Abstract
Small molecule histone deacetylase (HDAC) inhibitors block adverse cardiac remodeling in animal models of heart failure. The efficacious compounds target class I, class IIb and, to a lesser extent, class IIa HDACs. It is hypothesized that a selective inhibitor of a specific HDAC class (or an isoform within that class) will provide a favorable therapeutic window for the treatment of heart failure, although the optimal selectivity profile for such a compound remains unknown. Genetic studies have suggested that class I HDACs promote pathological cardiac remodeling, while class IIa HDACs are protective. In contrast, nothing is known about the function or regulation of class IIb HDACs in the heart. We developed assays to quantify catalytic activity of distinct HDAC classes in left and right ventricular cardiac tissue from animal models of hypertensive heart disease. Class I and IIa HDAC activity was elevated in some but not all diseased tissues. In contrast, catalytic activity of the class IIb HDAC, HDAC6, was consistently increased in stressed myocardium, but not in a model of physiologic hypertrophy. HDAC6 catalytic activity was also induced by diverse extracellular stimuli in cultured cardiac myocytes and fibroblasts. These findings suggest an unforeseen role for HDAC6 in the heart, and highlight the need for pre-clinical evaluation of HDAC6-selective inhibitors to determine whether this HDAC isoform is pathological or protective in the setting of cardiovascular disease.
Keywords: Histone deacetylase, hypertension, heart failure
1. Introduction
When the mammalian heart is subjected to increased afterload due to hypertension, the response is often characterized by myocyte hypertrophy, interstitial fibrosis and ventricular dysfunction [1]. Uncontrolled systemic or pulmonary arterial hypertension frequently leads to left ventricular (LV) or right ventricular (RV) heart failure, respectively. It is estimated that over 5 million adults in the U.S. suffer from heart failure, and that ~75% of these patients had antecedent hypertension [2]. Given the high mortality rate in patients with heart failure [2], there is a vital need for novel therapeutic approaches.
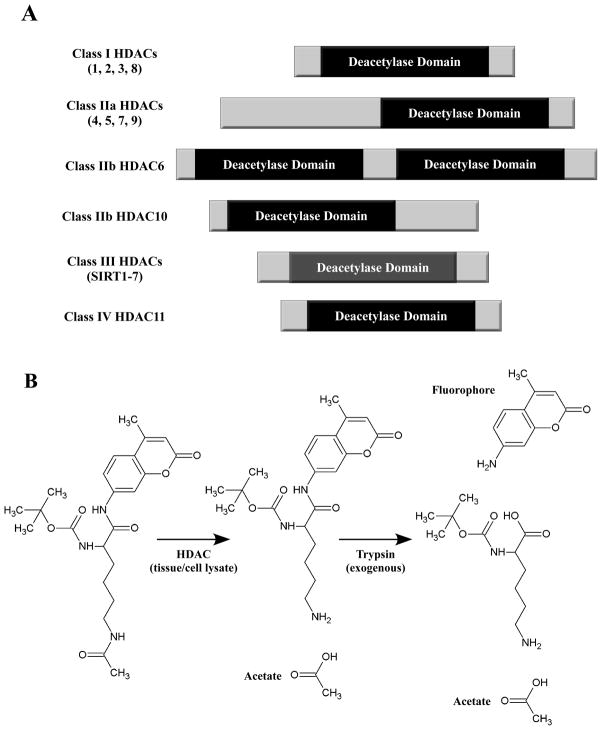
Small molecule inhibitors of histone deacetylases (HDACs) have shown efficacy in multiple animal models of heart failure [3–5]. HDACs catalyze removal of acetyl groups from ε-amino groups of lysine residues in a variety of proteins. HDACs have mainly been studied in the context of chromatin, where they serve an epigenetic function by deacetylating nucleosomal histones and altering the electrostatic properties of chromatin to favor gene repression. However, it is now clear that HDACs also deacetylate many non-histone proteins, including transcription factors, mitochondrial proteins and cytoskeletal proteins [6, 7]. The 18 HDACs are encoded by distinct genes and are grouped into four classes on the basis of similarity to yeast transcriptional repressors (Fig. 1A). Class I HDACs (HDACs 1, 2, 3 and 8) are related to yeast RPD3, class II HDACs (HDACs 4, 5, 6, 7, 9 and 10) to yeast HDA1, and class III HDACs (SirT1 – 7) to yeast Sir2. Class II HDACs are further divided into two subclasses, IIa (HDACs 4, 5, 7 and 9) and IIb (HDACs 6 and 10). HDAC11 falls into a fourth class [8].
Fig. 1.
Schematic representations of HDACs and an HDAC enzymatic assay. (A) HDACs are categorized into four distinct classes. Class II HDACs are further divided into two subclasses, IIa and IIb. Class III HDACs are also known as sirtuins. Class I, II and IV HDACs harbor zinc-dependent catalytic domains, while class III HDACs use NAD+ as a co-factor. (B) Example HDAC enzymatic assay. The substrate consists of lysine with three modifications: acetylation of the ε-amine, a protecting group on the α-amine, and conjugation of the carboxyl by the fluorophore 7-amino-4-methylcoumarin (AMC). HDAC activity from cell or tissue extracts removes the ε-acetyl group. Following incubation of the samples with substrate, trypsin is added, which is active only against deacetylated substrate. Trypsin releases AMC, resulting in increased fluorescence emission at 460 nm.
HDAC inhibitors block cardiac hypertrophy, fibrosis and inflammation in various rodent models, including myocardial infarction (MI) [9–12], transverse aortic constriction (TAC) [13, 14] and adrenergic excess [15]. Recently, HDAC inhibitors were shown to block LV hypertrophy in response to chronic systemic hypertension induced by the aldosterone mimetic, deoxycorticosterone acetate (DOCA) [16], and in spontaneously hypertensive rats (SHR) [17]. HDAC inhibitors also appear to reduce RV hypertrophy in response to increased afterload [18]. All of the compounds used in these pre-clinical studies were non-selective, pan-HDAC inhibitors, and thus it has not been possible to ascribe the beneficial effects to inhibition of a particular HDAC class or an isoform within a given class.
To address the possible involvement of specific HDAC classes in hypertension-induced cardiac remodeling, we established enzymatic assays that enable quantification of class I, IIa or IIb HDAC catalytic activity in cardiac tissue homogenates. These assays revealed an elevation in the catalytic activity of the class IIb HDAC, HDAC6, in multiple models of LV and RV pressure overload, including DOCA-treated rats, spontaneously hypertensive heart failure (SHHF) rats and rats with RV hypertrophy induced by chronic hypoxia, but not in hypertrophic hearts of exercised animals, demonstrating the specificity of HDAC6 activation to pathologic events. Studies with cultured cardiac myocytes and fibroblasts demonstrated that HDAC6 activity is induced by G-protein coupled receptor (GPCR) signaling in myocytes and cytokine receptor signaling in fibroblasts. Alterations in class I and class IIa HDAC activity were less consistent. These results suggest a role for HDAC6 in the heart and provide rationale for evaluation of the function of this HDAC isoform in the models of cardiovascular disease.
