Summary
Cell adhesion by classical cadherins is mediated by dimerization of their EC1 domains through the “swapping” of N-terminal β-strands. We use molecular simulations, measurements of binding affinities, and x-ray crystallography to provide a detailed picture of the structural and energetic factors that control the adhesive dimerization of cadherins. We show that strand swapping in EC1 is driven by conformational strain in cadherin monomers which arises from the anchoring of their short N-terminal strand at one end by the conserved Trp2 and at the other by ligation to Ca2+ ions. We also demonstrate that a conserved pro-pro motif functions to avoid the formation of an overly tight interface where affinity differences between different cadherins, crucial at the cellular level, are lost. We use these findings to design site-directed mutations which transform a monomeric EC2-EC3 domain cadherin construct, into a strand-swapped dimer.
Introduction
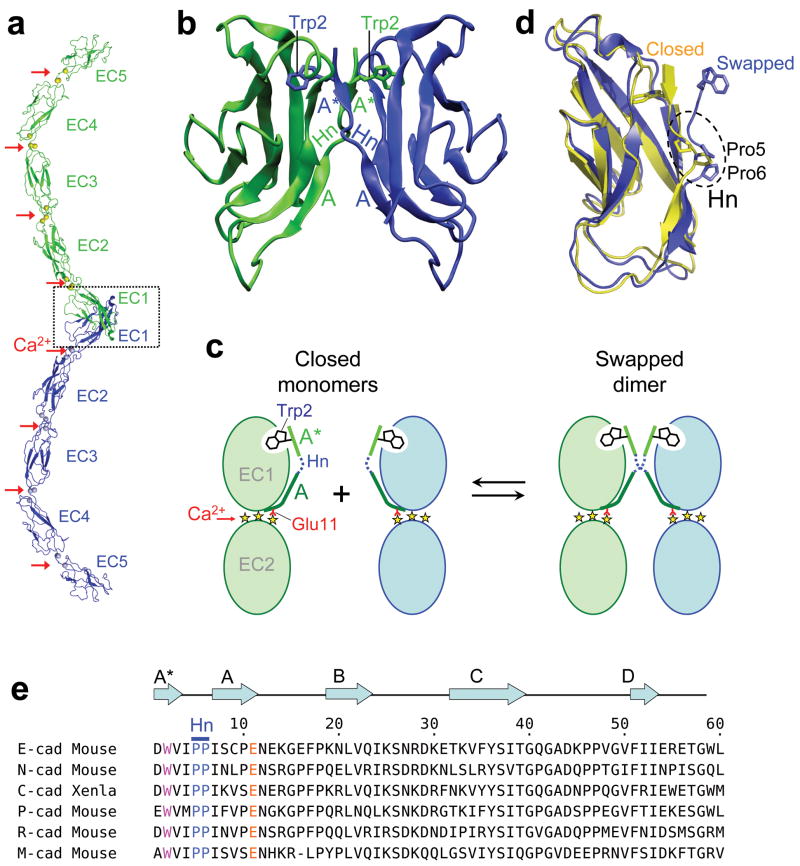
Cadherins constitute a large family of cell-cell adhesion proteins that are represented in both vertebrates and invertebrates 1,2. The “classical” type I and type II cadherins are found only in vertebrates and contain an extracellular region consisting of a tandem repeat of five extracellular cadherin immunoglobulin-like domains (EC1-EC5), that extend from the cell surface (Fig. 1A). Cadherin ectodomains bind between cells through the interaction of their EC1 domains which exchange, or swap, their N-terminal β-strand (the A* strand). Conserved anchor residues – Trp2 in type I cadherins or Trp2 and Trp4 in type II – dock into a complementary pocket in the partner molecule 3-6. The A* strand, which comprises residues 1-3, represents the N-terminal segment of a strand that, in type I cadherins spans residues 1-10 and includes a break at residues 4-6 due to the presence of prolines at positions 5 and 6, which provides a hinge that mediates conformational changes necessary for strand swapping (Fig. 1). Following our previous analysis we denote residues 7-10 as the A strand and residues 110 as the A*/A strand (Fig. 1b) 7.
Figure 1.
Dimerization by strand swapping in classical cadherins. (a) Ribbon representation of the strand swapped dimer of the entire type I C-cadherin ectodomain 3. The three Ca2+ bound at each interdomain region are indicated by red arrows. The dashed box indicates the region of the adhesive contact, which is encompassed entirely within the EC1 domain. (b) Cartoon representation of the strand swapped adhesive interface between EC1 domains of the type I E-cadherin 4. The A* and A strands are labeled, the hinge region is indicated (Hn) and the side chains of the swapped Trp2 are represented. (c) Schematic diagram of the strand swapping process. Two monomers are schematically represented in their closed conformation on the left and as a strand swapped dimer on the right. Only EC1 and EC2 domains are represented, and the three yellow stars between the domains denote bound calcium ions. The A* strand is colored in light green, the A strand in dark green and the hinge region in dashed dark blue. Trp2 and Glu11 (red line) are also indicated. (d) Superposed ribbon representations of the E-cadherin EC1 domain in its closed (in yellow) and swapped (in blue) conformations. The hinge region and the two prolines it contains are indicated. (e) Alignment of the N-terminal sequence of type I cadherins. Conserved residues Trp2, Glu11, Pro5 and Pro6 are colored. Secondary structure elements and the hinge region are indicated above the alignment. Xenla is an abbreviation for Xenopus laevis.
A considerable body of evidence demonstrates that cell-cell adhesive specificity is determined by the identity of the EC1 domain which contains the cadherin-cadherin binding interface 5,6,8-11. Cadherins within the same subfamily (eg., type I) are very similar in sequence and in structure, yet the small differences between them are sufficient to drive cell patterning behavior 12,13. For example, the difference in the binding affinities between N- and E-cadherin is on the order of 1 kcal.mol-1 12. This difference, which is conserved among species 12, is a crucial determinant of cell-cell binding specificity. Understanding the relationship between cadherin sequence, structure and binding energetics is thus a problem of considerable biological importance. Cadherin binding affinities are determined in part by the fact that formation of the EC1-EC1 interface involves β-strand swapping. An inherent feature of strand swapping or, more generally, of the domain swapping phenomenon, is that “closed” monomeric conformations act as competitive inhibitors of dimer formation, thus lowering affinities even when the dimer interface has the characteristics of high affinity complexes, e.g. large interfacial buried surface areas 13.
Two issues are addressed in this work. First, we consider how cadherins are designed to achieve strand swapping. Second, we show that both E- and N-cadherin and type II cadherins have undergone negative design so as to avoid the formation of a tight dimer interface that ablates functionally important differences in binding affinity. Our findings allow us to elucidate basic mechanisms of cadherin design and provide novel insights as to the possible evolutionary mechanisms that underlie the structure and function of this important protein family. In this regard, we also consider the properties of T-cadherin whose EC domains are very similar to those of classical cadherins but that forms a dimer, the X dimer, mediated by an interface that does not involve strand swapping 14.
