Abstract
Functional regulation of ligand-activated receptors is driven by alterations in the conformational dynamics of the protein upon ligand binding. Differential hydrogen/deuterium exchange (HDX) coupled with mass spectrometry has emerged as a rapid and sensitive approach for characterization of perturbations in conformational dynamics of proteins following ligand binding. While this technique is sensitive to detecting ligand interactions and alterations in receptor dynamics, it also can provide important mechanistic insights into ligand regulation. For example, HDX has been used to determine a novel mechanism of ligand activation of the nuclear receptor peroxisome proliferator activated receptor-γ, perform detailed analyses of binding modes of ligands within the ligand-binding pocket of two estrogen receptor isoforms, providing insight into selectivity, and helped classify different types of estrogen receptor-α ligands by correlating their pharmacology with the way they interact with the receptor based solely on hierarchical clustering of receptor HDX signatures. Beyond small-molecule–receptor interactions, this technique has also been applied to study protein–protein complexes, such as mapping antibody–antigen interactions. In this article, we summarize the current state of the differential HDX approaches and the future outlook. We summarize how HDX analysis of protein–ligand interactions has had an impact on biology and drug discovery.
Keywords: GPCR, HDX, ligand, MS, nuclear receptor, screening
Many pharmacological drugs target proteins that are receptors for small-molecule ligands. These ligands bind to and alter the structure of the protein target, resulting in modulation of its function. Protein targets of high interest to biology and the pharmaceutical industry include the nuclear receptor (NR) superfamily of ligand-dependent transcription factors, the G protein-coupled receptor (GPCR) family of seven-transmembrane receptors, and protein kinases, members of a large protein family that comprise the kinome. In addition to being receptors for small-molecule ligands, proteins can function directly as drugs such as monoclonal anti bodies or soluble decoy receptors, which target and bind to a host protein to neutralize its function or to deliver a warhead. In all cases, modulation of the structure and dynamics of the protein or protein target impacts its function and provides an opportunity for pharma ceutical intervention. Methods for characterizing the structure and dynamics of protein–ligand and protein–target complexes are therefore of considerable interest. Atomic structures solved with x-ray crystallography have played a key role in understanding protein structure–function relationships and the technology has had a profound impact on biology and the drug discovery process [1]. The advantages of x-ray crystallography are undisputed; however, atomic structures do not readily reveal information pertaining to the dynamics of the protein, although limited information may be obtained [2]. Long-range allosteric communication across protein–protein (or domain–domain) interaction surfaces can also be difficult to detect or infer from static crystal structures. Unfortunately, some proteins or protein states (i.e., the unliganded or apo-state) can be resistant to crystal formation.
Over the last two decades, solution phase amide hydrogen/deuterium exchange (HDX) coupled with mass spectrometry (MS) has become a valuable complement to x-ray crystallography in the determination of protein structure and dynamics. In this article, we describe the current state-of-the-art HDX MS methodology employed to study protein–ligand interactions and we highlight several examples of how the approach has been applied to the characterization of protein–ligand interactions that are critical to our understanding of biology as well as being of high interest to the pharmaceutical industry. It is important to note that many reviews have been published on the topic of HDX MS [3-6]. Many of these reviews have described the broad applications of the technology and compared various experimental approaches. This article, however, focuses specifically on the application of differential HDX to probe ligand interactions with their target proteins (where the ligand is either a small molecule or another protein).
Hydrogen/deuterium exchange MS
Amide hydrogens exchange with solvent hydrogen or deuterium through acid-, base- and water-catalyzed reactions. Measuring the rate of exchange for these amides provides detailed information about their surrounding environment [3]. Englander et al.’s intensive nuclear magnetic resonance (NMR) studies accumulated enough data to predict ‘intrinsic’ amide hydrogen exchange rates for small peptides of a given sequence when exposed to heavy water at a specific pH and temperature, assuming the peptide adopts a random coil conformation [7]. At pH 7.0 and room temperature, any amide hydrogen in a random coil conformation will exchange on the order of milliseconds to seconds. This same amide hydrogen within a folded protein may exhibit a decrease in exchange rate as great as 108. This slowing of amide hydrogen exchange rate is called the ‘protection factor’.
In a protein, amide hydrogen exchange rates are influenced by hydrogen bonding and, as such, measurement of this physical property is an excellent method to probe changes in protein structure and dynamics. However, in order to measure amide exchange rates, several factors need to be considered. First, amide hydrogen exchange is sensitive to pH and temperature. At room temperature, a minimum exchange rate is observed around pH 2.7 with an increase of an order of magnitude per pH unit away from the exchange rate minima. Second, temperature can reduce or increase the exchange rate by a factor of 10 per 22°C. Thus, precise control of experimental pH and temperature are therefore critical in experimental design.
Following exposure of the protein target to deuterium (on-exchange), two approaches can be employed to obtain localized HDX data: the ‘bottom-up’ approach of digesting the target protein with a protease, or the ‘top-down’ approach employing fragmentation technology such as electron-capture dissociation (ECD) or electron-transfer dissociation (ETD) that avoids randomization of deuterons within the protein in the gas phase. The ‘top-down’ approach is discussed in more detail in the ‘Emerging technology: instrumentation developments’ section later. In the ‘bottom-up’ approach, spatial resolution is obtained from the digestion of the intact deuterated protein following on-exchange into smaller proteolytic fragment peptides. Since the on-exchange precedes the digestion step, the measured extent of HDX in each peptide reflects the average rate of incorporation of deuterium across all amides within that sequence of the intact protein. The protein is usually digested with low specificity acid-stable proteases, such as pepsin, to generate a collection of small peptic peptides, many of which may overlap in sequence. In the absence of overlapping peptides, the spatial resolution of the HDX measurement is limited by the size of each proteolytic peptide minus the two amino terminal residues of each peptide (the new amino terminus of the proteolytic peptide is no longer an amide and the exchange behavior of the first amide within the new peptide is influenced by the presence of the free amino terminus). With overlapping peptides, sublocalization of amide HDX kinetics can be obtained via subtractive analysis [8,9]. For overlapping peptide regions that share the same start or end position, the accepted approach is to assume that the nonoverlapping segment would contain the difference between the two measured percent deuterium (%D) values. This approach makes it possible to obtain more localized HDX data, sometimes resolved to a single amide, and recently software developments have automated this process (software developments to support HDX studies are discussed in detail later) [10]. The assignment of amino acid sequence to specific proteolytic peptides can be performed manually, but is typically assisted by use of database correlation algorithms such as Mascot or SEQUEST.
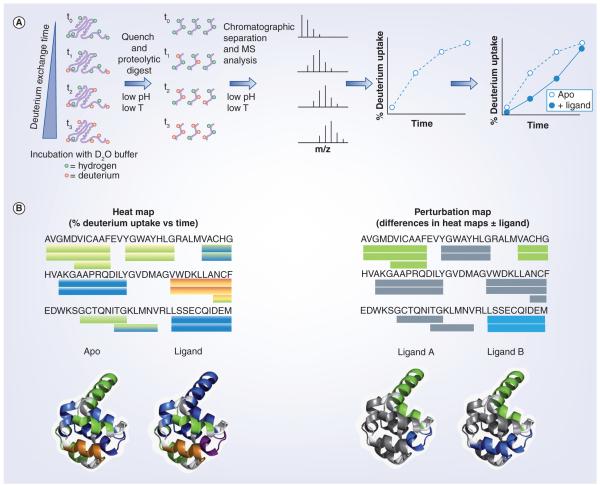
The increase in mass of a protein following solution phase amide hydrogen exchange with deuterium places MS as the ideal technique for measuring the rate of exchange over time [3,11,12]. Figure 1 shows a common workflow for the HDX MS experiment. Following each period of on-exchange, the reaction must be stopped or quenched to accurately preserve the level of deuterium incorporation until the digestion and mass spectrometric analysis can be completed. It is important that any further on-exchange and D/H back-exchange (loss of deuterium) with protic solvent be minimized. Even with a reduction of pH to 2.7 and temperature to 1°C, the experiment requires a sub-20 min time period in which the digestion, chromatographic separation and mass analysis must take place to avoid significant loss of the deuterium label. These experimental constraints prohibit the use of traditional proteomics methods that utilize off-line trypsin digestion and long shallow or multistage gradient liquid chromatography (LC) MS experiments.
Figure 1. Hydrogen/deuterium exchange experimental workflow.
(A) From left to right: incubation of target protein (± ligand) is performed for a series of predetermined time points. At the end point of the H/D exchange time, the reaction is ‘quenched’ with the addition of cold acidic solution and the protein is digested with enzyme. The percent deuterium incorporation into each peptide at each timepoint is measured with liquid chromatography electrospray ionization MS. Deuterium uptake curves are then plotted for the ‘apo’ sample, or a comparison between apo and ligand can be made. (B) Deuterium uptake information is often displayed using color gradients under a protein sequence or overlaid onto 3D crystal structure images. Differences in deuterium uptake can also be displayed in this manner to compare different ligands, as shown to the right.
D2O: Deuterium oxide; MS: Mass spectrometry; T: Temperature.
