Abstract
The human brm (hbrm) protein (homologue of the Drosophila melanogaster brahma and Saccharomyces cervisiae SNF-2 proteins) is part of a polypeptide complex believed to regulate chromatin conformation. We have shown that the hbrm protein is cleaved in NB4 leukemic cells after induction of apoptosis by UV-irradiation, DNA damaging agents, or staurosporine. Because hbrm is found only in the nucleus, we have investigated the nature of the proteases that may regulate the degradation of this protein during apoptosis. In an in vitro assay, the hbrm protein could not be cleaved by caspase-3, -7, or -6, the “effector” caspases generally believed to carry out the cleavage of nuclear protein substrates. In contrast, we find that cathepsin G, a granule enzyme found in NB4 cells, cleaves hbrm in a pattern similar to that observed in vivo during apoptosis. In addition, a peptide inhibitor of cathepsin G blocks hbrm cleavage during apoptosis but does not block activation of caspases or cleavage of the nuclear protein polyADP ribose polymerase (PARP). Although localized in granules and in the Golgi complex in untreated cells, cathepsin G becomes diffusely distributed during apoptosis. Cleavage by cathepsin G removes a 20-kDa fragment containing a bromodomain from the carboxyl terminus of hbrm. This cleavage disrupts the association between hbrm and the nuclear matrix; the 160-kDa hbrm cleavage fragment is less tightly associated with the nuclear matrix than full-length hbrm.
The human brm (hbrm) and BRG-1 (brm-related gene 1) proteins are homologs of the Drosophila melanogaster brahma and Saccharomyces cervisiae SNF proteins (1–9). The brahma and SNF proteins have been shown to regulate the transcription of genes necessary for development, including the Drosophilia genes bicoid and fushi-tarazu (2, 5). Brahma and SNF are components of a large protein complex believed to alter the structure of chromatin, allowing other transcription factors to gain access to promoter DNA (6). Recent work (10) has demonstrated that the hbrm complex can form a repressive complex with the retinoblastoma (Rb) protein alone or with the Rb protein and histone deacetylase. These repressor complexes arrest cells in the G1 phase of the cell cycle and inhibit the transcription of the genes encoding cyclin A and cyclin E. BRG-1 can associate with cyclin E and alters the ability of this protein to induce growth arrest (11). Therefore, the brm and BRG-1 protein complexes play a significant role in regulating DNA structure, transcriptional activation and repression, and the cell cycle.
To study the possible role of the hbrm protein in differentiation of hematopoietic cells, we initially examined the levels of hbrm during the differentiation of NB4 cells. NB4 cells are leukemic precursor cells that will differentiate into monocytes when treated with phorbol esters or into neutrophils when treated with all-trans retinoic acid (12). Differentiation of NB4 cells induced by treatment with 1 nM phorbol ester had no effect on the level of hbrm. However, treatment with 1 μM phorbol ester, which induces differentiation and apoptosis, led to marked decreases in hbrm. Because of this result, we checked other inducers of apoptosis and determined that induction of apoptosis in NB4 cells by UV-irradiation or chemotherapeutic agents led to cleavage and degradation of hbrm.
Based on this observation, we have attempted to discern whether hbrm is cleaved by the caspases known to cleave other nuclear proteins during apoptosis (caspase-3, -6, and -7). However, none of these caspases were able to cleave hbrm in vitro. We find instead that cathepsin G, which is found in granules (specialized lysosomes produced during neutrophil differentiation) in NB4 leukemic cells, is capable of cleaving hbrm. Immunofluorescence demonstrates that the induction of apoptosis in NB-4 cells causes the release of cathepsin G from granules to the nucleus where hbrm is cleaved.
Because cells from hbrm knockout mice undergo UV radiation-induced apoptosis more readily than cells from normal mice (12), this would suggest that the presence of brm could provide some degree of resistance to UV radiation-induced apoptosis. Cells that have the machinery to degrade brm might, therefore, be more sensitive to UV radiation-induced apoptosis. This suggestion agrees with the observation that NB4 cells undergo rapid UV- induced apoptosis when compared to other epithelial or fibroblast cells.
