Abstract
Hematopoietic malignancies are frequently associated with DNA hypomethylation but the molecular mechanisms involved in tumor formation remain poorly understood. Here we report that mice lacking Lsh develop leukemia associated with DNA hypomethylation and oncogene activation. Lsh is a member of the SNF2 chromatin remodeling family and is required for de novo methylation of genomic DNA. Mice that received Lsh deficient hematopoietic progenitors showed severe impairment of hematopoiesis, suggesting that Lsh is necessary for normal hematopoiesis. A subset of mice developed erythroleukemia, a tumor that does not spontaneously occur in mice. Tumor tissues were CpG hypomethylated and showed a modest elevation of the transcription factor PU.1, an oncogene that is crucial for Friend virus induced erythroleukemia. Analysis of Lsh−/− hematopoietic progenitors revealed widespread DNA hypomethylation at repetitive sequences and hypomethylation at specific retroviral elements within the PU.1 gene. Wild type cells showed Lsh and Dnmt3b binding at the retroviral elements located within the PU.1 gene. On the other hand, Lsh deficient cells had no detectable Dnmt3b association suggesting that Lsh is necessary for recruitment of Dnmt3b to its target. Furthermore, Lsh−/− hematopoietic precursors showed impaired suppression of retroviral elements in the PU.1 gene, an increase of PU.1 transcripts and protein levels. Thus DNA hypomethylation caused by Lsh depletion is linked to transcriptional upregulation of retroviral elements and oncogenes such as PU.1 which in turn may promote the development of erythroleukemia in mice.
Keywords: DNA methylation, chromatin, Lsh, SNF2, leukemia, hematopoiesis
Introduction
Epigenetic alterations have been demonstrated to be involved in tumorigenesis.1–3 Abnormal posttranslational histone modifications as well as aberrant CpG methylation patterns have been associated with human cancer. These abnormal chromatin states may lead to genomic instability, enhanced mutations rates, loss of imprinting, oncogene overexpression or silencing of tumor suppressor genes.
Human tumors are frequently associated with global reduction of DNA methylation.2–4 The cause for loss of CpG methylation remain unclear and it is still not known whether genomic hypomethylation causes or is a consequence of human cancer. However, since demethylating agents are currently in therapeutic use,4 it is important to determine if and how genomic hypomethylation affects tumorigenesis.
Previously, the role of DNA methylation in tumor development has been studied using transgenic mice carrying mutant forms of the DNA methyltransferase (Dnmt) family.5–11 Dnmt1 is present at replication forks and contributes to maintenance of methylation pattern whereas Dnmt3a and Dnmt3b perform de novo methylation and are crucial for silencing of retroviral transgenes.12 Mutations of Dnmt1, Dnmt3a and 3b lead to defects in embryogenesis including loss of imprinting, defects in x-inactivation and deregulation of gene expression suggesting a crucial role for DNA methylation during development.13 With respect to tumor formation, DNA hypomethylation can protect as well as promote depending on the genetic mouse model chosen.5–11 For example, decreased Dnmt1 activity was shown to inhibit intestinal tumor formation in ApcMin/+ mice or colorectal adenocarcinomas in Mlh1−/− mice.5–8 In these models, hypermethylation of tumor suppressor genes could be involved and therefore a defect in methylation may lead to tumor protection. In contrast, lymphomagenesis was increased in Dnmt1 deficient mice and chromosomal gains were associated with tumor development, suggesting a protective role for DNA methylation in genome stability.9 Thus depending on the origin of the tumor, its cell type specificity and the introduction of additional genetic mutations, tumor promotion as well as delay of onset were reported, suggesting a complex role for DNA methylation in cancer development.
To explore further the functional link between genomic hypomethylation and cancer, we studied the development of hematopoietic neoplasms using the Lsh−/− mouse model.14,15 Lsh is a member of the SNF2 family of chromatin remodeling proteins, and is a major epigenetic regulator in mice.16–20 Snf2 homologues can change nucleosomal positioning in vitro and therefore alter the access of DNA binding factors or chromatin modifying enzymes in vivo.21 Lsh is directly involved in the process of de novo methylation and can associate with Dnmt3a and Dnm3b.22 Deletion of Lsh in mice leads to perturbed heterochromatin structure, particularly reducing DNA methylation.16,19 As biological consequences of perturbed heterochromatin, we observed reactivation of endogenous retroviral elements, defects of imprinting at selected sites and abnormal mitosis.18,23,24 Since Lsh−/− mice died shortly after birth, tumor development cannot be studied using adult Lsh−/− mice.15 In this manner, we generated radiation chimera in order to investigate the formation of hematopoietic tumors in a normal environment using C57BL/6J mice as recipients. In addition, a mutant p53 allele was introduced into the Lsh deficient background since mice with a deletion of the p53 gene develop hematopoietic neoplasms at a high frequency.25 Here we show that deletion of Lsh induces the development of leukemia associated with hypomethylation and increased expression of the PU.1 gene, encoding a transcription factor that is frequently involved in the development of erythroleukemia.26,27