2. Materials and methods
2.1 Reagents
Reagents were purchased from the indicated vendors. HDAC inhibitors: apicidin (Enzo), SAHA (ChemieTek), nicotimamide (Sigma), MGCD0103 (Sai Advantium), Trichostatin A (TSA; Sigma). Agonists: phenylephrine (PE; Sigma), endothelin-1 (ET-1; Sigma), isoproterenol (ISO; Sigma), norepinephrine (NE; Sigma), prostaglandin F2α (PGF2α; EMD4 Biosciences), IL-1β (R&D Systems). Synthetic HDAC substrates: class I HDAC substrate (custom synthesis by Genscript), class IIa HDAC substrate (I-1985; Bachem), class I/IIb substrate (I-1875; Bachem). SU5416, Trypsin, Triton X-100, and DMSO were obtained from Sigma. 7-Amino-4-methylcoumarin (AMC; Alfa Aesar).
2.2 HDAC activity assays
Typically, assays were run simultaneously for class I, IIa, and IIb HDAC activities, using parallel sets of sample aliquots. Each of the HDAC substrates was based on ε-N-acylated lysine, derivatized on the carboxyl group with AMC [19]. Subsequent to deacylation by HDAC activity, trypsin was used to release AMC, resulting in a significant increase in fluorescence.
Tissue extracts were prepared in PBS (pH 7.4) containing 0.5% Triton X-100, 300 mM NaCl and protease/phosphatase inhibitor cocktail (Thermo Fisher) using a Bullet Blender homogenizer (Next Advance). Cultured cells were suspended in the same buffer and sonicated prior to clarification by centrifugation. Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Fisher). Cell and tissue extracts were diluted into PBS buffer in 100 μL total volumes in 96-well plate (60 μg ventricular protein/well; 15 μg neonatal rat ventricular myocyte [NRVM]/adult rat ventricular fibroblast [ARVF] protein/well; 30 μg adult rat ventricular myocyte [ARVM] protein/well); Where indicated, exogenous HDAC inhibitors (typically 1 μL of 100× DMSO stock solutions) or corresponding volumes of DMSO were added, followed by 30 – 60 minutes incubation at 37° C. Substrates were added (5 μL of 1 mM DMSO stock solutions), and the plates were returned to the 37°C incubator for 2 – 3 hours. Finally, developer/stop solution was added (50 μL per well of PBS with 1.5% Triton X-100, 3 μM TSA, and 0.75 mg/mL trypsin), with another 20 minute incubation at 37°C. AMC fluorescence was measured using a BioTek Synergy 2 plate reader, with excitation and emission filters of 360 nm and 460 nm, respectively (each with bandwidth 40 nm), along with a 400 nm dichroic top mirror. Background signals from buffer blanks were subtracted, and data were normalized as needed using appropriate controls.
Tissue extracts were often slightly colored, which variably quenched the AMC fluorescence. This issue was resolved by including an additional “reference” set of sample aliquots and buffer blank, which received identical aliquots of AMC (5 μL of 80 μM in DMSO) instead of substrate. The reference AMC signals were used to calculate quenching ratios relative to the AMC buffer blank signal. Each raw fluorescence value from the substrate-containing samples was divided by the corresponding quenching ratio, followed by subtraction of the appropriate substrate background.
GraphPad Prism software was used to generate graphs and analyze data. ANOVA with Bonferroni’s post test (p<0.05) was used to determine statistical differences between groups.
2.3 Enzyme kinetics
HDAC1, HDAC2, HDAC4 and HDAC6 recombinant enzymes were purchased from BPS Bioscience (San Diego, CA). Enzyme titrations were conducted to determine the conditions for each enzyme to be within the linear detection range of the assay. Enzyme efficiency (kcat/Km) was determined by incubating recombinant HDACs with titrated substrates, 5μM – 160μM, for 120 minutes at 37°C under standard assay conditions. A standard curve of the fluorescent product, AMC, was also run under these conditions to determine a conversation factor to express fluorescent values as moles of substrate produced. Raw fluorescent values were converted to a rate expressed as moles of substrate converted per second per mg of enzyme. This rate was plotted against substrate concentration and was fit to the Michaelis-Menten equation using GraphPad Prism 5 (La Jolla, CA). The best-fit line produced the Km (μM) and Vmax (moles sec−1 mg−1) of each enzyme/substrate pair. kcat (sec−1) was determined by dividing Vmax by the concentration of enzyme active site per mg of enzyme. kcat/Km, expressed in [sec−1 M−1], was determined for each enzyme on each substrate and represents the catalytic efficiency of the enzyme on the substrate.
2.4 Animal models
All animal experiments were conducted in accordance with the National Institutes Health ‘Guide for the Care and Use of Laboratory Animals’, and were approved by the Institutional Animal Care and Use Committee at the University of Colorado Denver. For RV hypertrophy, the hypoxia-induced pulmonary hypertension (PH) model was used. Male Sprague-Dawley (SD) rats weighing 250–280 grams (Charles River Labs; n = 8) were placed in a hypobaric chamber to simulate an altitude of 18,000 feet above sea level and create a hypoxic environment (10% 02) [20]. Normoxic control rats (n = 6) were maintained in chambers simulating sea level (21% 02). Some rats received a single subcutaneous injection of a VEGF receptor inhibitor, SU5416, at 20 mg/kg immediately prior to housing in the hypobaric chamber (n = 8). SU5416 has been shown to exacerbate pulmonary vascular lesions and aggravate PH [21, 22]. For LV remodeling, the DOCA-salt hypertension model was used. Male SD rats weighing 230–250 grams (n = 9) were subjected to unilateral nephrectomy followed one week later by implantation of a medical grade silastic pellet impregnated with 100 mg of deoxycorticosterone acetate (DOCA; Sigma), and given tap water containing 0.9% NaCl and 0.2% KCl, as previously described [23, 24]. Control animals (n = 8) were subjected to unilateral nephrectomy and received a silastic pellet containing vehicle, and were given regular tap water. Spontaneously hypertensive heart failure (SHHF) rats and the voluntary cage wheel running model of physiological cardiac hypertrophy in mice have been described previously [25, 26]. Male SHHF rats were from the McCune colony at the University of Colorado Boulder (young, n = 8; non-failing, n = 7, failing, n = 7). Young SHHF rats show no signs of heart failure and are generally accepted as optimal controls for determining how signaling and gene regulatory events are altered as the model progresses to heart failure [27]. For exercise studies, 3 to 4 month-old female C57BL/6 mice were used (Taconic; n = 15 per group).