Materials and Methods
Cell Culture.
NB4 cells and COS-7 cells were grown in RPMI medium supplemented with 10% bovine calf serum at 37°C in 4.9% CO2. HeLa cells were grown in DMEM supplemented with 10% bovine calf serum. HeLa-S3 cells were grown in Joklik's MEM (Sigma) plus 10% bovine calf serum. COS-7 cells were transfected with cathepsin G expression plasmid by using the Lipofectamine-Plus protocol (Life Technologies). Cathepsin G expression plasmids have been described previously (13).
Preparation of Whole Cell Lysates, Cytosol, and Nuclei from NB4 and HeLa Cells; Incubation of HeLa Nuclei with Caspases and Cathepsins.
NB4 cells were irradiated with 100 J/m2 UV in a minimal volume of PBS and then placed back in media plus bovine calf serum for various times. For whole-cell lysates, cells were harvested, washed with PBS, then lysed, and used for Western blots as previously described (14). NB4 cytosol and nuclei were prepared by rapid detergent lysis as described (15). HeLa nuclei were prepared as described previously (16.). Nuclei were then resuspended in homogenization buffer for incubation with cytosol or with purified caspases, or cathepsins. Purified active caspases were purchased from PharMingen; and cathepsins were purchased from Oncogene. For preparation of detergent free cytosols, cells were homogenized as described above for HeLa nuclei and then nuclei removed by centrifugation for 3 min at 3, 000 × g. The remaining supernatant was designated cell cytosol. Ni-nitrilotriacetic acid agarose was purchased from Qiagen, and 6-His ubiquitin-tagged proteins were purified as suggested by the manufacturer's protocols.
Western Blots.
Antibodies to the amino terminus of hbrm have been described (12). Antibodies to polyADP ribose polymerase (PARP) were purchased from Upstate Biotechnology (Lake Placid, NY), antibodies to DNA-dependent kinase p350 were purchased from PharMingen, and antibodies to NuMA (nuclear mitotic apparatus) protein were purchased from Calbiochem. Western blots were performed as described (14).
Staining NB4 Cells for Cathepsin G.
NB4 cells were incubated for 2 h in 50 μM LysoTracker (Molecular Probes, Eugene, OR), suspended in PBS containing 1% BSA, and then spun onto a microscope slide in a cytocentrifuge. Cells were then fixed, stained with anti-cathepsin G antibodies, and mounted in 4′,6-diamidino-2-phenylindole (DAPI)-containing medium (Vector Laboratories) as previously described for K562 cells (17).
Results and Discussion
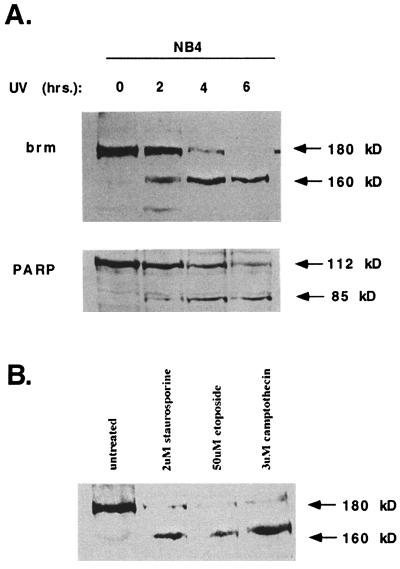
Treatment of NB-4 cells with UV-irradiation causes hbrm and PARP cleavage (Fig. 1A) and DNA fragmentation (data not shown). PARP is a known substrate of apoptotic caspases (18). The 180-kDa hbrm protein is cleaved into a smaller 160-kDa product at a rate almost identical to that of PARP cleavage. Treatment of NB4 cells with DNA damaging agents (etoposide or camptothecin) or staurosporine also induced apoptosis and cleavage of hbrm to the 160-kDa fragment (Fig. 1B). Anti-hbrm antibodies raised against the amino terminus of hbrm are able to detect the 160-kDa hbrm fragment (Fig. 1), whereas anti-hbrm antibodies raised against the carboxyl terminus of hbrm will not recognize this 160-kDa fragment (data not shown). These results indicate that the carboxyl terminus of hbrm has been removed by the apoptotic cleavage.