Results
Lsh is essential for normal hematopoiesis
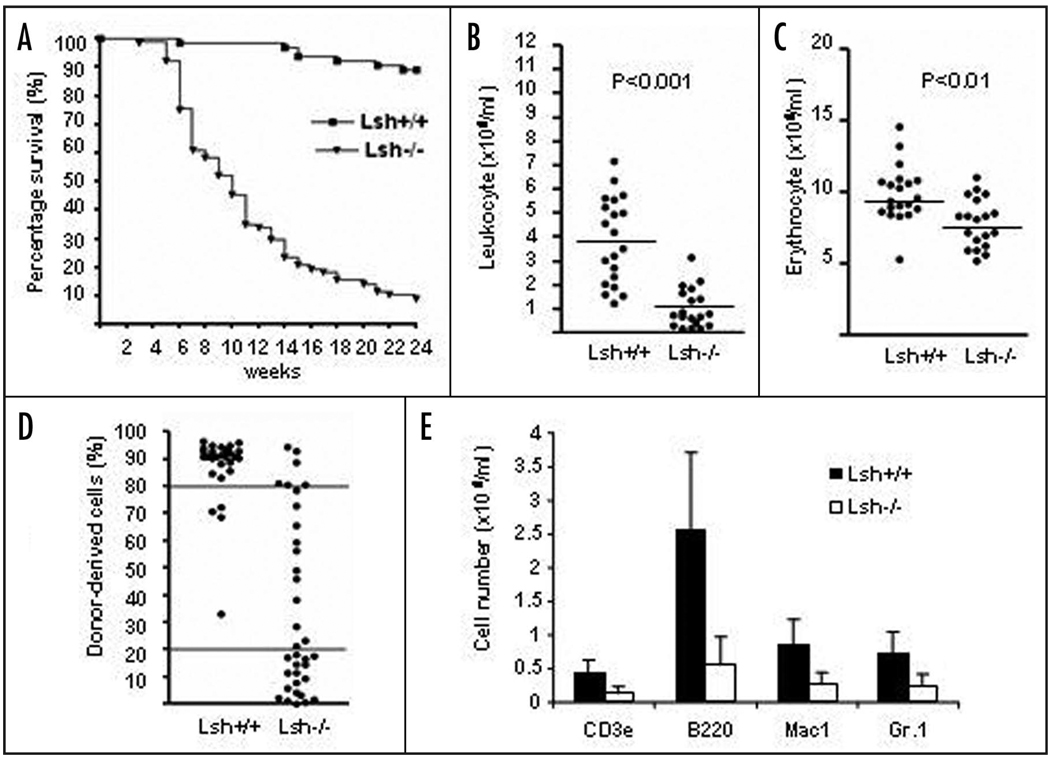
Since Lsh deletion is perinatal lethal we performed hematopoietic reconstitution experiments to investigate the role of Lsh in hematopoietic diseases.15 Hematopoietic precursor cells from fetal liver (day 14.5 to 16.5 gestation) derived from Lsh−/− or wild type littermates were transplanted into wild type lethally irradiated hosts. Mice that received Lsh−/− fetal liver cell transplants showed significantly reduced survival rates with 90% dying within 24 weeks (Fig. 1A). p53 depletion could not rescue the phenotype since mice transplanted with Lsh−/−p53−/− fetal liver cells died at a similar rate (Suppl. Fig. 1A). Peripheral leukocyte numbers were reduced about 70% in mice receiving Lsh−/− (Fig. 1B) or Lsh−/−p53−/− fetal liver cells (Suppl. Fig. 1B). Erythrocyte numbers in mice transplanted with Lsh−/− donor cells declined about 20% in comparison with controls that received wild type donor cells (Fig. 1C) and the hematocrit was reduced by a similar amount starting at three weeks after transplantation (Suppl. Fig. 1C). FACS (fluorescence-activated cell sorting) analysis of peripheral blood cells using antibodies against donor CD45.2+ allowed for monitoring of the fate of donor cells in the CD45.1+ expressing recipient strain C57BL/6J. As shown in Figure 1D, leukocytes that were derived from Lsh−/− derived donor cells were significantly decreased (in average 64% for Lsh−/− derived precursors versus 15% for Lsh+/+) indicating a reduced engraftment rate of Lsh−/− transplanted cells in comparison with Lsh+/+ transplanted cells. p53 mutations did not rescue the phenotype since Lsh−/−p53−/− transplanted cells showed a similar failure of engraftment (Suppl. Fig. 1D). FACS analysis for the expression of lineage specific markers and calculation of the absolute numbers for donor derived cells revealed a reduction of several lineages in the absence of Lsh such as reduced T cells (as marked by CD3+), B cells (B220+), neutrophils (GR1+) or Mac1+ positive cells (macrophages/ neutrophils) (Fig. 1E). Analysis of CD45.2+ donor derived cells in the bone marrow confirmed a relative decrease in B220+, GR1+ and Mac1+ precursors in the absence of Lsh (Suppl. Fig. 1E). Since CD45.2 is expressed on all lineages of hematopoietic precursors, FACS analysis using erythrocyte specific cell surface markers could also assess the proportion of donor derived immature erythrocytes in the bone marrow (note: CD45 is not expressed on mature erythrocytes in the peripheral blood). The relative increase of TER119+ (expressed on immature and mature erythrocytes) and transferrin receptor (CD71, high levels on developing erythroid cells) expressing cells suggested a maturation block in the erythroid lineage since peripheral blood displayed reduced numbers of circulating mature erythrocytes in mice transplanted with Lsh−/− fetal liver donor cells (Fig. 1C). Full pathology and histopathology of mice receiving Lsh−/− fetal liver cells revealed widespread signs of inflammation, including bacterial colonies in multiple tissues such as skin, colon, lung, nasal cavity and muscle (Table 1). Therefore, infection was the cause for mortality in 17/37 animals transplanted with Lsh depleted donor cells. This was consistent with the observation of a reduced number of white blood cells leading to a defect in immune response. Five animals with Lsh deficiency died because of anemia, which was consistent with reduced peripheral erythrocyte numbers. In summary, the data indicated that Lsh deficient hematopoietic progenitors show an intrinsic defect in hematopoiesis leading in part to a failure of host defense and early death of the animals.
Figure 1.
Impaired hematopoiesis in mice transplanted with Lsh−/− derived embryonal liver cells. (A) Survival curve of mice transplanted with fetal liver cells derived from Lsh−/− and Lsh+/+ embryos. Survival of mice was observed during a period of 24 weeks. Lsh−/− n = 77, Lsh+/+ n = 64. (B and C) Leukocytes were counted after lysis of erythrocytes using ACK buffer. Erythrocytes were counted from whole peripheral blood. Each dot represents an individual animal. The average cell number for different genotypes is represented by the horizontal line. (D) Reduced engraftment efficiency in Lsh deficient fetal liver transplanted mice. The engraftment in peripheral blood was determined by flow cytometry using antibodies to CD45.1+ and CD45.2+ at about ten weeks post-transplantation to discriminate between donor and host cells. Horizontal lines visualize arbitrarily the 20% and 80% engraftment efficiency. (E) FACS analysis of peripheral blood of reconstituted animals using distinct antibodies against the indicated cell surface antigens. The donor developed hematopoietic subsets are expressed as absolute cell number.
Table 1.
Cause for mortality in mice transplanted with Lsh−/− and Lsh−/−p53−/− donor cells
| Lsh+/+ (n = 10) | Lsh−/− (n = 15) | Lsh−/−p53−/− (n = 22) | Lsh+/+p53−/− (n = 12) | |
|---|---|---|---|---|
| Anemia | 0% | 27% (n = 4) | 5% (n = 1) | 0% |
| Infection/Inflammation | 10% (n = 1) | 60% (n = 9) | 36% (n = 8) | 0% |
| Pneumonia | 5% (n = 1) | |||
| Typhocolitis | 13% (n = 5) | 14% (n = 3) | ||
| Dermatitis | 20% (n = 3) | 14% (n = 3) | ||
| Myositis | 7% (n = 1) | 5% (n = 1) | ||
| Otitis media | 10% (n = 1) | |||
| Other* | 90% (n = 9) | 13% (n = 2) | 59% (n = 13) | 100% (n = 12) |
Other reasons include tumors and undetermined causes of death.