2.5 Cells and culture
Neonatal rat ventricular myocytes (NRVMs) were prepared from hearts of 1 – 3 day-old SD rats, as previously described [28]. Cells were cultured overnight on 10-cm plates coated with gelatin (Sigma; 0.2%) in DMEM containing fetal bovine serum (FBS, 10%), L-glutamine (2 mM), and penicillin-streptomycin. After overnight culture, cells were washed with serum-free medium and maintained in DMEM supplemented with Neutridoma-SP (Roche Applied Science), which contains albumin, insulin, transferrin, and other defined organic and inorganic compounds. Adult rat ventricular myocytes (ARVMs) and adult rat ventricular fibroblasts (ARVFs) were obtained from female SD rats, as described previously [29]. ARVMs were plated at a density of 100 to 150 cells/mm2 on laminin-coated 60-mm plates and maintained in serum-free DMEM supplemented with albumin (2 mg/ml), 2,3-butanedione monoxime (1 mg/ml), L-carnitine (2 mM), creatine (5 mM), penicillin-streptomycin (100 μg/ml), triiodothyronine (1 pM), and taurine (5 mM). ARVFs were cultured on 10-cm plates in DMEM containing FBS (10%), L-glutamine (2 mM) and gentamicin (0.1 mg/ml). ARVFs were serum-starved for 48 hrs in DMEM containing BSA (0.1%) prior to stimulation with FBS (10%) or IL-1β (10ng/ml) for an additional 48 hrs.
2.6 Adenovirus production and quantitative PCR for rat HDAC6 and HDAC10
Recombinant adenoviruses encoding short hairpin (sh) RNAs to rat HDAC6 and rat HDAC10 were prepared using the BLOCK-it™ Adenoviral RNAi Expression System (Invitrogen). Oligonucleotide targeting sequences are shown in Table 1. Top and bottom strands were annealed and ligated into the pENTR/U6 vector (Invitrogen). Positive sub-clones were recombined with the pAd/BLOCK-it™–Dest vector (Invitrogen) and then transfected into 293A cells using Fugene 6 (Roche). Virus was grown, amplified and recovered from 293A cells via multiple freeze/thaw cycles. Cell lysate containing viral particles was used for subsequent studies.
Table 1.
HDAC6 and HDAC10 shRNA targeting sequences and Q-PCR primers.
| Target | Top Strand shRNA | Bottom Strand shRNA |
|---|---|---|
| HDAC6 | 5′-CAC CGC ACC TAT GAT TCC GTT TAT CCG AAG ATA AAC GGA ATC ATA GGT GC-3′ | 5′-AAA AGC ACC TAT GAT TCC GTT TAT CTT CGG ATA AAC GGA ATC ATA GGT GC-3′ |
| HDAC10 | 5′-CAC CGC TTT AGT GGA AGA GGA ATC CCG AAG GAT TCC TCT TCC ACT AAA GC-3′ | 5′-AAA AGC TTT AGT GGA AGA GGA ATC CTT CGG GAT TCC TCT TCC ACT AAA GC-3′ |
| Target | Forward Q-PCR Primer | Reverse Q-PCR Primer |
| HDAC6 | 5′-GGG CGT CAG TGG CTC ACT CC-3′ | 5′-TCT GGG CGC TTG CAC AAG GT-3′ |
| HDAC10 | 5′-TGC TGT GTT GGA GTG CCC TGG-3′ | 5′-GCA GAT TGG GGG CGT AGG GC-3′ |
| 18S | 5′-GCC GCT AGA GGT GAA ATT CTT A-3′ | 5′-CTT TCG CTC TGG TCC GTC TT-3′ |
NRVMs were infected with shRNA encoding adenoviruses the day after plating, and total RNA was harvested using TRI Reagent™ (Ambion) 48 hrs after infection. All RNA samples were diluted to 100 ng/μl, and 5 μl (500 ng) of RNA was converted to cDNA using the Verso™ cDNA Synthesis Kit (Thermo Scientific). Quantitative PCR (Q-PCR) was performed using Absolute™ QPCR SYBR Green ROX mix (Thermo Scientific) on an ABI-7300 qPCR instrument (Applied Biosystems). Q-PCR primers for HDAC6 (XM_228753.5) and HDAC10 (NM_001035000.1) are shown in Table 1. Relative transcript levels were determined by measuring Ct values off of a stand curve made from serial dilutions of pooled cDNA. Additional NRVMs infected with shRNA adenoviruses were harvested for protein extracts, which were used in HDAC activity assays.
3. Results
3.1 Catalytic activity of specific HDAC classes in the heart
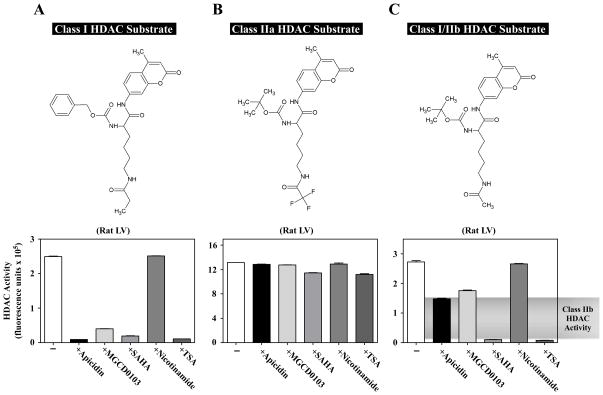
HDAC catalytic activity can be quantified using fluorogenic substrates containing acetyl-lysine moieties [30]. Upon deacetylation by HDACs, the substrates become susceptible to cleavage by trypsin, which releases the fluorophore, 7-amino-4-methylcoumarin (AMC) [31] (Fig. 1B). These substrates can be used to measure activity of recombinant HDACs or endogenous HDACs from tissue homogenates. The substrate shown in Fig. 1B is deacetylated by class I and class IIb HDACs [19]. By modifying the ε-acyl group and the α-amino protecting group, it is possible to create a substrate that is selectively deacetylated by class I HDACs [19] (Fig. 2A). Furthermore, a substrate having a trifluoroacetyl group on the ε-amine is specific for class IIa HDAC enzymes [32, 33] (Fig. 2B). Kinetic studies with recombinant forms of representative members of each HDAC class confirmed the selectivity of these substrates (Table 2). Specifically, HDAC1 and HDAC2 had the highest catalytic efficiency (kcat/Km) for the class I substrate, while HDAC4 and HDAC6 had higher kcat/Km values for the class IIa and Class I/IIb substrates, respectively.
Fig. 2.
