure 3A and 3B) [p < 0.05 for all PARP1 siRNA or Olaparib treatments]. As with our analysis of ERG-mediated transcription, knockdown of either ATM or XRCC4 did not have an effect on ERG-mediated invasion (Figure 3A and B). Treatment of stable RWPE cells stably overexpressing ETV1 (Tomlins et al., 2007a) with PARP1 or DNA-PKcs siRNA (Figure S2J) or small molecule inhibitors also led to a significant reduction in invasion (p < 0.01 for all PARP1 or DNA-PKcs treatments) (Figure 3C). Importantly, however, invasion of two negative control models, the ETS rearrangement-negative cell line PC3 and RWPE cells overexpressing an alternative prostate cancer gene fusion, SLC45A3-BRAF, was not affected by inhibiting either enzyme (Figure 3D and Figure S2K). To determine if the observed effect on cell invasion was due to cytotoxity, we performed chemosensitivity assays with both Olaparib and NU7026. Neither Olaparib nor NU7026 had an effect on the in vitro cell proliferation rate of any of the cell lines tested, suggesting that the reduction in cell invasion is not due to changes in cell proliferation (Figure S3A). In fact, the EC50 for both drugs was well beyond the dose shown to block transcription and invasion (Figure S3B and S3C).
Figure 3. ERG-mediated invasion requires engagement of PARP1 and DNA-PKcs.
(A) RWPE cells were infected with ERG adenovirus and treated with indicated siRNAs or different doses of the DNA-PKcs inhibitor, NU7026, or the small molecule PARP1 inhibitor, Olaparib, for 48 hours prior to plating cells in matrigel-coated Boyden chambers. After another 48 hours, cell invasion quantified.
(B) As in (A), except VCaP cells.
(C) As in (A), except stable RWPE cells transduced with ETV1 lentivirus.
(D) As in (A), except PC3 or RWPE-SLC45A3-BRAF cells.
Representative of three independent experiments.
Representative photomicrographs of invaded cells shown (lower Boyden chamber stained with crystal violet). For all experiments mean +/− SEM shown, * p < 0.05, ** p < 0.01. See also Figure S3.
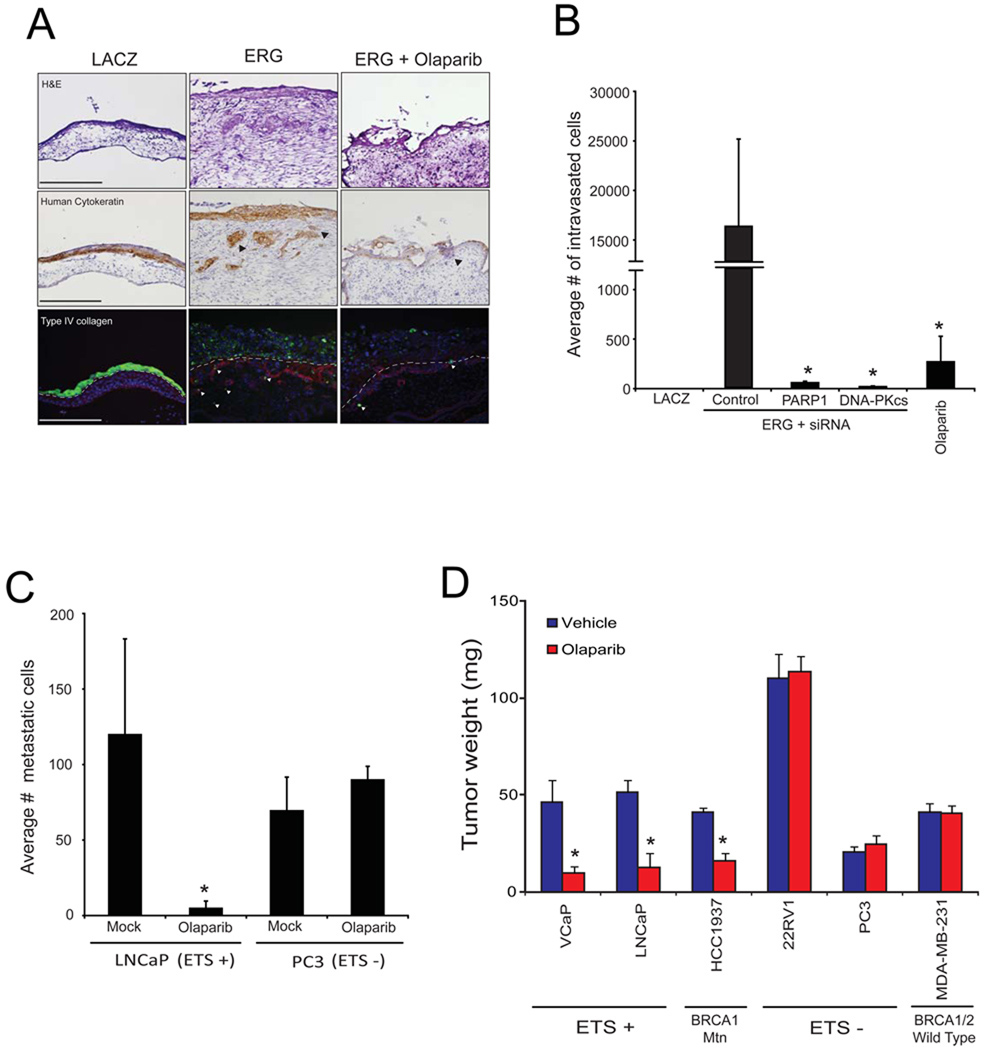
We next sought to define the role of PARP1 in ERG-mediated invasion and intravasation in vivo. To do this, we implanted cells onto the upper chorioallantoic membrane (CAM) of a fertilized chicken embryo and analyzed the relative number of cells that invade and intravasate into the vasculature of the lower CAM three days after implantation (Kim et al., 1998). In this assay, Olaparib blocked both ERG-mediated invasion and intravasation (p < 0.1) (Figure 4A and 4B). Because increased expression of EZH2 alone is sufficient to drive metastasis in several different cell systems, we monitored EZH2 mRNA expression and found that EZH2 expression was downregulated following either PARP1 or DNA-PKcs inhibition (Figure S4A), suggesting that mechanistically, PARP inhibition disrupts ERG-mediated invasion and intravasation by inhibiting ERG-mediated transcriptional activation of invasion-associated genes such as EZH2. More importantly, however, our models suggested that therapeutic disruption of either ERG-interacting enzymes (PARP1 or DNA-PKcs) inhibits the metastatic spread of prostate cancers harboring ETS gene fusions.
