Abstract
Misfolding and aggregation of amyloid beta (Aβ)-40 peptide play key roles in the development of Alzheimer's disease (AD). However, very little is known about the molecular mechanisms underlying these molecular processes. We developed a novel experimental approach that can directly probe aggregation-prone states of proteins and their interactions. In this approach, the proteins are anchored to the surface of the AFM substrate (mica) and the probe, and the interaction between anchored molecules is measured in the approach-retraction cycles. We used dynamic force spectroscopy (DFS) to measure the stability of transiently formed dimers. One of the major findings from DFS analysis of α-synuclein (α-Syn) is that dimeric complexes formed by misfolded α-Syn protein are very stable and dissociate over a range of seconds. This differs markedly from the dynamics of monomers, which occurs on a microsecond-nanosecond time scale. Here we applied the same approach to quantitatively characterize interactions of Aβ-40 peptides in a broad range of pH values. These studies showed that misfolded dimers are characterized by the lifetimes in the range of seconds. This value depends on pH and varies between 2.7 s for pH 2.7 and 0.1 s for pH 7, indicating that the aggregation properties of Aβ-40 are modulated by the environmental conditions. The analysis of the contour lengths revealed the existence of various pathways for dimer dissociation, suggesting that dimers with different conformations are formed. These structural variations result in different aggregation pathways, leading to different types of oligomers and higher order aggregates, including fibrils.
Protein misfolding and self-assembly into various morphologic aggregates is a wide spread phenomenon in development of various neurodegenerative disorders such as Alzheimer's and Parkinson's diseases (1, 2). The aggregation properties of amyloid beta (Aβ) peptide, a key protein for Alzheimer's disease, have been studied using various methods (3–7). The current model for amyloidogenic peptides dissects the aggregation kinetics into two main phases. Historically, self-assembly kinetics were observed initially in the earlier work of Hofrichter et al. (8), in which the gelation phenomenon of purified deoxyhemoglobin was investigated. The authors proposed a model that dissected the growth kinetics of fibrils into two phases. The first phase is the nucleation process. During this phase, a critical oligomer of a particular size (nucleus) is formed. The nucleus undergoes a thermodynamically favorable elongation process in which monomers are added via consecutive steps. This model, applied to the aggregation of deoxyhemoglobin, suggested the size of the nucleus to be as large as 30 monomeric units (8). Jarrett and Lansbury applied this model to analyze the aggregation of amyloids that followed a similar kinetic profile (9). It was recently shown that growth of amyloid plaques in vivo follows the same model (10). Both in vivo and in vitro studies indicate a considerably long lag period during which the formation of stable nuclei occurs. This period is considered a key step in the process of amyloid growth. However, a number of important questions arise. What are these nuclei? How large are they? How long do they live? Answering these questions is important, as soluble oligomeric assemblies of Aβ rather than fibrils are neurotoxic (11) and the dimer is the smallest synaptotoxic species (12, 13).
Progress has been made recently in understanding the early stages of aggregation and misfolding of proteins. The ionization mass spectrometry (ISI-MS) method was developed to characterize oligomeric species of Aβ peptides (3). This ISI-MS using study shows that oligomerization of Aβ-40 and Aβ-42 follow two different aggregation pathways with tetramers and hexamers as potential nuclei. Importantly, in both cases, dimers are the building blocks. Spectroscopic analysis of covalently cross-linked Aβ-42 oligomers (4, 14) showed that the structure changes of Aβ-42 occur rapidly in the transition from monomer to dimer and then proceed gradually for higher oligomers. A combined approach utilizing several methods to study entire aggregation kinetics has been recently proposed (15). The proposed kinetic model suggests that the β-LGA monomers are converted into dimers and tetramers in the early stages of aggregation and these species, primarily tetramers, constitute a reservoir of intermediates used for the later stages of the aggregation process. A common feature exists in the studies performed on two different types of amyloidogenic proteins – the conversion of monomers into dimers, followed by their assembly into tetramers. No trimers are detected in both studies. These findings suggest that dimers may contain a property that makes their accumulation preferable compared to the use of monomers as building materials for the growth of oligomers. Recently, a novel approach to characterize misfolded states and dimeric forms of the amyloidogenic proteins was proposed (16, 17). In this technique, the interaction between the proteins is measured by AFM force spectroscopy with significant rupture forces, suggesting that misfolded proteins form dimeric states. This approach was tested initially with the use of three different proteins, α-synuclein, lysozyme and amyloid β peptide (16). Further improvement of the protein immobilization methodology made it possible to provide the characterization of misfolded dimers at the single molecule level (18). The dynamic force spectroscopy (DFS) approach was used to characterize interactions of α-synuclein (α-Syn) (19, 20). One of the major findings from DFS analysis is that dimeric complexes formed by misfolded α-Syn protein are very stable and dissociate over a range of seconds. This differs markedly from the dynamics of monomers, which occurs on a microsecond-nanosecond time scale. This finding, along with other recent publications (3, 15), suggests that dimerization is the key step of the self-assembly process.