Quantification of class I, IIa and IIb HDAC catalytic activity in cardiac tissue homogenate. Rat LV extract was incubated with different HDAC substrates in the absence or presence of individual HDAC inhibitors: apicidin (3 μM), MGCD0103 (10 μM), SAHA (10 μM), nicotinamide (1 mM), or TSA (1 μM). Apicidin and MGCD0103 are class I-specific inhibitors. SAHA and TSA inhibit class I and IIb enzymes, and nicotinamide inhibits the sirtuins (class III). (A) Class I-selective substrate and corresponding HDAC activity data. In this case, apicidin and MGCD0103 behaved similar to the classical pan-inhibitors, SAHA and TSA. (B) Class IIa substrate and corresponding HDAC activity data. None of the inhibitors had a significant impact on the measured activity, confirming that this activity was not from class I, IIb, or III HDAC enzymes. (C) Class I/IIb substrate and corresponding activity data. Note that class I inhibitors lowered the activity by about 50%, revealing class IIb activity. All of the activity with this substrate was inhibited by SAHA and TSA, while nicotinamide had little effect.
Table 2.
Recombinant enzyme efficiency on class specific substrates as measured by kcat/Km.
| Enzyme | HDAC Class |
kcat/Km [sec−1 M−1]
|
||
|---|---|---|---|---|
| Class I Substrate | Class IIa Substrate | Class I/IIb Substrate | ||
| rHDAC 1 | Class I | 2425 | 189 | 742 |
| rHDAC 2 | Class I | 6139 | 750 | 2275 |
| rHDAC 4 | Class IIa | 0 | 21979 | 0 |
| rHDAC 6 | Class IIb | 45 | 200 | 6375 |
Validation studies were performed to determine whether these substrates could be used to quantify class-specific HDAC activity in cardiac tissue. As shown in Fig. 2A, HDAC activity was readily detected in rat LV tissue extract. As expected, this activity represented class I HDACs, as evidenced by the sensitivity of the assay to the class I HDAC inhibitors, apicidin and MGCD0103, as well as the pan-HDAC inhibitors, SAHA and TSA. In contrast, activity measured with this substrate was insensitive to nicotinamide (Fig. 2A), which inhibits class III HDACs [34].
A robust signal was also obtained using the class IIa HDAC substrate and rat LV extract (Fig. 2B). As predicted based on prior studies [35], this activity was largely resistant to class I and classical pan-HDAC inhibitors. The use of the class I/IIb substrate in combination with class I-specific inhibitors revealed the presence of class IIb HDAC activity in the rat LV (Fig. 2C).
3.2 Altered HDAC catalytic activity in models of pathological cardiac remodeling
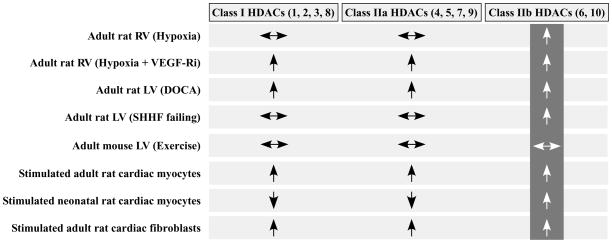
Experiments were performed to determine whether HDAC activity is changed in rodent models of cardiac remodeling induced by hypertension. The extent of cardiac hypertrophy and other model characteristics are shown in Table 3. In a three week model of pulmonary hypertension (PH) induced by chronic hypoxia, only class IIb HDAC catalytic activity was significantly increased in the RV (Fig. 3A). VEGF receptor inhibition in combination with chronic hypoxia is known to exacerbate pulmonary vascular remodeling and promote apoptosis and interstitial fibrosis in the RV [21, 22, 36]. In this more severe disease model, elevations in class I, IIa and IIb HDAC activity were detected in the RV (Fig. 3A).
Table 3.
Cardiac hypertrophy and hypertension in rodent models.
| MODEL | RV/LV | HW/BW | HW/BrainW | LV/TL | MAP | RVSP |
|---|---|---|---|---|---|---|
| Hypoxia | ↑ 94% | ↔ | ↔ | ↑ 83% | ||
| Hypoxia + SU5416 | ↑ 103% | ↔ | ↔ | ↑ 85% | ||
| DOCA | ↔ | ↑ 34% | ↑ 43% | ↑ 40% | ||
| SHHF (non-failing) | ↑ 8%* | ↑ 27%* | ↑ 8%* | |||
| SHHF (failing) | ↑ 49%* | ↑ 84%* | ↑ 60%* | |||
| Exercise (7-days) | ↑ 17% | |||||
| Exercise (21-days) | ↑ 23% |
DOCA, deoxycorticosterone acetate; SHHF, spontaneously hypertensive heart failure; RV, right ventricle; LV, left ventricle; HW, heart weight; BW, body weight; BrainW, brain weight; TL, tibia length; MAP, mean arterial pressure; RVSP, right ventricular systolic pressure.
Compared to young SHHF rats.
Fig. 3.
Elevated cardiac class IIb HDAC activity in animal models. Class I and IIa activities were measured using the appropriate substrates (see Fig. 2A and 2B). Class IIb activity was measured using the I/IIb substrate in the presence of 3 μM apicidin (see Fig. 2C). (A) HDAC activities in RV extracts prepared from rats exposed to normoxia (n = 6), hypoxia (n = 8), or hypoxia + SU5416 (n = 8). These are the only samples for which class I HDAC activity was determined using the class I/IIb substrate, as opposed to the class I HDAC-specific substrate; activity was defined based on apicidin sensitivity. (B) HDAC activities were measured in LV extracts prepared from rats with hypertension due to unilateral nephrectomy plus a DOCA pellet (n = 9) or normotensive controls (n = 8) that were subjected to nephrectomy and a sham pellet. (C) HDAC activities in LV extracts prepared from SHHF rats of different ages (young, n = 8; non-failing and failing, n = 7 each). (D) HDAC activities in LV extracts prepared from mice that were kept sedentary or were exercised (voluntary wheel running) for 7 or 21 days to induce physiological cardiac hypertrophy (n = 15 per group). Values represent averages +SEM. *P < 0.05 versus respective control.
In a rat model of systemic hypertension induced by the aldosterone mimetic, DOCA, combined with unilateral nephrectomy, the levels of class I, IIa and IIb HDAC catalytic activity were all significantly increased in pressure overloaded LVs. Class IIb HDAC activity was also increased in failing LVs from SHHF rats, which have severe systemic hypertension (Fig. 3C), but not in younger SHHF animals, which had cardiac hypertrophy but not heart failure. Interestingly, in this latter group of animals, a significant decrease in LV class I and IIa HDAC activity was noted.
To begin to address whether altered HDAC activity is a general response to cardiac growth signals, catalytic activity of the individual HDAC classes was quantified in hearts of mice with physiological cardiac hypertrophy induced by 7 or 21 days of voluntary cage wheel running (see Table 3). As shown in Fig. 3D, no significant changes in cardiac HDAC activity were found in this model of exercise-induced hypertrophy.