Lsh depletion leads to development of neoplasms
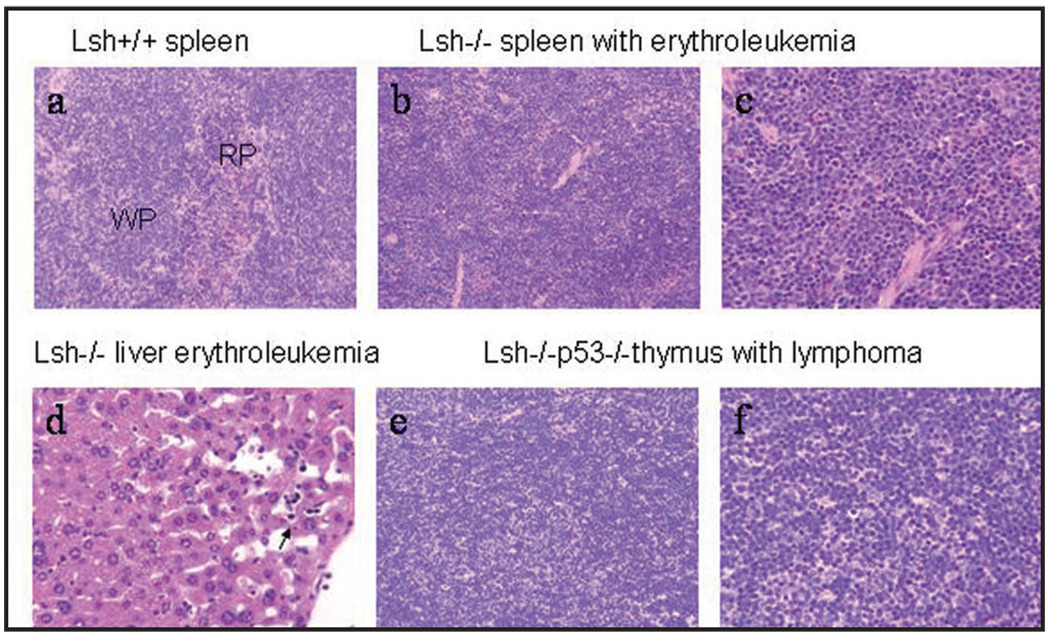
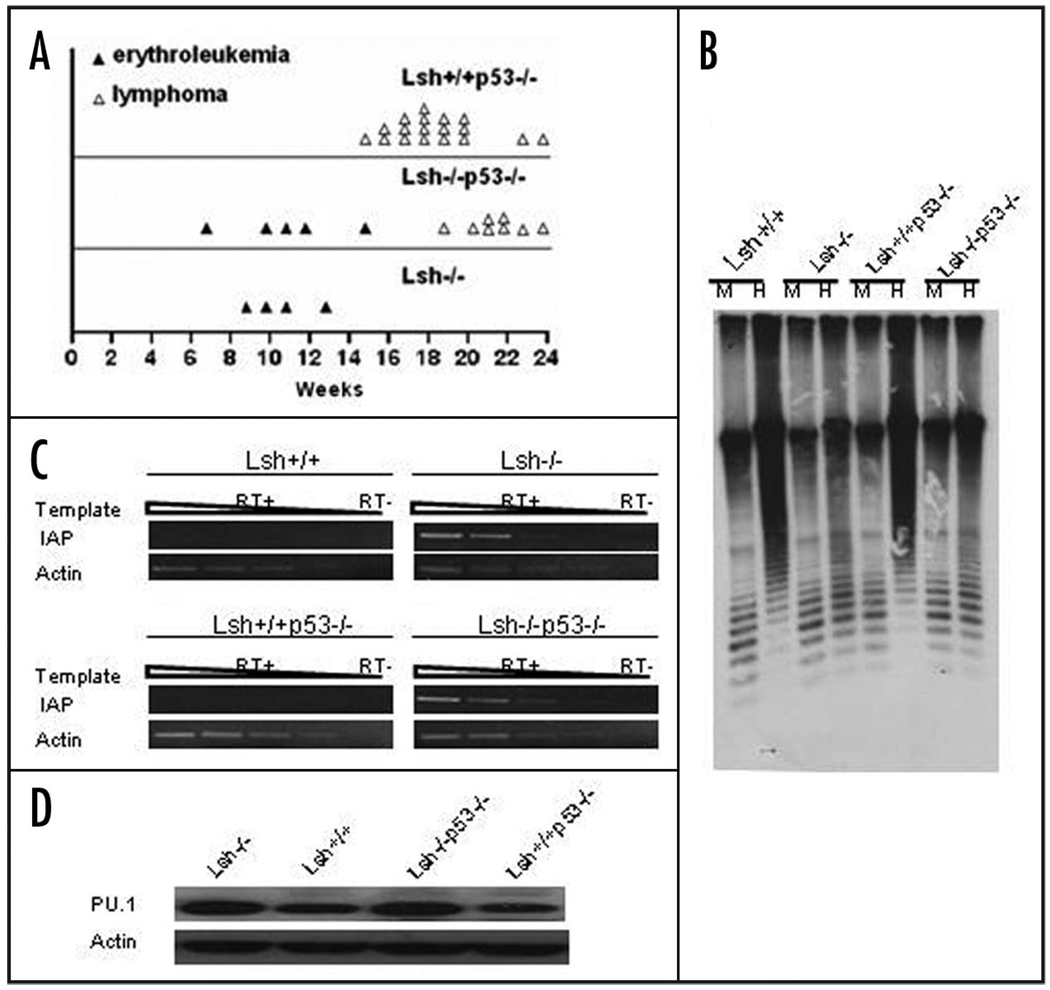
Though, most animals with a deletion of Lsh showed signs of infections, fifteen animals died for other reasons. Full pathology and histopathology revealed that twelve animals had developed hematopoietic neoplasms (Table 1 and Fig. 2). Eight were classified as lymphoma which was expected, because mice carrying a p53 mutation are prone to T cell lymphomas and all lymphomas were exclusively found in animals transplanted with Lsh−/−p53−/− donor cells. Surprisingly, four animals receiving Lsh−/− donor cells were diagnosed with erythroleukemia based on histopathology of the spleen, anemia, splenomegaly and erythroleukemic cells in the liver or bone marrow (Fig. 2). Erythroleukemia does not occur spontaneously in mice but is inducible by the Friend erythroleukemia virus. Since murine erythroleukemia originates in the spleen, the spleens of additional mice transplanted with Lsh−/− donor cells were evaluated pathologically. Altogether about 9% of all examined spleens transplanted with Lsh deficient donor cells (n = 9 out of 102 total) were diagnosed with erythroleukemia compared with none of the spleens from mice receiving wild type donor cells (n = 29) (Table 2). More than half of all tumorigenic animals transplanted with Lsh deficient fetal liver cells developed signs of abnormal erythropoiesis (14 out of 22 animals). There was no evidence that p53 mutation was promoting the development of erythroleukemia in contrast to a recent report.28 An effect of Lsh depletion on p53 induced lymphoma development could not be assessed since most mice receiving Lsh−/− donor cells died by infection or erythroleukemia before the onset of lymphoma which occurs after approximately 15 weeks (Fig. 3A).
Figure 2.
Histopathological analysis of erythroleukemia and lymphoma in fetal liver cell transplanted mice. (a) Normal Spleen from mice receiving Lsh+/+ fetal liver cells. Lymphoid tissue in white pulp (wp) and normal extramedullary hematopoiesis in red pulp (rp) (H&E 20× objective). (b) Spleen with erythroleukemia from mice transplanted with Lsh−/− fetal liver cells. Effacement of white pulp by expansion of erythropoiesis originating in the red pulp. This hematopoietic neoplasm occurred in mice transplanted with Lsh−/− and Lsh−/− p53−/− fetal liver cells (H&E, 20× objective). (c) Spleen with erythroleukemia from mice transplanted with Lsh−/− fetal liver cells. Many erythrocyte precursors (cells with prominent nucleoli) as well as numerous metarubricytes (dark nuclei) (H&E, 40× objective). (d) Liver erythroleukemia from mice transplanted with Lsh−/− fetal liver cells. Circulating metarubricytes in hepatic sinusoids can be seen (arrow) (H&E, 40× objective). (e) T cell lymphoma from mice transplanted with Lsh−/−p53−/− fetal liver cells (H&E, 20×, objective). (f) Thymus with sheets of lymphoblasts from mice transplanted with Lsh−/−p53−/− fetal liver cells. (H&E, 40× objective).
Table 2.
Occurrence of hematopoietic neoplasm in mice transplanted with Lsh−/− and Lsh−/−p53−/− donor cells
| Lsh+/+ (n = 17) | Lsh−/− (n = 58) | Lsh−/−p53−/− (n = 44) | Lsh+/+p53−/− (n = 12) | |
|---|---|---|---|---|
| Erythroleukemia | 0% | 7% (n = 4) | 11% (n = 5) | 0% |
| Atypic erythroid hyperplasia | 0% | 7% (n = 4) | 2% (n = 1) | 0% |
| Lymphoma/Thymoma | 0% | 0% | 18% (n = 8) | 100% (n = 12) |
| No neoplasm | 100% | 86% (n = 50) | 68% (n = 30) | 0% |
Figure 3.