Figure 1.
The brm and PARP proteins are degraded in NB4 cells undergoing apoptosis. (A) NB4 cells were treated with 100 J/m2 of UV and then lysed at the times after treatment indicated above the lanes. Cell lysates were used for Western blotting with anti-brm and anti-PARP antibodies. (B) Cells were treated for 2 h with etoposide, camptothecin, or staurosporine at the indicated concentrations and then lysed for Western blotting.
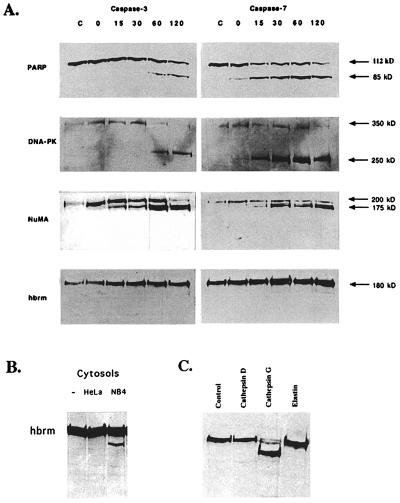
It is generally believed that nuclear apoptotic substrates like hbrm are cleaved by “effector” caspases, such as caspase-3 or caspase-7, which are activated in the final steps of apoptosis and enter the nuclei to cleave protein substrates. To determine whether caspase-3 or caspase-7 was able to cleave the hbrm protein, isolated HeLa nuclei were incubated with either active purified caspase-3 or caspase-7. The nuclei were then repelleted and used to make nuclear protein extracts. The nuclear extracts were used for Western blots with antibodies to three proteins known to be cleaved in vivo during apoptosis: PARP, DNA-dependent kinase (DNA-PK) 350-kDa subunit, and the NuMA protein, as well as for hbrm Westerns. Although caspase-7 cuts PARP and DNA-PK p350 more rapidly than caspase-3, both caspase-3 and caspase-7 cut all three nuclear apoptotic substrates (Fig. 2A). However, the hbrm protein was not cleaved by either caspase-3 or caspase-7 (or caspase-6, unpublished data). Although this is an in vitro system, this suggests that hbrm is cleaved by another protease, which is either capable of entering the nucleus during apoptosis, or is found in the nucleus.
Figure 2.
Incubation of HeLa cell nuclei with purified, active caspases and cathepsins. (A) HeLa cell nuclei were prepared and resuspended in 100 μl of homogenization buffer as described in Materials and Methods. Then 100 ng of purified, active caspase-3 or caspase-7 was added to each 100-μl sample, and the samples were incubated at 30°C for the times indicated above the lanes. Control samples were incubated 2 h at 30°C without added caspase. Immediately after incubation, nuclei were repelleted and then lysed in a small volume of Nonidet P-40 lysis buffer for 30 min at 4°C. The Nonidet P-40 lysates were cleared of DNA by spinning 10 min at 10,000 × g and then used for Western blotting. (B) HeLa nuclei were incubated at 30°C for 2 h in buffer, HeLa cytosol, or NB4 cytosol, as indicated above the lanes. (C) Nuclei were incubated with buffer, purified cathepsin G, cathepsin D, or elastin as indicated.
Because the hbrm protein is cleaved during apoptosis in NB4 cells, but is not cleaved by the usual effector caspases, it seemed possible that hbrm was cleaved by a protease specific to NB4 cells. To test this hypothesis, HeLa nuclei were incubated with detergent-free cytosol from untreated NB4 cells or HeLa cells. Fig. 2B shows that only the NB4 cytosol contained an activity that cleaved the hbrm protein. Proteases other than caspases have been implicated in the execution of apoptosis in NB4 cells or neutrophils, including cathepsins (19, 20), although cathepsins have not previously been shown to cleave nuclear substrates. To see whether neutrophil proteases had any effect on hbrm cleavage, HeLa nuclei were incubated with purified cathepsin D, cathepsin G, or elastin. Only cathepsin G was able to cleave hbrm (Fig. 2C); cathepsin G almost completely reduced the hbrm protein to a 160-kDa fragment, very similar in size to that produced by NB4 cytosol and observed in vivo during apoptosis. To verify that cathepsin G was present in the NB4 cytosol and was responsible for hbrm cleavage, NB4 cytosol was depleted of cathepsin G by repeated immunoprecipitation; the resulting depleted cytosol was much less active on hbrm when incubated with HeLa nuclei (unpublished data).