Here, we have applied the AFM force spectroscopy methodology to test whether the interaction of Aβ-40 peptide follows the same model. We used DFS to measure the stability of transiently formed Aβ-40 dimers. The aggregation properties of Aβ-40 depend on structural properties of Aβ-40 that are modulated by the environmental conditions (21–23). Therefore, we performed DFS analysis in a range of pH. These experiments showed that similar to α-Syn protein, Aβ-40 forms dimers with lifetimes in the range of seconds. The analysis of the contour lengths revealed variations in the interactions of Aβ-40. This suggests that dimers with different conformations are formed, which we propose are capable of leading to diverse aggregation pathways of Aβ-40.
MATERIALS AND METHODS
Synthesis of cysteine modified Aβ-40
The procedure is described in detail in the supplement. Briefly, the cysteine modified N-terminus Aβ-40 (Cys-Aβ-40) was synthesized on a CEM Liberty microwave peptide synthesizer using a Val-HMPB Chemmatrix resin. Synthesized peptide was cleaved from the resin by stirring the peptide resin with a mixture of TFA/Thioanisole/Phenol/H2O/Dimethylsulfide/ Ethanedi-thiol/Triisopropylsilane. After cleavage, the peptide was dissolved in 100 uL TFA which was then diluted with 100 mL H2O and injected into a Vydac C4 semi-preparative RP-HPLC column for purification (see detail in SI). The purified peptide was characterized by ESI mass spectrometry and SDS-PAGE gel electrophoresis (Fig. S1).
Silatrane derivatized tetrahedral shaped (T-silatrane) molecule
The nanoscale tetrahedral shaped tripodal silatrane incorporating a chemically reactive terminal maleimide group (Fig. S2) has been synthesized as described in the supplement and used without extra purification.
Tip and mica surface modification
The general process for the cleaning and modification of the tip and mica surfaces was similar to the previously reported protocol (19, 20). Briefly, tips (MLCT, Veeco, Santa Barbara, CA) were cleaned by ethylene alcohol followed by UV illumination for 30 min and modified by maleimide polyethylene glycol silatrane (MAS) (18). The freshly cleaved mica surface was modified by 1-(3-aminopropyl) silatrane (APS) and then N-hydroxysuccinimide-polyethylene glycol-maleimide (NHS-PEGMAL) (MW = 3400 g/mol, Laysan Bio Incorporation, Arab, AL) was coupled to the amine group of APS in DMSO in a dry chamber for 3 hours. Both functionalization protocols yielded surfaces terminated with maleimide head groups capable of covalent bonding of Aβ-40 at the N-terminal cysteine. To accomplish this step, the functionalized tip and mica were incubated with ~20 nM solution of Aβ-40 diluted in HEPES buffer (pH 7) for 1 hour and then gently rinsed with the dilution buffer and NaCO3-NaHCO2 (pH 10) buffer. Prepared tips and mica were kept in HEPES buffer until needed.
Modification of Si-wafer with tetrahedral shaped tripodal silatrane (T-shaped, see Fig. S2)
N-type Si-wafer (NOVA electronic materials, Flower Mound, TX) was cleaned by standard cleaning process (24). Before the standard cleaning process, wafer was cleaned by piranha solution (sulfuric acid: hydrogen peroxide = 3:1) for 30 min and then rinsed with DI water several times. Pre-cleaned Si-wafer was immersed in SC1 cleaner (water: ammonium hydroxide: hydrogen peroxide = 5:1:1) and then SC2 cleaner (water: hydrogen chloride: hydrogen peroxide = 6:1:1) at ~70 °C for 15 min for each step. After each cleaning step, wafer was washed with DI water. Cleaned Si-wafer surface was modified by T-shaped silatrane. The molecule was dissolved at 1 mg/ml in DMSO as a stock solution and then diluted in DMSO/H2O (80:20) before use. The cleaned wafer (1×1 cm2 was covered by 30 μL of diluted T-shaped molecule (167 μM) and was kept in a closed chamber overnight. After modifying, the wafer was rinsed several times using DMSO and then DI water. On the modified Si-wafer surface, Aβ protein was immobilized through the process described above.
The working buffer solutions were prepared at five different pHs: pH 2.7 (20 mM glycine-HCl); pH 3.7 (10 mM sodium acetate); pH 5 (10 mM sodium acetate); pH 7 (20 mM HEPES); pH 9.8 (100 mM sodium carbonate and sodium bicarbonate), respectively. All buffers were adjusted to the ionic strength of 150 mM with NaCl.