Genomic hypomethylation, activation of retroviral elements and PU.1 elevation in tumor tissues. (A) Time point of erythroleukemia and thymoma/lymphoma development in mice transplanted with fetal liver cells derived from the indicated genotypes. (B) Southern blot analysis of minor satellite repeat region in genomic DNA from spleens of mice transplanted with fetal liver cells. Genomic DNA digested with either MspI (M) or HpaII (H) was probed with MR150. Spleen from Lsh−/− and Lsh−/−p53−/− recipient mice was diagnosed with erythroleukemia. (C) RT-PCR analysis for detection of IAP transcripts in spleens from Lsh−/− and Lsh−/−p53−/− recipient mice that were diagnosed with erythroleukemia and from control mice. (D) PU.1 protein levels were examined in spleens transplanted with Lsh−/− or Lsh−/−p53−/− fetal livers or Lsh+/+ and Lsh+/+p53−/− controls by Western blot analysis. Spleens from Lsh−/− and Lsh−/−p53−/− recipient mice were diagnosed with erythroleukemia.
Increases of PU.1 protein in the tumor
To characterize further the developing erythroleukemia, we examined global DNA methylation levels in the tumor tissue. As shown in Figure 3B, erythroleukemic spleen derived from mice transplanted with Lsh−/− as well as Lsh−/−p53−/− donor cells showed global DNA hypomethylation as demonstrated by higher digestibility at satellite sequences using the methylation sensitive restriction enzyme HpaII. Furthermore, expression of retroviral elements that are dispersed throughout the genome was analyzed using primers located within the repeat sequences. An activation of usually silenced retroviral elements such as IAP (intracisternal A particle) could be detected in Lsh depleted tumor samples (Fig. 3C) as has been previously reported for Dnmt1 or Lsh depleted tissues.18,29 Thus tumor development was associated with global DNA hypomethylation and derepression of endogenous retroviral repeats.
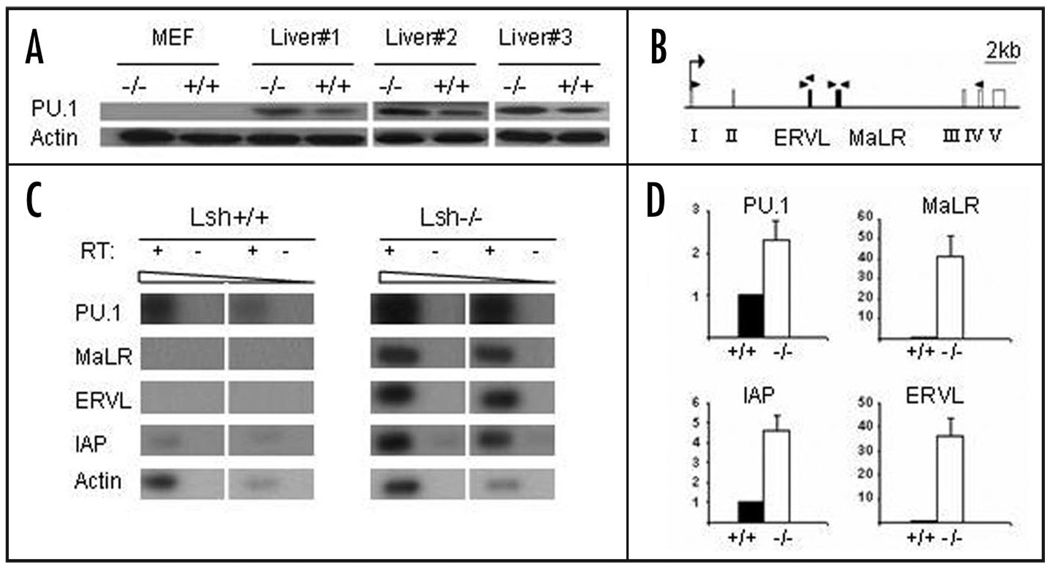
To further investigate the molecular mechanisms of erythroleukemia development, we considered the possibility that activation of PU.1 could play a role in tumor development. PU.1 is crucial for normal hematopoiesis and the PU.1 gene has been originally identified as an oncogene in the Friend virus erythroleukemia model.26,27 The PU.1 locus also known as Sfpi1 (Spleen focus forming virus Proviral Integration 1) is a common integration site of the Friend virus in murine erythroleukemia. The retroviral sequences brought into close proximity of the promoter region lead to PU.1 upregulation. PU.1 is a transcription factor that blocks terminal differentiation of erythroblasts and overexpression leads to expansion of the erythroid precursor pool.30,31,27 Though subsequent events are involved in tumorigenesis, overexpression of PU.1 alone can ultimately lead to the development of erythroleukemia in mice indicating that PU.1 can promote the disease.27 Because of the eminent role of PU.1 in erythroleukemia and since mice that had received Lsh−/− fetal liver transplants also showed a relative expansion of the erythrocyte precursor pool in the bone marrow, it prompted us to examine PU.1 expression levels in erythroleukemia. As shown in Figure 3D, Western analysis revealed a slight increase in PU.1 protein levels in tumor tissues of mice transplanted with Lsh−/− or Lsh−/−p53−/− donor cells compared to controls. In the Friend erythroleukemia model integration of the exogenous retrovirus leads to activation of the PU.1 gene. However, preliminary Southern analysis did not reveal any evidence for mobility and integration of endogenous retroviral elements into the PU.1 locus in tumor tissues (data not shown). Unexpectedly, we found a similar raise in PU.1 protein levels in Lsh depleted hematopoietic progenitors (Fig. 4A). To test whether this PU.1 increase was due to a transcriptional elevation in hematopoietic progenitors, Lin− Sca1+Kit+ hematopoietic stem cells were purified from fetal liver and were examined for PU.1 mRNA expression. A two to three-fold increase of PU.1 transcripts in the absence of Lsh was detected using RT-PCR analysis as compared to controls (Fig. 4C and D). This confirmed that the rise in PU.1 was transcriptional regulated and that it was not due to an altered composition of hematopoietic subsets in the liver. Thus the cause for PU.1 elevation was already present in progenitors and thus rather a preceding event than a consequence of tumor transformation.
Figure 4.
Increased PU.1 expression in Lsh−/− hematopoietic progenitors and reactivation of endogenous retroviral elements. (A) PU.1 protein levels were examined in extracts derived from MEFs or fetal livers from three different Lsh−/− or Lsh+/+ control mice by Western blot analysis. (B) Graph of the PU.1 locus showing the five exons (open boxes), the location of the two retroviral elements ERVL and MaLR (black boxes), the transcriptional start site (arrow) and the position of the primers used for RT-PCR analysis (black triangles). The primers used for detection of PU.1 transcripts were located in exon I and exon IV. The 5' and 3' primer for MaLR and the 5' primer for ERVL were located outside of the repeat region allowing specific detection of transcripts. (C) RT-PCR analysis for detection of PU.1 transcripts and ERVL and MaLR transcripts located in the PU.1 locus. RT-PCR analysis for IAP transcripts was used for comparison and actin as a control. RNA was prepared from Lin-Sca1+Kit+ purified progenitor cells derived from Lsh−/− or Lsh+/+ fetal liver. (D) Bar graph of semi-quantitative RT-PCR analysis for detection of PU.1, IAP, ERVL and MaLR. Number represents arbitrary units relative to actin. The data for PU.1 transcripts summarizes PCR analysis performed with RNA from three independent purified Lin−Sca1+Kit+ progenitor cells. The other graphs summarize results from two independent stem cell purifications.