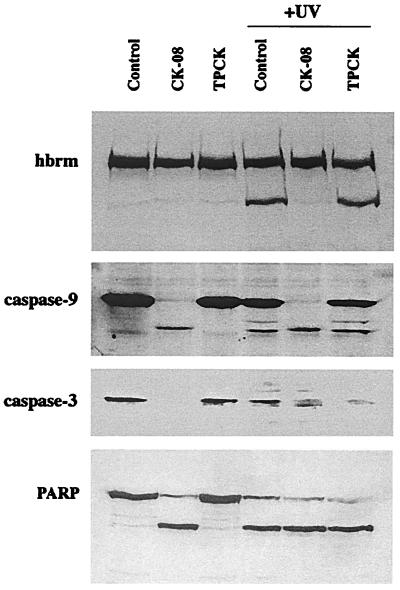
To determine whether cathepsin G was necessary for hbrm cleavage in vivo, NB4 cells were pretreated with either the specific cathepsin G inhibitor CK-08 (carbobenzoxy-glycyl-leucyl-phenylalanyl-chloromethyl ketone) or the more general serine protease inhibitor l-1-tosylamido-2-phenylethyl chloromethyl ketone. The cells were then UV-irradiated, and incubated a further 4 h. Cell sample lysates were then analyzed by Western blot (Fig. 3). As shown at the top of Fig. 3, the cathepsin G inhibitor CK-08 almost completely blocked hbrm cleavage in response to UV irradiation, whereas l-1-tosylamido-2-phenylethyl chloromethyl ketone had little effect. It is also clear that CK-08 does not act by blocking some upstream step of the apoptotic response, such as caspase activation. As shown in the lower part of Fig. 3, CK-08 not only fails to block caspase activation in response to UV-irradiation, but CK-08 itself, in contrast to l-1-tosylamido-2-phenylethyl chloromethyl ketone, is a powerful activator of caspases. Caspase-9, caspase-3 (Fig. 4), and caspase-7 (not shown) are all cleaved and activated in cells treated with CK-08 alone or with CK-08 followed by UV irradiation. The PARP protein is also cleaved from 112 kDa to 85 kDa in cells treated with CK-08, showing that activated caspases enter the nucleus in these cells (Fig. 3). These results indicate that CK-08 must either directly block cathepsin G cleavage of hbrm, or block some alternate apoptotic pathway that is dependent on cathepsin G activity and does not involve caspases. In view of the fact that purified cathepsin G does cleave hbrm in vitro in a manner similar to that observed in vivo, the former possibility seems more likely.
Figure 3.
Effect of serine protease inhibitors on cleavage of hbrm and activation of caspases. NB4 cells were left untreated, pretreated for 30 min with 5 μM cathepsin G inhibitor CK-08 (Z-Gly-Leu-Phe-CK), or pretreated for 30 min with the more general serine protease inhibitor l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK) as indicated above the lanes. Untreated and inhibitor-treated samples were then UV-irradiated and incubated a further 4 h (with protease inhibitors added back to the appropriate samples). Whole cell lysates were then prepared as described in Materials and Methods and used for Western blot analysis with antibodies to the proteins indicated at left.
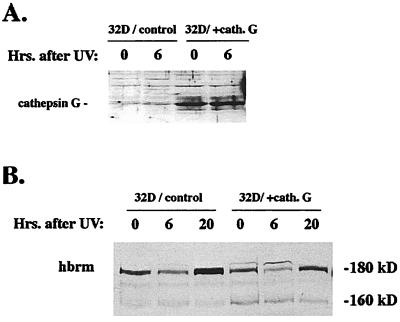
Figure 4.
