Abstract
The oxidation of protein cysteine residues represents significant posttranslational modifications that impact a wide variety of signal transduction cascades and diverse biological processes. Oxidation of cysteines occurs through reactions with reactive oxygen as well as nitrogen species. These oxidative events can lead to irreversible modifications, such as the formation of sulfonic acids, or manifest as reversible modifications such as the conjugation of glutathione with the cysteine moiety, a process termed S-glutathionylation (also referred to as S-glutathiolation, or protein mixed disulfides). Similarly, S-nitrosothiols can also react with the thiol group in a process known as S-nitrosylation (or S-nitrosation). It is the latter two events that have recently come to the forefront of cellular biology through their ability to reversibly impact numerous cellular processes. Herein we describe two protocols for the detection of S-glutathionylated or S-nitrosylated proteins in situ. The protocol for the detection of S-glutathionylated proteins relies on the catalytic specificity of glutaredoxin-1 for the reduction of S-glutathionylated proteins. The protocol for the detection of S-nitrosylated proteins represents a modification of the previously described biotin switch protocol, which relies on ascorbate in the presence of chelators to decompose S-nitrosylated proteins. These techniques can be applied in situ to elucidate which compartments in tissues are affected in diseased states whose underlying pathologies are thought to represent a redox imbalance.
1. INTRODUCTION
Cysteines with low pKa values ranging from 4 to 5 as dictated by the charges and the electrophilic nature of surrounding amino acids are described as being “reactive” with regard to their susceptibility to undergo oxidative modifications. The pKa of most cysteines within a protein’s structure is 8.5 and not considered to be highly susceptible to oxidative modification and are thus considered “nonreactive” (Janssen-Heininger et al., 2008; Meng et al., 2002). In nature, free cysteines are found to be conserved within the primary sequence in numerous classes of proteins throughout species, suggesting that there may be other roles intended for cysteine residues beyond metal coordination and structural disulfide formation. Notably, there are numerous redox-based modifications applicable to cysteine residues, including hydroxylation (SOH), nitrosylation (SNO), glutathionylation (SSG), and the formation of inter/intramolecular disulfides (S–S), with each modification having the potential to modify not only protein structure but also function in a widely diverse fashion (Giles et al., 2003; Jacob et al., 2003; Ji et al., 1999; Kim et al., 2002; Mallis and Thomas, 2000).
2. PROTEIN S-GLUTATHIONYLATION
Protein S-glutathionylation represents the conjugation of the low molecular weight antioxidant molecule glutathione to cysteines within a protein. As described, during oxidative stress, cysteines are among the most vulnerable with regard to oxidative modifications. As an antioxidant molecule, GSH is present within cells at millimolar concentrations (1–10 mM) and it is believed that conjugation of GSH to oxidized cysteines serves a protective mechanism in the prevention of overoxidation. More recently, however, protein S-glutathionylation has been compared to O-phosphorylation with regard to its impact on protein structure and function, and its ability to reversibly impact signaling pathways (Dalle-Donne et al., 2007; Ghezzi, 2005; Holmgren et al., 2005). This comparison is further bolstered by the observation that protein S-glutathionylation occurs not only in response to overt oxidative injury but also in pathophysiological states, and in settings where ratios of GSH to oxidized GSH (GSSG) are low (i.e., 100:1 vs. 3:1). Further, the heterogeneity of cysteine reactivity with oxidants imparts a unique specificity toward protein S-glutathionylation formation. Finally, there exist mechanisms by which S-glutathionylation formation and resolution are exquisitely regulated (Dalle-Donne et al., 2007, 2008; Gallogly and Mieyal, 2007; Ghezzi, 2005; Shelton et al., 2005).