PU.1 increases are associated with hypomethylation at specific sites of the PU.1 gene
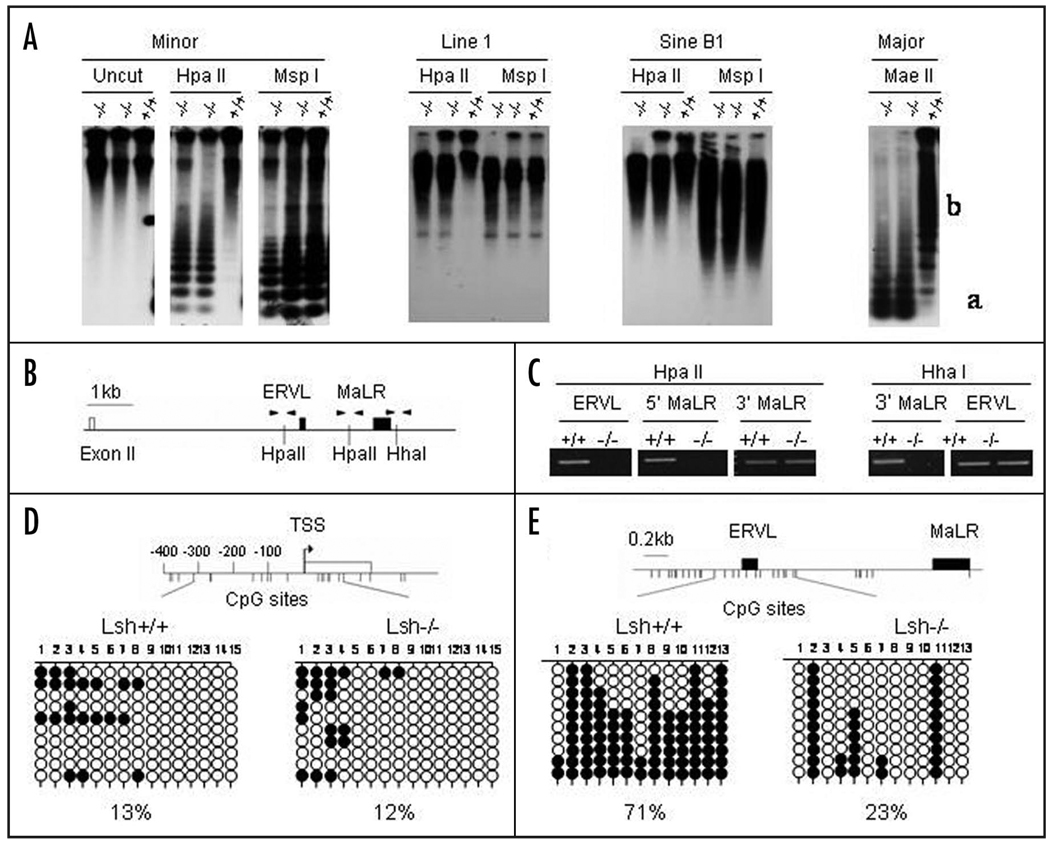
To study the mechanism for the increase in PU.1 transcription in Lsh depleted progenitors, we examined the DNA methylation pattern in hematopoietic precursors. First, we analyzed DNA methylation at repetitive sequences using Southern analysis. As shown in Figure 5A, embryonal liver precursors derived from Lsh−/− embryos were highly sensitive to Hpa II digestion (or Mae II) digestion and revealed DNA hypomethylation at minor and major satellite sequences. In addition, Line 1 and Sine B1 elements that are located throughout the genome were highly sensitive to Hpa II digestion (Fig. 5A). The results indicated widespread DNA hypomethylation in hematopoietic precursor cells in the absence of Lsh.
Figure 5.
Genomic hypomethylation in hematopoietic precursors at retroviral elements located in the PU.1 locus. (A) Southern analysis of genomic DNA derived from fetal liver of Lsh−/− embryos or Lsh+/+ embryos using indicated methylation sensitive restriction enzymes HpaII or MaeII or MspI as control. For detection of minor, major satellites or Line1 and Sine B1 elements the indicated probes were used. (B) Graph of exon II of the PU.1 gene and the retroviral elements ERVL and MaLR located 3' of exon II. The position of the restriction enzyme sites HpaII and HhaI that are used in the methylation sensitive PCR analysis is indicated as well as the primers (black triangles) upstream of ERVL (ERVL), between ERVL and MaLR (5' MaLR) and downstream of MaLR (3' MaLR). (C) Methylation sensitive PCR analysis of two HpaII sites and one HhaI site located around the retroviral elements in the PU.1 locus. Genomic DNA derived from Lsh−/− or Lsh+/+ fetal liver was digested with the indicated restriction enzymes and subjected to PCR analysis. HpaII digested DNA and primers for detection of HpaII sites served as control for HhaI analysis and vice versa. (D) Bisulphite sequencing analysis of the PU.1 promoter using genomic DNA derived from Lsh−/− or Lsh+/+ fetal liver samples. Each line represents sequencing result of individual clones. The 15 circles (unmethylated open circle and methylated black circle) represent 15 CpG sites at the PU.1 promoter as illustrated in the graph above. (E) Bisulphite sequencing analysis of genomic DNA derived from Lsh−/− or Lsh+/+ fetal liver samples. Each line represents a sequencing result of individual clones. The 13 circles represent 13 CpG sites located around ERVL and MaLR elements of the PU.1 locus as illustrated in the graph above.
Next, we used methylation sensitive PCR analysis to address the question whether the PU.1 gene too showed DNA hypomethylation in the absence of Lsh. Utilizing the methylation sensitive restriction enzymes Hpa II or HhaI we analyzed first the methylation status at two retroviral elements located between exon 2 and 3 of the PU.1 gene (Fig. 5B). The two retroviral elements with long terminal repeats (LTR), ERVL (Endogenous RetroVirus Like) and MaLR (Mammalian Apparent LTR-Retrotransposon), were the only retroviral elements detected in the PU.1 locus using the Repeatmasker Program (including in the search a region 16000 bp upstream of the transcriptional start site). Whereas successful amplification around the HpaII sites indicated CpG methylation in wild type samples, Lsh−/− samples revealed loss of DNA methylation (Fig. 5C). PCR analysis using primer pairs surrounding a HhaI site served as a control for equal input of DNA. Likewise, successful amplification of a fragment around a HhaI site after HhaI digestion indicated DNA methylation in wild type, but showed hypomethylation in Lsh−/− samples. Thus Lsh depletion leads to reduced CpG methylation at selected sites within the PU.1 locus.
To confirm the evidence of hypomethylation within the PU.1 locus and also to analyze the methylation status at the promoter region, bisulphite sequencing was performed. Previous reports had identified a 500 base pair region including 350 bp upstream of the transcriptional start site that was sufficient to confer a tissue specific expression pattern using reporter assays.31 Therefore we examined the methylation status of CpG sites within the PU.1 promoter (Fig. 5D). Wild type cells as well as Lsh depleted cells showed very little DNA methylation (13% and 12% respectively) suggesting that the promoter region was not affected by DNA methylation. This result is in accordance with previous reports that find little DNA methylation at promoter regions in normal tissue.13,32,33 In contrast, significant methylation differences were detected at the two retroviral elements, ERVL and MaLR, located between exon 2 and 3 of the PU.1 gene (Fig. 5E). Retroviral elements are usually methylated in the genome and it has been hypothesized that CpG methylation is a crucial epigenetic mechanism to silence these parasitic elements.13,29 As shown in Figure 5E, whereas wild type samples were methylated about 71% at the examined CpG sites located around the ERVL and MaLR, Lsh−/− samples showed a reduction in methylation to 23%.
To search for functional consequences of DNA hypomethylation, we designed primers to detect transcripts of these specific retroviral repeats by RT-PCR analysis (Fig. 4B). Wild type samples of Lin− Sca1+Kit+ progenitors showed no detectable transcripts, in contrast, Lsh depletion resulted in reactivation of both ERVL and MaLR elements in purified hematopoietic progenitor cells (Fig. 4C and D).