Single molecule force spectroscopy
The force-distance curve (FDC) of Aβ was measured at four different pH values (pH 2.7, 3.7, 5, and 7) with MFP3D AFM (Asylum Research, Santa Barbara, CA) at room temperature. The spring constant of tips (MLCT, Veeco) was 30–60 pN/nm which was calibrated with the thermal noise analysis method. We applied 100 pN force (trigger) at the contact. At high retracting velocities (1 μm/s and above), the tip was kept at the surface for 0.3 s (dwell time). The retracting velocity varied from 50 nm/s to 3 μm/s with 7~8 discrete steps corresponding to 1,000~200,000 pN/s of apparent loading rate. FDCs were collected more than 2,000 times per each retracting velocity on 5~7 different batches. The selected FDCs were analyzed using the worm-like chain (WLC) model with Igor Pro 6.01 software.
Data analysis
The contour length analysis was performed with the WLC model for flexible linkers (25) as described in previous publications (18, 19, 26, 27). These papers also describe specifics for DFS methods used in this paper. While the individual FDCs were fitted to WLC model ( Fp=¼kBT(1-x/L)−2−1/4+x/L, where F is rupture force; p is persistence length; kBT is thermal energy; L is contour length), the persistence length was allowed to be varied for the best fitting curve and evaluated as a variable parameter along with contour length. The WLC fitting procedure as a part of the software was provided by the MFP 3D manufacturer (Asylum Research). The example of such a fit is shown in Fig. S6A. The persistence length was an adjustable parameter with a mean value 0.16 ± 0.1 nm in the distribution histogram (Fig. S6B). Based on the parameters of the DFS result, the conceptual simplified energy landscape was constructed on the reaction coordinate (28).
RESULTS
1. Experimental design
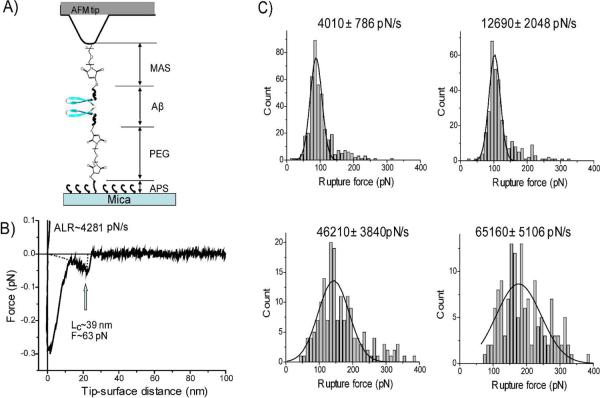
Figure 1A provides the schematics for our experimental approach. We used a variant of Aβ-40 peptide with cysteine at the N-terminus to specifically immobilize proteins at the tip and the mica surface functionalized by maleimide using appropriate linkers (18–20, 27). This immobilization approach was selected because the N-terminus of Aβ-40 is not involved in fibril formation (6, 29, 30). Aβ-40 was covalently attached to the maleimide-terminated surface at its N-terminal cysteine and the interaction between the protein molecules was measured by multiple approach-retraction cycles. The cysteine sulfhydril group was attached to the mica surface via a polyethylene glycol linker (PEG; 19–26 nm long). A shorter PEG spacer of maleimide silatrane (MAS) provided immobilization of Aβ-40 to the AFM using a one-step modification procedure (18). A freshly prepared ~20 nM working solution of peptide was used in each experiment and 0.25 mM TCEP was added to minimize the dimerization of the peptide via S-S bond formation. Note that the concentration of the protein was more than three orders of magnitude less than the protein concentration used in the aggregation experiments in vitro. At these conditions we obtained a sparse distribution of the peptide on the surface minimizing the double rupture events (26).
Figure 1.
(A) Schematic experimental system of SMFS. AFM tip and mica surface were functionalized by MAS/Aβ-40 and APS/PEG/Aβ-40, respectively. In the schematic, Aβs are shown as β-hairpin structures in dimeric form. (B) A representative force-distance curve measured at pH 5 with apparent loading rate (ALR) 4280 pN/s. (C) The histograms of the rupture force distribution measured at pH 5 at ALR 4000 pN/s, 12690 pN/s, 46210 pN/s, 65160 pN/s. The most probable rupture force (Fr) for each loading rate was ~88 pN, ~102 pN, ~143 pN, and ~177 pN, respectively, which were calculated by Gaussian fitting.
2. Interactions between Aβ-40 at pH 5
A typical force distance curve for the rupture events of the experiments performed at pH 5 is shown in Fig. 1B. pH 5 corresponds to conditions at which Aβ-40 is mostly neutral (pI 5.4). The first large peak of the FDC corresponds to the short-range non-specific interactions typically appearing in AFM force measurement experiments (20, 27). The second peak is separated from the initial peak by a specific distance, similar to prior studies (19, 20, 26). This distance is defined primarily by the length of the flexible linkers and unstructured segments of the peptide. The peak shown in Fig. 1B corresponds to the contour length Lc~39 nm which is in the range of expected values (Fig. 1A and Table S1). The force curves similar to the one in this figure were acquired by multiple probing over various positions of the tip over the surface. The DFS analysis required the system pulling with various rates. Therefore, the probing experiments were performed in a broad range of loading rates (0.1 nN/s ~ 100 nN/s). To generate the DFS spectrum, more than 25,000 force curves were collected. The mean yield of the rupture events was 6.5 % and they all were analyzed. In the control experiments without peptides non-specific rupture events appeared at short contour lengths (< 30 nm) and the forces were distributed over a broad range (20 pN~1.5 nN). The yield of such event was less than 1%. The most probable rupture force (Fr) for a selected pulling rate was calculated from the Gaussian fitting of the histograms of rupture force distribution (Fig. 1C).