Figure 4. ERG-mediated invasion, intravasation and metastasis requires PARP1 activity in vivo.
(A) CAM invasion assay performed using stable RWPE-LACZ, or RWPE-ERG cells labeled with microspheres (green fluorescence emission) and treated with or without a single dose of Olaparib [40mg/kg] as indicated. Seventy two hours after implantation, the upper CAM was harvested. Frozen sections were created and stained for hematoxylin and eosin (top row), human-specific cytokeratin (immunohistochemistry, middle row) or chicken-specific Type IV collagen (red immunofluorescence, bottom row). Arrows indicate cells invaded through the upper CAM. Representative images are shown. Scale bars are 200µM.
(B) CAM intravasation assay performed using stable RWPE-ERG cells pre-treated with siRNA as indicated. Alternatively, RWPE-ERG cells were implanted and treated with a single dose of Olaparib immediately after implantation [40mg/kg]. Seventy two hours after implantation, the lower CAM was harvested. Total DNA was isolated from the lower CAM and qPCR was performed using human-specific ALU PCR primers. Total cell number was determined by comparing to a standard curve created using varying amounts of RWPE cells as input.
(C) Liver metastasis in chicken embryos was assessed 8 days following implantation of either LNCaP (ETV1 rearrangement) or PC3 (no ETS rearrangement) cells onto the upper CAM. Animals were injected every other day with Olaparib [40mg/kg] prior to harvesting chicken livers. Total cell number was then quantified by qPCR as in (B).
(D) ETS positive (VCaP and LNCaP), ETS negative (PC3 and 22RV1) prostate cancer cells as well as BRCA1 mutant (HCC1937) and BRCA1/2 WT (MDA-MB-231) breast cancer cells were implanted onto the upper CAM. These cell line xenografts were then treated with 40mg/kg Olaparib every other day for 8 days. Tumors (non-invaded cells remaining on the upper CAM) were collected and weighed. Average tumor weight is shown.
For all experiments mean +/− SEM shown, * p < 0.05, ** p < 0.01.
See also Figure S4.
To test this postulate, we analyzed the metastatic potential of an ETS positive (LNCaP) and an ETS negative (PC3) cell line. As shown in Figure 4C, Olaparib treatment blocked the formation of liver metastases from LNCaP (p = 0.01), but not PC3 cells. Importantly, we also noticed that over the extended treatment period the ETS-positive tumors were significantly smaller than the ETS-negative tumors (Figure 4D) with p < 0.05 for VCaP and p < 0.01 for LNCaP. This suggests that PARP1 could play a role in the long term maintenance of ETS-positive cancer cell survival. Because it appeared that the long term survival of ETS overexpressing tumors can be diminished by treatment with Olaparib, we sought to compare the magnitude of effect to that of a clinically validated model. Therefore, we xenografted HCC1937 (BRCA1 mutant) and MDA-MB-231 (BRCA1/2 WT), following Olaparib treatment a significant effect was observed on the BRCA1 mutant HCC1937 tumors, while no measurable effect was observed in MDA-MB-231 tumors. Surprisingly, the magnitude of effect observed in the HCC1937 cells was equivalent to the magnitude of effect observed in the two ETS-positive cell line xenografts (Figure 4D).