We address questions of cadherin design through an integrated computational and experimental approach. Molecular dynamics (MD) simulations and earlier structural bioinformatics analysis 7 are first used to provide a hypothesis as to the basic mechanism used by cadherins to achieve strand swapping; specifically that strain in the short A*/A strand in the closed monomer conformation resulting from anchorage at one end by the conserved Trp2 and at the other by a Ca2+-Glu11 ion pair, provides a driving force for strand expulsion and swapping. To test this hypothesis, we carried out binding affinity measurements on mutant proteins from mouse designed to either relieve or increase strain in the monomer. Measurements using analytical ultracentrifugation (AUC) yielded results in good agreement with our predictions. Using the molecular principles uncovered in these experiments, we were able to design mutations that conferred the adhesive strand-swap dimerization properties of EC1 domains on a naturally monomeric non-swapping EC2 domain.
A second discovery that emerged from this work resulted from experiments aimed at determining the role of the pro-pro motif conserved in type I cadherin N-termini at positions 5 and 6. We found that the mutating Pro5 and/or Pro6 into alanine increased the dimerization affinity of E-cadherin by almost two orders of magnitude. Moreover, these mutations abolished affinity differences between E- and N-cadherin. To identify the source of this effect we determined the crystal structure of the E-cadherin EC1-2 P5A P6A mutant which revealed a new type of strand-swapped cadherin interface. It appears then that the pro-pro motif in type I cadherins functions to prevent the formation of this mutant β-interface that, if formed, would remove cell-cell adhesion specificity.
The results presented here provide a clear picture of the molecular design principles used by classic cadherins to achieve cell-cell adhesive specificity. Strand swapping provides a basis for dimerizing with high specificity and low affinity 13 but to achieve this property evolutionary forces have had to design against a potentially competing high affinity dimer conformation. The driving force for swapping involves generating strain in the monomer through a novel mechanism involving an elongated N-terminal β-strand that is fixed at both ends by evolutionarily conserved anchor points.
Results
Ca 2+ binding promotes swapping of the A*/A strand
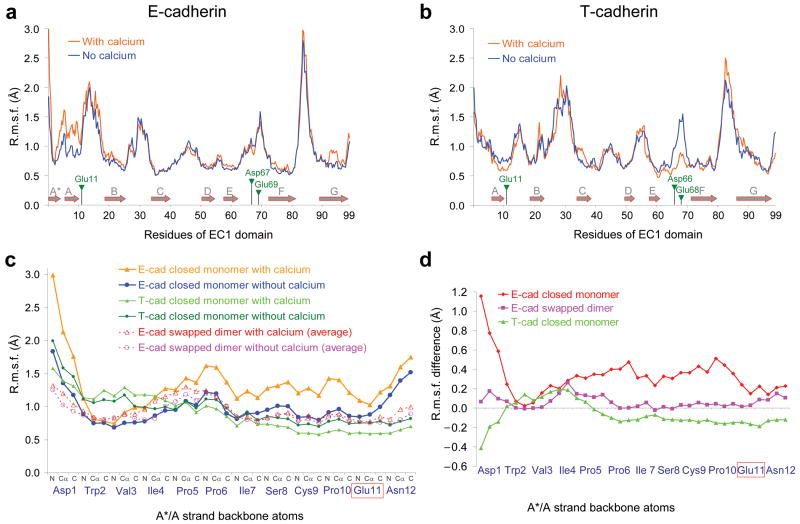
Molecular Dynamics (MD) simulations were carried out on the closed monomer of E-cadherin, of the closely related N-cadherin, and on the monomer of T-cadherin, which forms an X-dimer but does not strand swap 14. Crystal structures are available for E- and T- cadherin monomers while the MD simulations for N-cadherin were carried out on a homology model (see Supplementary methods). We reasoned that differences between T-cadherin and the classical cadherins E- and N-cadherins might illuminate features unique to the A*/A strand swapping reaction. Figure 2a and 2b report Root Mean-Square Fluctuations (R.m.s.f.) of backbone atoms for the EC1 domain of respectively, E- and T-cadherin, obtained from the simulation runs of their EC1-2 domains in explicit solvent. The data for N-cadherin are shown in Supplementary Figure 1.
Figure 2.
Root Mean-Square Fluctuations (R.m.s.f., a measure of the average atomic mobility) of backbone atoms (N, Cα and C atoms) during the MD simulations. (a and b) R.m.s.f. of the EC1 domain backbone atoms of (a) the closed E-cadherin monomer and (b) the closed T-cadherin monomer during MD simulations in the presence (orange trace) or the absence of Ca2+ (blue trace). Secondary structure elements are represented by grey arrows and the positions of the Ca2+-binding residues are indicated by green triangles. (c) and (d) zoom in on the N-terminal fragment of the EC1 domain (residues 1-12). (c) R.m.s.f. of the backbone atoms during three sets of MD simulations: the closed E-cadherin monomer, the swapped E-cadherin dimer and the closed T-cadherin monomer, each time either in the presence or the absence of Ca2+. For the case of the E-cadherin swapped dimer simulations with or without Ca2+, the average R.m.s.f. of the two A*/A strands contained in the dimer (one per protomer) is represented. The color code is indicated in the top right panel on the graph. (d) Difference between the R.m.s.f. in the presence of Ca2+ and in the absence of Ca2+ for the three systems described in (c).
With the exception of the A*/A strand, the results obtained for all three molecules are quite similar: Ca2+ ions have little effect on the mobility of most of the β-strands and the loops containing Ca2+-coordinating residues exhibit an increased mobility in the absence of Ca2+ (Fig. 2a and 2b, Supplementary Fig. 2). However the A*/A strands of E- and T-cadherin are affected differently by the presence of Ca2+. As can be clearly seen in Figure 2c, there is almost no effect of Ca2+ on the mobility of the A*/A strand in T-cadherin, except for a slight reduction of the R.m.s.f. close to the Ca2+ binding residue Glu11. In contrast, the A*/A strand of E-cadherin exhibits a marked increase in mobility when Ca2+ ions are bound. The simulations on N-cadherin do not reveal substantial Ca2+ effects on A*/A strand mobility (Supplementary Fig. 1). The difference between N- and E- cadherin, may be due to the use of a homology model for N-cadherin or to the length of the simulations which may be too short to see large conformational changes, but it is also possible that still undetermined factors are responsible for the strand-swapping properties of N-cadherin. Here we focus on E-cadherin for which the key result of the simulations is that the binding of Ca2+ ions to increases fluctuations in the region that swaps, behavior that is not observed for any other strand in any of the structures that were studied.