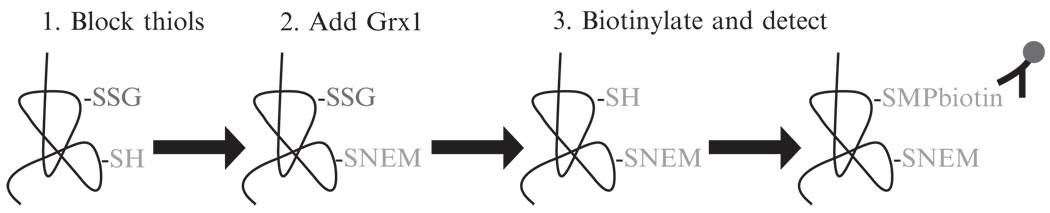
Glutaredoxins (Grx) are low molecular weight (9–14 kDa) enzymes of the oxidoreductase class of enzymes, and under physiological conditions play an important role in the reduction of S-glutathionylated proteins. It is worthy of mention that under conditions of oxidative stress when GSSG concentrations are increased, Grx causes increases in protein S-glutathionylation, instead of decreases. Of the mammalian isoforms, Grx1 is the most extensively described and characterized. Localized primarily in the cytoplasm, Grx1 compared to other thiol–disulfide oxidoreductases (i.e., Grx2 and thioredoxin (Trx)) is significantly more efficient in catalyzing the deglutathionylation of proteins. Despite its lower concentration within cells (1 µMcompared to 10 µM for Trx), Grx1 has a 10-fold lower Km value for ribonucleotide reductase and a 5000-fold higher kcat/Km for S-glutathionylated cysteines in vitro as compared to Trx (Chrestensen et al., 2000; Holmgren, 1976; Holmgren et al., 1978). Under physiological conditions where GSH/GSSG ratios are high, Grx1 functions through the monothiol mechanism in the reduction of mixed disulfides. With regard to cellular processes, Grx1 has been shown to be important in cellular differentiation, regulation of transcription factor activation, and apoptosis (Anathy et al., 2009; Pineda-Molina et al., 2001; Takashima et al., 1999). We have utilized the catalytic specificity of Grx1 to successfully detect S-glutathionylated proteins in paraffin embedded tissues in situ. The protocol is schematically depicted in Fig. 17.1, and described in the following section.
Figure 17.1.
Schematic representation of the Grx1-catalyzed cysteine derivatization protocol. Free thiols are blocked by alkylation with NEM (1), followed by Grx1-catalyzed reduction of S-glutathionylated proteins (2), and finally newly reduced cysteines are labeled using MPB and detected using streptavidin-conjugated fluorophores (3).
2.1. In situ detection of S-glutathionylated proteins
After dewaxing tissue samples in three changes of xylene, tissue is rehydrated in 100%, 95%, and 75% ethanol, and washed in one change of Tris-buffered saline (TBS). Free thiol groups are blocked with 40 mM N-ethylmaleimide (NEM) in buffer that contains 25 mM HEPES, pH 7.4, 0.1 mM EDTA, pH 8.0, 0.01 mM neocuproine, and 1% Triton for 30 min.
After three washes with TBS, S-glutathionylated cysteine groups are reduced by incubation with a reaction mix that contains recombinant Grx1 and the components necessary for its activity as follows: 13.5 µg/ml recombinant human Grx1, 35 µg/ml GSSG reductase, 1 mMGSH, 1 mM NADPH, 18 µmol EDTA, and 137 mM Tris–HCl, pH 8.0 for 30 min.
After three washes with TBS, newly reduced cysteine residues are labeled with 1 mM N-(3-maleimidylpropionyl) biocytin (MPB) dissolved in 25 mM HEPES, pH 7.4, 0.1 mM EDTA, pH 8.0, 0.01 mM neocuproine for 30 min.
Excess MPB is removed by three washes with TBS.
Tissue samples are incubated with 0.5 µg/ml streptavidin-conjugated fluorophores for 30 min. Nuclei are counter stained using appropriate concentrations of dye (Fig. 17.2 A and B).