ETS gene fusion prostate tumors are preferentially susceptible to PARP1 inhibition in vivo
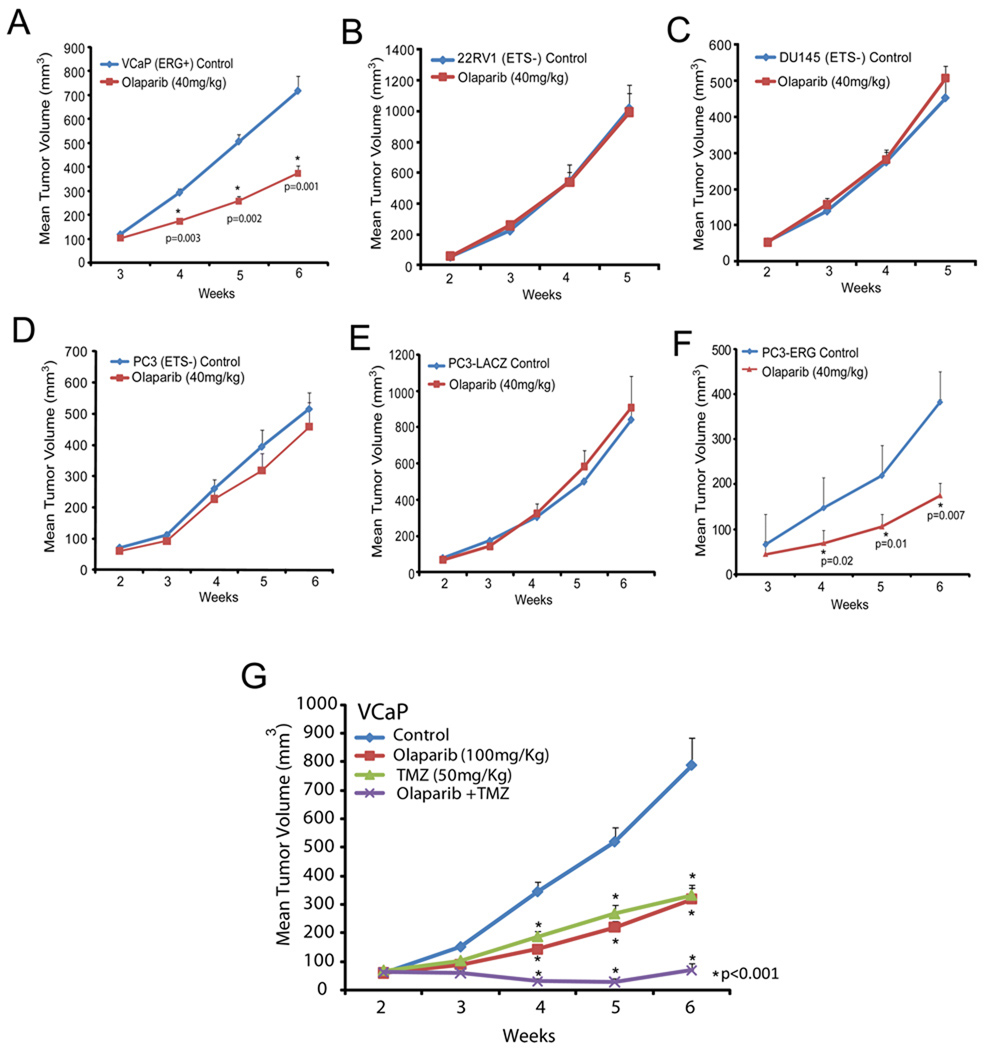
Based upon our in vivo data from the chicken CAM assay, we hypothesized that inhibition of PARP1 would inhibit ETS-positive prostate cancer growth in mouse xenograft models. Several PARP inhibitors have entered Phase I and Phase II clinical trials (Audeh et al., 2010; O'Shaughnessy et al., 2011; Tutt et al., 2010). One of these, Olaparib, was shown to be well tolerated in cancer patients with a minimal side effect profile (Fong et al., 2009). Thus, to first test our hypothesis, we implanted VCaP (ERG positive) or PC3-LACZ (ETS negative) cells and studied the impact of Olaparib (100mg/kg/day, IP) on xenograft growth. Importantly, we observed a significant reduction of tumor growth in the ETS-rearranged cell line relative to that of the vehicle control, but no change was observed in the ETS-negative control cell line (p = 0.002) suggesting preferential sensitivity of ETS-positive tumors (Figure S5A and S5B).
We then extended our experiment to analyze the effects of Olaparib on a panel of ETS-positive and ETS-negative prostate cancer cell lines including an isogenic model. Because this experiment intended to test the specificity of Olaparib-induced growth inhibtion for ERG-overexpressing prostate xenografts, we chose to use a dose similar to that used in previously published xenograft experiments (Rottenberg et al., 2008). Consistent with our hypothesis, this dose of Olaparib had a significant effect on VCaP cells (p = 0.001), but did not inhibit the growth of two additional ETS negative cell line xenografts (22RV1 or DU145) (Figure 5A, 5B and 5C). This experiment also demonstrated that the VCaP tumor growth response was dose-dependent. To then create an isogenic model, we overexpressed the primary TMPRSS2:ERG gene fusion product in the PC3 prostate cancer cell line (PC3-ERG). Western blotting confirmed protein overexpression and IP-Western confirmed the ERG:DNA-Pkcs and ERG:PARP1 interactions (Figure S5C and S5D). ChIP assays demonstrated that ERG binds to known target genes in PC3 cells (Figure S5E) and qPCR demonstrated ERG transcriptional activity (Figure S5F). Surprisingly, ERG overexpression led to a slightly reduced growth rate of PC3 cells relative to LACZ-overexpressing PC3 cells (Figure S5G). Consistent with the model that ETS-positive, but not ETS-negative prostate tumors are susceptible to PARP inhibition, overexpression of ERG was sufficient to significantly sensitize PC3 cells to PARP inhibition (p = 0.007) suggesting that ERG overexpression provides a selective mechanism for Olaparib-mediated growth inhibition (Figure 5D, 5E and 5F). Western blot and qPCR analysis of flash frozen, staged PC3-ERG tumors treated with or without drug for 4 hours confirmed inhibition of PARP1 activity and loss of ERG-target gene expression after treatment with Olaparib (Figure S5H–K). In line with the clinical observation that Olaparib is well tolerated at doses capable of inhibiting PARP activity (Fong et al., 2009), Olaparib treatments in our xenograft models did not lead to a significant decrease in total body weight and did not lead to liver toxicity as assessed by serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) (Figure S5L).