To determine the effect of Ca2+ coordination on the strand swapped dimer of E-cadherin, we performed similar MD simulations starting from the dimeric EC1-2 structure (PDB ID 2QVF15). In contrast to the closed monomer, the mobility of the A*/A strand in the strand swapped dimer remains largely unaffected by Ca2+ binding (Fig. 2c and 2d). Our results thus indicate that Ca2+ binding has a differential effect on the mobility of the closed monomer and swapped dimer that are likely to reflect a role in the energetics of dimerization. This conclusion is also consistent with the findings of Sotomayor and Schulten16 who carried out simulations on a monomeric but opened conformation of the C-cadherin ectodomain and found that in the presence of Ca2+, Trp2 remained mostly exposed to solvent while in the Ca2+-free system, Trp2 fluctuated between an exposed and partially buried conformation. Taken together, both sets of simulations suggest that Ca2+ binding leads to the destabilization of the A*/A strand in the closed monomer. In the next section we propose a hypothesis that explains the computational results and then use it as a basis for the design of mutants aimed at testing its validity.
Origins of conformational strain in the monomer A*/A strand
In previous work we have shown that the short A*/A strand in EC1 domains and the presence of a Trp residue at position 2 are associated with strand swapping 7. Our current simulations also reveal that, in E-cadherin, backbone atoms near Trp2 and Glu11 are quite immobile whereas backbone atoms linking these two sites exhibit a Ca2+-dependent increase in mobility (Fig. 2c and 2d, Supplementary Fig. 1). These observations lead us to the hypothesis that, in the closed monomer, the docking of the side chain of Trp2 and the ligation of Ca2+ by Glu11 anchor the A*/A strand at its N- and C-termini and that due to the relatively small number of intervening residues, fixing the strand at these two anchor points destabilizes the closed monomer. We suggest that the fluctuations seen in the simulations reflect Ca2+-dependent destabilization of the monomer and are an early indicator of conformational changes that take place on a longer time scale 17.
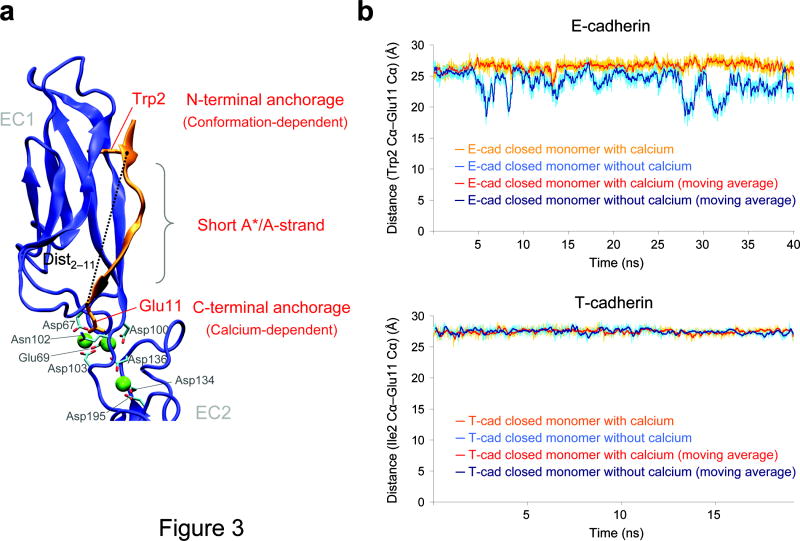
Our hypothesis about conformational strain in the monomer is supported by a structural analysis of the A*/A strand. Specifically, in simulations of the closed E-cadherin monomer, Ca2+-depletion results in a substantial shortening of the distance between the N- and C-termini of the strand, with the Trp2 Cα - Glu11 Cα distance (<dist2-11>) reduced from an average of 26.5 Å in the presence of Ca2+ to 23.9 Å in its absence of Ca2+ (Fig. 3). In contrast, in the swapped dimer, <dist2-11> = 27.7 Å, both in the presence and in the absence of Ca2+, Supplementary Fig. 3b). These results suggest that in the closed monomer, the presence of Ca2+ elongates the strand beyond its preferred length in the apo state, an effect not seen in the dimer. It seems reasonable to posit that the A*/A strand is better able to bridge the two anchor points in the strand-swapped dimer, where inter-molecular degrees of freedom at the interface might enable a relaxation of conformational strain that is not possible in the closed monomer. In this regard, no appreciable change in the length of the A*/A-strand is observed upon Ca2+ removal in the simulation of the non-swapping T-cadherin (27.5 Å and 27.4 Å for <dist2-11> in the presence and absence of Ca2+, respectively) (Fig. 3b).
Figure 3.
Distance between the N- and C-terminal anchorage points of the A*/A strand during the simulations of the closed E-cadherin monomer and T-cadherin. (a) Ribbon representation of the E-cadherin closed conformation. The distance used to monitor the “length” of the A*/A strand (dist2–11) is represented. The sources of the β-strand strain in our model are also indicated. (b) Distance between Trp2 Cα atom and Glu11 Cα atom during the simulations of E-cadherin closed monomer (top) and T-cadherin (bottom). In each graph the distance in the simulation in the presence and in the absence of Ca2+ are both represented together with their moving average (over 20 values) to facilitate the comparison. The color code is indicated in the panel on each graph.
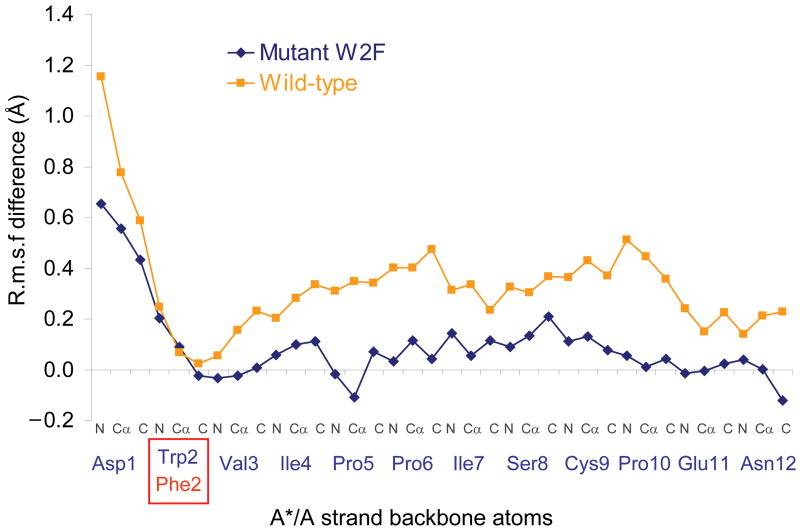
Our model implies that both anchor points are required simultaneously to induce strain in the A*/A strand which would, otherwise, be able to assume a more relaxed conformation. In order to further computationally test this hypothesis, we carried out simulations on the closed conformation of the E-cadherin W2F mutant. Note that in the apo state the A*/A strand in the W2F mutant undergoes larger fluctuations than wild type (Supplementary Fig. 4) but that in the Ca2+-bound state the fluctuations are of comparable magnitude. Figure 4 plots the difference in R.m.s.f. between the Ca2+-bound state and apo state of both the wild-type and the W2F mutant. It is clear from the figure that Ca2+ binding has only limited effect on the mobility of the A*/A strand in the mutant. These results suggest a coupling in the wild type protein between the N- and C-termini, which are located more than 25 Å apart. This coupling is absent in the W2F mutant which we interpret as a reduction in constraints due to the loss of a hydrogen bond between the NH group of Trp2 side chain and the carboxyl group of Asp90 and to the smaller Phe2 side chain which poses fewer conformational constraints than the indole ring of Trp2 (Supplementary Fig. 5 and Supplementary Fig. 4). The simulations also show that the W2F mutant <dist2-11> ≈ 25.5Ǻ, in both the Ca2+ bound and Ca2+ free states so that the Ca2+ induced elongation of the mutant A*/A strand has disappeared (Supplementary Fig. 3a).