Measuring the percent deuterium exchange for each of the proteolytic peptides is achieved by calculating the number average mass-to-charge ratio (m/z) value of the peptide ion isotope cluster (centroid approach), or through fitting of theoretical isotopic distributions to experimental data. For the centroid approach, the percent deuterium incorporation can be calculated by subtracting the number average m/z value for the nondeuterated control from the number average m/z value of the deuterated peptide ion. The measured Δm/z value is then corrected to account for the number of exchangeable amides, charge state of the ion, the D2O concentration of the reaction and corrected for back-exchange. The %D incorporation can then be plotted as a function of time (s) as shown in Figure 1. In addition to the qualitative ‘fingerprint’ obtained from differential HDX data, the work of Englander has demonstrated how the results from the HDX method can be reported as a quantitative change in the protection factor [13]. The relationship between protection factor (Pf), measured exchange (kex), intrinsic exchange (kref) is shown in Equation 1.
| (1) |
Sample processing & data acquisition
The first HDX MS experiments to include protease digestion were reported by Smith and Zhang in the early 1990s [14]. As illustrated in Figure 1A, the processing of the sample during the HDX experiment is comprised of four main steps, all of which require careful control of pH and temperature: incubation with D2O buffer (on-exchange); quenching of the reaction with cold acidic solution (to stop further on-exchange and minimize off-exchange [loss of deuterium label]); exposure of the target protein to protease to facilitate enzymatic digestion; and chromatographic separation coupled with mass analysis. As emphasized previously, in order to minimize D/H back-exchange, quenching of the reaction with cold acidic solution and chromatographic separation coupled with mass analysis should be performed at a low temperature (1°C) and be maintained around pH 2.7.
The quench step allows the opportunity to denature the target protein prior to exposure to protease to facilitate efficient proteolysis. The two most common denaturants employed in HDX studies are urea and guanidine hydrochloride. Often, it is desirable to chemically reduce all intramolecular disulphide bonds at this quench step and this is commonly achieved through the addition of tris(2-carboxyethyl)phosphine (TCEP). The use of progressive proteolysis, multiple enzymes, optimized denaturant conditions and TCEP to improve digestion and analysis were described in detail by Woods and Hamuro in 2001 [5].
The most common enzyme used in HDX experiments to date is pepsin; however two alternative acid-stable proteases (fungal protease XIII and XVIII) have been in use since 2003 [15]. All three of these enzymes perform well when covalently coupled to a stationary phase and packed into columns for use as online immobilized enzyme reactors [16]. The immobilization of the enzyme ensures that the enzyme is not injected into the LC MS system, which can lead to a reduction in dynamic range (the enzyme may overload the capacity of the sample trap) and system robustness. Optimization of methods to produce immobilized pepsin columns have been described by Busby et al. [17].
In certain ligand screening applications, it may be beneficial to remove the ligand of interest prior to digestion and analysis. One such example would be the removal of excess DNA or oligodeoxynucleotide (ODN) from protein/DNA complexes. An online system for the removal of ODN prior to MS analysis was described by the Gross laboratory in 2008 [18].
The automated processing of samples for HDX MS experiments has received significant attention over the last 10 years. The automation of the HDX MS experiment was first described by Woods and Hamuro in 2001 with the description of a modified autosampler that thawed frozen samples from manually generated on-exchange time points and injected them into a cooled HPLC system housed within a refrigerator [5]. This system remains in use today. The advantage of this technique is that the period of on-exchange is under manual control and is therefore highly flexible. The decoupling of the manual on-exchange and automated injection steps may also be considered a disadvantage for the processing of large numbers of samples; complete automation with this approach is not possible. The complete automation of the entire HDX MS experiment was first described by Hamuro et al. in 2003 and was based upon the use of CTC Analytics (Zwingen, Switzerland) HTS PAL liquid-handling robots [19]. Subsequent coupling of this automated platform to high-resolution MS systems was described by Chalmers et al. in 2006 [20]. The liquid-handling robots processes all the replicate on-exchange events and controls the quench, online digestion and LC MS analysis [21]. This approach has expanded and has found use in a growing number of laboratories [22]. The system has been integrated with a custom ultra-performance liquid chromatography pump capable of high-pressure chromatography at 1°C [23] and has been configured to operate with a dual-column HPLC system to increase the rate of data acquisition [22]. The precision and reproducibility of these automated HDX MS systems have been evaluated [22] and the results demonstrate that the experiments can be performed with a precision suitable for compound screening studies [24].
The measurement of HDX rates on a millisecond time scale is best performed with a pulsed label or quenched-flow apparatus [25-27]. Rist et al. described a quench flow HDX system capable of measuring on-exchange time points between 100 ms and 30 s [26]. In an alternative approach, Chalmers et al. demonstrated the ability to perform automated on-exchange in the 130 ms timescale by elimination of the movement of samples, and relying on aspiration of air gaps between target protein, on-exchange buffer and quench solution [20].
Data processing & analysis
The starting point of the HDX experiment is the identification of the protoelytic peptides generated from the digestion of the protein of interest with LC MS/MS. The conditions of the quench event and the choice of enzyme are optimized to provide the highest attainable sequence coverage. The methods used for peptide identification are often the same as traditional proteomics workflows for the identification of peptide product ion spectra. Such workflows rely on database search engines such as SEQUEST and/or Mascot [28,29].
As mentioned previously, the enzyme pepsin is commonly used in HDX experiments, due to both its ability to function at low pH and its broad specificity. Common enzymes used in proteomics experiments, such as trypsin, are not active at low pH. Recent work from Hamuro et al. has evaluated the specificity of pepsin across many hundreds of experiments [30]. This paper provides the percent probability of a pepsin cleavage event for all combinations of amino acid pairs. For example, the highest probability cleavage event was between the amino acid pair F-F (85%). Importantly it was demonstrated that no cleavage was observed between R-X and P-X residues (where X is any amino acid). The detection of a pepsin peptide that breaks these Hamuro ‘forbidden cleavage’ rules should therefore be treated with caution, especially if assignment is based solely upon measured mass and not product ion data. To accommodate the broad specificity of pepsin it is therefore recommended to conduct a ‘no-enzyme’ database search for HDX studies. The high computational expense usually associated with no enzymes searches is not an issue with HDX studies as the databases are small, often containing only a single protein.
As with any proteomics experiment, it is important to establish false discovery rates in peptide assignments. One way to address this is the application of a target–decoy search strategy, where the original ‘target’ protein sequence is combined with a ‘decoy’ sequence. This decoy sequence can be generated in various ways, such as reversing the sequence or the generation of a random database of equal size and amino acid composition. The concatenated ‘target’ and ‘decoy’ database is used for the MS/MS search. The false-positive rate can then be estimated by doubling the number of matches to the decoy database [31]. These approaches are all combined to establish a final set of peptides for HDX data analysis with software applications such as Scaffold (Proteome Software, Portland, OR, USA).
As shown in Figure 1, the typical HDX experiment consists of multiple samples that are analyzed at each of a series of discrete on-exchange time points. The HDX data for each time point are acquired with multiple replicate experiments in order to allow the researcher to determine the variance within the data. Each protein within the sample will generate many proteolytic peptides, each of which need to be identified as described earlier and the extent of deuterium incorporation across all the time points and replicates must be determined. All the data must then be stored and made available for statistical analysis and cross comparison with other HDX datasets. One of the most demanding problems remaining is how to process and manage the data that are generated in a typical HDX experiment on a timescale that is equivalent to the rate of data generation. Although methods for data analysis have progressed in recent years, it remains significantly quicker to acquire the data than it is to process and validate it in sufficient detail that the results can be considered ready for publication or report generation. Compounding the problems that arise from the size and complexity of the datasets are the different types of mass spectrometers that are used to perform the mass analysis. For example, the approach for analyzing low-resolution MS data differ significantly from that for the analysis of high-resolution data.
Here, we outline the approaches that are currently available for HDX MS data analysis. The simplest approach for the extraction of deuterium content is to measure the weighted m/z average of each peptide distribution over the experimental time course and use this to determine its rate of deuterium exchange [14]. Associating exchange rate data to the regions of the intact protein provides information related to the dynamics of the investigated protein. The software MagTran [32] provided a means to accomplish these calculations and allow the calculation of the peptide ion centroid m/z values. Alternate approaches aim to extract information directly from measured isotopic envelopes. One such approach uses theoretical isotopic distributions to determine the %D incorporation by comparing multiple theoretical profiles to the measured isotopic distribution. Determination of the best fit is performed with least-squares regression [33-36]. Others have implemented a maximum entropy method for the determination of deuterium content [37]. The software HX-Express uses the widths of the peak envelopes to extract deuterium content from spectral data for the examination of EX1-type kinetics [38,39].
Tools have been produced that provide a simple user interface, automation capabilities and visualization components for data analysis and interrogation. The Deuterator [40] and HD Desktop [10] combined existing and novel approaches into a convenient web-based user interface. This software was designed to include visualization tools that are able to process data on a project-wide scale. A similar approach was taken for the subsequent development of the web-based MSTools software package [41]. Hydra offers equivalent attributes and provides new features such as support for the analysis of product ion data and deconvolving isotopic distributions [42]. HD Desktop and Hydra operate from a common file format for MS data, mzXML [43], in an effort to support a broad range of users. The standalone proprietary solutions DXMS [5] and HD Express (ExSAR Corp., NJ, USA, unpublished) have been built to facilitate the extraction of deuterium content. The software package TOF2H provides multiple tools for the management and interrogation of LC-MALDI TOF MS HDX data [44].
The starting point for many of these applications is a list of MS/MS-identified peptides as described previously; however, this limits the approach to obtaining HDX data only for peptides in the starting list. The software Hexicon attempts to increase sequence coverage by performing an in silico digest of the intact protein sequence, and searches for the resultant peptides in the spectral data in regions lacking coverage in the list of peptides originating from MS/MS experiments [45]. The software allows the user to fully automate the extraction of deuterium content. A solution that utilizes both MS and MS/MS for peptide identification from fourier transform ion cyclotron resonance MS data has been described by the Marshall group [46].