Endogenous murine brm cleavage is enhanced by over-expression of human cathepsin G in murine 32D cells. (A) Murine 32D cells stably transfected with either empty vector or vector expressing human cathepsin G were UV-irradiated and then lysed at the times after irradiation indicated above the lanes. Cell lysates were then used for Western blot analysis with anti-cathepsin G antibodies. (B) 32D lysates from the same experiment shown in A were used for Western blot analysis with anti-hbrm antibodies. Positions of full-length endogenous murine brm (180 kDa) and cleavage product (160 kDa) are indicated.
To confirm that expression of cathepsin G affects hbrm cleavage in vivo, hbrm cleavage was examined in murine myeloid 32D cell lines (21) stably transfected with either empty vector or vector expressing human cathepsin G (Fig. 4A). The 32D cells, like other IL-3-dependent cells, will not undergo complete apoptosis unless starved for IL-3 (22); in the presence of IL-3, 32D cells subjected to UV-irradiation will enter the early stages of apoptosis, then recover. Fig. 4B shows endogenous mouse brm levels in 32D cells after UV-irradiation but without IL-3 starvation. The 32D cells transfected with human cathepsin G clearly have lower levels of full-length endogenous brm (180 kDa) and higher levels of brm cleavage product (160 kDa); this is apparent under all conditions.
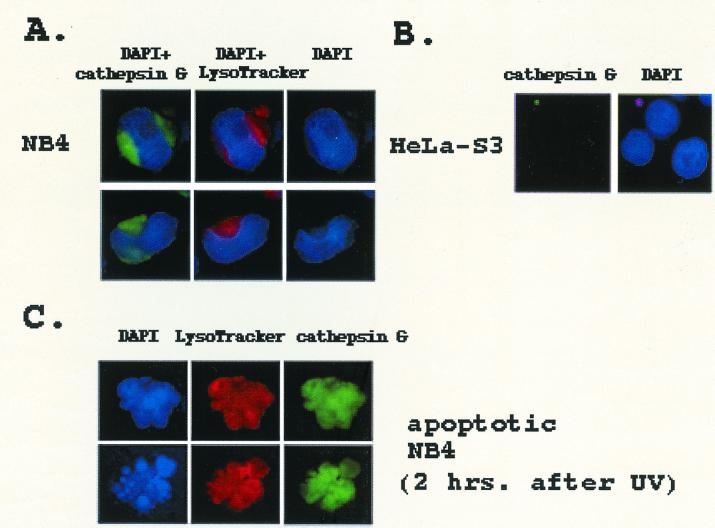
If cathepsin G actually cleaves hbrm in vivo, it must be released from the granules and have access to nuclear proteins. To observe the subcellular distribution of cathepsin G in NB4 cells, the cells were spun onto slides, then costained by using anti-cathepsin G antibodies and LysoTracker (LysoTracker is a florescent cell-permeable peptide that accumulates selectively in acidic organelles such as lysosomes). As shown in Fig. 5A, the anti-cathepsin G antibodies stain specific structures in the NB4 cells very intensely. The cap-like structure, which is stained by the anti-cathepsin G antibodies but not by LysoTracker, is probably the golgi apparatus. The smaller circular areas are almost certainly granules because LysoTracker peptide also accumulates in them (see Fig. 5A). As demonstrated by the DAPI staining, NB4 cells contain very large nuclei and a relatively small volume of cytoplasm. As a control, HeLa-S3 cells, which grow in suspension and are similar in size and shape to NB4 cells, were also stained with the anti-cathepsin G antibodies. As seen in Fig. 5B, almost no staining of HeLa-S3 cells by anti-cathepsin G antibodies is observed. To determine whether cathepsin G undergoes any change in subcellular location during apoptosis, NB4 cells were UV-irradiated. After 1 h LysoTracker was added to the irradiated cells; after one additional hour the cells were spun onto slides and fixed. The cells were stained by using the anti-cathepsin G antibodies, then mounted with media containing DAPI. Apoptotic cells were identified by their irregular shape and condensed chromatin. As demonstrated by the clumped DAPI staining, Fig. 5C shows two apoptotic NB4 cells. In comparison to Fig. 5A, the pattern of both cathepsin G and LysoTracker staining is clearly different, becoming more diffuse and associated with the nucleus. This change in subcellular distribution 2 h after irradiation supports the idea that granule structure is disrupted during apoptosis, and granule proteins, such as cathepsin G, can move into the nucleus and cleave nuclear proteins.