Figure 4.
Modulation of the effect of Ca2+ on the A*/A strand mobility by the W2F mutation. The graph presents the difference between the R.m.s.f. observed in presence of Ca2+ and the R.m.s.f. observed in absence of Ca2+ for the E-cadherin wild-type (blue) and W2F mutant (orange) closed monomer sets of simulations.
To summarize, our simulations suggest the presence of conformational strain in the Ca2+ bound closed monomer of E-cadherin that is due to anchoring of the short A*/A strand at both end. The evidence is based a) on enhanced fluctuations in the A*/A strand that we interpret as early evidence of conformational change, b) reduced fluctuations when either of the two anchor points are removed (mutating Trp to Phe or removing Ca2+), c) the pronounced shortening of the strand in the Ca2+ bound state of the monomer that is not seen in the dimer, in T-cadherin, or in the W2F mutant of E-cadherin. These observations do not by themselves point to a single underlying source of the strain which may well be due to subtle effects involving inter-atomic interactions and/or conformational entropy whose identification is beyond the resolution of the simulations. However what emerges clearly from the simulations on E-cadherin is that the presence of two anchor points induces strain in the closed monomer conformation and a corresponding elongation of the strand that would otherwise prefer to have a shorter distance between the two termini. The mechanism we have proposed here is consistent with the simulation data and its basic premises can be tested experimentally. Specifically, if it is correct, mutations designed to release the strain in the monomer should lower the binding affinity for dimerization whereas mutations that increase strain should have the opposite effect. These expectations provide the rationale for the experiments described in the following sections.
Strain modulating mutations affectbinding affinities as predicted
Table 1 reports dissociation constants (KD) for two-domain constructs (EC1-EC2) of wild type and mutant E-cadherins determined by AUC. The KD for wild type E-cadherin is 96.5 ± 10.6 μM and the effect of the various mutations can be assessed relative to this value. Our simulations suggest that the W2F mutant should decrease strain in the monomer and, as expected, its KD is significantly increased relative to wild type, to 246.5 ± 2.1 μM (p-value < 0.0001, see Methods). Moreover, our model suggests that increasing the length of the A* strand should increase the KD and indeed mutants where one or two alanines are inserted between residues 2 and 3 in the A* strand increase the KD relative to wild-type (p-values < 0.0001), to 1517 ± 726.2 μM and 195 ± 8.6 μM, respectively. We speculate that the insertion of one alanine results in a higher KD than the insertion of two alanines, because adding a single residue to the N-terminal β-strand is likely to result in a radical change in the orientation of Trp2, whereas a two residues insertion retains the register of the strand.
Table 1.
Dissociation constants for the homodimerization of E-cadherin EC1-EC2, E-cadherin EC2-EC3, N-cadherin EC1-EC2 and Cadherin-11 EC1-EC2 domains constructs measured by analytical ultracentrifugation at 25°C. Data are represented as mean ± standard deviation.
| Protein | Mean KD (in μM) | Number of AUC measurements |
|---|---|---|
| E-cadherin EC1-2 constructs | ||
| E-cad12 WT | 96.5 ± 10.6 | 4 |
| E-cad12 E11D | 71.2 ± 12.2 | 5 |
| E-cad12 W2F | 246.5 ± 2.1 | 2 |
| E-cad12 Ins A betw. 2-3 | 1517 ± 726.2 | 2 |
| E-cad12 Ins AA betw. 2-3 | 195 ± 8.6 | 2 |
| E-cad12 P5A P6A | 3.7 ± 0.1 | 3 |
| E-cad12 P5S P6S | 2.9 ± 0.04 | 2 |
| E-cad12 P5G P6G | 2.7 ± 1.68 | 2 |
| E-cad12 P5A | 2.9 ± 2.1 | 2 |
| E-cad12 P6A | 4.8 ± 1.65 | 2 |
| E-cadherin EC2-3 constructs | ||
| E-cad23 WT | monomer | 2 |
| E-cad23 F108W | monomer | 2 |
| E-cad23 ΔTQE | monomer | 3 |
| E-cad23 F108W, ΔTQE | 46.5 ± 0.9 | 3 |
| E-cad23 F108W, ΔTQE, A193I | monomer | 2 |
| E-cad23 F108W, ΔTQE, A205I | monomer | 2 |
| N-cadherin EC1-2 constructs | ||
| N-cad12 WT | 25.8 ± 1.5 | 4 |
| N-cad12 P5A P6A | 3.6 ± 0.2 | 2 |
| Cadherin-11 EC1-2 constructs | ||
| Cad-1112 WT | 25.2 ± 4.3 | 2 |
| Cad-1112 F8A | 3.7 ± 0.2 | 2 |
Another possible interpretation of our results is that the various mutants we have generated form an X-dimer 15 so that the various binding affinities are not those of the strand-swapped adhesive interface. The X-dimer involves an interface located in the EC1-EC2 linker region and is seen when strand swapping is precluded, for example in the W2A mutant. However, the KD for the X-dimer is about 800-900 μM and, with one exception, all of the mutants we report dimerize with significantly lower KDs and thus do not form this interface. The only mutant whose binding affinity is compatible with that of an X-dimer is E-cadherin with two alanines inserted between residues 2 and 3 (KD = 1517 μM). If this mutant forms an X-dimer this would not affect the evidence that the insertion significantly lowers affinities since dimerization through its strand-swapped interface would have to involve an even lower affinity than that of the X-dimer.
In order to design a mutant with increased strain, we reasoned that shortening the Glu11 side chain while preserving the Ca2+ binding site would pull the A*/A strand in the direction of the bound Ca2+ ion thus increasing the distance between the two anchor points. This effect was achieved by replacing Glu11 with Asp (the E11D mutant). As can be seen in Table 1, despite the conservative nature of the mutation, a slight but significant decrease in KD is observed (p-value = 0.0013).