3.3 Signal-dependent changes in HDAC catalytic activity in cultured cardiac cells
Hypertension triggers release of humoral factors such as norepinephrine (NE) and endothelin-1 (ET-1) that promote growth of the two major cell types in the heart, cardiac myocytes and cardiac fibroblasts [37]. Experiments were performed with purified populations of myocytes or fibroblasts from rat hearts to determine whether HDAC activity is directly altered by extracellular stimuli that activate these cells. As shown in Fig. 4A, treatment of cultured NRVMs with NE, which induces cardiomyocyte hypertrophy through stimulation of both α- and β-adrenergic receptors, caused a dramatic increase in class IIb HDAC activity. This increase was recapitulated with phenylephrine (PE), which selectively agonizes α-ARs, but not with the β-AR selective agonist, isoproterenol (ISO). In a second study, NE also stimulated class IIb HDAC activity in NRVMs, as did prostaglandin F2α (PGF2α) and ET-1, which are also potent hypertrophic agonists (Fig. 4B). These results suggest that class IIb HDAC activity is induced by agonists targeting G-protein coupled receptors (GPCRs) that couple to Gαq (PE, PGF2α, ET-1), but not Gαs (ISO).
Fig. 4.
Extracellular stimuli induce class IIb HDAC catalytic activity in cultured cardiac myocytes and fibroblasts. HDAC activities were measured as described for Figure 3. (A) NRVMs were left untreated (−) or stimulated for 72 hrs with NE (10 μM), PE (10 μM) or ISO (1 μM), then harvested for HDAC activity measurements. (B) NRVMs were cultured in the absence (−) or presence of NE (10 μM), PGF2α (10 μM), or ET-1 (25 nM) for 72 hrs prior to measuring HDAC activity in lysates. (C) ARVMs were treated with NE (10 μM), PE (20 μM), PGF2α (10 μM) or ET-1 (25 nM) for 72 hrs and harvested for HDAC activity assays. (D) ARVFs were treated with FBS (10%) or IL-1β (10ng/ml) for 48 hrs prior to harvesting cells for HDAC activity assays. Values for A – D represent averages (n = 3 per condition) +SEM. *P < 0.05 versus respective control.
NE, PE and PGF2α also induced class IIb HDAC catalytic activity in cultured adult rat ventricular myocytes (ARVMs) (Fig. 4C). However, in contrast to what was observed with NRVMs, PE and PGF2α treatment of ARVMs additionally led to elevations in class I and IIa HDAC activity, and NE stimulated class I HDAC activity. The reason for these discrepancies between ARVMs and NRVMs is unknown, but may reflect developmental stage-specific differences in signaling in cardiomyocytes, as has been described for cardiac α- and β-ARs [38]. ET-1 failed to alter HDAC activity in ARVMs, possibly due to a lack of function ET receptors in our myocyte preparations.
HDAC activity was subsequently measured in extracts from cultured adult rat cardiac fibroblasts (ARVFs) exposed to fetal bovine serum (FBS), which stimulates fibroblast proliferation and migration. Alternatively, cells were treated with interleukin-1β (IL-1β), which promotes cardiac fibroblast migration, but not proliferation. As shown in Fig. 4D, FBS treatment stimulated class I, IIa and IIb activity in ARVFs, while IL-1β selectively promoted activity of class I and IIb HDACs. These results suggest that distinct HDACs can respond to signaling events governing cardiac fibroblast proliferation and/or migration.
3.4 Altered HDAC6 activity and expression
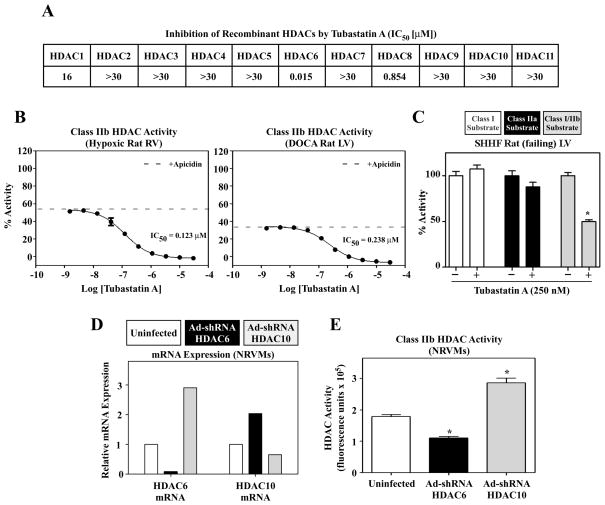
A summary of the enzymatic activity of HDAC classes in cardiac tissue and cells is shown in Fig. 5. Only class IIb HDAC catalytic activity was consistently elevated in all of the pathologic models tested. There are two mammalian class IIb HDACs, HDAC6 and HDAC10. To begin to address whether the elevated activity observed in these studies was due to increases in HDAC6 and/or HDAC10, dose-response studies were performed with RV and LV homogenates and a small molecule HDAC inhibitor, Tubastatin A. Tubastatin A was shown to inhibit recombinant HDAC6 with a half-maximal inhibitory concentration (IC50) of 15 nM, while failing to inhibit HDAC10 at concentrations of up to 30,000 nM (Fig. 6A) [39]. As shown in Fig. 6B, class IIb HDAC activity from hypertrophic RV and LV samples was inhibited by Tubastatin A in a concentration-dependent manner (IC50 = 123 nM and 238 nM for RV and LV, respectively), suggesting that the cardiac activity targeting this substrate is primarily derived from HDAC6 as opposed to HDAC10.
Fig. 5.
Summary of cardiac HDAC activities. General increase in activity (↑), decrease in activity (↓), or activity unchanged (↔) compared to respective controls.
Fig. 6.
HDAC6 activity in the heart. (A) Half-maximal inhibitory concentration (IC50) of Tubastatin A for each recombinant HDAC isoform [39]. (B) Tubastatin A titration of class IIb HDAC activity. RV or LV protein extracts from rats with hypoxia-induced PH or DOCA-induced systemic hypertension, respectively, were exposed to apicidin to inhibit class I HDACs prior to addition of increasing concentrations of Tubastatin A. Class I/IIb substrate (Fig. 2C) was used. Three replicates were used per Tubastatin concentration. The dashed line indicates the level of inhibition due to apicidin alone; remaining HDAC activity is class IIb. Tubastatin A inhibits all the IIb activity from RV and LV with high potency. These results implicate HDAC6 as the sole source of the activity, because Tubastatin A does not significantly inhibit HDAC10. (C) LV protein extract from failing SHHF rats was exposed to 250 nM Tubastatin A and HDAC activity was measured using class I, class IIa or class I/IIb HDAC substrates (n = 4 per condition). Tubastatin A only inhibited deacetylation of the class I/IIb HDAC substrate, demonstrating that HDAC6 deacetylates this substrate but not the class I and class IIa substrates. (D and E) NRVMs were infected with adenoviruses (n = 3 plates/condition) encoding shRNA against rat HDAC6 (Ad-shRNA HDAC6; black shading) or rat HDAC10 (Ad-shRNA HDAC10; grey shading). After 48 hrs of infection, total RNA was prepared and levels of HDAC6 and HDAC10 mRNA transcripts were analyzed by Q-PCR. Protein was prepared from parallel plates (n = 3 per condition) for HDAC activity measurements. The Class IIb HDAC activity pattern (E) approximates that of HDAC6 mRNA expression (D).