Figure 5. Inhibition of PARP1 alters ETS positive, but not ETS negative cell line xenograft growth.
(A–F) Specificity screen for ETS positive and ETS negative tumor cell line xenografts. Cell lines were injected subcutaneously and grown until tumors were palpable. Xenografted mice then received I.P. injections of Olaparib 40mg/kg as indicated 5 days/week. Caliper measurements were taken weekly. ETS positive cell line xenografts were (A) VCaP (ERG rearrangement) and (F) PC3-ERG cells and the ETS negative xenografts are (B) 22RV1, (C) DU145 and (D, E) PC3-Control/-LACZ, respectively.
(G) Mice xenografted with VCaP cells were treated as in as in (A) except with 100mg/kg Olaparib and/or 50 mg/kg Temozolomide (TMZ) as indicated. Olaparib was administered I.P. five days per week. TMZ was administered in two 5 day cycles with the first occurring during week 3 and the second occurring during week 5.
For all experiments mean +/− SEM shown, p < 0.05, ** p < 0.01 unless indicated.
See also Figure S5.
Given the specific effect of Olaparib on ERG-positive cell line xenograft growth (Figure S5M), we extended our analysis with the use of a model of primary human prostate tumors maintained in serial xenografts (Li et al., 2008). We identified one ERG-positive (MDA-PCa-133), one ETV1-positive (MDA-PCa-2b–T) (FISH-confirmed (Tomlins et al., 2007a)) and one ETS-negative (MDA-PCa-118b) model for this experiment by assessing relative levels of ETS gene expression by qPCR (Figure S5N). Functional ETS gene status was assessed by testing the relative expression of several ETS target genes including EZH2, DNMT3a, ZNF100, PBX1, ZNF618, PLA1A and PLAT, between the models (Figure S5O). As shown in Figure S5P and S5Q, low dose Olaparib altered the growth of both the ERG and ETV1 overexpressing primary human prostate cancer xenografts [MDA-PCa-133: p = 0.05 (day 8) and p < 0.01 (day 12, 16, 20, 24, 28 and 32); MDA-PCa-2b–T: p < 0.01 (day 8, 12 and 16)], but had no effect on the ETS-negative primary human xenograft model (Figure S5R). In all cases, Olaparib did not have an observable effect on total body weight (Figure S5S–S5AA).
Because we were able to validate the hypothesis that Olaparib specifically alters the growth of either ETS overexpressing or BRCA1/2 deficient cell lines, we sought to extend the treatment regimen to identify combination therapies that enhance the magnitude of inhibition without causing significant toxicity. Recently, an alkylating agent called temozolomide (TMZ) has been shown to potentiate the effects of other PARP inhibitors in several cancer xenograft models (Donawho et al., 2007; Liu et al., 2008; Palma et al., 2009) as well as caused a complete or partial response in some patients enrolled in a Phase II trial for metastatic breast cancer (Isakoff S J, 2010). As expected, the combination treatment resulted in a significant growth reduction of VCaP tumors that was maintained over the entire 6 weeks (p < 0.001 for all combination treatments) (Figure 5G). Even with the combination therapy, at this dose range, no overt toxicity such as excessive weight loss was observed (Figure S5AB). This suggested that the addition of PARP inhibitor therapy to existing chemotherapeutic regimens will help enhance the overall effect for ETS positive tumors.
ETS transcription factors drive DNA double strand break formation
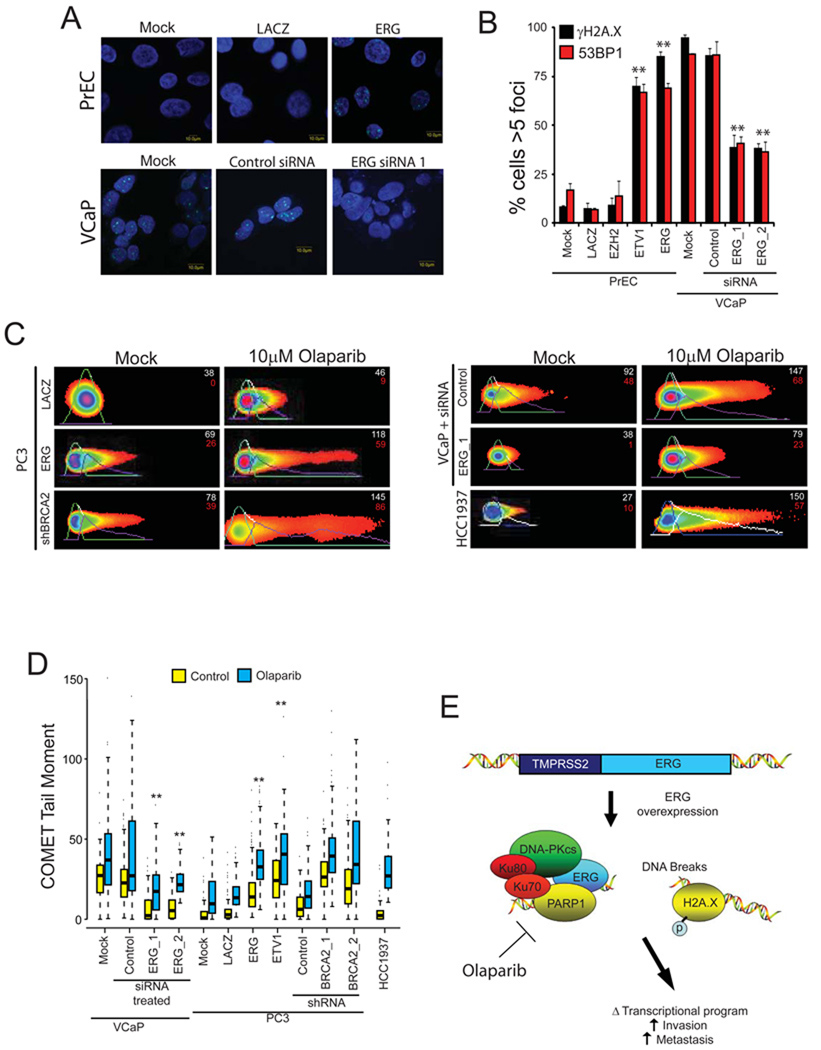