Inducing strand swapping in a non-swapping EC2 domain
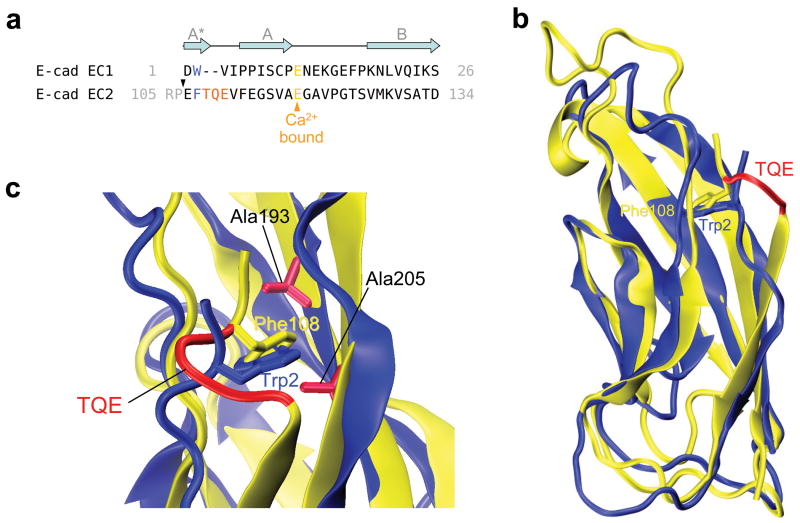
EC2 domains differ from EC1 domains in that they have a Phe instead of a Trp at position 2 and they have two additional residues in their A-strands 7. The expectation based on simulation results is that these two factors reduce strain in the A/A* strand and thus inhibit swapping. Indeed isolated EC2 domains are monomeric in solution 18. We produced mutant EC2-EC3 domain constructs which contained one or two of the swapping determinants. This was accomplished with mutants where Phe108 (equivalent to Trp2 in EC1 domains) was replaced with a Trp and/or where the A*/A strand was shortened by three residues. Specifically, Thr109, Gln110 and Glu111 that form a bulge in the A* strand (Fig. 5) were deleted. The relevant KDs are reported in Table 1. Consistent with the results for a single EC2 domain construct 18 wild-type EC2-3 does not dimerize in solution (Table 1). Similarly, both the F108W and the Δ109-111 mutants are also monomeric. However the combined F108W, Δ109-111 mutant is dimeric in solution with a KD of 46.5 μM which is lower than that of the wild type EC1-EC2 construct.
Figure 5.
Comparison of E-cadherin EC1 and EC2 domains. (a) Sequence alignment of the mouse E-cadherin EC1 and EC2 N-terminal residues. The grey arrows represent the secondary structure elements, and the black triangle above the EC2 sequence indicates the beginning of the EC2-3 construct used. Trp2 and homologous residue Phe108 are highlighted in blue, and the deleted TQE residues in red. (b) Structural alignment of E-cadherin EC1 (in blue) and EC2 (in yellow) domains. The side chains of Trp2 and Phe108 are shown, and the deleted residues are colored in red in the EC2 structure. (c) Detail of the hydrophobic pocket. Residues Ala193 and Ala205 mutated to Ile in the pocket-filling mutants are represented.
To confirm that the EC2-3 double mutant dimerizes via strand swapping, two additional mutants were designed with the goal of filling the Trp2 binding pocket thus abrogating strand swapping. To this end the pocket-lining residues Ala193 and Ala205, were separately mutated to isoleucine (Fig. 5c). Equivalent mutations in the EC1 domain (A78M and A80M) were shown to abrogate binding through strand swapping 19. As shown in Table 1, these mutants are monomeric.
Pro5 and Pro6 mutants form a new non-specific interface
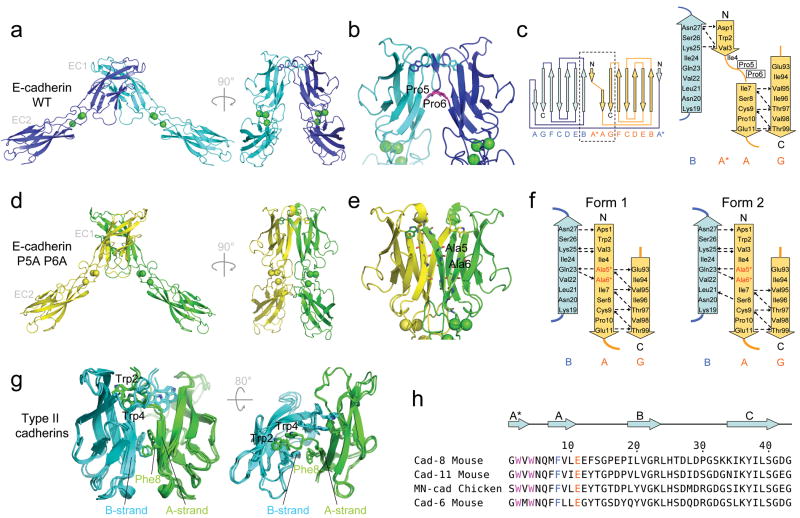
In order to test the role of the pro-pro motif in the hinge region, we measured the KDs of several E-cadherin EC1-2 constructs where Pro5 and/or Pro6 were mutated. Contrary to expectations, mutating Pro5 and/or Pro6 significantly increases binding affinity in E-cadherin (Table 1). Furthermore, the binding affinity of the mutants does not depend on the identity of the new side chain or on whether one or both prolines are mutated (Table 1).
The explanation for this intriguing finding is evident from the 1.8 Å crystal structure we determined for the EC1-EC2 construct of the E-cadherin P5A P6A mutant (Table 2). Strand-swapping occurs, however the dimer interface is quite different than observed in the wild type protein: the break at positions 4-6 in the A*/A strand has disappeared and the backbone of mutated residues Ala5 and Ala6 now engages in hydrogen bonds with the adjacent B-strand of the dimer mate molecule (Fig. 6). In this way, the A*/A strand becomes continuous from residue 1 to residue 11 (Fig. 6c, 6d, 6e, 6f and Supplementary Fig. 6). In the wild type proteins, Pro5 and Pro6 prevent the formation of this long “β-strand interface” since their backbone nitrogen cannot play the role of a hydrogen bond donor.
Table 2.
Data collection and refinement statistics
| Ecad EC1-2 P5AP6A | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 111.2, 92.6, 79.5 |
| a, b, g (°) | 90.0, 92.3, 90.0 |
| Resolution (Å) | 1.8 (1.86-1.8)* |
| Rsym or Rmerge | 6.3 (66.4) |
| I / sI | 24.6 (2.4) |
| Completeness (%) | 98.9 (98.1) |
| Redundancy | 3.8 (3.8) |
| Refinement | |
| Resolution (Å) | 1.8 |
| No. reflections | 69883 |
| Rwork / Rfree | 16.5/19.9 |
| No. atoms | |
| Protein | 3332 |
| Ligand/ion | 6 |
| Water | 903 |
| B-factors | |
| Protein | 32.8 |
| Ligand/ion | 17.7 |
| Water | 41.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.34 |
Values in parentheses are for highest-resolution shell.
Figure 6.