Figure 5.
Staining of NB4 and HeLa-S3 cells with anti- cathepsin G antibodies or LysoTracker. (A) NB4 cells were incubated for 1 h at 37°C in media containing 50 μM LysoTracker peptide and then spun onto slides by using a cytocentrifuge. After fixation in neutral-buffered formalin followed by washing, the cells were incubated first with anti-cathepsin G antibodies and then with FITC-labeled secondary antibodies. Cells were prepared for viewing under the microscope by adding mounting media containing DAPI and coverslips. Photographs show DAPI plus cathepsin G staining, DAPI plus LysoTracker, DAPI alone, cathepsin G alone, or LysoTracker alone, as indicated. (B) HeLa-S3 cells were prepared as in A but were not incubated with LysoTracker. (C) NB4 cells were UV-irradiated as described in Materials and Methods; 1 h after irradiation, 50 μM LysoTracker was added to the cell media. After an additional hour of incubation, cells were spun onto a glass slide. After fixing and staining with anti-cathepsin G antibodies, cells were mounted by using DAPI-containing medium. Cell shape and chromatin condensation was used to distinguish apoptotic from nonapoptotic cells.
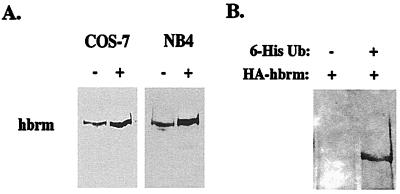
Although cathepsin G appears to be involved in hbrm cleavage in NB4 cells, cathepsin G is normally expressed only in certain myeloid cells (neutrophils, macrophages, mast cells, and their precursors). This raises the question of changes in hbrm levels observed in other cell types. Fig. 6 presents evidence that hbrm can be degraded by proteasome activity. The specific proteasome inhibitor lactacystin causes an increase in hbrm protein levels in both NB4 and COS-7 cells (Fig. 6A), suggesting a basal rate of hbrm ubiquitination and degradation exists in both cell types. To verify that hbrm is ubiquinated, COS-7 cells were cotransfected with plasmids expressing HA-tagged hbrm and 6-His-tagged ubiquitin. Lysates from the transfected cells were then incubated with Ni-nitrilotriacetic acid agarose, and bound proteins eluted and used for Western blot analysis with anti-HA antibodies. Hbrm bound to the Ni-nitriloacetic acid agarose but only when the HA-tagged hbrm plasmid was cotransfected with the 6-His-ubiquitin plasmid (Fig. 6B). The results described above suggest that hbrm degradation in cells without cathepsin G, and perhaps the hbrm degradation in NB4 cells that occurs after cleavage by cathepsin G, is mediated by the proteasome. The level of hbrm ubiquitinylation does not appear to increase after UV-irradiation (our unpublished data); however, this does not necessarily indicate that the ubiqutin pathway is unimportant in hbrm degradation. Other groups (24, 25) have proposed proteasome-mediated apoptotic degradation but have not shown detectable increases in ubiquiated substrate during apoptosis. It is possible that apoptosis results in increased activity of both ubiquinating enzymes and proteasome proteases, causing more rapid protein degradation without detectable increases in ubiquinated protein.
Figure 6.