To explore potential mechanisms of ETS-specific therapeutic response to these inhibitors of DNA repair, we assessed total levels of DNA double strand breaks in vitro. We hypothesized that constitutive overexpression of ERG may lead to an increased susceptibility to DNA damage. Thus, we first assessed the total levels of a histone mark of DNA double strand breaks called γ-H2A.X in Olaparib-treated versus untreated VCaP cells. Surprisingly, the untreated cells had a high level of γ-H2A.X foci (Figure 6A) leading us to test the hypothesis that overexpression of ETS genes induces DNA double strand breaks. Overexpression of ETS genes in primary prostate epithelial (PrEC) cells induced >5 γ-H2A.X foci in greater than 75% of the analyzed cells while the control genes LACZ and EZH2 had no effect (Figure 6A–B). Quantitative PCR confirmed overexpression (Figure S6A–C). Likewise, other ETS genes were also capable of inducing γ-H2A.X foci in several different prostate cell lines (Figure S6D–E). To then confirm that ERG induces γ-H2A.X in an endogenous setting, we depleted ERG from VCaP cells by RNA interference (Figure S6I) and found significantly decreased γ-H2A.X foci, p = 7.16 × 10−3 and p = 1.36 × 10−3 for two independent siRNAs, respectively (Figure 6A–B). While γ-H2A.X foci represent an early mark of DNA damage recognition, 53BP1 is present only in the later stages of repair (Bennett and Harper, 2008). As such, in the presence of a DNA damage response, we expected to observe an increase in 53BP1 foci formation. Indeed, the ETS genes also induced 53BP1 foci formation (Figure 6A–B and Figure S6D).
Figure 6. ETS transcription factors induce DNA damage which is potentiated by PARP inhibition.
(A) γ-H2A.X immunofluorescence staining shows that ERG induces the formation of γ-H2A.X foci. Top row, benign prostate epithelial cells(PrEC) were infected with lentiviruses expressing LACZ or ERG. Bottom row, VCaP cells treated with control siRNA or ERG siRNA.
(B) Quantification of γ-H2A.X and 53BP1 immunofluorescence staining in PrEC or VCaP cells. For all experiments mean +/− SEM shown, * p < 0.05, ** p < 0.01.
(C) ETS overexpression or BRCA2 knockdown (with shRNA) induces DNA damage as assessed by neutral COMET assay in VCaP cells. Cells were treated with or without 10µM Olaparib for 48 hours. Cells with DNA damage have an extended “tail moment” of fragmented DNA shown in red. Relative tail length is shown in white. Representative images showing quantification of head and tail height, length and fluorescence intensity are shown (as indicated).
(D) Quantification of average COMET tail moments following treatment as noted in the box plot. Statistical tests were performed using the two-way ANOVA test (described in S.O.M.) to determine if the increase in DNA damage in Olaparib-treated ETS overexpressing cells (PC3-ERG, PC3-ETV1 and VCaP) was statistically greater than the increase observed in Olaparib-treated control cells with low ETS expression (PrEC-LACZ, PC3-LACZ or VCaP treated with ERG siRNA) as indicated in the text. Similar statistical tests were used to compare the increase in BRCA2 shRNA expressing cells to PC3 cells transduced with control shRNA ** p < 0.01
(E) Proposed model to therapeutically target ETS gene fusions via their interacting enzyme, PARP1.
All bar graphs are shown with +/− SEM unless otherwise indicated.
See also Figure S6.
After demonstrating that ETS gene overexpression drives the accumulation of markers of DNA double strand breaks, we sought to confirm the presence of DNA double strand breaks by directly analyzing cellular DNA for fragmentation using the COMET assay. As with the γ-H2A.X and 53BP1 foci formation assays, in PC3 cells, ERG or ETV1 overexpression was sufficient to induce significantly longer and brighter tails than those observed in controls (p < 0.01 for both ETS genes) and treatment with either ERG siRNA led to a reduction in relative level of DNA double strand breaks (p < 0.01) (Figure 6C–D).