Structure of the E-cadherin P5A P6A mutant swapped dimer and comparison with the wild type swapped dimer. (a) Ribbon representation of E-cadherin wild-type swapped dimer crystal structure with a close-up view of the swapped interface in (b). Bound calcium ions are represented as green spheres. (c) Topology diagram of the wild-type E-cadherin swapped dimer. The dashed box on the complete diagram on the left indicates the interface region of which a close-up view is shown on the right. (d) Ribbon representation of the E-cadherin P5A P6A mutant crystal structure, with a close-up view of the new swapped interface in (e). Electron density of the new feature formed by the A*/A strand at the mutant interface is shown in Supplementary Figure 6. The mutated residues 5 and 6 are indicated in panels (b, e). (f) Topology diagram at the interface region in the two forms of the E-cadherin P5A P6A mutant swapped dimer observed in the crystal structure. (g) Ribbon representation of the available type II swapped dimer structures superposed (cadherin-11 (2A4C.pdb), cadherin-8 (1ZXK.pdb) and MN-cadherin (1ZVN) 5). Phe8 and the two strands (A and B strands from apposed protomers) between which it intercalates are indicated. (h) Alignment of N-terminal sequence of type II cadherins. Conserved residues Trp2, Trp4, Phe8 and Glu11 are colored, and secondary structure elements are indicated above the alignment.
The high affinity of the newly identified mutant “β-strand interface” is related to its increased surface area relative to the wild type protein. The total surface buried in this interface (2238.8 and 1953.9 Å2 for the two crystal forms) is substantially larger than in the wild type (1834.1 Å2 in 2QVF.pdb 15), and includes a larger hydrophobic surface (1201.0 and 1100.5 Å2 as compared to 1008.5 Å2 in wild type E-cadherin). Since most of the additional hydrophobic contacts involve residues that do not swap (at the bottom of the EC1 domain, Supplementary Fig. 7), their effect is not reduced by the disruption of equivalent contacts in the monomer 13.
In order to determine whether the mutant β-interface was formed by other type I cadherins, we produced the P5A P6A double mutant of the N-cadherin EC1-2 domain and measured its KD by AUC. Similar to what was observed in E-cadherin, the mutation resulted in a significantly increased binding affinity for N-cadherin. Remarkably, despite the significant difference in KD between the wild-type proteins, the KDs of the double mutant of E- and N-cadherin are essentially identical. Thus, the consequence of tighter binding has been a loss of affinity differences between these two cadherins.
This effect is seen dramatically in type II cadherins which, despite the absence of a pro-pro-motif in their A*/A strand (Fig. 6h), do not form an extended intermolecular β-sheet at their swapped interface 5. Crystal structures of type II swapped dimers reveal why: In all available structures the A- and B-strands from opposite protomers, which form the long intermolecular β-sheet in the type I E-cadherin P5A P6A mutant, are kept apart by the presence of bulky side chains 5, particularly the highly conserved Phe8 (Fig. 6g). In order to test this structural inference, we measured the KDs for wild-type and F8A EC1-2 constructs of the type II cadherin-11 (Table 1). Phe8 is located in the middle of the large hydrophobic cluster below the swapped interface, in a region that engages in binding but does not swap 5. Since the F8A mutant should reduce the buried hydrophobic area of the dimer, a possible effect would be a reduction in binding affinity. Strikingly, we observe the opposite: cadherin-11 F8A mutant binds significantly more tightly than wild-type protein (Table 1), suggesting that cadherin-11 F8A forms a β-strand interface similar to the one observed in E-cadherin P5A P6A mutants. That the binding affinities are the same (Table 1) strongly supports this hypothesis.
Discussion
It has been well established through structural 3-5, biophysical 20,21, in vitro 22 and in vivo 5,9,19 mutagenesis studies that the adhesive dimerization interface formed between cadherins involves the swapping of N-terminal β-strands between their EC1 domains. We have previously described how strand swapping enables the formation of large and specific inter-protein interfaces that have relatively low binding affinities 13. At the cellular level we have shown that the small differences in these affinities can result in highly specific cell-cell interactions 12,13. Thus, the fine tuning of cadherin dimerization energetics, which is achieved through β-strand swapping, is an essential feature of cell sorting behavior. In this study we discuss the structural and energetic principles underlying the swapping process.
Since, by definition, strand swapping implies the replacement of one set of interactions in two monomers with an equivalent set in a dimer, there must be some additional factor that overcomes the entropy loss associated with dimerization that allows swapping to occur. In a previous study we took a bioinformatics approach to identify sequence and structural determinants of the swapping process and found that the conserved Trp at position 2 and a shortened A*/A strand were unique features of EC1 domains that were most likely to be important determinants of strand swap binding 7. However the physical mechanisms underlying these observations were unclear. One fundamental question concerns the role of the conserved Trp2 which is generally assumed to play a central role in dimerization by anchoring the swapped strand into a hydrophobic pocket on the partner molecule. But in energetic terms this explains very little since the same hydrophobic interactions should be present in the monomer as well so that there is no a priori reason to suggest that it prefers either monomer or dimer. Moreover, for the same reason, it has not been clear why any change (i.e. the W2F mutant) would affect binding affinities.
The role of Ca2+ ions in the swapping process has also been of uncertain origin. It is known that Ca2+ ions that are bound in inter-domain linker regions are essential for cadherin mediated cell adhesion, but this has been attributed primarily to the rigidification of the entire ectodomain. Cadherin rigidification by Ca2+ appears to be necessary to facilitate the preference for trans (cell-to-cell) dimerization over cis dimerization (between cadherins from the same cell) 23-27. However it has become clear that bound Ca2+ ions also play a direct role in the swapping process 28,16,29.
Our results suggest that these various observations can be explained in the context of a single mechanism characterized by four molecular features of critical functional importance. First, the swapped strand requires an anchor residue(s) near the N-terminus, Trp 2 in type I cadherins, and Trps 2 and 4 in type II cadherins. Second, the swapping A*/A strand is anchored at the C-terminal end through bidentate ligation of the conserved Glu11 side chain to two of the three Ca2+ ions in the linker between the EC1 and EC2 domains. Third, conformational strain is produced in the shortened A*/A strand by this dual ligation mediated by Trp anchor residue(s) at the N-terminal and Ca2+ ligation at the C-terminal end of the swapping A*/A strand. This strain, which arises only in the context of the monomer, provides the driving force that favors strand swap dimerization over the nearly structurally identical monomer. Fourth, the pro-pro motif in the A*/A strand of type I cadherins, and the Phe8 side chain of type II cadherins, serve to insure that the swapping strands of a cadherin pair cannot form a cross-dimer continuous hydrogen-bonded β-sheet, which our results show to yield unnaturally tight cadherin dimer complexes that lack binding specificity. This latter observation was surprising since, based on precedent from other systems 30-34, and consistent with a recent suggestion 4, it was expected that the prolines would introduce strain in the monomer so that strand swapping to the dimer form would be enhanced. Each aspect of our model has been confirmed through binding affinity measurements using site-directed mutants which also show how the various factors are tightly coupled.