The specificity of Tubastatin A and the HDAC substrates was confirmed using LV homogenate from SHHF rats with heart failure. As shown in Fig. 6C, Tubastatin failed to inhibit deacetylation of the class I and class IIa HDAC substrates, but reduced deacetylation of the class I/IIb substrate by ~50%.
To further address whether the class IIb HDAC activity measured in cardiac cells is derived from HDAC6 and/or HDAC10, recombinant adenoviruses were constructed to express short hairpin (sh) RNAs to knockdown expression of endogenous rat HDAC6 or HDAC10 in cardiac myocytes. As shown in Fig. 6D, HDAC6 mRNA was efficiently reduced in NRVMs infected with Ad-shHDAC6. Interestingly, this led to reciprocal upregulation of HDAC10 mRNA expression. Likewise, Ad-shHDAC10 reduced HDAC10 mRNA expression but led to a dramatic, compensatory increase in HDAC6 expression. Importantly, the changes in HDAC6 expression correlated precisely with deacetylation of the class IIb HDAC substrate (Fig. 6E), further suggesting that HDAC6 is primarily responsible for the observed enzymatic activity in our assays.
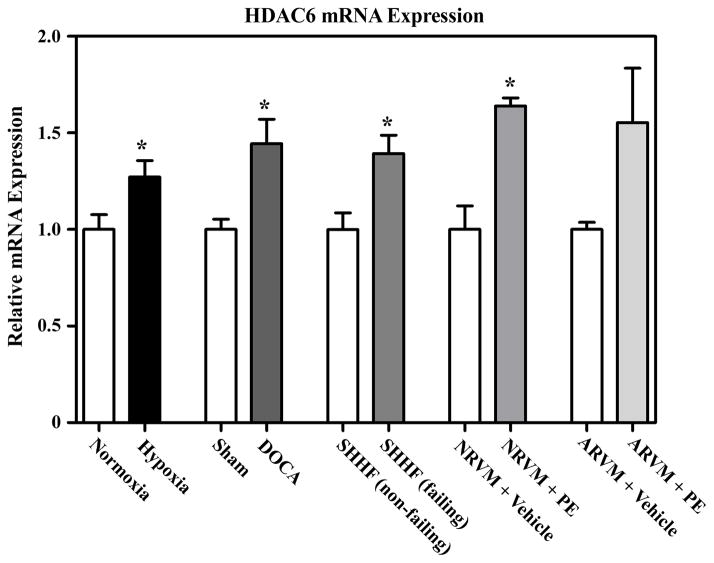
Quantitative PCR analysis was performed to determine whether the changes in HDAC6 activity observed in cardiac cells correlated with increased expression of this HDAC isoform. As shown in Fig. 7, HDAC6 mRNA transcripts were modestly elevated in hypertrophied ventricles from rats subjected to chronic hypoxia or DOCA salt, as well as in LVs from SHHF rats with cardiac hypertrophy and heart failure. Additionally, HDAC6 mRNA levels were increased by PE stimulation in NRVMs and ARVMs, although the increase in ARVMs failed to reach statistical significance. These results suggest that the enhanced HDAC6 catalytic activity observed in response to hypertrophic signals is due, in part, to elevated expression of this HDAC isoform.
Fig. 7.
Increased HDAC6 mRNA expression during cardiac hypertrophy. HDAC6 mRNA levels were measured by Q-PCR using RV RNA from the rat hypoxia model or LV RNA from the DOCA rat or SHHF rat models. Normoxia (n = 6), Hypoxia (n = 8); Sham (n = 8), DOCA (n = 9); non-failing SHHF (n = 8), failing SHHF (n = 7). HDAC6 mRNA was also quantified in unstimulated NRVMs or ARVMs or cells stimulated with PE (10 μM) for 72 hrs. n = 3 per condition. Values represent averages +SEM. *P < 0.05 versus respective controls.
4. Discussion
The major finding reported here is that HDAC6 catalytic activity is increased in response to cardiac stress signals. The results highlight the need for loss-of-function studies to determine whether HDAC6 serves a protective or pathological role in the heart.
Broad-spectrum, ‘pan’ HDAC inhibitors are efficacious in animal models of pathological cardiac remodeling. However, since these compounds are associated with toxicities such as thrombocytopenia, nausea and fatigue [40], many in the field remain skeptical of the prospects of translating these pre-clinical findings to better therapies for heart failure patients. Knowledge of the cardiac function(s) of distinct HDAC isoforms, including HDAC6, should facilitate development of safer, isoform-selective HDAC inhibitors for chronic indications such as heart failure.
We report an unbiased comparison of the catalytic activity of distinct HDAC classes in the heart. Global HDAC activity was shown to be increased in hypertrophic SHR rat hearts [17], in hearts of transgenic mice with cardiac hypertrophy due to overexpression of homeodomain protein (Hop) [15], and in a model of cardiac ischemia-reperfusion injury [9]. Kee et al. used sequential immunoprecipitation-HDAC activity assays to provide evidence that HDAC2, but not HDAC1, is activated in the heart in response to hypertrophic stimuli, including pressure overload due to aortic constriction [41]. Another study reported rapid induction of HDAC3 catalytic activity in NRVMs treated with lipopolysaccharide (LPS) [42]. Conversely, HDAC catalytic activity was reduced in NRVMs exposed to hypoxic conditions [43]. Our inability to detect changes in class I HDAC activity in certain samples (e.g. SHHF hearts and stimulated NRVMs) could be a reflection of complexities related to kinetics of HDAC activation. Indeed, the report by Kee et al. noted only transient activation of HDAC2 following aortic constriction, and this activation preceded hypertrophic growth of the heart [41]. The studies by other groups did not assess activity of class IIb HDACs in the heart.
It should be emphasized that increased catalytic activity of an HDAC isoform is not requisite to establish the enzyme as a valid drug target. For example, a class I HDAC could be aberrantly recruited to the promoter region of a protective gene in response to cardiac stress signals, and de-repression of this gene with an HDAC inhibitor would thus provide benefit to the heart. This concept is exemplified by the anti-oncogenic actions of HDAC inhibitors, which are largely mediated by derepression of pro-apoptotic and anti-proliferative genes (e.g., cyclin-dependent kinase inhibitor, p21) [44]. There are two marketed HDAC inhibitors for cutaneous T cell lymphoma, and many ongoing clinical trials of HDAC inhibitors for other hematological malignancies, as well as solid tumors [45, 46]. Nevertheless, the degree to which the activity of distinct HDAC isoforms is altered in various cancers is largely unknown. As such, with regard to the current study, the absence of detectable increases in cardiac class I HDAC activity in most of the models tested should not be viewed as invalidation of members of this HDAC class as drug targets for heart failure.