Olaparib potentiates ETS-induced DNA damage
After finding that aberrantly expressed ETS transcription factors drive the accumulation of DNA double strand breaks, we hypothesized that by having a baseline level of DNA damage, ETS positive cancers may be specifically susceptible to accumulating DNA damage following inhibition of the interacting DNA repair enzyme PARP1. To test this hypothesis, we analyzed VCaP cells treated with Olaparib for 48 hours. Olaparib-treated VCaP cells had a very high level of γ-H2A.X foci (Figure S6F). Importantly, by depleting endogenous ERG using either of two independent siRNAs (confirmed in Figure S6I), we were able to reverse the gross increase in γ-H2A.X. Similar increases in foci were observed in PC3-ERG cells or PC3 cells with BRCA2 knockdown, but not in the control cells (Figure S6F). Knockdown efficiency was confirmed by qPCR (Figure S6G). Quantification of the relative levels of DNA double strand breaks demonstrated that, while there was an increase in the tail moment of all Olaparib-treated cell lines, Olaparib caused a significantly greater increase in the tail moments of ERG-positive cells than controls (p < 0.001, two-way ANOVA) (Figure 6C–D). In conjunction with this observation, both ERG siRNAs led to a significant reduction in DNA damage following Olaparib and blocked the synergistic increase of DNA damage observed with Olaparib (p = 0.001 for ERG siRNA_1 and p < 0.001 for ERG siRNA_2, two-way ANOVA). Consistent with this observation, the BRCA1 mutant HCC1937 breast cells and PC3 cells with BRCA2 shRNA underwent a significant increase in the total levels of DNA damage when treated with Olaparib. Taken together, this data demonstrates that similar to BRCA1/2-deficent tumors, ETS-positive, but not ETS-negative, prostate cancer models are susceptible to PARP inhibition through the increased incidence of DNA double strand breaks (Figure 6E).
To discriminate between mechanisms of ERG-potentiated DNA damage, we performed the COMET assays after 0.5, 1, 24 and 48 hours exposure to Olaparib. Surprisingly, the potentiated DNA damage was observed in PC3-ERG cells relative to PC3-LACZ cells as early as 30 minutes after treatment (Figure S6H) (p = 0.002 at 30 minutes, two-way ANOVA). This suggested that the mechanism of ERG-induced potentiation is independent of the genes regulated by PARP1-mediated transcriptional activation. Focused expression analysis of genes involved in DNA damage repair pathways demonstrated that no significant change in any of repair genes analyzed, suggesting that the DNA damage phenomena is independent of changes to ERG-regulated gene expression (Figure S6I). To analyze the role of repair pathways directly, we tested the postulate that downregulation of a protein critical for the execution of non-homologous end joining (NHEJ) pathway such as XRCC4 would lead to a synergistic induction of DNA damage in a homologous recombination (HR) deficient cell. Treatment of HR-deficient HCC1937 cells with siRNA confirmed a greater increase in DNA damage following XRCC4 knockdown (NHEJ) than by XRCC3 knockdown (HR) (p < 0.05, one-way ANOVA). In contrast, the synergistic induction of DNA damage following XRCC4 or XRCC3 knockdown was not observed in PC3-ERG cells as compared to PC3-LACZ cells (Figure S6J and S6K). This suggested that ERG overexpression does not completely block either of these double strand break repair pathways. This was further confirmed by HR-efficiency assays which demonstrated that HR is not significantly altered by ERG overexpression (Figure S6L).
DISCUSSION
In this study, we discovered that the ETS gene fusion product, ERG, physically interacts with the enzymes PARP1 and DNA-PKcs. Both PARP1 and DNA-PKcs are required for ERG-mediated transcription and cell invasion, suggesting that both of these co-factors are necessary for ERG-mediated prostate cancer progression. Moreover, therapeutic inhibition of PARP1 preferentially sensitized ETS-overexpressing prostate cancer xenografts compared to ETS negative xenografts. Thus, similar to the successful paradigm of targeting the BCR-ABL gene fusion in CML with the small molecule kinase inhibitor imatinib meyslate (Druker et al., 2001), one could envision targeting the ETS-PARP1 axis in prostate cancer and possibly other ETS gene fusion-dependent cancers. While directly inhibiting transcription factors, such as ERG, may be difficult, blocking the function of regulatory co-factors, such as PARP1, is more feasible, and may represent a viable treatment paradigm in cancer therapy.
In particular, PARP1 represents a very promising therapeutic target. Based on its role in base excision repair, PARP1 has been explored in both preclinical and clinical settings as a target in tumors with deficiencies in double-stranded DNA repair, such as mutations in BRCA1 and BRCA2 (Bryant et al., 2005; Farmer et al., 2005). In these cancers, the inhibition of PARP1 in cells with an inherent defect in homologous repair results in stalled replication forks and subsequent cell death (Bryant et al., 2005; Farmer et al., 2005). An initial Phase I trial of the PARP inhibitor Olaparib has suggested an excellent therapeutic response in patients with BRCA1/2 deficient tumors from multiple organ sites with most patients experiencing a large reduction in total tumor volume (Fong et al., 2009). However, most cancers do not harbor BRCA1/2 mutations; only 5–6% of all breast cancers are associated with an inherited BRCA1/2 gene mutation (Malone et al., 1998) and only 3% of prostate tumors from an Ashkenazi Jewish population of 832 patients were BRCA1/2-deficient (Gallagher et al., 2010).