Looking forward, there are many challenges that still exist for analysis of HDX data. The near-future HDX data analysis will allow for more fully automated analysis, further reducing the data-processing time. Software will provide more support for detection of post-translational modifications (PTMs), which has become a growing need in the field, as well as comparing HDX kinetics for the protein or the same peptide region with and without the PTM. It has now been shown that alternative fragmentation processes from ECD and ETD can be used to investigate deuterium content from HDX studies at single amide resolution [47,48]. These techniques are poised to change the current landscape by providing site-localized HDX data from regions containing PTMs, allowing single amide resolution of deuterium content, as well as facilitating ‘top-down’ experiments. We also envisage that a computational platform that provides near-real-time analysis of complete HDX experiments will be forthcoming. Such a platform will allow quick changes in experimental conditions and design to be performed and would facilitate screening of protein–ligand interactions. A true end-to-end high-throughput HDX workflow that combines these advances will reduce cost per experiment and enable HDX to become a more widely adopted technique.
Emerging technologies: instrumentation developments
One of the most exciting areas of research in recent years has focused on the possibility that ECD [49] and ETD [50] may provide the ability to measure HDX kinetics with a spatial resolution of a single amide. The gas-phase fragmentation of protein and peptide ions without any intramolecular rearrangement of hydrogen or deuterium atoms would allow the HDX kinetics for each amide to be established. At present, the only technique available for the direct measurement of individual amide exchange kinetics is NMR spectroscopy. However, NMR analysis requires significant amounts of protein compared with the equivalent HDX MS experiment and is typically limited to the analysis of small proteins.
Nearly a decade has passed since the first description of HDX experiments in combination with ECD [51] and the issue of the ‘scrambling’ of the solution phase HDX data during gas-phase fragmentation remains the subject of some debate. However, a number of recent publications have made a compelling case that, despite some valid concerns, the experiment is indeed feasible [47,48,52,53]. The publication by Rand and Jørgensen in 2007 characterizing a series of synthetic peptides provided a simple reference system for researchers to determine the degree of scrambling induced during a particular MS/MS experiment [54]. The analysis of control peptides during these sublocalization experiments is predicted to become an established requirement of method development and validation prior to publication. Subsequent publications from the same group have demonstrated that single amide HDX data correlated well with reference NMR data [47].
The development of ‘top-down’ MS following HDX has progressed alongside the use of ECD/ETD. The removal of the proteolysis step from the experiment is an ambitious goal and good progress has been made with small (<20 kDa) proteins [55]. The progression of ‘top-down’ HDX MS/MS experiments was the focus of a recent review and will not be covered in detail here [4]. However, it is possible that the limiting factor of top-down HDX experiments will be the ability to transfer protein ions into the gas phase and fragment the parent ions into a sufficient number of product ions. It is nontrivial to perform the isolation and fragmentation of protein ions above 40 kDa with comprehensive product ion sequence coverage. By contrast, the use of proteolysis in HDX research allows multiprotein complexes in excess of 100 kDa to be analyzed routinely. These complexes can include upwards of three protein components, small-molecule ligands and DNA [56]. Thus, the ideal experimental design at this point in time would appear to be the use of ECD/ETD to sublocalize amide exchange kinetics within peptide fragments generated via proteolysis, or so-called ‘middle-down experiments.’
The use of alternative mass separation technologies and alternative scan functions has offered important options for HDX studies. The recent availability of a commercial ion mobility-TOF MS instrument has a number of potential benefits to the HDX MS experiment. The introduction of an additional dimension of ion separation prior to TOF MS analysis increases the peak capacity of the experiment and should aid in HDX studies of very large proteins or the study of complex multicomponent samples [57,58]. In addition, the traveling wave ion guide allows for gas-phase HDX experiments [59]. In the traditional HDX experiment, a mass spectrometer is used to measure a shift in m/z upon incubation with deuterium. A recent publication from Percy and Schriemer reevaluated this experimental design [60]. Taking advantage of an established method, they describe the use of a mass spectrometer operating in a multiple reaction monitoring (MRM) mode to measure the mass shift perturbations. The key advantage of the experiment arises from an increase in sensitivity and a simplified data analysis workflow that can take advantage of commercial software optimized for small molecule quantification applications. The approach may be limited to a small number of MRM transitions (and therefore peptides); however, it appears ideally suited for ligand screening applications and may simplify and expand the use of the HDX MS experiment.
Emerging technologies: new methods
The increasing size of the protein complexes that are being characterized with HDX has lead a number of researchers to propose the isotopic depletion of one (or more) of the proteins of interest [51,61]. It has been demonstrated that depletion of 13C (and 15N) isotopes during protein expression can significantly reduce the complexity of the mass spectrum and increase the signal/noise ratio of the ions through consolidation of ion current into a single isotope [61]. It was also demonstrated how the use of isotope depletion aided the identification of peptide ions where mass conflicts occur, assuming the peptides cannot be separated during the chromatography step [61].
The efficient digestion of the target protein is an important step in the ‘bottom-up’ HDX experiment; however, to preserve deuterium label on the target protein, the proteolysis has to be performed in a short period of time at low temperature and pH. If the protein is not digested efficiently under these conditions, the sensitivity of the experiment is reduced and the sequence coverage of the protein will be compromised. To improve the efficiency of the digestion step, a high-pressure digestion apparatus compatible with the HDX experiment has been introduced by the Gross laboratory [62]. As the HDX MS method moves to larger protein complexes and more challenging targets, improved methods for the efficient digestion of the protein will be required. Conditions that denature the target protein without causing precipitation or aggregation are critical to enable efficient digestion and maintain sensitivity. This is particularly evident as the technique is applied to the characterization of transmembrane proteins such as GPCRs. Zhang et al. demonstrate that maintenance of appropriate detergent in the on-exchange and quench buffer when performing HDX on β2-adrenergic receptor (a seven-transmembrane GPCR) is required to prevent loss of protein target to precipitation or aggregation [63]. In this study, HDX analysis was performed with buffers containing the detergent n-dodecyl-β-D-maltoside and cholesterol hemisuccinate. While this was encouraging, the ability to study proteins in solution with a wide range of detergents would be of benefit. To address this need, a recent publication has described a system for the removal of detergents such as Brij®58, PEG-4000, Tween®20 and Triton X-100™ in a system compatible with HDX experiments [64].
As an alternate approach, the insertion of the protein of interest within phospholipid bilayer nanodisks prior to HDX analysis has also been described by Engen’s group [65]. The method is demonstrated with the HDX analysis of the 94 kDa transmembrane protein γ-glutamyl-carboxylase (GGCX). HDX is performed on the intact nanodisk complex that is comprised of membrane scaffold protein (MSP), GGCX and lipid. Following the HDX on-exchange time period, the nanodisk is disrupted with cholate and the lipid is sequestered with ZrO2-coated silica beads. The 2:1 molar excess of MSP to GGCX was not detrimental to the detection of GGCX peptides in the UPLC MS step. The authors were able to obtain the HDX data from 71 peptides that spanned 45% of the GGCX protein sequence.
Alternate approaches
All the experiments detailed so far follow the traditional HDX workflow outlined in Figure 1. There are additional MS-based techniques that take advantage of protein deuterium exchange to discern information regarding protein folding and protein–ligand interactions [66]. The stability of unpurified proteins from rates of H/D exchange (SUPREX) technique monitors the global unfolding equilibrium of proteins with deuterium exchange to measure their thermodynamic global stability [67]. In contrast to the traditional HDX experiments, in the SUPREX experiment the period of D2O incubation is fixed and the concentration of a chemical denaturant is varied. The deuterium uptake of a protein is used to follow its denaturant-induced transition to the unfolded state. The denaturant concentration midpoint of this transition (C1/2) allows for the determination of the Gibbs energy (ΔGf) for the protein of interest. The comparison of the SUPREX transition’s C1/2 values in the presence and absence of ligand allows for the change in the free energy of folding (ΔΔGf), and thereby the dissociation constant (Kd), to be calculated [66]. Similar to the traditional HDX experiments, the SUPREX technique can be combined with a proteolysis step to explore localized regions of stability within proteins, which has been useful for analysis of multidomain proteins [68]. Protein–ligand interaction by mass spectrometry, titration, and H/D exchange (PLIMSTEX) is a similar technique to SUPREX; however ligand concentration, not denaturant, is varied while the on-exchange time is constant [69]. The experiment measures binding affinities and stoichiometries for protein–ligand complexes. Stability of proteins from rates of oxidation (SPROX) is a method for the detection of protein–drug interactions [70]. Although it is not based upon HDX, it is compatible with standard proteomic chromatography and it does provide for the identification of on- and off-target drug interactions, and as such is expected to be of interest to the readers of this article. A thorough comparison of the SUPREX, PLIMSTEX and SPROX methods are given in a recent review [66].
Application of HDX to protein–ligand interactions
The HDX technique is ideally suited to the characterization of interactions between protein targets and small molecules that bind to them. A typical HDX ligand screening application involves multiple differential HDX MS experiments to generate a ‘HDX fingerprint’ for each ligand. The differences between the fingerprints can allow for the clustering of ligands into groups that share a similar binding mode [71]. It is important to note that the differences in HDX between ligand-free (apo) and ligand-bound (holo) protein can also arise from direct interactions with the ligand within the ligand-binding pocket (LBP), as well as changes to the protein structure or dynamics remote to the binding site. This allostery may span domain:domain and protein:protein interaction surfaces, and while this presents challenges in the interpretation of HDX studies, this allosteric communication is not often detected with x-ray crystallography experiments.