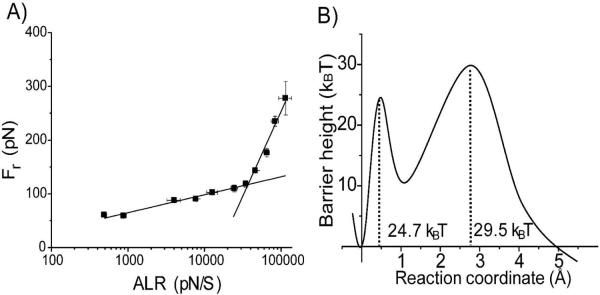
Figure 2A shows the plot of Fr obtained for different pulling rates versus the apparent loading rates on logarithmic scale. Each data point was obtained by averaging over 5 independent experiments. The data set was approximated by the Bell-Evans model (31–33) with two linear lines indicating that there were two transient states for the Aβ-40 dissociation at this condition. Two major parameters obtained from this plot, the intercept and the slope, were used to reconstruct the energy landscape profile for the dissociation of the dimer shown in Fig. 2B. The first (inner barrier) and second (outer barrier) transient states correspond to the higher and the lower slopes in Fig. 2 A, respectively. The transient states for this process are located at 0.3 Å and 2.6 Å from the ground state of the dimer. The inner barrier height (ΔG=24.7 kBT) and the dissociation lifetime (τin=0.009 s) were obtained from the off-rate constant (Koff). Similar analysis for the outer barrier provided the following numbers: ΔG=29.5 kBT and τout=1.13 s.
Figure 2.
Results of the dynamic force spectroscopy analysis for pH 5. (A) The dependence of Fr on logarithm of ALR. The solid lines represent the best fit of data points in two regimes by Bell's model. (B) Profile of the energy landscape calculated from the DFS plot above. The parameters are listed in Table 1.
3. Effect of pH on the Aβ-40 interactions
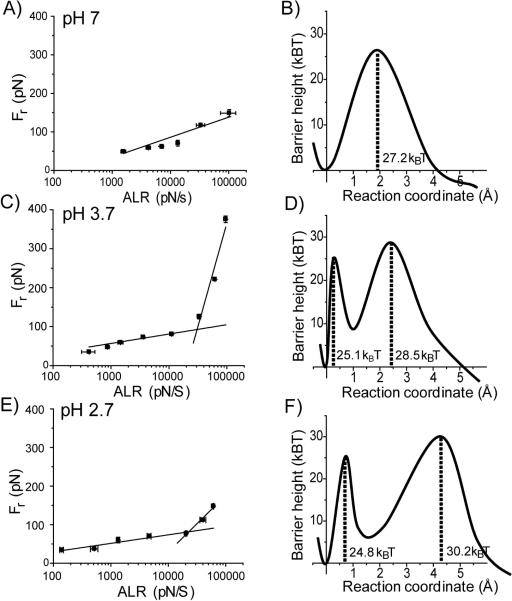
Similar DFS analysis was performed at pH values 7, 3.7 and 2.7. The results are shown in DFS plots and landscape energy profiles in Fig. 3. The DFS plot for pH 7 (Fig. 3A) is represented by a single line suggesting that there is only one transient state. The corresponding energy landscape profile is shown to the right (Fig. 3B). Figure 3C shows the DFS result obtained at pH 3.7. This plot has two slopes and the corresponding profile of the energy landscape with two barriers is shown in Fig. 3D. Note a close location of the inner barrier to the ground state of the dimer. The results of the DFS analysis for acidic conditions, pH 2.7, are shown in Fig. 3E and F. The two-slope plot (Fig. 3E) generates the profile of the energy landscape with two barriers (Fig. 3F). Note a relatively distant position of the outer barrier compared with other profiles. The values for the off-rate constants, lifetimes and positions of the barriers characterizing the Aβ-40 dissociation processes are summarized in Table 1. The lifetimes corresponding to the dimer dissociation (outer barrier height) vary between 2.7 s for pH 2.7 and 0.1 s for pH 7. The lifetimes for the inner barriers of pH 2.7, 3.7 and 5 are in the range of 10 ms indicating that the transient state of the inner barrier is ~100 times more dynamic than that of the outer barrier.
Figure 3.