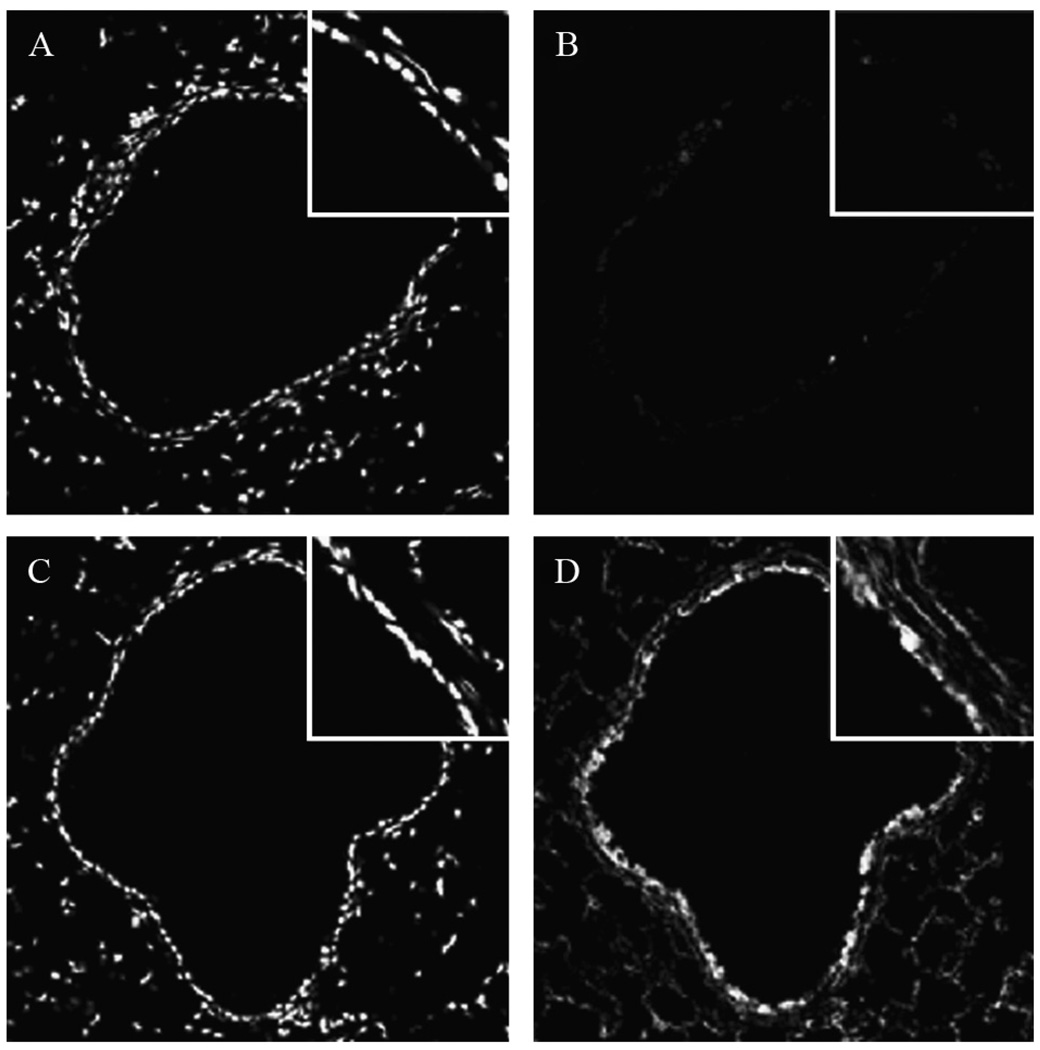
Figure 17.2.
Representative image of untreated paraffin embedded murine lungs stained for S-glutathionylated proteins using the Grx1-catalyzed derivatization protocol and visualization via confocal laser scanning microscopy (B and D). Panels A and C represent nuclear counter stains for the tissues evaluated in (B) and (D), respectively. Panel D: Following the protocol outlined herein, reactivity is seen primarily within the epithelial cells of the conducting airways (inset) with some parenchymal reactivity. Panel B: As a negative control, GSH was omitted from the reaction mix, resulting in a loss of Grx-1-catalyzed labeling. Magnification = 200×. Insets: zoom = 4×.
Prior to derivatization of tissues, all buffers are prepared fresh, and buffers containing MPB protected from direct exposure to light. The reaction mix containing Grx1 is prepared immediately prior to application. All steps are conducted at room temperature.
Following Grx1-catalyzed cysteine derivatization, samples are routinely analyzed by confocal microscopy. Use of confocal imaging is critical for optimal detection of S-glutathionylated proteins over potential background signals, and to elucidate tissue or cellular compartments that harbor Sglutathionylated proteins which would be impossible using wide field fluorescence. The specificity of Grx1 for S-glutathionylated proteins is highlighted by the competitive inhibition of detectable signal by incubating samples with 2.5 mg/ml S-glutathionylated bovine serum albumin (BSA), as compared to fully reduced BSA. Omission of Grx1 from the reaction mix should be an important reagent control as it highlights the requirement of the presence of Grx1 in the observed labeling reaction. Furthermore, omission of free GSH from the reaction mix completely abrogates detectable signal owing to its necessity in reducing catalytically active Grx1 (Fig. 17.2). As an additional negative control, samples should be incubated with dithiothreitol (DTT) prior to incubation with NEM, in order to assess the efficiency of blocking free cysteine residues. As a positive control, tissue sections can be incubated with 400 µM diamide and 1 mM GSH for 10 min prior to application of NEM (Aesif et al., 2009; Reynaert et al., 2006).