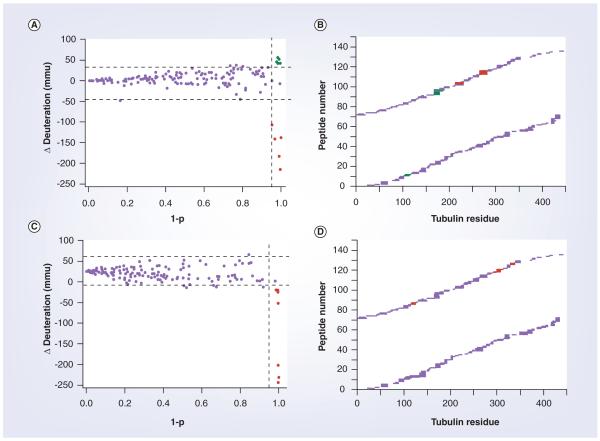
There are many examples of the application of HDX to study protein/small-molecule interactions. A recent study from the Schriemer laboratory discovered the unique binding site of laulimalide on microtubules using differential HDX [72]. The binding site for laulimalide had been determined to be nonoverlapping with the binding site for paclitaxel, but its location on the protein was unknown. Interestingly, but not unexpectedly, differential HDX of laulimalide-stabilized microtubules revealed extensive perturbations in amide exchange in regions throughout the protein, making it difficult to localize the binding site of the ligand using the differential HDX data alone. The authors proposed that HDX perturbations unique to the binding site of laulimalide could be determined by suppressing common allosteric effects of ligand binding to the paclitaxel site on microtubules. Thus, differential HDX of laulimalide-stabilized microtubules was performed in the presence and absence of docetaxel, a ligand that binds to the paclitaxel site and is not competitive with laulimalide binding. The reverse experiment was also performed, differential HDX of docetaxel-stabilized microtubules in the presence and absence of laulimalide. Comparison of these datasets resulted in defining a contiguous patch of solvent-accessible surface area on the microtubule that can be uniquely assigned as the laulimalide binding site as the allosteric effects of each ligand and their combination are identical under saturating conditions. Figure 2 shows the mass shift perturbation data from the study. In a unique approach, the authors plot the difference in HDX according to the statistical significance of the change (1-p), as opposed to a traditional Δ%D [72]. In these studies, it was determined that laulimalide binds to the exterior of the microtubule on β-tubulin in a region previously unknown to support ligand binding and well removed from the known paclitaxel binding site. These and other studies highlight the importance of HDX as a technique for mechanistic and structure–function studies of biologically and pharmacologically important proteins. As it would be impossible to address all HDX studies focused on protein–ligand interactions in one review, in the following section we summarize the impact HDX has had on three classes of proteins that are important to our understanding of biology and have significant commercial value to the pharmaceutical industry.
Figure 2. Mass shift perturbation data between singly and doubly ligated microtubules.
Mass shift perturbation data from analyses of doubly ligated microtubules relative to the singly ligated state. (A) Scatterplot of replication membrane scaffold protein data for a comparison of doubly ligated microtubules with laulimalide-stabilized microtubules, highlighting the perturbations unique to docetaxel. (B) Significant changes in (A) mapped to their locations in tubulin sequence. (C) Scatterplot of replication membrane scaffold protein data for a comparison of doubly ligated microtubules with docetaxel-stabilized microtubules, highlighting the perturbations unique to laulimalide. (D) Significant changes in (C) mapped to their locations in tubulin sequence. Each point in (A,C) represents the shift perturbation of a single peptide in millimass units. The horizontal dotted lines represent a ±2 standard deviation cutoff based on noise in ΔD measurements and the vertical dotted line represents a 1-p cutoff value of 0.95. Green represents positive mass shifts resulting from single ligation and red represents negative mass shifts resulting from single ligation. The sequence maps (B,D) are arranged with α-tubulin on the bottom and β-tubulin on the top.
mmu: Millimass unit.
Reproduced with permission from [71]. For more details, refer to the text or reference [71].
Nuclear receptors
Nuclear receptors comprise a superfamily of ligand-dependent transcription factors, including receptors for thyroid hormones, steroid hormones, retinoids, fatty acids and sterols, as well as receptors with no currently known ligand (termed orphan receptors). In the late 1980s, the identification of the canonical domain structure and conserved sequence of members of this superfamily led to an intense effort to identify additional members of this superfamily. The conserved domain structure of NRs consists of four major functional domains. The variable amino-terminal (A/B) domain is followed by the central, highly conserved DNA-binding domain (DBD), which contains two zinc fingers (C domain). Adjacent to the C domain is the hinge region (D domain), followed by a carboxy-terminal ligand-binding domain (LBD; E domain). Several nuclear receptors, such as the estrogen receptor, contain an additional C-terminal F domain whose role is poorly defined. NRs bind to specific DNA sequences typically in the promoter region of target genes and these sequences are referred to as response elements. The LBDs of the nuclear receptors are multifunctional and have a secondary domain structure that is characteristic of all NRs. Small pharmacological compounds bind within the LBP of the LBD and this interaction with ligand alters the dynamics and conformation of the receptor. The human genome contains 48 NRs and these proteins can form an array of complexes where the central NR is present as either a monomer, homodimer or heterodimer and bound with coregulatory proteins (coactivators such as steroid receptor coactivator [SRC]-2, or co repressors such as nuclear receptor corepressor [NCoR] or silencing mediator of retinoid and thyroid hormone receptors [SMRT]), and DNA [56,73]. The transcriptional output of the NR complex is modulated by binding of small-molecule ligands (agonists, partial agonists, antagonists and inverse agonists) that induce conformational changes in the receptor that alter interactions with coregulatory proteins involved in chromatin remodeling or facilitate regulatory PTMs of the complex. Such ligands have already been successfully designed for approximately 23 NRs and several of these ligands are used clinically to treat diseases such as osteoporosis, Type 2 diabetes and various cancers.
The combinatorial regulatory mechanism of NRs makes them ideally suited for HDX MS studies. HDX can be used to profile the changes in dynamics upon ligand binding, addition of PTMs or binding of coregulatory proteins and DNA. HDX characterization of ligand binding can provide insights into binding mode and can aid compound optimization, whereas HDX analysis of NR interaction with coreceptor (heterodimer partner), coregulatory protein and DNA can provide a mechanistic insight into how these complexes modulate transcriptional output of specific target genes. Many of the 48 human nuclear receptors have been studied by HDX. Several of these receptors are of significant therapeutic interest and some of these studies are reviewed in the next section.
Retinoid, fatty acid, oxysterol & vitamin A NRs
The retinoid X receptor (RXR) is in the NR2B subfamily of NRs and is the common heterodimer partner (coreceptor) for many NR1 subfamily nuclear receptors, including the vitamin D receptor (VDR), the peroxisome proliferator-activated receptor (PPAR), the thyroid hormone receptor, the liver X receptor, the farnesoid X receptor and the constitutive androstane receptor [74]. It seems fitting, therefore, that this common heterodimer partner was among the first NRs to be studied with HDX by the Deinzer laboratory [75]. RXR is activated through binding of 9-cis retinoic acid and the changes to the HDX signature upon ligand binding were profiled, along with a comparison to the atomic structure of the RXRα LBD [75]. Importantly, this work identified changes in dynamics upon ligand binding within regions of RXR LBD that could not be predicted from the x-ray RXR: 9-cis retinoic acid cocrystal structure. This work was followed in 2007 by a comparison of the HDX behavior of RXR in complex with two synthetic antagonists [76]. More recent work from the Renfrow laboratory has profiled the HDX behavior of RXR in complex with 9-cis retinoic acid and a 13-mer RXR coactivator peptide, glutamate receptor-interacting protein (GRIP)-1 [77].
The RXR heterodimer partner PPARγ is the master regulator of adipocyte differentiation and the molecular target of the antidiabetic thiazolidinedione (glitazones or thiazolidinediones [TZDs]), ligands such as rosiglitazone (Avandia™) and pioglitazone (Actos™). The glitazones are agonists of PPARγ and strong insulin sensitizers; however, several well-publicized adverse effects have been associated with the use of these drugs, including cardiac events, weight gain and increased fluid retention [78]. Development of second generation PPARγ modulators, so-called selective PPARγ modulators, suggested that it might be possible to dissociate the insulin-sensitizing effects from the weight-promoting activity and fluid retention. Thus, understanding the mechanism of activation of PPARγ is therefore of significant interest and has been the subject of decades of research. The HDX behavior of PPARγ has been profiled by a number of researchers [21,79-82]. In 2006, Hamuro et al. published a HDX study investigating changes in the dynamics of the PPARγ LBD following binding of four synthetic ligands, including two full agonists (rosiglitazone and GW1929), a non-TZD partial agonist (nTZDpa) and an antagonist known to covalently bind to the receptor (GW9662) [82]. Importantly, this study demonstrated that all four compounds reduced the dynamics of the receptor, but that only the full agonists reduced the rate of HDX in a region of the LBD referred to as ‘activation function-2’ or AF-2, which is a 3D surface that forms a hydrophobic cleft and includes helix 3–4 loop and helix 12 (H12). AF2 had been shown to be a critical structural component involved in the activation of most, if not all, NRs and it has been shown to be the primary binding site for coactivator and corepressor on NRs [73]. The observation that partial agonists do not stabilize AF2 of PPARγ suggested an alternative mode of action for these compounds compared with full agonists; however, the specific mechanism was still unclear. These studies were extended beyond synthetic ligands, where HDX was used to probe the interaction of the receptor with several putative endogenous ligands and results from this study were consistent with observations for synthetic ligands, suggesting the existence of putative endogenous partial agonists [21].