The mechanism(s) for HDAC6 activation in response to cardiac stress signaling is unknown. Preliminary kinetic studies with NRVMs suggest that prolonged stimulation of the cells is required for HDAC6 activation, which may reflect a need for new protein synthesis (D.D. Lemon and T.A. McKinsey, data not shown). We hypothesize that induction of HDAC6 catalytic activity in the heart is due in part to increased HDAC6 expression, and also occurs through post-translational activation of the protein. With regard to HDAC6 expression, modest increases in HDAC6 mRNA expression were observed in hypertrophic hearts and stimulated cardiomyocytes (Fig. 7); we have failed to identify a commercially available antibody that efficiently recognizes rat HDAC6 protein. Multiple post-translational mechanisms for regulation of HDAC6 activity have been described in other cell types. For example, association of TPPP/p25 (tubulin polymerization-promoting protein/p25) [47], CYLD [48], IIp45 [49] or Tau [50] with HDAC6 has been shown to inhibit its catalytic activity, as has direct phosphorylation or acetylation of HDAC6 by epidermal growth factor receptor or p300, respectively [51, 52]. Conversely, phosphorylation of HDAC6 by glycogen synthase kinase-3β was shown to activate HDAC6 catalytic activity in neurons [53].
Elucidation of the function of HDAC6 in the heart awaits loss-of-function analyses. In this regard, HDAC6 knockout mice are viable and fertile, and thus assessment of the response of these mice to increased ventricular afterload should be particularly enlightening [54]. Furthermore, the availability of selective pharmacological inhibitors of HDAC6, such as Tubastatin A, may provide an alternative ‘chemical genetic’ approach for determining the role of HDAC6 catalytic activity in models of pathological cardiac remodeling. HDAC6 has been implicated in diverse processes, including regulation of autophagy, cytoskeletal dynamics and cytokine signaling [55], and thus it seems likely that this enzyme will play important roles in the heart. To facilitate drug development, it will be essential to establish whether these functions of HDAC6 are operative in the heart and, if so, whether they are beneficial and/or deleterious to the myocardium in the setting of hypertension.
Acknowledgments
We thank A.P. Kozikowski (U. of Illinois Chicago) for Tubastatin A and B.G. Reid (U. of Colorado Denver) for advice regarding enzyme kinetics. The authors acknowledge NIH grant HL50560 to LAL and the American Heart Association for support of EC.
Footnotes
Disclosures
No conflicts of interest exist for the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yip GW, Fung JW, Tan YT, Sanderson JE. Hypertension and heart failure: a dysfunction of systole, diastole or both? J Hum Hypertens. 2009;23:295–306. doi: 10.1038/jhh.2008.141. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De SG, et al. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 3.Berry JM, Cao DJ, Rothermel BA, Hill JA. Histone deacetylase inhibition in the treatment of heart disease. Expert Opin Drug Saf. 2008;7:53–67. doi: 10.1517/14740338.7.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Bush EW, McKinsey TA. Protein acetylation in the cardiorenal axis: the promise of histone deacetylase inhibitors. Circ Res. 2010;106:272–84. doi: 10.1161/CIRCRESAHA.109.209338. [DOI] [PubMed] [Google Scholar]
- 5.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norris KL, Lee JY, Yao TP. Acetylation goes global: the emergence of acetylation biology. Sci Signal. 2009;2:e76. doi: 10.1126/scisignal.297pe76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith KT, Workman JL. Introducing the acetylome. Nat Biotechnol. 2009;27:917–9. doi: 10.1038/nbt1009-917. [DOI] [PubMed] [Google Scholar]
- 8.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Granger A, Abdullah I, Huebner F, Stout A, Wang T, Huebner T, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 2008;22:3549–60. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang LX, Zhao Y, Cheng G, Guo TL, Chin YE, Liu PY, et al. Targeted deletion of NF-kappaB p50 diminishes the cardioprotection of histone deacetylase inhibition. Am J Physiol Heart Circ Physiol. 2010;298:H2154–H2163. doi: 10.1152/ajpheart.01015.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao TC, Cheng G, Zhang LX, Tseng YT, Padbury JF. Inhibition of histone deacetylases triggers pharmacologic preconditioning effects against myocardial ischemic injury. Cardiovasc Res. 2007;76:473–81. doi: 10.1016/j.cardiores.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Zhao TC, Zhang LX, Cheng G, Liu JT. gp-91 mediates histone deacetylase inhibition-induced cardioprotection. Biochim Biophys Acta. 2010;1803:872–80. doi: 10.1016/j.bbamcr.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–9. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 14.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–88. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man YW, Mackay J, et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003;112:863–71. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer A, Fenning A, Lim J, Le GT, Reid RC, Halili MA, et al. Antifibrotic activity of an inhibitor of histone deacetylases in DOCA-salt hypertensive rats. Br J Pharmacol. 2010;159:1408–17. doi: 10.1111/j.1476-5381.2010.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardinale JP, Sriramula S, Pariaut R, Guggilam A, Mariappan N, Elks CM, et al. HDAC inhibition attenuates inflammatory, hypertrophic, and hypertensive responses in spontaneously hypertensive rats. Hypertension. 2010;56:437–44. doi: 10.1161/HYPERTENSIONAHA.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho YK, Eom GH, Kee HJ, Kim HS, Choi WY, Nam KI, et al. Sodium valproate, a histone deacetylase inhibitor, but not captopril, prevents right ventricular hypertrophy in rats. Circ J. 2010;74:760–70. doi: 10.1253/circj.cj-09-0580. [DOI] [PubMed] [Google Scholar]
- 19.Heltweg B, Dequiedt F, Marshall BL, Brauch C, Yoshida M, Nishino N, et al. Subtype selective substrates for histone deacetylases. J Med Chem. 2004;47:5235–43. doi: 10.1021/jm0497592. [DOI] [PubMed] [Google Scholar]
- 20.Crossno JT, Jr, Garat CV, Reusch JE, Morris KG, Dempsey EC, McMurtry IF, et al. Rosiglitazone attenuates hypoxia-induced pulmonary arterial remodeling. Am J Physiol Lung Cell Mol Physiol. 2007;292:L885–L897. doi: 10.1152/ajplung.00258.2006. [DOI] [PubMed] [Google Scholar]
- 21.Bogaard HJ, Natarajan R, Henderson SC, Long CS, Kraskauskas D, Smithson L, et al. Chronic pulmonary artery pressure elevation is insufficient to explain right heart failure. Circulation. 2009;120:1951–60. doi: 10.1161/CIRCULATIONAHA.109.883843. [DOI] [PubMed] [Google Scholar]