The term “conformational strain” as used here is necessarily ambiguous because it does not define specific interactions that cause the strain. Strain can be distributed over many degrees of freedom and attempts to partition energies into individual contributions are unlikely to be meaningful. What can be said with certainty is that our simulations on E-cadherin indicate that the A*/A strand is unusually mobile when Ca2+ is bound in the closed monomer and that this is associated with fixed anchor points at both ends and with an elongation of the strand. The simulation results then suggest that the presence of both anchors combined with a short strand destabilize the monomer. The success of this model in explaining the mutagenesis data, and in suggesting the design of an EC2-EC3 constructs that swaps, supports its validity, but of course there may yet to be other swapping determinants still to be discovered. One possibility that in principle might drive swapping is that the interactions made by the A*/A strand in the dimer are different than in the monomer, and energetically more favorable. However a precise and reliable comparison of the two conformations is difficult since there is no crystal structure available for the native closed monomer. Indeed, all crystal structures of the closed conformation have been obtained via mutations that prevent strand swapping and potentially locally affect the structure 15,23,25. What can be said is that the observed contacts established by the A*/A strand residues in the available structures of the closed monomer are nearly identical to those seen in the swapped dimer crystal structure so that differences, if they do exist, are very small.
Although our simulations and experiments have focused primarily on E-cadherin, and, to a lesser extent N-cadherin (for which well-behaved mutants are hard to obtain), the swapping determinants we have identified are common to all other type I cadherins 7. Moreover, Trp2, a shortened A*/A strand and Glu11 are all conserved in EC1 domains of type II and desmosomal cadherins suggesting that each of these sub-families exploits the same structural design principles. Similarly, we have shown that both type I and type II cadherins are designed so as to avoid the formation of the mutant β-interface again suggesting the generality of the findings reported in this work. Our study does not consider the source of the difference in binding affinity among cadherins within the same sub-family which, as we have recently discussed, is a crucial determinant of cell-cell binding specificity 12. However since strain in the A*/A strand in the monomer appears to provide the major driving force for dimerization, it is likely that subtle differences in the conformational energetics of this region of the protein will prove to be important. Indeed, the difference in the behavior of the A*/A strand seen in our simulations of N- and E-cadherin suggest the existence of additional factors that still need to be identified.
An important finding of this study is the identification of the mutant β-interface which is characterized by a KD of about 2-4 μM for type I E- and N-cadherins and the type II cadherin-11. This represents significantly stronger association than that of the wild-type proteins but, as we have discussed, the resulting loss of differences in affinity between N- and E-cadherin would be expected to remove cell-cell adhesive specificity.
Finally, the fact that a small number of mutations in a natively monomeric non-swapping EC2 domain, designed based on the structure/function principles elaborated here for cadherins, can induce dimerization via strand swapping raises the possibility that we have in some way mimicked an evolutionary process. We have recently shown that, when strand swapping is inhibited, classical cadherins dimerize through an interface formed in the calcium binding region located between EC1 and EC2 15. In classical cadherins this “X-dimer” functions as a binding intermediate on the path to strand swapping, but in T-cadherin it corresponds to the cell-cell adhesive interface. We have suggested that, earlier in their evolution, some branches of the cadherin superfamily may have used (or may still use) this interface for their primary mode of adhesion. If this is indeed the case, the path from the X-dimer to a strand swapped dimer may well have involved sequence and structure modifications in the A/A* strand similar to those described in this work.
Experimental Procedures
DNA constructs and protein production
The following Mouse E-cadherin constructs were used: EC1-2 wild-type (residues 1213, UNP residues 157369), EC1-2 W2F, EC1-2 E11D, EC1-2 P5A P6A, EC1-2 P5G P6G, EC1-2 P5S P6S, EC1-2 P5A, EC1-2 P6A, EC2-3 wild-type (residues 107323), EC2-3 F108W, EC2-3 del-TQE (residues 109111 of the wild-type sequence were deleted), EC2-3 F108W del-TQE, EC2-3 F108W del-TQE A193I and EC2-3 F108W del-TQE A205I. The following Mouse N-cadherin constructs were used: EC1-2 wild type (residues 1215, UNP residues 160374). The following Mouse cadherin-11 constructs were used: EC1-2 wild type (residues 1215). Mutations were prepared from the wild-type EC1-2 and EC2-3 constructs using the QuickChange site-directed mutagenesis method (Stratagene Inc.). All proteins were produced using the prokaryotic pSMT3 SUMO–His-tag fusion system and expressed in E.coli BL21 (DE3) cells. The proteins were purified by affinity chromatography on a nickel chelating column, ion exchange on a MonoQ column, and size exclusion. The tag was removed by overnight digestion with the SUMO-specific protease Ulp1. The proteins were concentrated to 10 mg.ml-1 in 20 mM TrisHCl, pH 8.0, 150 mM NaCl, and 10mM CaCl2.
Crystallization and structure determination
Crystals of E-cadherin EC1-2 P5A P6A mutant were grown in 10% (w/v) PEG8000, 10% (v/v) ethylene glycol, 0.1M HEPES, pH 7.5. Crystals were cryoprotected with the mother liquor supplemented with 20% (v/v) ethylene glycol before flash-cooling in liquid nitrogen. Diffraction data were collected on a single frozen crystal at the X4A beam line of the National Synchrotron Light Source, Brookhaven National Laboratory, and processed with the HKL program suite35. The structure was solved by molecular replacement using the structure of mouse E-cadherin EC1-2 (PDB ID 1FF525) as a search model with MOLREP36. Manual rebuilding was done using COOT37 and refinement using REFMAC38. Data collection and Refinement statistics are summarized in Table 2. All residues in the refined model are in allowed (98.4% in favoured) regions of the Ramachandran plot, as analyzed by MOLPROBITY39. Coordinates have been deposited in the Protein Data Bank with a deposition code 3QRB.
Analytic Ultracentrifugation
All purified proteins were dialyzed in the following buffer overnight at 4C: TRIS 20 mM, NaCl 150 mM, 10 mM CaCl2, 10 mM TCEP pH 8.0. Samples were analyzed by sedimentation equilibrium measurements at 25°C for 20 hours at 23500g, then 10 hours at 35200g and 10 hours at 49100g. All analyses were performed using a Beckman XL-A/I ultracentrifuge equipped with a Ti50An rotor. Three concentrations were used for each measurement (0.77, 0.50, and 0.27 mg.ml-1). Raw data were collected using both UV detection at 280 nm and interference measurement at 660 nm. Both interference data and UV data for the three concentrations were used simultaneously in global fittings. Calculations were done using the HeteroAnalysis software, version 1.1.0.28 from the Analytical Ultracentrifugation Facility at University of Connecticut (http://www.biotech/uconn.edu/auf/). For each protein, the experiment was repeated a number of times (N) and the average KD was obtained.
Statistical analysis
Statistical significance of the difference between the distribution of the KDs measured by AUC for the wild-type and for the different mutants was tested using a two-sided t-test with an alpha level of 0.05, in all cases where a p-value is reported in the text. The sample size for each mutant is indicated in Table 1.
Structures used as starting points for the different MD simulations
(see Supplementary Methods).