Results of the dynamic force spectroscopy analysis for different pH. Each pair consists of the DFS plot and the corresponding profile of the energy landscape. pH 7 (A and B), pH 3.7 (C and D), and pH 2.7 (E and F). In the energy landscapes, the first valley is for the bound state and the second is for the intermediate and the last is for the dissociation state of Aβ-40 dimers. The potential barrier heights were calculated using Koff (see the text) extracted from the linear fitting of Bell's model in the plot of Fr vs. ln [ALR]. All parameters used for energy landscapes were listed on table 1.
Table 1.
Characteristic parameters of DFS results. Each xβ1,2 and Koff1,2 are the outer and inner potential barrier location and dissociation ratio, respectively. The lifetime (τ) was calculated by τ~1/Koff and the standard deviation (SD) was calculated from several data sets measured at each pH.
| pH | Xβ1 (pm) | Koff1 (s−1) | Lifetime (s) | Xβ2 (pm) | Koff2 (s−1) | Lifetime (ms) |
|---|---|---|---|---|---|---|
| 2.7 | 427±21 | 0.4±0.2 | 2.7±1.1 | 60±10 | 104±21 | 10±2 |
| 3.7 | 244±26 | 2.5±0.8 | 0.4±0.1 | 20±6 | 78±12 | 13±2 |
| 5 | 265±27 | 0.9±0.2 | 1.1±0.3 | 28±5 | 114±12 | 9±1 |
| 7 | 182±11 | 9.4±1.5 | 0.1±0.01 |
4. Modes of interaction in Aβ-40 dimers
The force curves in the AFM force spectroscopy experiments, in addition to the rupture force values, provide another important value, the contour length Lc (Fig. 1B). According to Fig. 1A and B, the contour length primarily comprises the length of flexible linker and the N-terminal segment of the peptide not involved in dimer formation. Therefore, subtracting the lengths of the linkers from the experimentally measured contour length yields the length of the N-terminal segment of the peptide preceding the structured segment of Aβ-40 involved into the interpeptide interaction. We tested and applied this approach in our recent work on the localization of interacting segments in misfolded α-synuclein(19, 20).
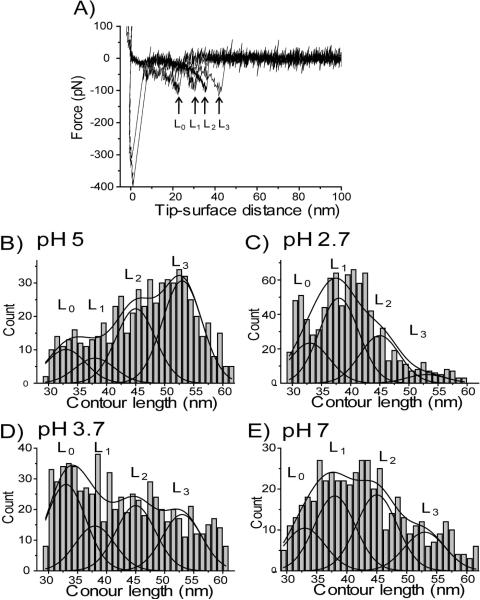
Figure 4A shows a set of four force curves obtained at pH 5. These force curves have very close rupture force values (~120 pN) but differ in the rupture position between ~25 nm (L0) and ~45 nm (L3). The contour lengths measured from hundreds of rupture events were assembled and shown as a histogram in Fig. 4B. The contour lengths vary between 30 nm and 60 nm. Similar data were obtained for pH 2.7, 3.7 and 7, as shown in Figs. 4 C, D and E respectively. The histograms at each pH are broad but profiles are different. For example, the representative rupture events for pH 5 are those with lengths between 49 nm and 56 nm, whereas probing at pH 2.7 has the most representative lengths between 36 nm and 43 nm. The contribution of flexible linkers to the contour lengths is 26 (± 4) nm (Fig. 1A) (see supplemental materials). The error in this value is due to the heterogeneity of the PEG linker and rupture force. Subtracting this value from the total range of contour lengths yields a range between 8 nm and 38 nm, which represents the variability of the N-terminal segment of Aβ-40 peptide including the dimeric structure length. Such broad variability suggests that the conformation of the peptide in the complexes is not constant. Rather the Aβ-40 adopts a set of conformations. To confirm this assumption and to exclude the potential effect of the linkers, we performed similar probing experiments with the use of very short linkers. The major problem with the use of short linkers is the adhesion at small distances. To minimize this problem we used tetrahedral shaped silatrane linkers (T- silatrane) with tripodal silatrane arms (Fig. S2). This molecule has three arms with silatrane end groups enabling the immobilization of the construct on the surface. The forth arm contains a chemically reactive maleimide group that provides covalent coupling with the cysteine of the Aβ-40 peptide. For the modification of the silicon substrate, T-silatrane was mixed with similar tetrahedral molecules containing silatrane in all arms. The use of the second molecule allowed us to decrease the surface density of active maleimide groups and hence the number of non-specific events. The results of the contour length measurements for pH 5 and 2.7 are shown in Fig. S5. The distribution of the lengths is broad with a range between 10 nm and 45 nm. This is a similar range of contour lengths as described above, given the fact that the contribution of the linkers to these values is ~10 nm.