3. PROTEIN S-NITROSYLATION
Nitric oxide (NO) is an important signaling molecule that exerts many of its effects through the posttranslational modification S-nitrosylation. S-Nitrosylation is the oxidation of cysteine thiols by NO-derived species such as N2O3, but also by transnitrosylation from low molecular weight nitrosothiols such as S-nitrosylated glutathione (GSNO) or nitrosylated proteins (Stamler et al., 2001). To date, a large number of targets for S-nitrosylation have been identified and linked to functional consequences. Importantly, protein regulation by S-nitrosylation has been coupled to physiological stimuli that involve both receptor-mediated activation of nitric oxide synthases (NOS), as well as stimulus-induced denitrosylation (Benhar et al., 2009; Gow et al., 2002; Hess et al., 2005). Multiple mechanisms for denitrosylation exist, including SNO decomposition by various nonenzymatic compounds, ascorbate, and two enzymatic pathways, namely Trx1 and GSNO reductase (Benhar et al., 2009).
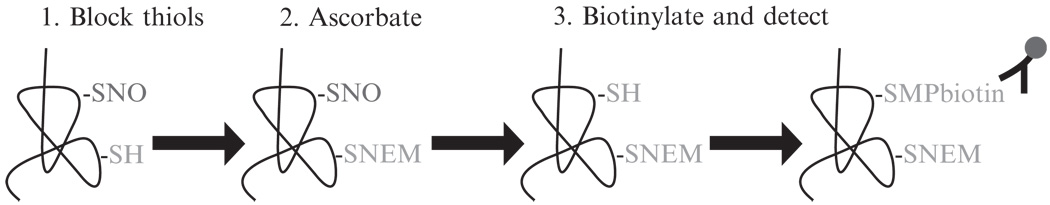
Sample preparation and storage for the determination of nitrosothiol content and protein nitrosylation requires great care since these moieties are readily decomposed by a myriad of factors. Furthermore, only recently have technological advances been made to facilitate research into this redox-dependent posttranslational modification. For instance, an antibody was developed against nitrosocysteine that has been used among others to visualize protein S-nitrosylation in various tissues (Gow et al., 2002). S-Nitrosylation of specific proteins can be achieved by measuring the release of NO by these proteins using chemiluminescence, or by the biotin-switch protocol as described by Jaffrey and Snyder (2001) and subsequently published variations thereof. Finally, we have successfully adapted this biotin derivatization procedure to detect protein S-nitrosylation in situ (Ckless et al., 2004). The protocol is schematically depicted in Fig. 17.3 and described in the following section.
Figure 17.3.
Schematic representation of the protocol for detecting S-nitrosylated proteins in situ. Free thiols are blocked by alkylation with NEM (1), S-nitrosylated cysteines are reduced using ascorbate (2), and finally newly reduced cysteines are labeled using MPB and detected using streptavidin-conjugated fluorophores (3).
3.1. In situ detection of S-nitrosylated proteins
After dewaxing tissue sections in three changes of xylene, tissue is rehydrated in 100%, 95%, and 75% ethanol, and washed three times with phosphate-buffered saline (PBS) containing 0.4 mM EDTA and 0.04 mM neocuproine. Next, free thiol groups are blocked with 40 mM NEM in PBS containing 0.4 mM EDTA, 0.04 mM neocuproine, and 2.5% SDS for 30 min.
After removal of blocking solution and three washes with PBS containing 0.4 mM EDTA and 0.04 mM neocuproine, sections are incubated with 1 mM sodium ascorbate in PBS for 15 min to reduce S-nitrosylated proteins.
Newly reduced cysteine residues are then labeled with 0.1 mM MPB in PBS for 30 min.
Excess MPB is removed by three washes with PBS containing 0.4 mM EDTA and 0.04 mM neocuproine.
Tissues are next incubated for 30 min with 0.5 µg/ml of a fluorophore-labeled streptavidin in order to visualize biotinylated proteins and nuclei are counterstained with an appropriate dye.
All procedures prior to labeling with MPB must be performed under protection from direct light and all reagents must be dissolved freshly before their use. All incubation steps in the protocol occur at room temperature.