Lsh associates with retroviral elements and controls association of Dnmt3b
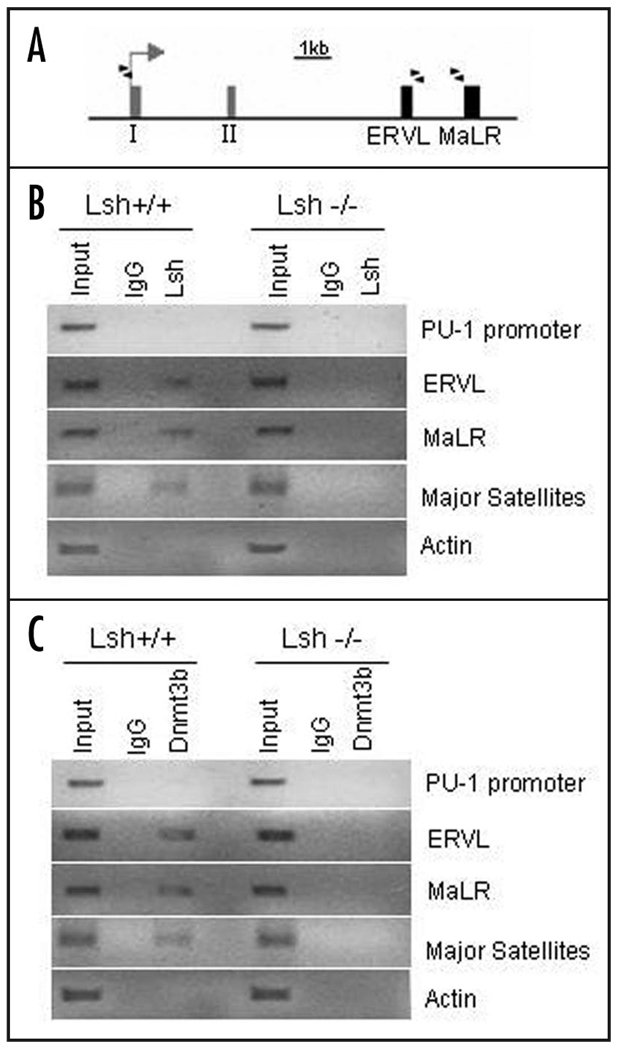
In order to address the question whether Lsh is directly involved in DNA methylation at the ERVL and MaLR element of the PU.1 gene we performed ChIPs analysis. Using chromatin derived from wild type embryonal liver cells we first examined binding of Lsh to distinct sites of the PU.1 gene (Fig. 6A and B). Whereas association of Lsh to the promoter elements was undetectable, Lsh showed binding to ERVL and MaLR elements within the PU.1 locus. In contrast, using chromatin derived from Lsh−/− embryonal liver cells did not show any precipitation using anti-Lsh antibodies (similar to IgG controls). As further controls, Lsh was also found associated with major satellite sequences being consistent with the observation that Lsh controls DNA methylation levels at major satellite repeats (Fig. 5A). In contrast, there was no detectable association of Lsh with the housekeeping actin gene (Fig. 6B). The correlation of Lsh binding with DNA methylation levels suggests a direct involvement of Lsh in the regulation of DNA methylation at the PU.1 gene.
Figure 6.
Lsh associates with specific sites of the PU.1 gene and regulates Dnmt3b binding to the PU.1 gene. (A) Graph of the PU.1 locus showing the first two exons (grey boxes), the location of the two retroviral elements ERVL and MaLR (black boxes), the transcriptional start site (arrow) and the position of the primers used for ChIPs analysis (black triangles). (B) ChIP analysis was performed using chromatin extracts of fetal liver derived from Lsh−/− and Lsh+/+ embryos and using specific antiserum against Lsh or IgG control. PCR analysis was performed for detection of binding to regions in the PU.1 promoter, the ERVL and MaLR elements. PCR analysis for major satellite or actin sequences served as positive and negative controls, respectively. The figure shows an inversion of an ethidium bromide image after agarose gel electrophoresis. (C) ChIP analysis was performed from chromatin extracts of fetal liver derived from Lsh−/− and Lsh+/+ embryos as in (B) using specific antiserum against Dnmt3b and species matched IgG control.
Next, we asked whether the presence of Lsh could affect Dnmt3b binding, since we had previously reported co-precipitation of Lsh and Dnmt3b suggesting a close interaction of the two proteins.22,39 As shown in Figure 6C, in wild type cells Dnmt3b associated with the retroviral elements ERVL and MaLR, as well as the major satellite sequence (serving as positive control) and there was no detectable Dnmt3b binding to the PU.1 promoter or the actin gene. In contrast, Lsh deficient cells showed greatly reduced Dnmt3b binding to PU.1 retroviral elements. Thus Dnmt3 and Lsh binding correlated well with DNA methylation levels and the results suggested that Dnmt3b binding is controlled by the presence of Lsh.
In summary, Lsh depletion resulted in genome wide DNA hypomethylation as well as specific hypomethylation at retroviral elements located in the PU.1 locus. This was associated with transcriptional activation of those endogenous retroviral elements and a slight increase in mRNA and protein levels of the tumor promoting factor PU.1.
Discussion
Here we report hematopoietic defects as well as erythroleukemia development in mice transplanted with Lsh−/− donor cells. Lsh depletion reduces Dnmt3b binding and DNA methylation at the PU.1 gene in hematopoietic stem cells. The rise in PU.1 transcripts and the slight rise in PU.1 protein level may then contribute to the development of erythroleukemia in the absence of Lsh.
Previous models of DNA hypomethylation and hematopoietic malignancies have utilized a hypomorphic Dnmt1 allele. Attenuation of Dnmt1 activity is sufficient to induce neoplasms, however, in contrast to our work these neoplasms are of T cell origin.9 By the time lymphomas were developing in Dnmt1 attenuated mice (4 to 8 months), most Lsh−/− recipients had already died because of failed engraftment, reduced immune defense, or erythroleukemia, thus excluding an evaluation of Lsh deletion on lymphomagenesis. On the other hand, erythroleukemias were not observed in Dnmt1 attenuated mouse models, indicating significant differences in the molecular actions of Lsh and Dnmt1. Whereas Dnmt1 depletion leads to substantial alteration of imprints, loss of Lsh showed limited effects (so far only one imprinted locus was affected). While the Lsh−/− model leads to a decrease in CpG methylation of about 50% resulting in retroviral activation, the hypomorphic Dnmt1 allele shows only moderate loss of methylation and as a consequence no enhanced retroviral activity.9 Furthermore, Dnmt1 shows multiple interactions with transcriptional repressors and tumor suppressor genes (e.g., Rb and HDAC) suggesting aberrant transcriptional regulation with Dnmt1 attenuation.13,15 Thus the exploration of molecular pathways using distinct genetic models is crucial for determining how genomic hypomethylation is involved in tumorigenesis.