Figure 4.
(A) The representative force curves for rupture events with different rupture lengths obtained for pH 5. All force curves were obtained at the same ALR regime (~34 nN/s) and correspond to rupture forces ~120 pN. (B–E) Histograms of the contour length distributions measured at pH 5 (B), 2.7 (C), 3.7 (D), and 7(E). Each histogram is approximated with four Gaussian curves with centers located around 33 nm (L0), 38 nm (L1), 45 nm (L2), and 53 nm (L3), respectively. The data were collected for the pulling rate range between 0.1 nN/s and 10 nN/s
DISCUSSION
Stability of Dimers of Aβ-40 and molecular mechanisms of early stages of aggregation
The single molecule force spectroscopy analysis revealed a number of novel properties of Aβ-40 in misfolded states. The DFS experiments demonstrate that the transiently formed dimers are characterized by lifetimes in the range of seconds, regardless of the pH. It is important to compare this value with the lifetime of misfolded conformations of the monomer peptides. When Aβ peptides are monomers, their conformational dynamics occurs in the range of 10−6 − 10−9 s (34). Some of these transient states existing in the nanosecond-microsecond time scale are misfolded conformations. When two misfolded monomers associate to form a dimer, their lifetime increases significantly, reaching the value of several seconds. Molecular dynamics (MD) simulations of oligomeric structure of Aβ (25–35) revealed that dimerization and formation of stabile β-sheet dimers takes place in nanosecond time scale (35). Furthermore, MD simulations of ologomeric Aβ (17–42) showed that a double layer of β-hairpins is energetically favored (36). Thus, the dimerization of misfolded proteins leads to the stabilization of the misfolded states of Aβ peptides. However, the dimers show different properties depending on the pH. The DFS plot for pH 7 (Fig. 3A) is represented by one line, yielding a profile of the energy landscape with one barrier (Fig. 3B). The barrier height is 27.2 kBT, which corresponds to a lifetime of 0.1 s. The dimer dissociation pathways in acidic conditions (pH 5, 3.7 and 2.7) are more complex because the DFS graphs have two slopes corresponding to two potential barriers in the energy landscapes. The inner barriers for these conditions have very close heights, corresponding to lifetimes in the range of 10 ms. The heights of outer barriers are larger and correspond to lifetimes in the range between 0.43 s (pH 3.7) and 2.7 s (pH 2.7).
Two major conclusions emerge from these findings. Compared to the monomeric proteins, the misfolded conformation of Aβ peptides in the dimeric state is 106 times more stable. This finding suggests that Aβ peptide dimers have a different conformation compared to the monomers. This is in agreement with the recent publication of Ono et al (4), in which a dramatic change of the secondary structure was observed between monomer and dimer of photo-induced cross-linked Aβ-40. Although further oligomerizations increase the β-sheet content relative to the random coil, these are gradual processes compared to that in the monomer to dimer transition. Given this conformational change preceding or along with dimerization (4, 37), our results suggest that the misfolded dimers have large lifetimes most likely through interactions with β-hairpins.
The increased stability of the dimers suggests the following mechanism for the aggregation process: Aβ peptides in the monomeric state are very dynamic occurring in the nano- to microsecond scale (34, 38), which enables Aβ to adopt various conformations including misfolded states. When two monomers make a dimer, it remains in the misfolded state 106–109 times longer. Therefore, the probability of aggregation with the participation of dimers is 106–109 times higher compared to that with monomers. For example, the probability of the formation of tetramers with the assembly of two dimers will be 106 times higher compared to the formation of trimers via the interaction of a dimer and a monomer. As a result, the preferred mechanism for aggregation is via dimers, rather than monomers. This assumption is in agreement with the recent publication of Bernstein et al (3). Quantitative analysis using the ISI-MS method showed that oligomers with only even numbers of monomers, dimers and tetramers for Aβ-40 peptide are formed.
Additional support for the hypothesis of a critical role of Aβ peptide dimers in the aggregation kinetics is provided by recent publications (4, 39). These papers showed that stabilization of Aβ peptide dimers by crosslinking or mutation resulted in a rapid increase of the aggregation rate with a short lag phase. Additionally, a publication (40) in which the hairpin conformation of Aβ peptide monomers was stabilized by intramolecular disulfide bonding, demonstrated that the Aβ peptide oligomers assembled with stabilized β-sheets led to an increase in their toxicity compared to the control samples. Furthermore, Shankar et al showed that Aβ dimers are the abundant species and have a high neurotoxicity through the analysis of the Aβ peptides isolated from brain (12). This suggests an important biological role for Aβ dimers.