Following the biotin switch protocol to label protein S-nitrosylation, tissues are analyzed by confocal microscopy. Use of confocal imaging is critical to optimal detection of S-nitrosylated proteins over potential background signal, and to elucidate tissue or cellular compartments that harbor S-nitrosylated proteins, which would be impossible to achieve using wide field fluorescence. As a negative control, omission of the ascorbate decomposition step should be performed, which, in the case of optimal blocking of reduced cysteines, should result in the absence of labeling. Alternatively, S-nitrosothiols can be decomposed with 1 mM ascorbate, or subjected UV-mediated prephotolysis at 335 nm (Forrester et al., 2009) prior to blocking. These steps should result in the absence of labeling. As a positive control, tissue sections can be incubated with a nitrosothiol such as GSNO or l-CysNO (100–500 µM) prior to the application of NEM.
4. SUMMARY
The protocols presented herein represent robust avenues for the in situ detection of S-glutathionylated and S-nitrosylated proteins in tissues. They are, however, not without potential and long debatable pitfalls, as neither protocol absolutely excludes the possibility of false positivity due to the reduction of intermolecular disulfides, or other oxidative modifications: The labeling and detection of newly generated thiols depend on the specificity of ascorbate and human Grx1 toward S-nitrosylated and S-glutathionylated proteins, respectively. As screening tools, however, these assays and adaptations thereof that undoubtedly will be developed in the future will provide valuable insights into which tissue or organ compartments are affected by reversible cysteine oxidations.
REFERENCES
- Aesif SW, et al. In situ analysis of protein S-glutathionylation in lung tissue using glutaredoxin-1-catalyzed cysteine derivatization. Am. J. Pathol. 2009;175:36–45. doi: 10.2353/ajpath.2009.080736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anathy V, et al. Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J. Cell Biol. 2009;184:241–252. doi: 10.1083/jcb.200807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, et al. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- Chrestensen CA, et al. Acute cadmium exposure inactivates thioltransferase (Glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. J. Biol. Chem. 2000;275:26556–26565. doi: 10.1074/jbc.M004097200. [DOI] [PubMed] [Google Scholar]
- Ckless K, et al. In situ detection and visualization of S-nitrosylated proteins following chemical derivatization: Identification of Ran GTPase as a target for S-nitrosylation. Nitric Oxide. 2004;11:216–227. doi: 10.1016/j.niox.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, et al. S-glutathionylation in protein redox regulation. Free Radic. Biol. Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, et al. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxid. Redox Signal. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- Forrester MT, et al. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ghezzi P. Regulation of protein function by glutathionylation. Free Radic. Res. 2005;39:573–580. doi: 10.1080/10715760500072172. [DOI] [PubMed] [Google Scholar]
- Giles GI, et al. Evaluation of sulfur, selenium and tellurium catalysts with antioxidant potential. Org. Biomol. Chem. 2003;1:4317–4322. doi: 10.1039/b308117f. [DOI] [PubMed] [Google Scholar]
- Gow AJ, et al. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- Hess DT, et al. Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. USA. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A, et al. Thiroedoxin from Escherichia coli. Radioimmunological and enzymatic determinations in wild type cells and mutants defective in phage T7 DNA replication. J. Biol. Chem. 1978;253:430–436. [PubMed] [Google Scholar]
- Holmgren A, et al. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- Jacob C, et al. Sulfur and selenium: the role of oxidation state in protein structure and function. Angew. Chem. Int. Ed. Engl. 2003;42:4742–4758. doi: 10.1002/anie.200300573. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- Janssen-Heininger YM, et al. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic. Biol. Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, et al. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch. Biochem. Biophys. 1999;362:67–78. doi: 10.1006/abbi.1998.1013. [DOI] [PubMed] [Google Scholar]
- Kim SO, et al. OxyR: A molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- Mallis RJ, Thomas JA. Effect of S-nitrosothiols on cellular glutathione and reactive protein sulfhydryls. Arch. Biochem. Biophys. 2000;383:60–69. doi: 10.1006/abbi.2000.2048. [DOI] [PubMed] [Google Scholar]
- Meng TC, et al. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, et al. Glutathionylation of the p50 subunit of NF-kappaB: A mechanism for redox-induced inhibition of DNA binding. Biochemistry. 2001;40:14134–14142. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- Reynaert NL, et al. In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim. Biophys. Acta. 2006;1760:380–387. doi: 10.1016/j.bbagen.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Shelton MD, et al. Glutaredoxin: Role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- Stamler JS, et al. Nitrosylation. The prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Takashima Y, et al. Differential expression of glutaredoxin and thioredoxin during monocytic differentiation. Immunol. Lett. 1999;68:397–401. doi: 10.1016/s0165-2478(99)00087-5. [DOI] [PubMed] [Google Scholar]