Our data support a link between PU.1 increase and erythroleukemia development as has been previously demonstrated in the PU.1 transgenic animal model. It had been demonstrated that overexpression of PU.1 alone is sufficient to promote erythroleukemia.27,28 PU.1, an Ets family transcription factor, is expressed in hematopoietic lineages including erythroblasts.30 During normal hematopoiesis PU.1 is turned off in erythrocyte precursors. However, continuous overexpression of PU.1 blocks terminal differentiation and leads to an expansion of the erythrocyte precursor pool. This is consistent with our observation of a relative increase of erythrocyte precursors in the bone marrow of mice transplanted with Lsh−/− progenitors. Subsequent mutational events are thought to be required for transformation of the expanded erythrocyte precursor pool.34 Based on the Friend virus model and transgenic mice data, PU.1 elevation in the absence of Lsh may play a role in leukemia development. However, additional mechanisms that are linked to DNA hypomethylation may also participate such as a failure to maintain all genomic imprints,35 chromosomal segregation defects due to perturbed centromeric heterochromatin,9,36 increases in mutations due to CpG hypomethylation37 and the transcriptional deregulation of other genes possibly involved in tumorigenesis.38–40
Lsh is involved in de novo methylation and can associate with Dnmt3b.22,39,40 Here we report binding of Lsh to retroviral elements located in the PU.1 gene and show that the presence of Lsh is important for association of Dnmt3b with these retroviral elements. This is consistent with the hypothesis that Lsh may regulate in part DNA methylation by controlling access of Dnmt3b to appropriate target sites.39 The two retroviral elements of the PU.1 gene, ERVL and MaLR, that showed differential methylation after Lsh depletion belong to the family of degenerate long-terminal repeat (LTR) transposons. 41,42 LTR elements make up 4–8% of the mammalian genome and are probably derived from past retroviruses. Many LTR elements lack a reverse transcriptase encoding region, have lost the ability to transpose, and are only 300 to 500 bp in size with a putative TATA box. The LTRs of the Moloney MuLV have been demonstrated to contain strong enhancer activity and insertion of this viral LTR up to 300 kb distal to the c-myc gene can lead to c-myc overexpression.42,43 DNA methylation is known to affect the LTR activity of retroviral elements in the mammalian genome.29,44 We observed de-methylation and transcriptional reactivation of the MaLR and ERVL elements at the PU.1 locus. This suggests the possibility that the re-activated LTR enhancers may also affect the PU.1 promoter. There are many examples for transcriptional control of mammalian genes by nearby integrated LTRs that are also controlled by DNA methylation such as the oncomodulin gene, the nocturnin gene and the agouti locus.44–47 Alternatively, the alteration in DNA methylation levels at the retroviral elements may subsequently affect other histone modifications and spread along the gene. Thus it may facilitate Pol II initiation at the promoter region or Pol II elongation through the gene body and therefore enhance PU.1 transcript levels.
In summary, genomic hypomethylation after Lsh deletion can cause a dual risk; on the one hand it may lead to severe hematopoietic defects with the danger of reduced immune responses and on the other hand, it may create an enhanced risk of developing hematopoietic neoplasm. Whether demethylating agents currently used in the clinic or developed in the future can reactivate silenced tumor suppressor genes and predispose patients to an additional cancer risk by oncogene activation is an important issue. Only future studies that address the contribution of epigenetics to tumorigenesis using suitable epigenetic models, will further clarify the risks and benefits of such treatments.
Materials and Methods
Mouse strains
Lsh heterozygous animals were maintained on a mixed C57BL/6J×129/SvJ background.15 Male heterozygous p53-deficient 129/Sv-TrpmlTyj mice, generated by germline disruption of the p53 gene (Jackson Laboratories) were crossed with Lsh+/− mice to generate Lsh+/−p53+/− hybrids and subsequently Lsh−/−p53−/− embryos. NCI-Frederick is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the “Guide for the Care and Use of Laboratory Animals” (National Research Council; 1996; National Academy Press; Washington DC).
Fetal liver cell transplantation
About 3 × 106 fetal liver cells (day 14.5 to 16.5 of gestation) were intravenously injected into C57BL/6 (CD45.1+) recipient mice that had been exposed to 950 Rad of total body irradiation. After 8 to 16 weeks the peripheral blood or the bone marrow was analyzed by FACS. Hematopoietic reconstitution by donor and host cells was determined in recipient mice by two-color FACS analysis using monoclonal antibodies A-20 (CD45.1+) and 104 (CD45.2+) cells.
Antibodies/FACS analysis
Flow cytometry analysis was performed on single-cell suspensions of fetal liver, thymus, spleen, peripheral blood and bone marrow prepared by standard methods. After blocking the FcR with antibody 2.4 G2, cells were stained with fluorescein isothiocyanate (FITC)—or phycoerythrin (PE)—conjugated antibodies and analyzed on a Coulter Epics XL-MCL cytometer. The data was collected and analyzed using the Coulter System II software provided by the manufacturer. Scatter-gating for size were used to exclude dead cells from the analysis. Monoclonal antibodies (MAb) RA3-6B2 (B220), 500A2(CD3e), RB6-8C5 (Gr-1), M1/70 (CD11b/Mac-1), C2(CD71), R6-60.2(IgM), A20(Ly5.2), 104(Ly5.1), and TER-119 were used according to the instructions of the supplier (Pharmingen).
Histopathology analysis
Mice were euthanized with CO2 and complete necropsy examinations performed. Tissues were fixed in 10% neutral buffered formalin, paraffin-embedded, sectioned into 5 µm sections, and stained with hematoxylin and eosin for pathology evaluation.
Purification of progenitors
Fetal liver cells were suspended at a density of 1 × 106 cells/100 µL in PBS with 0.1% BSA (Sigma). To exclude terminally differentiated hematopoietic cells, the cells were incubated with a cocktail of antibodies specific for the following lineage markers: CD3ε, CD4, CD8, B220, Gr-1 and TER119 (B–D Pharmingen, San Diego, CA). After incubation for 30 min at 4°C, cells were washed twice and resuspended in media at a density of 108 cells/mL. Magnetic beads coated with anti-rat immunoglobulin G (IgG; Dynal, New Hyde Park, NY, USA) were incubated with cells at a concentration of 20 beads/cell at 4°C on a rotating platform for 45 minutes. The lineage-positive (Lin+) cells were removed using a magnetic particle concentrator, and the resulting Lin-negative (Lin−) cell population was stained for further FACS. Lin− were first incubated with FcR (2.4 G2)-blocking antibodies for 10 minutes at 4°C, washed twice, and then incubated with the following cocktail of directly conjugated monoclonal antibodies: phycoerythrin-conjugated anti-c-Kit (2B8), phycoerythrin-Cy5 anti-IL-7Rα (A7R34) (BD pharmingen, San Diego, CA), allophycocyanin-conjugated anti-Sca-1 (Ly-6A/E) monoclonal antibodies (eBioscience, San Diego, CA). Throughout staining, the cells were kept at 4°C, and the antibodies were used at a concentration of 0.5 µg/1 × 106 cells. Cells were then sorted by high-speed sorter (MoFlo; Cytomation) for the IL-7Rα−/c-Kit+/Sca-1+ cell population identified as HSCs.
Western blot analysis
Protein extracts were prepared from day 15.5 fetal liver and from spleens of reconstituted mice. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and electrotransfered to an Immobilon membrane (Millipore). The blotted membrane was incubated with an affinity-purified rabbit anti-PU.1 antibody (T21, sc-352 Santa Cruz), or anti-actin antibody. The blots were incubated with HRP-coupled secondary antibodies. Immunoreactive proteins were detected using an enhanced chemiluminescence visualization system (Amersham).
RT-PCR analysis
Reverse transcription was performed on 1 µg of total RNA using the iScript™ cDNA Synthesis Kit (BIO-RAD). Control reactions were prepared in parallel omitting reverse transcriptase. PCR analysis was performed in serial dilutions (1:3) for the detection of transcripts. The primers used for detection of PU.1, IAP, ERVL, MaLR or actin are listed in Supplemental Table 1. The PCR products were separated on agarose gels, blotted and hybridized with 32P-radiolabeled internal probes. The radioactive blots were scanned using a Phosphoimager and the data standardized to actin.