While PARP inhibitors can exploit the DNA repair defects of BRCA deficient tumors to induce cell death, we now demonstrate that they can also inhibit ETS gene fusion protein activity by preventing ETS transcriptional activity, inhibiting ETS-associated invasion, and enhancing ETS-mediated DNA damage. Future studies will help determine if, as with AR-mediated transcription (Haffner et al., 2010), ETS-mediated transcription is directly coupled the induction of DNA damage. Importantly, the potentiation of ETS-induced DNA damage by PARP inhibition is of particular clinical interest; analogous to the “synthetic lethality” resulting from PARP inhibition in BRCA1/2-deficient tumors. By suggesting that cancers driven by specific oncogenic transcription factors may respond to PARP inhibition independent of BRCA1/2 status, our data supports the notion that multiple tumor subtypes will be susceptible to PARP pharmacotherapy. It is important to note that the company Sanofi-BiPAR recently released a press report that their phase III trial assessing the addition of their next generation PARP inhibitor, Iniparib, to a gemcitabine-carboplatin regimen for patients with metastatic triple-negative breast cancer, was negative for an overall survival benefit (http://sanofi-aventis.mediaroom.com/index.php?s=43&item=310). This is in direct contrast to the recently reported phase II trial showing that the addition of Iniparib approximately doubled overall survival in this setting (O'Shaughnessy et al., 2011), and some questions about specificity have been raised (Carey and Sharpless, 2011). Nonetheless, these results highlight the importance of target selection; it is expected that ongoing phase III trials assessing chemotherapy with or without PARP inhibitor in BRCA-mutant cancers (instead of a non-specific triple-negative breast cancer population) will be positive, because the patient population is selected based on the presence of the PARP inhibitor target – BRCA mutation (Ellisen, 2011). Here, we have shown that Olaparib very specifically, and in a dose dependent manner, delays tumor growth of ETS positive, but not ETS negative prostate cancer xenografts.
By exploiting the ETS:PARP1 interaction to selectively target ETS-overexpressing xenografts, our studies significantly expand the total population of patients who could benefit from PARP inhibition. Consequently, the data presented here also have implications on the design of subsequent clinical trials which will follow the recently reported Phase I trial of Olaparib (Fong et al., 2009). While most trials will undoubtedly be designed to target and subtype BRCA-deficient tumors, trials designed in organ sites that are also known to harbor aberrantly expressed ETS genes, such as breast, melanoma, Ewing’s sarcoma and especially prostate, should also subtype the disease by ETS status. Based on the data presented here, ETS-positive tumors are expected to respond with a higher probability to PARP inhibition than ETS-negative tumors, potentially making ETS status an important predictive biomarker. In line with the observation that PARP inhibitors can significantly increase the mean overall survival of patients with triple negative breast cancer when added onto an existing regimen, our data suggests that the best design for a clinical trial in hormone-refractory metastatic prostate cancer will be to add PARP inhibitors in combination with chemotherapeutics known to potentiate the effects of PARP inhibition such as temozolomide.
Finally, the observation that gene fusions which drive the gross overexpression of ETS genes also induce DNA double strand break formation provides additional mechanistic insight into how ETS gene fusions drive cancer progression. Specifically, by causing DNA double-strand breaks, ETS gene overexpression may also play a role in the gradual evolution of genomic rearrangements. This finding may explain why recurrent ETS gene fusions may have been difficult to discover, because they simply lead to the accumulation of additional gene fusions only some of which will drive disease progression. In fact, this model may partially explain the clinical behavior of prostate cancers which lie dormant for years only to spontaneously become extremely aggressive.
Experimental Procedures
Xenograft and Primary Human Xenograft Models
For human prostate cancer xenografts, written informed consent was obtained before sample acquisition and the sample was processed according to a protocol approved by The University of Texas, MD Anderson Cancer Center institutional review board. All procedures were approved by the University of Michigan’s University Committee on Use and Care of Animals (UCUCA).
Accession Number
Expression profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Santa Clara, CA) according to the manufacturer's protocol and described previously (Tomlins et al., 2007b). Data accession number: GSE27723.
Significance
While genomic rearrangements leading to ETS gene overexpression occur in about 50% of prostate cancers, transcription factors like the ETS genes have been notoriously difficult to target therapeutically. Here, we show that the ETS:PARP1 interaction axis may represent a target for therapeutic intervention in cancers with ETS gene fusions. Our study also suggests that inhibition of co-factors necessary for function may represent a promising paradigm of treatment for malignancies driven by oncogenic transcription factors. Furthermore, our study motivates the assessment of ETS gene fusions as a potential biomarker of response in future clinical trials incorporating PARP inhibitors into the treatment of prostate cancer and other ETS fusion-positive malignancies.
Supplementary Material
Acknowledgements
We thank F. Alt, D. Ferguson, T. Wilson, R. Mani and J. R. Prensner for discussions of the data, K.Suleman, A. Ayuningtyas, J. Granger, G. Hao, X. Jiang, X. Jing, B. Laxman, B. Russo and A. Fullen for their technical support and A. Kim, K. Giles for critically reading the manuscript. We would also like to thank S.J. Weiss for assistance in performing CAM assays. A.M.C. is an American Cancer Society Research Professor. A.M.C. is supported by a Burroughs Welcome Foundation Award in Clinical Translational Research and the Doris Duke Charitable Foundation Clinical Scientist Award. B.A. is supported by Genentech Foundation Postdoctoral Fellowship and Young Investigator Award from Expedition Inspiration. F.Y.F. is supported by a Physician Research Training Award from the Department of Defense (PC094231).This work was supported in part by the National Institutes of Health (R01CA132874 to A.M.C.), Prostate SPORE P50CA69568 to K.J.P. and A.M.C., and the Early Detection Research Network (UO1 CA113913 to A.M.C.) and the Department of Defense (PC051081 to A.M.C.). We also received support from the Prostate Cancer Foundation (F.Y.F., C.R.M., S.A.T., N.M.N., K.J.P. and A.M.C.).