Molecular dynamics simulations
All molecular dynamics simulations were performed with Gromacs-3.2.1 40, using the GROMOS force field. In all cases proteins were embedded in a triclinic box filled with SPC water molecules so that no atom of the protein is closer than 10 Å from the box boundaries. In the eight closed monomer simulations (E-cadherin wild type, N-cadherin wild type, E-cadherin W2F mutant and T-cadherin wild type, each time in the presence or the absence of Ca2+), the simulation box size was approximately 32 × 64 × 80 Å, and each system contained a little more than 41000 atoms. In the two E-cadherin swapped dimer simulations (in the presence and in the absence of Ca2+), the size of the simulation box was approximately 48 × 66 × 130 Å, and each system contained around 83500 atoms. In all the systems, the N-terminal terminus (which correspond to the actual N-terminus in solution) was positively charged (ammonium group), and the C-terminal was negatively charged (carboxyl group). We ensured global charge neutrality of the different systems by adding counterions to the box as needed. Periodic boundary conditions were assumed in all cases. A uniform integration step of 1 fs was used for all types of interactions, throughout all simulations except those of N-cadherin closed monomer where an integration step of 2 fs was used. A cutoff of 12 Å was used for Van der Waals interactions, and electrostatic interactions were calculated with the particle mesh technique for Ewald sums (PME), also with a cutoff of 12 Å. A Berendsen thermostat was used to maintain a constant temperature of 300K, and a constant pressure of 1 atm was maintained using a Berendsen barostat.
All simulations started with an equilibration of the system consisting of four heating steps of 250 ps each, with the water molecules free to move and protein heavy atoms constrained. The production phase was then run and frames were recorded at 0.1 ps intervals. The production phase lasted 40ns for each of the wild type E-cadherin closed monomer simulation (with and without Ca2+), 20 ns for each of the T-cadherin corresponding simulations (with and without Ca2+), 21 ns for each of the W2F mutant E-cadherin closed monomer simulations (with and without Ca2+) and 18 ns and 15 ns for the E-cadherin swapped dimer simulation with and without Ca2+, respectively.
Root mean square fluctuations of the backbone atoms (i.e. C, Cα and N atoms) were computed using the g_rmsf module of Gromacs. In a first step, each frame of the trajectory was superposed with the first frame via a structural alignment of the protein backbone.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB-0918535 (to B.H.H.) and National Institutes of Health Grant R01 GM062270–07 (to L.S.).
Footnotes
Author Contributions: J.V. performed and analyzed the simulations; J.V. and S.P. designed the mutants; F.B. produced all the wild-type and mutant proteins; G.A. performed and analyzed the AUC experiments; X.J. determined and refined the crystal structures; J.V., L.S. and B.H. designed experiments, analyzed data and wrote the manuscript.
Accession codes. Coordinates of the E-cadherin EC1-2 P5A P6A mutant structure have been deposited in the Protein Data Bank with a deposition code 3QRB.
References
- 1.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–72. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 2.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–69. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–13. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 4.Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373:401–11. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel SD, et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–68. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Harrison OJ, et al. The extracellular architecture of adherens juntions revealed by crystal structures of type I cadherins. Structure. 2010 doi: 10.1016/j.str.2010.11.016. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:952–66. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nose A, Tsuji K, Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell. 1990;61:147–55. doi: 10.1016/0092-8674(90)90222-z. [DOI] [PubMed] [Google Scholar]
- 9.Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–16. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- 10.Shan WS, Koch A, Murray J, Colman DR, Shapiro L. The adhesive binding site of cadherins revisited. Biophys Chem. 1999;82:157–63. doi: 10.1016/s0301-4622(99)00115-5. [DOI] [PubMed] [Google Scholar]
- 11.Shan W, et al. The minimal essential unit for cadherin-mediated intercellular adhesion comprises extracellular domains 1 and 2. J Biol Chem. 2004;279:55914–23. doi: 10.1074/jbc.M407827200. [DOI] [PubMed] [Google Scholar]
- 12.Katsamba P, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci U S A. 2009;106:11594–9. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–6. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciatto C, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol. 2010;17:339–47. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison OJ, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–57. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sotomayor M, Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys J. 2008;94:4621–33. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miloushev VZ, et al. Dynamic properties of a type II cadherin adhesive domain: implications for the mechanism of strand-swapping of classical cadherins. Structure. 2008;16:1195–205. doi: 10.1016/j.str.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad A, Housley NA, Pedigo S. Thermodynamic stability of domain 2 of epithelial cadherin. Biochemistry. 2004;43:8055–66. doi: 10.1021/bi049693c. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–63. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 20.Sivasankar S, Zhang Y, Nelson WJ, Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–81. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci U S A. 2009;106:109–14. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison OJ, Corps EM, Kilshaw PJ. Cadherin adhesion depends on a salt bridge at the N-terminus. J Cell Sci. 2005;118:4123–30. doi: 10.1242/jcs.02539. [DOI] [PubMed] [Google Scholar]
- 23.Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–4. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 24.Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223:1019–26. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 25.Pertz O, et al. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–47. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cailliez F, Lavery R. Cadherin mechanics and complexation: the importance of calcium binding. Biophys J. 2005;89:3895–903. doi: 10.1529/biophysj.105.067322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro L, Weis WI. Structure and Biochemistry of Cadherins and Catenins. Cold Spring Harb Perpect Biol. 2009;1 doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haussinger D, et al. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison OJ, Corps EM, Berge T, Kilshaw PJ. The mechanism of cell adhesion by classical cadherins: the role of domain 1. J Cell Sci. 2005;118:711–21. doi: 10.1242/jcs.01665. [DOI] [PubMed] [Google Scholar]
- 30.Green SM, Gittis AG, Meeker AK, Lattman EE. One-step evolution of a dimer from a monomeric protein. Nat Struct Biol. 1995;2:746–51. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 31.Murray AJ, Head JG, Barker JJ, Brady RL. Engineering an intertwined form of CD2 for stability and assembly. Nat Struct Biol. 1998;5:778–82. doi: 10.1038/1816. [DOI] [PubMed] [Google Scholar]
- 32.Kelley BS, Chang LC, Bewley CA. Engineering an obligate domain-swapped dimer of cyanovirin-N with enhanced anti-HIV activity. J Am Chem Soc. 2002;124:3210–1. doi: 10.1021/ja025537m. [DOI] [PubMed] [Google Scholar]
- 33.Simeoni F, Masotti L, Neyroz P. Structural role of the proline residues of the beta-hinge region of p13suc1 as revealed by site-directed mutagenesis and fluorescence studies. Biochemistry. 2001;40:8030–42. doi: 10.1021/bi002812d. [DOI] [PubMed] [Google Scholar]
- 34.Barrientos LG, Gronenborn AM. The domain-swapped dimer of cyanovirin-N contains two sets of oligosaccharide binding sites in solution. Biochem Biophys Res Commun. 2002;298:598–602. doi: 10.1016/s0006-291x(02)02489-0. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–5. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindahl E, Hess B, Van Der Spoel D. GROMACS: A package for molecular simulation and trajectory analysis. J Mol Mod. 2001;7:306–317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.