These publications were followed by a more extensive study performed by Bruning et al. that combined the use of HDX with x-ray crystallography and cell-based transcriptional assays to further investigate the mechanism of partial agonist activation of PPARγ [79]. This work profiled the structural dynamics of the receptor with a series of compounds that exhibited a gradient of transcriptional activity from partial to full activation. In all cases, the backbone structure of PPARγ obtained from the x-ray crystallography experiments was unchanged for each of the ligand complexes, suggesting no obvious conformational change induced by this array of ligands. As such, these co-crystal structures provided little insight into the mechanism associated with different transcriptional activities of each ligand class, but they did provide an atomic resolution structure detailing the precise orientation of the ligands within the binding pocket. Interestingly, HDX analysis revealed major differences between all of the compounds profiled in terms of localized alterations in dynamics, and compounds with similar binding modes had similar HDX characteristics. Comparing the two datasets, a complete picture of the changes in the receptor dynamics following ligand binding was obtained. Combining these findings with the cell-based data, the researchers proposed a model for partial agonist activation of PPARγ that was independent of involvement of H12 (AF2-independent). It was proposed that ligands that do not utilize H12 engage other regions of the LBP such as the β-sheet region of the receptor (located on the opposite side of the LBP from H12) and this engagement compensates for a lack of interaction with H12 to support high-affinity binding. Of particular interest in this study was the analysis of two regioisomers, MRL20 and MRL24. MRL20 was shown to have full agonist activity in cellular transaction assays whereas MRL24 afforded partial agonist activity. The x-ray co-crystal structures showed that these two highly similar compounds had opposite orientations (binding modes) in the ligand pocket. The acid functionality of MRL20 was shown to be interacting with H12, consistent with its full agonist activity, and the acid functionality of MRL24 was shown to be interacting with the β-sheet, consistent with its partial agonist activity. Even with this level of structural detail, this study still did not fully define the insulin sensitization efficacy of PPARγ partial agonists.
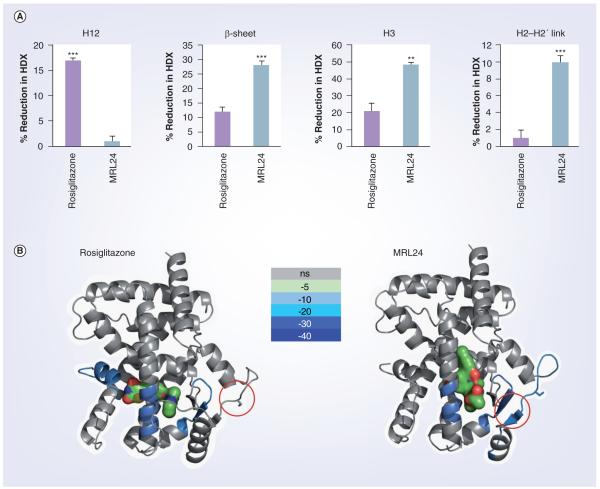
The role of the β-sheet region of the PPARγ LBD was recently highlighted in a landmark publication demonstrating that PPARγ is a substrate of cdk5 [81]. The single site of cdk5-dependent phosphorylation was shown to be serine 273 (S273) which is located in the H2-H2′ region of the receptor and in close proximity to the β-sheet region shown to be strongly stabilized by partial agonists. The HDX data for this region of the receptor are shown in Figure 3. The Spiegelman group demonstrated that phosphory lation of S273 alone was capable of dysregulating expression of adipokines, which are genes involved in insulin sensitization [81]. Interestingly, the extent of phosphorylation was inversely proportional to the expression levels of adiponectin and adipsin. More importantly, treatment with agonist in vitro or in cells afforded a dose-dependent blockage of this PTM. To their surprise, the partial agonist MRL24 was more potent at blocking the PTM than the full agonist rosiglitazone (controls demonstrated that the compounds do not directly inhibit cdk5). Regardless of level of PTM state of the receptor, cdk5 was coprecipitated with PPARγ, suggesting that the kinase binds to PPARγ, but ligand binding alters the kinases ability to interact and modify S273. Subsequent studies revealed that the insulin sensitivity activity in rodents and in humans correlated with the level of this PTM. HDX studies showed that the full agonist rosiglitazone did not stabilize this H2-H2′ link region containing S273 or the β-sheet. By contrast, the partial agonist MRL24 stabilized both the β-sheet and the H2-H2′ link region. It was therefore proposed that the alterations in the dynamics of the receptor, as determined by HDX, modulate the extent of cdk5 phosphorylation. In turn, the phosphorylation state of the receptor influences the transcriptional output of this NR complex. This study is a powerful example of how perturbations in the dynamics of this receptor, even when subtle, can reflect profound changes in the biology of this important transcriptional regulator.
Figure 3. Differential hydrogen/deuterium exchange coupled with mass spectrometry data for peroxisome proliferator-activated receptor γ-ligand binding domain ± rosiglitazone and MRL24.
The HDX data shown correspond to four regions of interest: helix 3 (IRIFQGCQF), the β-sheet region (ISEGQGFMTRE), helix 12 (QEIYKDLY) and the helix 2–2′ link region containing the site of CDK5 phosphorylation (KTTDKSPFVIYDM). (A) Histograms showing the percent reduction in HDX for each peptide region. Values are calculated relative to the measured %D value for apo peroxisome proliferator-activated receptor (PPAR)γ-LBD (n = 4; error bars are standard error of the mean; p < 0.01, p < 0.001). (B) HDX data for the four peptides of interest are plotted over the structures of PPARγ-LBD bound with rosiglitazone (left: PDB:2PRG) and MRL24 (right: PDB:2Q5P). Percent reduction in HDX relative to apo receptor is colored according to the key. Red circle indicates Ser273 residue of PPARγ.
H12: Helix 12; HDX: Hydrogen/deuterium exchange; LBD: Ligand-binding domain; ns: Not significant.
Reproduced with permission from [79].
It is surprising that it was not until 2008 that the first crystal structure of a full-length NR was published. This structure high-lighted the atomic nature of the PPARγ/RXRα heterodimer in complex with ligand and DNA (PDB:3DZU) [80]. This fascinating structure supported many theories and provided a unique insight into the structure of the heterodimer complex. For example, this structure revealed that the β-sheet region of PPARγ was in close proximity to the DNA-binding domain of RXRα, suggesting possible cross-domain interactions. As highlighted previously, potent insulin sensitizers alter the dynamics of this region of the receptor and it is possible that these ligands modulate the interaction of the RXRα DBD with DNA. This study used HDX to complement the x-ray crystallography data to provide supporting insight into this and other domain:domain interactions within the intact heterodimer.
There are a number of orphan NRs for which no endogenous ligand has been identified or there is still debate as to the identity of an endogenous ligand. The retinoic acid receptor-related orphan receptors (RORs) are an example of the latter. Identification of ligands that potentially regulate the activity of RORs has been met with controversy. Elegant x-ray crystallographic studies provided significant insight into the nature of a putative physiological ligand. The resolution of the x-ray structure of the RORα LBD revealed the presence of cholesterol in the binding pocket. A second crystallographic study established that in addition to cholesterol, cholesterol sulfate could be bound within the LBD of RORα. Although the idea that RORα functioned as a receptor for cholesterol and cholesterol sulfate was attractive, it was still unclear whether cholesterol or derivatives of cholesterol were truly physiological ligands for RORα. Wang et al. recently demonstrated that 7-oxygenated sterols function as high-affinity ligands for both RORα and RORγ with Ki values in the range of 10–20 nmol/l, as determined by radioligand-binding assays [83]. These data indicated that 7-oxygenated sterols (7α-OHC, 7β-OHC and 7-ketocholeseterol) bind RORα and RORγ with higher affinity than cholesterol sulfate, whereas cholesterol binding is barely detectable. More importantly, this study demonstrated that addition of a 7-oxygenated sterol to apo RORα perturbed the HDX dynamics of the receptor. This study demonstrates a role for HDX in deorphanizing orphan NRs.
The majority of NR HDX studies performed to date have focused on the analysis of the receptor ligand-binding domain. The dynamics of the full-length PPAR:RXR heterodimer complex were detailed for the first time alongside the publication of the x-ray structure of the complex as already described [80]. More recent work described a study of intact full-length vitamin D receptor (VDR) in complex with full-length RXR and three ligands of interest, 1α25-dihydroxyvitamin D3 (active form of VD3), alfacidol and ED-71 [56,84]. The data presented in this study illustrate how the ligand-induced changes to HDX behavior for the LBD of VDR are similar to those induced within the full-length heterodimer complex. These data further highlight the relevance of earlier NR HDX experiments performed on the LBDs alone.
Steroid & hormone receptors
Human glucocorticoid receptor functions as a homodimer, binds cortisol, and is involved in the regulation of glucose homeostasis, lipid metabolism and inflammation [85]. In 2006, the HDX profiles of the receptor in complex with the anti-inflammatory agonist dexamethasone and the antagonist RU-486 were detailed [86]. These data are supplemented with HDX data from complexes of glucocorticoid receptor with corepressor (NCoR) and coactivator (TIF2/SRC2) peptides. In agreement with previously obtained x-ray structures, the agonist showed reduced dynamics of H12 when compared with the antagonist.
Since the activation and transcriptional output of NRs is driven by changes in structure and dynamics of the receptor upon ligand binding, and since these changes are reflected in a differential HDX experiment, it is logical to suspect that it may be possible to predict the pharmacological properties of NR ligands directly from HDX data. This hypothesis was put to the test in a 2008 paper by Dai et al. that focused on the prediction of the pharmacology of ligands for the estrogen receptor (ER) [71]. Ligands profiled included the human estrogen (estradiol, a full agonist) and several selective ER modulators including raloxifene (a compound used to prevent osteoporosis) and 4-hydroxytamoxifen (the active metabolite of the anticancer drug tamoxifen). Seven of these known compounds were profiled with HDX and classified into three distinct groups. Then, in a blinded study, four unknown compounds were profiled with HDX and assigned to one of these three groups. Only the antagonist ICI 182780 and one of the four unknown compounds were incorrectly assigned. Although the study is limited in the number of compounds (both in the training set and the blinded compound set), the data do support the hypothesis that the HDX profile can, at least in certain cases, predict the pharmacology of ER ligands. A follow-up manuscript to this work described a comparison between the ERα and ERβ HDX profiles of 12 ER ligands [87].