Structure of misfolded dimers
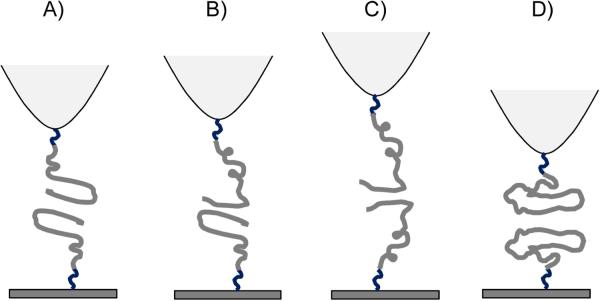
The measurements of the contour lengths (Fig. 4A) provide an insight into the structure of misfolded dimers of Aβ-40. Although the range of contour lengths is rather broad, the distribution is not smooth and can be divided into four groups, L0, L1, L2 and L3, as indicated in the figure (Fig. 4B–E). In fibrils, Aβ chains interact with each other via β-hairpin structures, therefore, we assumed the presence of similar structures during probing experiments. The estimate for the contour length for this interaction produced L1 value. Values L2 and L3 correspond to hypothetical interactions of transiently opened hairpin structures. These are schematically shown in Fig. 5. The model for the Aβ-40 dimer (Fig. 5A) is assembled by two Aβ-40 molecules adopting the structure found in fibrils. Once again, monomers interact via β-hairpin and residues D1-Y10 of the N terminus are non-structured. The rupture of such a dimer should occur at the contour length 38(±4) nm, which includes the length of the linkers (26±4 nm) and unstructured N-terminal segments of the peptides (7~8 nm) (41) and two hairpin structures (~4 nm) (29). The overall length correlates very well with the L1 value in Fig. 4B–E. Fig. 5B shows the model in which the dimer is formed by the Aβ-40 peptides in two different conformations. One confirmation is the monomer with a hairpin structure that interacts with the C-terminal segment of a second Aβ-40 which is half of the hairpin structure. The rupture of this construct occurs at contour lengths of 45 ± 4 nm which is larger compared to the previous model due to extended length of N-terminal segment in one of the constructs. This overall length is in agreement with the L2 value. Similarly, the model in Fig. 5C corresponds to the assembly of two Aβ-40 molecules through their C-terminal regions with extended unstructured N-terminal regions contributing ~23 nm to the contour lengths of the linkers (26±4 nm). The overall expected contour length for the rupture of such a construct is 53 ± 4 nm, which is close to L3.
Figure 5.
Schematics of four intermediate dimeric structures. (A) The dimer structure is stabilized by the interaction between β-hairpins -. (B) The dimer structure stabilized by the interaction between β-hairpins and the C-terminal segment of the peptide. (C) The dimer is stabilized by the interaction of the C-terminal segments of the peptide. (D) The interaction between collapsed conformers of Aβ 40 monomer.
To explain the appearance of rupture events with the lengths as small as L0, we hypothesize that the dimers can adopt a collapsed conformation in which the residues A2-F4 of the N-terminal region are involved with the stabilization of the dimer. Such a model is supported by molecular dynamics simulation (34) showing that N-terminal residues of Aβ-40 are involved with the formation of oligomer assembly. This, however, is in disagreement with Takeda et al (42). The structure of the dimer with interactions between collapsed structures is sketched schematically in Fig. 5D. The rupture of such a dimer occurs after stretching of linkers, providing the expected contour length of 33 ±4 nm which is close to the L0 value.
It is important to apply this model to understanding the effect of pH on the conformation of Aβ dimers. The data shown in Fig. 4B indicates that at pH 5, conformers L2 and L3 are the most predominant species suggesting that at these conditions the stabilization of dimers by models B and C is rather favorable. In contrast, at pH 2.7 (Fig. 4C) the complexes A and B comprise the most preferable states for the dimers with a few events with conformation C. This difference can be explained by electrostatic repulsion that is stronger at acidic pH compared with pH 5 that is close to the pI value for Aβ-40.
Additional insight into the structure of the transiently formed dimers is provided by the values of the rupture forces. The rupture forces of Aβ-40 dimers were in the range of 60 ~100 pN at the loading rate of 4 ~20 nN/s (200 nm~1 um/s) at pH 7. We can compare this value with the data on the AFM unfolding of titin Ig domains that mainly consists of β-sheet structures (43). The unfolding forces obtained under the loading rate range of 200~400 nm/s were 170~180 pN, which is about three times larger (~60 pN) than the rupture force for Aβ-40 obtained at the same loading rate. At the same time, unfolding of α-helical structure of T4 lysozyme required considerably less force, ~60 pN compared to ~100 pN for Aβ-40, at the loading rate of 1 m/s (44). This comparison suggests that the conformation of Aβ-40 dimers is less stable than β-sheets, but more stable than α-helical structures. This conclusion is in agreement with the structural analysis of Aβ-40 (4, 6), and suggests that unstructured monomers undergo significant conformational changes while they are oligomerized.