The proteasome is involved in hbrm degradation. (A) NB4 cells or COS-7 cells were treated with 20 μM lactacystin for 4 h or left untreated. Cells were then lysed for Western analysis. (B) COS-7 cells were transfected with either HA-tagged hbrm plasmid plus empty vector or with HA-tagged hbrm plasmid plus vector expressing 6-His Ubiquitin, as indicated above the lanes. Both cell samples were lysed, incubated with Ni-nitrilotriacetic acid agarose for binding of 6-His-tagged proteins, and then bound proteins eluted with 150 mM EDTA. Eluted proteins were used for Western blot analysis with anti-HA tag antibodies.
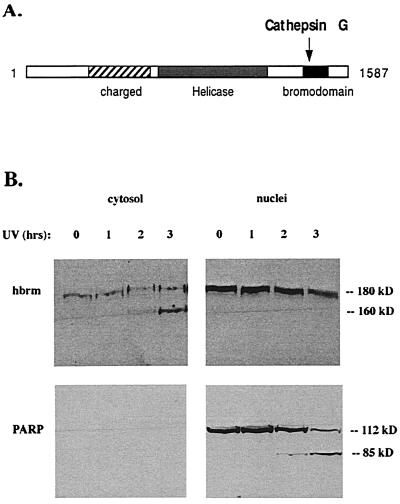
Because both cathepsin G and the proteasome appear to be involved in hbrm cleavage in NB4 cells, the question of what role each plays in the in vivo degradation of cathepsin G arises. The region of the carboxyl terminus removed by cathepsin G cleavage contains a bromodomain (Fig. 7A), which is believed to mediate protein-protein interaction (9). This suggests that hbrm cleavage may disrupt normal interactions between hbrm and other nuclear proteins. To determine whether cathepsin G cleavage affects the association of the hbrm protein with nuclear structures, subcellular fractionation experiments were used to examine the degree to which full-length hbrm as opposed to the 160-kDa hbrm fragment was able to remain associated with the nucleus. NB4 cells were irradiated with UV, and then at various times afterward they were fractionated into cytosol and nuclei by rapid detergent lysis as described in Materials and Methods (Fig. 7B). In contrast to full-length 180-kDa hbrm, where the majority of protein remained tightly associated with the nucleus, all detectable 160-kDa hbrm fragment was found in the cytosol (Fig. 7B). This is in contrast to the PARP 85-kDa apoptotic cleavage fragment, which remains in the nucleus. These results clearly indicate that cleavage of ≈20 kDa from the carboxyl terminus of hbrm results in the disruption of an interaction between hbrm and some other nuclear protein, leading to loss of tight association with the nucleus. It also may result in increased accessibility to the proteasome, facilitating further apoptotic degradation. It is reasonable to suggest that hbrm must be associated with other nuclear proteins in the chromatin to perform its normal function, and this model is supported by studies of BRG-1 association with chromatin (15). The BRG-1 protein is highly homologous to hbrm, and when lymphocytes are stimulated by using either ionomycin or phorbol 12-myristate 13-acetate, BRG-1 becomes more tightly associated with the chromatin.
Figure 7.
The 160-kDa hbrm cleavage fragment is less strongly associated with other nuclear proteins than full-length hbrm. (A) Diagram of hbrm domains and cathepsin G cleavage site. (B) NB4 cells were irradiated with UV, and then at various times after irradation the cells were fractionated into cytosol and nuclei by rapid detergent lysis (see Materials and Methods). Both fractions were used for Western blot analysis with anti-hbrm and anti-PARP antibodies.
In summary, NB4 cells respond to both UV irradiation and staurosporine treatment with rapid cleavage of the carboxyl-terminal 20 kDa of the hbrm protein containing a bromodomain, resulting in the loss of tight association with nuclear structures. Unlike most nuclear apoptotic substrates, hbrm is not cleaved by caspase-3, -6, or -7. The serine protease cathepsin G cleaves hbrm in vitro, and an inhibitor of cathepsin G, but not other serine protease inhibitors, can block UV-induced cleavage of hbrm in vivo. Induced over- expression of human cathepsin G in murine 32D cells enhances the level of hbrm cleavage observed after UV-irradiation. These results suggest that hbrm is cleaved in NB4 cells by cathepsin G released from cell granules during apoptosis. We have also observed that cathepsin G will cleave the PARP, NuMA, and DNA-PK p350 proteins in vitro (unpublished data). However, unlike the results observed with hbrm, the digestion pattern is dissimilar to that observed in vivo. The in vivo digestion pattern of these proteins resembles instead that generated in vitro by caspases. In addition, in vivo cleavage of PARP is not blocked by the cathepsin G inhibitor CK-08 (Fig. 3). For these reasons, we believe the nuclear apoptotic substrates PARP, NuMA, and DNA-PK p350, in contrast to hbrm, are cleaved in vivo by caspase-3 or -7.