Protein kinases
Protein kinases are responsible for the addition of phosphate to serine, threonine and tyrosine residues of proteins. This reversible phosphorylation is a key regulatory step in many signal transduction pathways. As such, the study of the structure and function of these enzymes is of paramount importance to deciphering cell signaling and the role these enzymes play in physiology. Importantly, aberrant signaling by kinases is often associated with the pathogenesis of disease; thus, a detailed understanding of the structure–function relationship of these proteins is critical to tackling diseases such as cancer, neurodegeneration and immune disorders. A few examples of the application of HDX to the study of kinases and interactions with ligands are presented in the next section.
The chimeric oncoprotein Bcr-Abl has been implicated in the onset of chronic myelogenous leukemia (CML) and other leukemias. Bcr-Abl is a constitutively active protein kinase, and resistance to small-molecule ATP competitive inhibitors of the Abl kinase domain (such as imatinib) has become a serious impediment to the successful therapeutic treatment of CML. In studies to further understand the role of protein dynamics in the regulation of the Abl kinase domain, the HDX behavior of Abl SH2 and SH3 domains has been characterized [88-90]. In more recent work, a series of known imatinib-resistant mutants were compared with the wild-type protein [91]. It was demonstrated that the T315I mutant induced conformational changes when compared with wild type; however, no changes in HDX profile were detected for the Y253H and E255V mutants. In order to overcome the resistance of Bcr-Abl kinase mutants to inhibition with ATP competitive ligands, efforts have been refocused on the development of allosteric inhibitors of Abl to be used in cooperation with ATP competitive inhibitors. These allosteric inhibitors have recently been characterized with HDX and the data compared with the x-ray structure, leading to new insights into the mechanism of action of these allosteric inhibitors [92].
Hydrogen/deuterium exchange studies have probed the effect of mutations on the dynamics on another kinase of importance, the receptor tryrosine kinase KIT. Gajiwala et al. have detailed the HDX behavior of wild-type KIT and KIT mutants associated with gastrointestinal stromal tumors [93]. The same group have also profiled the dynamics of wild-type KIT in complex with imatinib and sunitinib [94].
G protein-coupled receptors
The GPCRs belong to a large family of cell surface receptors that are the molecular targets of many marketed therapeutics [95,96]. All GPCRs share a seven-transmembrane α-helical structure and they function as integrators of extracellular stimuli into intracellular signal transduction pathways. The hydrophobic nature of the transmembrane regions of GPCRs presents a formidable challenge for structural biologists trying to understand mechanism of action of modulators of these receptors. Structural characterization of GPCRs is also complicated by the extensive PTM state of the receptor. There are heterogeneous sites of N-glycosylation, sites of phosphorylation, intramolecular disulphide linkages and sites of palmitoylation. Although the x-ray structure of rhodopsin was first solved in 2000 [97], it was not until 2007 that the first ligand-diffusible GPCR structures were reported [98-100]. Crystallization of the β2-adrenergic receptor (β2-AR) was achieved via protein engineering where the highly dynamic third intracellular loop (ICL3 located between TM5 and TM6) was replaced with T4 lysozyme [99,100], or by formation of a protein complex with a monoclonal antibody bound to the third intracellular loop [100]. The first HDX MS analysis of a GPCR followed and was published in 2010 [63]. Working with a construct of β2-AR that contained a more native ICL3 sequence, Zhang et al. were able to determine HDX MS-compatible conditions that allowed the dynamics of the receptor in complex with the inverse agonist carazolol to be profiled [63]. It was observed that all the transmembrane helices were resistant to exchange; however, the ICL regions were highly dynamic. Although not the first HDX manuscript to profile a membrane protein [101-104], this study paves the way for the analysis of β2-AR in complex with other ligands such as full and partial agonists and antagonists. More importantly, the study paves the way to facilitate HDX analysis of the many other important GPCRs, as well as studying the interaction of the receptor with G proteins, arrestins and GPCR kinases. These studies will help dissect the structural dynamics important to functional selectivity of ligands that drive biased GPCR signaling.
Can HDX be used for small-molecule screening?
One desirable application of the HDX technique would be to screen chemical libraries containing small molecules, natural products or fragments for chemical entities that impart a desired HDX fingerprint on the target protein. Although an attractive prospect, the use of HDX as a screening tool is problematic and has yet to be demonstrated at a throughput capable of screening even a simple library of a few thousand compounds. However, a number of recent papers offer two approaches that may provide opportunities in the future. If the desired HDX behavior can be distinguished from the ligand-free protein at a single on-exchange time point, as is the case for PPARγ, then as many as 10–20 compounds can be screened within a few hours [21]. This approach was validated with PPAR modulators with previously determined HDX signatures, and was then used to provide the first HDX fingerprints for a number of putative endogenous ligands of the receptor. The limit to the throughput of this method is the requirement for a chromatographic step of sufficient length to separate the peptide of interest from any interfering peptides. The use of the MRM approach already described would allow for a more rapid chromatography step and also provide for a simplified data analysis workflow.
A workflow and performance evaluation for automated HDX was evaluated in a recent manuscript from the Griffin laboratory [24]. In this work, the precision of the automated system was evaluated over a period of 8 months. The manuscript also shows a heat map containing statistically significant changes to the HDX behavior of VDR upon ligand binding (Figure 4). The data provided in this manuscript demonstrate the feasibility of HDX for small-molecule screening applications. This is an area predicted to see significant progress in the next couple of years.
Figure 4. Hydrogen/deuterium exchange heat map obtained from the analysis of vitamin D receptor ligand-binding domain in complex with 87 ligands of interest.
Data are clustered and ordered according to Wards method (see [24] for details). Changes with Tukey-adjusted p-values <0.05 are colored according to the key. p-values >0.05 are colored gray.
Reproduced with permission from [24].
Challenges to the development of the HDX technique
Although there have been many successful applications of HDX covered in this article, no technique is without its limitations. The reader may want to consider the following points when contemplating the use of HDX MS. It should be noted that it is often unclear if a change in HDX rate is a result of a direct interaction (e.g., formation of a new H-bond), or the result of an allosteric effect causing a perturbation of the hydrogen-bonding network at a site remote from the site of ligand binding. Care must also be taken to ensure that the ligand concentration is sufficiently above its Kd to ensure binding. Problems may arise from the precipitation of DNA at a low pH, leading to a loss of system robustness. Finally, if no peptide can be detected that spans a region of interest within a protein, then no HDX data can be obtained. Despite these limitations, HDX MS experiments clearly provide a valuable addition to many research programs around the world.
Expert commentary
The field of HDX MS continues to evolve at a rapid pace. Building on the foundations of the early work from the likes of Englander, Smith, Woods and numerous others, the technique has become widely adopted by the MS community. The last 10 years have seen significant advancements in the HDX MS method. The experiment has been automated and many software applications are available for the analysis and management of HDX MS datasets.
As the method has matured, the application of HDX to the drug discovery process has expanded. HDX MS is now routinely applied in our laboratory in support of the medicinal chemistry efforts of our collaborators in the pharmaceutical industry. Other academic groups have partnered with industry to characterize therapeutic antibodies, and the internal use of the HDX method in the pharmaceutical industry can be expected to continue to grow.
The expansion of the size of the datasets that are being generated now allows researchers to undertake large-scale mechanism of action projects, such as those performed on the NRs. These large-scale projects will provide new insight into the regulation of key pharmaceutical targets and further increase the impact of the method on the understanding of the associated biological systems. Through a number of recent studies, it has become apparent that minor changes in HDX behavior can have significant impacts on the biology and pharmacology of the systems being studied [81,91]. Methods for determining the statistical significance of these minor changes in HDX data are therefore of importance and will undoubtedly receive more prominence in future HDX studies.
Five-year view
As we look back on the development of the HDX method, the future will surely provide some exciting developments and provide many key insights into the pharmacology and biology of important drug targets. Consistent with early efforts in the field, the work of Englander continues to set a high standard and provide direction to the field, particularly in the treatment of HDX datasets. It might be expected that future HDX screening applications will report data not simply as a change in %D, but as a more quantitative description of a change in the protection factor (ΔPf) of individual amides. Such single-amide-resolution HDX will be made possible by the use of ECD/ETD for peptide ion fragmentation. The size of the protein complexes amenable to HDX MS experiments is expected to increase, with data for complexes in excess of 200 kDa being reported. It is expected that the average HDX manuscript will no longer be a single experiment with an isolated protein, but will comprise an evaluation of many tens, or even hundreds of datasets in the context of full-length mutiprotein complexes.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, National Institute of Mental Health (Grant U54-MH074404) and the National Institute of General Medical Sciences (Grant R01-GM084041).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Blundell TL, Patel S. High-throughput X-ray crystallography for drug discovery. Curr. Opin. Pharmacol. 2004;4(5):490–496. doi: 10.1016/j.coph.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Kohn JE, Afonine PV, Ruscio JZ, Adams PD, Head-Gordon T. Evidence of functional protein dynamics from X-ray crystallographic ensembles. PLoS Comput. Biol. 2010;6(8):pii–e1000911. doi: 10.1371/journal.pcbi.1000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Englander SW. Hydrogen exchange and mass spectrometry: a historical perspective. J. Am. Soc. Mass Spectrom. 2006;17(11):1481–1489. doi: 10.1016/j.jasms.2006.06.006. •• A historical perspective of the development of hydrogen/deuterium exchange (HDX) mass spectrometry experiments.