A. Chinnaiyan serves on the advisory board of Gen-Probe; and is a co-inventor on a patent filed by the University of Michigan covering the diagnostic and therapeutic field of use for ETS fusions in prostate cancer. The diagnostic field of use has been licensed to Gen-Probe, Inc. Gen-Probe did not play a role in the design and conduct of this study, in the collection, analysis, or interpretation of the data, or in the preparation, review, or approval of the article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.C.B., F.Y.F. and A.M.C. designed the overall project and directed the experimental studies. B.A., N.M.N., and K.J.P. coordinated the xenograft/primary prostate cancer xenograft experiments. J.S., W.Y. and R.M. developed tissue microarrays and carried out pathology evaluation. A.K.Y., Y.L., V.B., J.C.B., K.S.J.E., and S.V. carried out proteomics analysis. I.A.A. carried out and analyzed CAM assays. R.J.L. and C.A.M. performed statistical analysis. J.C.B., B.A., Y.L., S.P., H.L., Q.C., J.Y., N.P., X.W., X.C., A.S. J.Y., and S.A.T., performed experimental studies. J.C.B, B.A., M.H., K.J.P., F.Y.F. and A.M.C. interpreted the data. J.C.B, S.A.T., F.Y.F. and A.M.C. wrote the paper.
References
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nature structural & molecular biology. 2008;15:20–22. doi: 10.1038/nsmb0108-20. [DOI] [PubMed] [Google Scholar]
- Brenner JC, Chinnaiyan AM. Translocations in epithelial cancers. Biochimica et biophysica acta 1796. 2009:201–215. doi: 10.1016/j.bbcan.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Carey LA, Sharpless NE. PARP and cancer--if it's broke, don't fix it. The New England journal of medicine. 2011;364:277–279. doi: 10.1056/NEJMe1012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nature genetics. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE., Jr Transcription factors as targets for cancer therapy. Nature reviews. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. The New England journal of medicine. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Ellisen LW. PARP Inhibitors in Cancer Therapy: Promise, Progress and Puzzles. Cancer cell. 2011;19:165–167. doi: 10.1016/j.ccr.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Gallagher DJ, Gaudet MM, Pal P, Kirchhoff T, Balistreri L, Vora K, Bhatia J, Stadler Z, Fine SW, Reuter V, et al. Germline BRCA Mutations Denote a Clinicopathologic Subset of Prostate Cancer. Clin Cancer Res. 2010;16:2115–2121. doi: 10.1158/1078-0432.CCR-09-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Molecular cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al. Androgen-induced TOP2B–mediated double-strand breaks and prostate cancer gene rearrangements. Nature genetics. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer research. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- Hermans KG, van der Korput HA, van Marion R, van de Wijngaart DJ, Ziel-van der Made A, Dits NF, Boormans JL, van der Kwast TH, van Dekken H, Bangma CH, et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer research. 2008;68:7541–7549. doi: 10.1158/0008-5472.CAN-07-5930. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, OB, Tung NM, Gelman RS, Giranda VL, Bernhard KM, Habin KR, Ellisen LW, Winer EP, Goss PE. A phase II trial of the PARP inhibitor veliparib (ABT888) and temozolomide for metastatic breast cancer. Journal of Clinical Oncology; Paper presented at: American Society of Clinical Oncology; Chicago, IL. 2010. [Google Scholar]
- Jane-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, Baker AC, Nazarian RM, Vijayendran KG, Sellers WR, et al. An oncogenic role for ETV1 in melanoma. Cancer research. 70:2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, Baker AC, Nazarian RM, Vijayendran KG, Sellers WR, et al. An oncogenic role for ETV1 in melanoma. Cancer research. 2010;70:2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon IS, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- Kim J, Yu W, Kovalski K, Ossowski L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell. 1998;94:353–362. doi: 10.1016/s0092-8674(00)81478-6. [DOI] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nature genetics. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nature reviews. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer research. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, Sikes C, Multani AS, Efstathiou E, Lopez A, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. The Journal of clinical investigation. 2008;118:2697–2710. doi: 10.1172/JCI33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Shi Y, Guan R, Donawho C, Luo Y, Palma J, Zhu GD, Johnson EF, Rodriguez LE, Ghoreishi-Haack N, et al. Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol Cancer Res. 2008;6:1621–1629. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- Malone KE, Daling JR, Thompson JD, O'Brien CA, Francisco LV, Ostrander EA. BRCA1 mutations and breast cancer in the general population: analyses in women before age 35 years and in women before age 45 years with first-degree family history. Jama. 1998;279:922–929. doi: 10.1001/jama.279.12.922. [DOI] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science (New York, NY. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. The New England journal of medicine. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- Palma JP, Wang YC, Rodriguez LE, Montgomery D, Ellis PA, Bukofzer G, Niquette A, Liu X, Shi Y, Lasko L, et al. ABT-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res. 2009;15:7277–7290. doi: 10.1158/1078-0432.CCR-09-1245. [DOI] [PubMed] [Google Scholar]
- Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia (New York, NY. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. The American journal of surgical pathology. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, Barrette TR, Ghosh D, Varambally S, Chinnaiyan AM. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia (New York, NY. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M, Zevenhoven J, Lau A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff SA, Buijs A, Behm FG, Rubnitz JE, Raimondi SC, Hancock ML, Chan GC, Pui CH, Grosveld G, Downing JR. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nature genetics. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Molecular cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007a;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia (New York, NY. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007b;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer research. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (New York, NY. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer research. 2003;63:3877–3882. [PubMed] [Google Scholar]
- Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer research. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? The Journal of cell biology. 2007;179:183–186. doi: 10.1083/jcb.200705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.