Methylation sensitive PCR
Genomic DNA was completely digested with HpaII or HhaI. Primers to analyze the methylation status of the retroviral repeats located in the PU.1 locus are listed in Supplemental Table 1. The PCR products were separated on agarose gels and stained with ethidium bromide. For every methylationsensitive PCR, the starting amount of template was adjusted using undigested DNA in the PCR reaction before enzymatic digestion.
Southern analysis
Genomic DNA was extracted from fetal liver (gestation day14.5) or adult spleen, digested accordingly with either Hpa II, MspI or Mae II, then separated by electrophoresis on 1.2% agarose gels and transferred by blotting on Nytran plus membranes (Schleicher & Schuell). Membranes were hybridized overnight at 42°C in hybridization buffer (Amersham) with 32P-labeled probes as published elsewhere.16 The hybridized membranes were washed twice with 6xSSC at 42°C for 15 minutes and visualized by autoradiography.
Bisulphite sequencing
Genomic DNA was subjected to bisulphite treatment using the CpGenome DNA modification kit (Chemicon International) according to the manufacturer’s instructions. Primers were designed using the Methprimer program and are listed in Supplemental Table 1. The PCR products were separated in agarose gels and purified using the QIAEX II gel extraction kit (Qiagen). Amplified fragments were subcloned into the pCR2.1-TOPO vector with the TOPO TA Cloning Kit (Invitrogen). Independent clones for each fragment were sequenced by using the M13 F or M 13 R primer.
Chromatin immunoprecipitation
For chromatin immunoprecipitations (ChIPs) fetal liver cells derived from Lsh−/− or Lsh+/+ embryos (day 14.5 gestation) were cross-linked with 1% formaldehyde, lysed and sonicated to generate DNA fragments (200–800 bp). Immunoprecipitation was performed using affinity purified rabbit anti-recombinant Lsh antiserum, Dnmt3b antibody (Alexis), or species specific IgG controls. After reversal of crosslinking, nucleic acids were prepared from the eluted complex and PCR analysis was performed, followed by agarose gel electrophoresis. Amplification conditions were as follows: 94°C for 4 min; 94°C for 1 min; 55°C for 1 min; 72°C for 1 min (35 cycles) and 72°C for 7 min. PCR primer pairs for ChIPs analysis are listed in Supplemental Table 1.
Supplementary Material
Acknowledgements
We are grateful to Rodney Wiles and Terry Stull for excellent technical animal assistance.
This project has been funded in whole or part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-C0-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
This research was supported by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Note
Supplementary materials can be found at: www.landesbioscience.com/supplement/FanEPI3-3-sup.pdf
References
- 1.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA. Epigenetics in carcinogenesis and cancer prevention. Ann N Y Acad Sci. 2003;983:213–219. doi: 10.1111/j.1749-6632.2003.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 3.Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 4.Villar-Garea A, Esteller M. DNA demethylating agents and chromatin-remodelling drugs: which, how and why? Curr Drug Metab. 2003;4:11–31. doi: 10.2174/1389200033336757. [DOI] [PubMed] [Google Scholar]
- 5.Laird PW, Jackson-Grusby L, Fazeli A, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 6.Cormier RT, Dove WF. Dnmt1N/+ reduces the net growth rate and multiplicity of intestinal adenomas in C57BL/6-multiple intestinal neoplasia (Min)/+ mice independently of p53 but demonstrates strong synergy with the modifier of Min 1(AKR) resistance allele. Cancer Res. 2000;60:3965–3970. [PubMed] [Google Scholar]
- 7.Trinh BN, Long TI, Nickel AE, Shibata D, Laird PW. DNA methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair. Mol Cell Biol. 2002;22:2906–2917. doi: 10.1128/MCB.22.9.2906-2917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eads CA, Nickel AE, Laird PW. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic mice. Cancer Res. 2002;62:1296–1299. [PubMed] [Google Scholar]
- 9.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 10.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 11.Lin H, Yamada Y, Nguyen S, et al. Suppression of Intestinal Neoplasia by Deletion of Dnmt3b. Mol Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Geiman TM, Muegge K. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci USA. 2000;97:4772–4777. doi: 10.1073/pnas.97.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta. 2001;1526:211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- 16.Dennis K, Fan T, Geiman TM, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Q, Huang J, Fan T, Zhu H, Muegge K. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. Embo J. 2003;22:5154–5162. doi: 10.1093/emboj/cdg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Fan T, Yan Q, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun LQ, Lee DW, Zhang Q, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muegge K. Lsh, a guardian of heterochromatin at repeat elements. Biochem Cell Biol. 2005;83:548–554. doi: 10.1139/o05-119. [DOI] [PubMed] [Google Scholar]
- 21.Fyodorov DV, Kadonaga JT. The many faces of chromatin remodeling: SWItching beyond transcription. Cell. 2001;106:523–525. doi: 10.1016/s0092-8674(01)00478-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Geiman TM, Xi S, et al. K Lsh is involved in de novo methylation of DNA. Embo J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K. Lsh controls silencing of the imprinted Cdkn1c gene. Development. 2005;132:635–644. doi: 10.1242/dev.01612. [DOI] [PubMed] [Google Scholar]
- 24.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosomesynapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 25.Attardi LD, Jacks T. The role of p53 in tumour suppression: lessons from mouse models. Cell Mol Life Sci. 1999;55:48–63. doi: 10.1007/s000180050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 27.Moreau-Gachelin F, Wendling F, Molina T, et al. A Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scolan EL, Wendling F, Barnache S, et al. Germ-line deletion of p53 reveals a multistage tumor progression in spi-1/PU.1 transgenic proerythroblasts. Oncogene. 2001;20:5484–5492. doi: 10.1038/sj.onc.1204708. [DOI] [PubMed] [Google Scholar]
- 29.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 30.Gupta P, Gurudutta GU, Verma YK, et al. PU.1: An ETS family transcription factor that regulates leukemogenesis besides normal hematopoiesis. Stem Cells Dev. 2006;15:606–617. doi: 10.1089/scd.2006.15.609. [DOI] [PubMed] [Google Scholar]
- 31.Chen HM, Ray-Gallet D, Zhang P, et al. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- 32.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 34.Kosmider O, Denis D, Lacout C, et al. Kit-activating mutations cooperate with Spi-1/PU.1 overexpression to promote tumorigenic progression during erythroleukemia in mice. Cancer Cell. 2005;8:467–478. doi: 10.1016/j.ccr.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 36.Fan T, Yan Q, Huang J, et al. Lsh deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res. 2003;63:4677–4683. [PubMed] [Google Scholar]
- 37.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 38.Ben-David Y, Giddens EB, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 39.Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci USA. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 42.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 43.Dudley JP, Mertz JA, Rajan I, Lozano M, Broussard DR. What retroviruses teach us about the involvement of c-Myc in leukemias and lymphomas. Leukemia. 2002;16:1086–1098. doi: 10.1038/sj.leu.2402451. [DOI] [PubMed] [Google Scholar]
- 44.Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994;8:1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 45.Rentsch JM, Hegersberg M, Banville D, Berchtold MW. The LTR promoter of the rat oncomodulin gene is regulated by cell-line specific accessibility in the LTR U3 region. Arch Biochem Biophys. 2006;447:68–79. doi: 10.1016/j.abb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gwynn B, Lueders K, Sands MS, Birkenmeier EH. Intracisternal A-particle element transposition into the murine β-glucuronidase gene correlates with loss of enzyme activity: a new model for β-glucuronidase deficiency in the C3H mouse. Mol Cell Biol. 1998;18:6474–6481. doi: 10.1128/mcb.18.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.