- 4.Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: is there a need for a top-down approach? Anal. Chem. 2009;81(19):7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woods VL, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell Biochem. Suppl. 2001;37:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 6.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 2009;81(19):7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai YW, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen-exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns-Hamuro LL, Hamuro Y, Kim JS, et al. Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14(12):2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Resing KA, Hoofnagle AN, Ahn NG. Modeling deuterium exchange behavior of ERK2 using pepsin mapping to probe secondary structure. J. Am. Soc. Mass Spectrom. 1999;10(8):685–702. doi: 10.1016/S1044-0305(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 10.Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD Desktop: an integrated platform for the analysis and visualization of H/D exchange data. J. Am. Soc. Mass Spectrom. 2009;20(4):601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1991;5(4):214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 12.Thevenon-Emeric G, Kozlowski J, Zhang Z, Smith DL. Determination of amide hydrogen exchange rates in peptides by mass spectrometry. Anal. Chem. 1992;64(20):2456–2458. doi: 10.1021/ac00044a027. [DOI] [PubMed] [Google Scholar]
- 13.Chetty PS, Mayne L, Lund-Katz S, Stranz D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl Acad. Sci. USA. 2009;106(45):19005–19010. doi: 10.1073/pnas.0909708106. • HDX analysis of apolipoprotein. Data analysis shows the amide exchange data plotted as log (protection factor).
- 14.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2(4):522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cravello L, Lascoux D, Forest E. Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins. Rapid Commun. Mass Spectrom. 2003;17(21):2387–2393. doi: 10.1002/rcm.1207. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol. Cell Proteomics. 2002;1(2):132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Busby SA, Chalmers MJ, Griffin PR. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. Int. J. Mass Spectrom. 2007;259(1–3):130–139. [Google Scholar]
- 18.Sperry JB, Wilcox JM, Gross ML. Strong anion exchange for studying protein-DNA interactions by H/D exchange mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19(6):887–890. doi: 10.1016/j.jasms.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J. Biomol. Tech. 2003;14(3):171–182. [PMC free article] [PubMed] [Google Scholar]
- 20.Chalmers MJ, Busby SA, Pascal BD, et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 2006;78(4):1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 21.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J. Biomol. Tech. 2007;18(4):194–204. [PMC free article] [PubMed] [Google Scholar]
- 22.Burkitt W, O’Connor G. Assessment of the repeatability and reproducibility of hydrogen/deuterium exchange mass spectrometry measurements. Rapid Commun. Mass Spectrom. 2008;22(23):3893–3901. doi: 10.1002/rcm.3794. [DOI] [PubMed] [Google Scholar]
- 23.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 2008;80(17):6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalmers MJ, Pascal BD, Willis S, et al. Methods for the analysis of high precision differential hydrogen-deuterium exchange data. Int. J. Mass Spectrom. 2010 doi: 10.1016/j.ijms.2010.08.002. DOI: 10.1016/j.ijms.2010.08.002 (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons DA, Konermann L. Characterization of transient protein folding intermediates during myoglobin reconstitution by time-resolved electrospray mass spectrometry with on-line isotopic pulse labeling. Biochemistry. 2002;41(6):1906–1914. doi: 10.1021/bi011697j. [DOI] [PubMed] [Google Scholar]
- 26.Rist W, Rodriguez F, Jørgensen TJD, Mayer MP. Analysis of subsecond protein dynamics by amide hydrogen exchange and mass spectrometry using a quenched-flow setup. Protein Sci. 2005;14(3):626–632. doi: 10.1110/ps.041098305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan J, Rintala-Dempsey AC, Li Y, Shaw GS, Konermann L. Folding kinetics of the S100A11 protein dimer studied by time-resolved electrospray mass spectrometry and pulsed hydrogen-deuterium exchange. Biochemistry. 2006;45(9):3005–3013. doi: 10.1021/bi052349a. [DOI] [PubMed] [Google Scholar]
- 28.Eng JK, Mccormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 29.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun. Mass Spectrom. 2008;22(7):1041–1046. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
- 31.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9(3):225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 33.Hotchko M, Anand GS, Komives EA, Ten Eyck LF. Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 2006;15(3):583–601. doi: 10.1110/ps.051774906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmblad M, Buijs J, Hakansson P. Automatic analysis of hydrogen/deuterium exchange mass spectra of peptides and proteins using calculations of isotopic distributions. J. Am. Soc. Mass Spectrom. 2001;12(11):1153–1162. doi: 10.1016/S1044-0305(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 35.Chik JK, Vande Graaf JL, Schriemer DC. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Anal. Chem. 2006;78(1):207–214. doi: 10.1021/ac050988l. [DOI] [PubMed] [Google Scholar]
- 36.Abzalimov RR, Kaltashov IA. Extraction of local hydrogen exchange data from HDX CAD MS measurements by deconvolution of isotopic distributions of fragment ions. J. Am. Soc. Mass Spectrom. 2006;17(11):1543–1551. doi: 10.1016/j.jasms.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Zhang ZQ, Guan SH, Marshall AG. Enhancement of the effective resolution of mass spectra of high-mass biomolecules by maximum entropy-based deconvolution to eliminate the isotopic natural abundance distribution. J. Am. Soc. Mass Spectrom. 1997;8(6):659–670. [Google Scholar]
- 38.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 2006;17(12):1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 39.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 2006;17(11):1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Pascal BD, Chalmers MJ, Busby SA, et al. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kavan D, Man P. MSTools – web based application for visualization and presentation of HXMS data. Int. J. Mass Spectrom. 2010 In Press. [Google Scholar]
- 42.Slysz GW, Baker CA, Bozsa BM, et al. Hydra: software for tailored processing of H/D exchange data from MS or tandem MS analyses. BMC Bioinformatics. 2009;10:162. doi: 10.1186/1471-2105-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedrioli PG, Eng JK, Hubley R, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004;22(11):1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 44.Nikamanon P, Pun E, Chou W, Koter MD, Gershon PD. ‘TOF2H’: a precision toolbox for rapid, high density/high coverage hydrogen-deuterium exchange mass spectrometry via an LC-MALDI approach, covering the data pipeline from spectral acquisition to HDX rate analysis. BMC Bioinformatics. 2008;9:387. doi: 10.1186/1471-2105-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lou X, Kirchner M, Renard BY, et al. Deuteration distribution estimation with improved sequence coverage for HX/MS experiments. Bioinformatics. 2010;26(12):1535–1541. doi: 10.1093/bioinformatics/btq165. [DOI] [PubMed] [Google Scholar]
- 46.Kazazic S, Zhang HM, Schaub TM, et al. Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 2010;21(4):550–558. doi: 10.1016/j.jasms.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rand KD, Zehl M, Jensen ON, Jørgensen TJD. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 2009;81(14):5577–5584. doi: 10.1021/ac9008447. •• Highlights the feasibility of the use of electron-transfer dissociation for single-amide resolution HDX measurements.
- 48.Zehl M, Rand KD, Jensen ON, Jørgensen TJ. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J. Am. Chem. Soc. 2008;130(51):17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 49.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120(13):3265–3266. [Google Scholar]
- 50.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl Acad. Sci. USA. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charlebois JP, Patrie SM, Kelleher NL. Electron capture dissociation and 13C,15N depletion for deuterium localization in intact proteins after solution-phase exchange. Anal. Chem. 2003;75(13):3263–3266. doi: 10.1021/ac020690k. [DOI] [PubMed] [Google Scholar]
- 52.Rand KD, Adams CM, Zubarev RA, Jørgensen TJ. Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 2008;130(4):1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 53.Hagman C, Tsybin YO, Hakansson P. Solution-phase deuterium/hydrogen exchange at a specific residue using nozzle-skimmer and electron capture dissociation mass spectrometry. Rapid Commun. Mass Spectrom. 2006;20(4):661–665. doi: 10.1002/rcm.2339. [DOI] [PubMed] [Google Scholar]
- 54.Rand KD, Jørgensen TJD. Development of a peptide probe for the occurrence of hydrogen (H-1/H-2) scrambling upon gas-phase fragmentation. Anal. Chem. 2007;79(22):8686–8693. doi: 10.1021/ac0710782. [DOI] [PubMed] [Google Scholar]
- 55.Pan J, Han J, Borchers CH, Konermann L. Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. J. Am. Chem. Soc. 2009;131(35):12801–12808. doi: 10.1021/ja904379w. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Chalmers MJ, Stayrook KR, et al. DNA binding alters coactivator interaction surfaces of the intact VDR–RXR complex. Nat. Struct. Mol. Biol. 2010 doi: 10.1038/nsmb.2046. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004;18(20):2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 58.Iacob RE, Murphy JP, 3rd, Engen JR. Ion mobility adds an additional dimension to mass spectrometric analysis of solution-phase hydrogen/deuterium exchange. Rapid Commun. Mass Spectrom. 2008;22(18):2898–2904. doi: 10.1002/rcm.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rand KD, Pringle SD, Murphy JP, 3rd, Fadgen KE, Brown J, Engen JR. Gasphase hydrogen/deuterium exchange in a traveling wave ion guide for the examination of protein conformations. Anal. Chem. 2009;81(24):10019–10028. doi: 10.1021/ac901897x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Percy AJ, Schriemer DC. MRM methods for high precision shift measurements in H/DX-MS. Int. J. Mass Spectrom. 2010 In Press. [Google Scholar]
- 61.Bou-Assaf GM, Chamoun JE, Emmett MR, Fajer PG, Marshall AG. Advantages of isotopic depletion of proteins for hydrogen/deuterium exchange experiments monitored by mass spectrometry. Anal. Chem. 2010;82(8):3293–3299. doi: 10.1021/ac100079z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones LM, Zhang H, Vidavsky I, Gross ML. Online, high-pressure digestion system for protein characterization by hydrogen/deuterium exchange and mass spectrometry. Anal. Chem. 2010;82(4):1171–1174. doi: 10.1021/ac902477u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Chien EYT, Chalmers MJ, et al. Dynamics of the β(2)-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal. Chem. 2010;82(3):1100–1108. doi: 10.1021/ac902484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rey M, Mrazek H, Pompach P, et al. Effective removal of nonionic detergents in protein mass spectrometry, hydrogen/deuterium exchange, and proteomics. Anal. Chem. 2010;82(12):5107–5116. doi: 10.1021/ac100171m. [DOI] [PubMed] [Google Scholar]
- 65.Hebling CM, Morgan CR, Stafford DW, Jorgenson JW, Rand KD, Engen JR. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal. Chem. 2010;82(13):5415–5419. doi: 10.1021/ac100962c. • Shows the use of nanodisks to enable the HDX analysis of integral membrane proteins. Such methods will be required for the HDX analysis of challenging targets such as G protein-coupled receptors.