The above discussion suggests that certain hematopoietic cells, such as neutrophils, may possess a unique method for the rapid inactivation of the hbrm protein in response to DNA damage caused by UV radiation or other agents. Possession of such a mechanism might make cells prone to rapid apoptosis after incurring DNA damage; cells from mice lacking the hbrm gene do in fact undergo a greater degree of apoptosis after UV-irradiation (17). In addition, the protein complex containing hbrm has been shown to form part of a repressor complex which inhibits transcription of the genes for cyclins E and A, thereby inducing growth arrest in the G1 phase of the cell cycle (9). In myeloid stem cells or leukemic cells, inactivation of hbrm might affect the activity of this repressor complex, thereby preventing growth-arrest in response to DNA damage. This in turn might ensure that cells containing DNA damage would undergo apoptosis rather than growth arrest.
Acknowledgments
We thank Marileila Varella-Garcia (supported by National Institutes of Health Grant CA46934) and Theresa Boomer for help with microscopy. This work was supported by National Institutes of Health Grant CA42533 (to A.S.K.).
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- DNA-PK
DNA-dependent kinase
- PARP
polyADP ribose polymerase
- hbrm
human brm
- BRG-1
brm-related gene 1
- NuMA
nuclear mitotic antigen
References
- 1.Khavare P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. Nature (London) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 2.Peterson C L, Herskowitz I. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 3.Hirschhorn J N, Brown S A, Clark C D, Winston F. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 4.Peterson C L, Tamkun J W. Trends Biochem Sci. 1995;20:143–147. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 5.Laurent B C, Carlson M. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, et al. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 7.Bazett-Jones D P, Coete J, Landel C C, Peterson C L, Workman J W. Mol Cell Biol. 1999;19:1470–1478. doi: 10.1128/mcb.19.2.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muchardt C, Yaniv M. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H S, Gaven M, Dahiyz A, Postigo A, Ma D, Luo R X, Harbour J W, Dean D C. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 10.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes J C, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z B, Uphoff C C, Lanotte M, Drexler H G. Leukemia. 1993;7:1817–1823. [PubMed] [Google Scholar]
- 13.Garwicz D, Lindmark A, Persson A M, Gullberg U. Blood. 1998;92:1415–1422. [PubMed] [Google Scholar]
- 14.Biggs J R, Kraft A S. J Biol Chem. 1999;274:36987–36994. doi: 10.1074/jbc.274.52.36987. [DOI] [PubMed] [Google Scholar]
- 15.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 17.Whalen A M, Galasinski S, Shapiro P S, Nahreini T S, Ahn N G. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen G M. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata R K, Chen S T, Weil S C. Hematol Pathol. 1993;7:225–238. [PubMed] [Google Scholar]
- 20.Brunk U T, Svensson I. Redox Rep. 1999;4:3–11. doi: 10.1179/135100099101534675. [DOI] [PubMed] [Google Scholar]
- 21.Garwicz D, Lindmark A, Gullberg U. J Biol Chem. 1995;270:28413–28418. doi: 10.1074/jbc.270.47.28413. [DOI] [PubMed] [Google Scholar]
- 22.Canman C E, Gilmer T M, Coutts S B, Kastan M B. Genes Dev. 1995;9:600–611. doi: 10.1101/gad.9.5.600. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Salvesen G. Biochem J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailand N, Falck J, Lukas C, Syljuasen K, Welker M, Bartok J, Lukas J. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Fang S, Jensen J, Wiessman A, Ashwell J D. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]