- 22.Taraseviciene-Stewart L, Kasahara Y, Alger L, Hirth P, Mc MG, Waltenberger J, et al. Inhibition of the VEGF receptor 2 combined with chronic hypoxia causes cell death-dependent pulmonary endothelial cell proliferation and severe pulmonary hypertension. FASEB J. 2001;15:427–38. doi: 10.1096/fj.00-0343com. [DOI] [PubMed] [Google Scholar]
- 23.Lange DL, Haywood JR, Hinojosa-Laborde C. Endothelin enhances and inhibits adrenal catecholamine release in deoxycorticosterone acetate-salt hypertensive rats. Hypertension. 2000;35:385–90. doi: 10.1161/01.hyp.35.1.385. [DOI] [PubMed] [Google Scholar]
- 24.Qualy JM, Westfall TC. Overflow of endogenous norepinephrine from PVH nucleus of DOCA-salt hypertensive rats. Am J Physiol. 1995;268:H1549–H1554. doi: 10.1152/ajpheart.1995.268.4.H1549. [DOI] [PubMed] [Google Scholar]
- 25.Konhilas JP, Watson PA, Maass A, Boucek DM, Horn T, Stauffer BL, et al. Exercise can prevent and reverse the severity of hypertrophic cardiomyopathy. Circ Res. 2006;98:540–8. doi: 10.1161/01.RES.0000205766.97556.00. [DOI] [PubMed] [Google Scholar]
- 26.Onodera T, Tamura T, Said S, McCune SA, Gerdes AM. Maladaptive remodeling of cardiac myocyte shape begins long before failure in hypertension. Hypertension. 1998;32:753–7. doi: 10.1161/01.hyp.32.4.753. [DOI] [PubMed] [Google Scholar]
- 27.Sack MN, Rader TA, Park S, Bastin J, McCune SA, Kelly DP. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996;94:2837–42. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 28.Palmer JN, Hartogensis WE, Patten M, Fortuin FD, Long CS. Interleukin-1 beta induces cardiac myocyte growth but inhibits cardiac fibroblast proliferation in culture. J Clin Invest. 1995;95:2555–64. doi: 10.1172/JCI117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satoh N, Suter TM, Liao R, Colucci WS. Chronic alpha-adrenergic receptor stimulation modulates the contractile phenotype of cardiac myocytes in vitro. Circulation. 2000;102:2249–54. doi: 10.1161/01.cir.102.18.2249. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann K, Brosch G, Loidl P, Jung M. A non-isotopic assay for histone deacetylase activity. Nucleic Acids Res. 1999;27:2057–8. doi: 10.1093/nar/27.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riester D, Wegener D, Hildmann C, Schwienhorst A. Members of the histone deacetylase superfamily differ in substrate specificity towards small synthetic substrates. Biochem Biophys Res Commun. 2004;324:1116–23. doi: 10.1016/j.bbrc.2004.09.155. [DOI] [PubMed] [Google Scholar]
- 32.Jones P, Altamura S, De FR, Gallinari P, Lahm A, Neddermann P, et al. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18:1814–9. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–40. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 35.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–43. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogaard HJ, Natarajan R, Mizuno S, Abbate A, Chang PJ, Chau VQ, et al. Adrenergic receptor blockade reverses right heart remodeling and dysfunction in pulmonary hypertensive rats. Am J Respir Crit Care Med. 2010;182:652–60. doi: 10.1164/rccm.201003-0335OC. [DOI] [PubMed] [Google Scholar]
- 37.Porter KE, Turner NA. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther. 2009;123:255–78. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Tobise K, Ishikawa Y, Holmer SR, Im MJ, Newell JB, Yoshie H, et al. Changes in type VI adenylyl cyclase isoform expression correlate with a decreased capacity for cAMP generation in the aging ventricle. Circ Res. 1994;74:596–603. doi: 10.1161/01.res.74.4.596. [DOI] [PubMed] [Google Scholar]
- 39.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–6. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15:3958–69. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
- 41.Kee HJ, Eom GH, Joung H, Shin S, Kim JR, Cho YK, et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ Res. 2008;103:1259–69. doi: 10.1161/01.RES.0000338570.27156.84. [DOI] [PubMed] [Google Scholar]
- 42.Zhu H, Shan L, Schiller PW, Mai A, Peng T. Histone deacetylase-3 activation promotes tumor necrosis factor-alpha (TNF-alpha) expression in cardiomyocytes during lipopolysaccharide stimulation. J Biol Chem. 2010;285:9429–36. doi: 10.1074/jbc.M109.071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw J, Zhang T, Rzeszutek M, Yurkova N, Baetz D, Davie JR, et al. Transcriptional silencing of the death gene BNIP3 by cooperative action of NF-kappaB and histone deacetylase 1 in ventricular myocytes. Circ Res. 2006;99:1347–54. doi: 10.1161/01.RES.0000251744.06138.50. [DOI] [PubMed] [Google Scholar]
- 44.Marks PA. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin Investig Drugs. 2010;19:1049–66. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;280:211–21. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Horwitz SM. The Emerging Role of Histone Deacetylase Inhibitors in Treating T-cell Lymphomas. Curr Hematol Malig Rep. 2010 doi: 10.1007/s11899-010-0067-5. [DOI] [PubMed] [Google Scholar]
- 47.Tokesi N, Lehotzky A, Horvath I, Szabo B, Olah J, Lau P, et al. TPPP/p25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J Biol Chem. 2010;285:17896–906. doi: 10.1074/jbc.M109.096578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickstrom SA, Masoumi KC, Khochbin S, Fassler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–44. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Song SW, Sun J, Bruner JM, Fuller GN, Zhang W. IIp45 inhibits cell migration through inhibition of HDAC6. J Biol Chem. 2010;285:3554–60. doi: 10.1074/jbc.M109.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez M, Santa-Maria I, Gomez de BE, Zhu X, Cuadros R, Cabrero JR, et al. Tau--an inhibitor of deacetylase HDAC6 function. J Neurochem. 2009;109:1756–66. doi: 10.1111/j.1471-4159.2009.06102.x. [DOI] [PubMed] [Google Scholar]
- 51.Deribe YL, Wild P, Chandrashaker A, Curak J, Schmidt MH, Kalaidzidis Y, et al. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci Signal. 2009;2:ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- 52.Han Y, Jeong HM, Jin YH, Kim YJ, Jeong HG, Yeo CY, et al. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2009;383:88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]
- 53.Chen S, Owens GC, Makarenkova H, Edelman DB. HDAC6 regulates mitochondrial transport in hippocampal neurons. PLoS One. 2010;5:e10848. doi: 10.1371/journal.pone.0010848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JY, Yao TP. Quality control autophagy: A joint effort of ubiquitin, protein deacetylase and actin cytoskeleton. Autophagy. 2010:6. doi: 10.4161/auto.6.4.11812. [DOI] [PMC free article] [PubMed] [Google Scholar]