- 66.Fitzgerald MC, West GM. Painting proteins with covalent labels: what’s in the picture? J. Am. Soc. Mass Spectrom. 2009;20(6):1193–1206. doi: 10.1016/j.jasms.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 67.Ghaemmaghami S, Fitzgerald MC, Oas TG. A quantitative, high-throughput screen for protein stability. Proc. Natl Acad. Sci. USA. 2000;97(15):8296–8301. doi: 10.1073/pnas.140111397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang L, Roulhac PL, Fitzgerald MC. H/D exchange and mass spectrometry-based method for biophysical analysis of multidomain proteins at the domain level. Anal. Chem. 2007;79(22):8728–8739. doi: 10.1021/ac071380a. [DOI] [PubMed] [Google Scholar]
- 69.Zhu MM, Rempel DL, Du Z, Gross ML. Quantification of protein-ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. J. Am. Chem. Soc. 2003;125(18):5252–5253. doi: 10.1021/ja029460d. [DOI] [PubMed] [Google Scholar]
- 70.West GM, Tucker CL, Xu T, et al. Quantitative proteomics approach for identifying protein–drug interactions in complex mixtures using protein stability measurements. Proc. Natl Acad. Sci. USA. 2010;107(20):9078–9082. doi: 10.1073/pnas.1000148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dai SY, Chalmers MJ, Bruning J, et al. Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc. Natl Acad. Sci. USA. 2008;105(20):7171–7176. doi: 10.1073/pnas.0710802105. • Demonstrates that HDX signatures can be used to predict the tissue selective pharmacology of a number of estrogen receptor ligands.
- 72.Bennett MJ, Barakat K, Huzil JT, Tuszynski J, Schriemer DC. Discovery and characterization of the laulimalide-microtubule binding mode by mass shift perturbation mapping. Chem. Biol. 2010;17(7):725–734. doi: 10.1016/j.chembiol.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 73.Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 2005;67:309–333. doi: 10.1146/annurev.physiol.66.032802.154710. [DOI] [PubMed] [Google Scholar]
- 74.Wright E, Busby SA, Weisecarver S, Vincent J, Griffin PR, Fernandez EJ. Helix 11 dynamics is critical for constitutive androstane receptor activity. Structure. 2011;19(1):37–44. doi: 10.1016/j.str.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan X, Broderick D, Leid ME, Schimerlik MI, Deinzer ML. Dynamics and ligand-induced solvent accessibility changes in human retinoid X receptor homodimer determined by hydrogen deuterium exchange and mass spectrometry. Biochemistry. 2004;43(4):909–917. doi: 10.1021/bi030183c. [DOI] [PubMed] [Google Scholar]
- 76.Yan X, Perez E, Leid M, Schimerlik MI, de Lera AR, Deinzer ML. Deuterium exchange and mass spectrometry reveal the interaction differences of two synthetic modulators of RXRα LBD. Protein Sci. 2007;16(11):2491–2501. doi: 10.1110/ps.073019707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xia G, Boerma LJ, Cox BD, et al. Structure, energetics and dynamics of binding coactivator peptide to human retinoid x receptor α ligand binding domain complex with 9-cis-retinoic acid. Biochemistry. 2010 doi: 10.1021/bi101288y. DOI: 10.1021/bi101288y. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lipscombe LL, Gomes T, Levesque LE, Hux JE, Juurlink DN, Alter DA. Thiazolidinediones and cardiovascular outcomes in older patients with diabetes. JAMA. 2007;298(22):2634–2643. doi: 10.1001/jama.298.22.2634. [DOI] [PubMed] [Google Scholar]
- 79.Bruning JB, Chalmers MJ, Prasad S, et al. Partial agonists activate PPARγ using a helix 12 independent mechanism. Structure. 2007;15(10):1258–1271. doi: 10.1016/j.str.2007.07.014. •• HDX is used to discover a novel mechanism for the activation of a nuclear receptor.
- 80.Chandra V, Huang P, Hamuro Y, et al. Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature. 2008:350–356. doi: 10.1038/nature07413. •• Describes the first x-ray structure and HDX data for a full-length nuclear receptor complex.
- 81.Choi JH, Banks AS, Estall JL, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARγ by Cdk5. Nature. 2010;466(7305):451–456. doi: 10.1038/nature09291. •• Peroxisome proliferator-activated receptor γ (PPARγ) is demonstrated to be a substrate of CDK5. Reversible phosphorylation of PPAR is corelated with genes linked to metabolic dysregulation.
- 82.Hamuro Y, Coales SJ, Morrow JA, et al. Hydrogen/deuterium-exchange (H/D-Ex) of PPARγ LBD in the presence of various modulators. Protein Sci. 2006;15(8):1883–1892. doi: 10.1110/ps.062103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang YJ, Kumar N, Solt LA, et al. Modulation of retinoic acid receptor-related orphan receptor α and γ activity by 7-oxygenated sterol ligands. J. Biol. Chem. 2010;285(7):5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, Chalmers MJ, Stayrook KR, et al. Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within the intact vitamin D receptor/retinoid x receptor heterodimer. Structure. 2010;18(10):1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicolaides NC, Galata Z, Kino T, Chrousos GP, Charmandari E. The human glucocorticoid receptor: molecular basis of biologic function. Steroids. 2010;75(1):1–12. doi: 10.1016/j.steroids.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frego L, Davidson W. Conformational changes of the glucocorticoid receptor ligand binding domain induced by ligand and cofactor binding, and the location of cofactor binding sites determined by hydrogen/deuterium exchange mass spectrometry. Protein Sci. 2006;15(4):722–730. doi: 10.1110/ps.051781406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dai SY, Burris TP, Dodge JA, et al. Unique ligand binding patterns between estrogen receptor α and β revealed by hydrogen-deuterium exchange. Biochemistry. 2009;48(40):9668–9676. doi: 10.1021/bi901149t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007;16(4):572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen S, O’Reilly LP, Smithgall TE, Engen JR. Tyrosine phosphorylation in the SH3 domain disrupts negative regulatory interactions within the c-Abl kinase core. J. Mol. Biol. 2008;383(2):414–423. doi: 10.1016/j.jmb.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen S, Dumitrescu TP, Smithgall TE, Engen JR. Abl N-terminal cap stabilization of SH3 domain dynamics. Biochemistry. 2008;47(21):5795–5803. doi: 10.1021/bi800446b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc. Natl Acad. Sci. USA. 2009;106(5):1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J, Adrian FJ, Jahnke W, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463(7280):501–506. doi: 10.1038/nature08675. •• HDX study of wild-type and drug-resistant Bcr-Abl mutants.
- 93.Gajiwala KS, Wu JC, Christensen J, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc. Natl Acad. Sci. USA. 2009;106(5):1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang HM, Yu X, Greig MJ, et al. Drug binding and resistance mechanism of KIT tyrosine kinase revealed by hydrogen/deuterium exchange FTICR mass spectrometry. Protein Sci. 2010;19(4):703–715. doi: 10.1002/pro.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.George SR, O’Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat. Rev. Drug Discov. 2002;1(10):808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 96.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009;8(1):41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 98.Cherezov V, Rosenbaum DM, Hanson MA, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosenbaum DM, Cherezov V, Hanson MA, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. •• First crystal structures of a diffusible ligand G protein-coupled receptor.
- 100.Rasmussen SG, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–387. doi: 10.1038/nature06325. •• First crystal structures of a diffusible ligand G protein-coupled receptor.
- 101.Busenlehner LS, Salomonsson L, Brzezinski P, Armstrong RN. Mapping protein dynamics in catalytic intermediates of the redox-driven proton pump cytochrome c oxidase. Proc. Natl Acad. Sci. USA. 2006;103(42):15398–15403. doi: 10.1073/pnas.0601451103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Busenlehner LS, Codreanu SG, Holm PJ, et al. Stress sensor triggers conformational response of the integral membrane protein microsomal glutathione transferase 1. Biochemistry. 2004;43(35):11145–11152. doi: 10.1021/bi048716k. [DOI] [PubMed] [Google Scholar]
- 103.Joh NH, Min A, Faham S, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453(7199):1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stelzer W, Poschner BC, Stalz H, Heck AJ, Langosch D. Sequence-specific conformational flexibility of SNARE transmembrane helices probed by hydrogen/deuterium exchange. Biophys. J. 2008;95(3):1326–1335. doi: 10.1529/biophysj.108.132928. [DOI] [PMC free article] [PubMed] [Google Scholar]