Altogether, the AFM force spectroscopy identified long lifetimes of misfolded Aβ-40 dimers, compared to the monomers which retain the misfolded state for very short periods of time. Additionally, the misfolded dimers are 106 times more stable. Note that qualitatively similar results for lifetimes were obtained previously from α-synuclein (19, 20, 26). Thus, the long lifetime of the Aβ-40 dimers is a fundamental property of this peptide and possible for other amyloidogeneic proteins. Based on these findings, we propose a model in which the formation of dimers is a mechanism by which aggregation prone misfolded proteins becomes highly stabilized. Due to its long lifetime, the probability that the dimer will have further interactions dramatically increases. Thus, the ability to form long-lived dimers is a fundamental property of misfolded proteins and triggers the protein aggregation process. This is an entirely novel view on the aggregation process that may lead to a paradigm shift in the development of diagnostic and therapeutic treatments for protein misfolding diseases, narrowing the search to approaches that can prevent dimer formation. Given the ability of AFM to detect the interaction between the proteins at the single molecule level, AFM force spectroscopy may be a potential diagnostic and therapeutic tool in the future. AFM in the imaging mode is a primary tool for the visualization of amyloid aggregates (45), so the combination of both AFM capabilities would provide important information on the mechanism of the aggregation process. However, a small size of monomeric Aβ peptides complicates unambiguous identification of all oligomeric species in the AFM scans. Further improvement in the AFM imaging methodology is needed for accomplishing this goal.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank A. Krasnoslobodtsev, J. Yu, A. Portillo and other Lyubchenko lab members for insightful discussions.
The work was supported by grants from DOE (DE-FG02-08ER64579), NATO (SfP 983204), and the Nebraska Research Initiative (NRI, all to YLL) and the NIH grant INBRE P20 RR016469 to SL.
ABBREVIATIONS
- AFM
Atomic Force Microscopy
- ALR
Apparent Loading Rate
- Aβ
Amyloid beta
- APS
1-(3-aminopropyl) silatrane
- α-Syn
α-Synuclein
- DFS
Dynamic Force Spectroscopy
- FDC
Force-Distance Curve
- MAS
Maleimide silatrane
- PEG
Polyethylene glycol
- TCEP
Tris(2-carboxyethyl)phosphine
- T-silatrane
Tetrahedral shaped tripodal silatrane molecule
- WLC model
Worm-Like Chain model
Footnotes
SUPPORTING INFORMATION AVAILABLE This section describes details for the synthesis of cysteine modified Aβ-40 and tetrahedral shaped tripodal silatrane (T-shaped) molecule, the results of contour length distribution measured with the use of the T-silatrane molecules, and the estimates for polydispersity of linker molecule. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nature Chemistry. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci U S A. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Evidence of fibril-like beta-sheet structures in a neurotoxic amyloid intermediate of Alzheimer's beta-amyloid. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Lee M. Observation of multi-step conformation switching in beta-amyloid peptide aggregation by fluorescence resonance energy transfer. Biochem Biophys Res Commun. 2004;316:393–397. doi: 10.1016/j.bbrc.2004.02.059. [DOI] [PubMed] [Google Scholar]
- 8.Hofrichter J, Ross PD, Eaton WA. Kinetics and mechanism of deoxyhemoglobin S gelation: a new approach to understanding sickle cell disease. Proc Natl Acad Sci U S A. 1974;71:4864–4868. doi: 10.1073/pnas.71.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarrett JT, Lansbury PT., Jr. Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 10.Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer's disease. Nature. 2008;451:720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshpande A, Mina E, Glabe C, Busciglio J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. J Neurosci. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 14.Bitan G, Teplow DB. Rapid photochemical cross-linking--a new tool for studies of metastable, amyloidogenic protein assemblies. Acc Chem Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 15.He X, Giurleo JT, Talaga DS. Role of small oligomers on the amyloidogenic aggregation free-energy landscape. J Mol Biol. 2009;395:134–154. doi: 10.1016/j.jmb.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAllister C, Karymov MA, Kawano Y, Lushnikov AY, Mikheikin A, Uversky VN, Lyubchenko YL. Protein interactions and misfolding analyzed by AFM force spectroscopy. J Mol Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Lyubchenko YL, Sherman S, Shlyakhtenko LS, Uversky VN. Nanoimaging for protein misfolding and related diseases. J Cell Biochem. 2006;99:52–70. doi: 10.1002/jcb.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kransnoslobodtsev AV, Shlyakhtenko LS, Ukraintsev E, Zaikova TO, Keana JF, Lyubchenko YL. Nanomedicine and protein misfolding diseases. Nanomedicine. 2005;1:300–305. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Lyubchenko YL. Early stages for Parkinson's development: alpha-synuclein misfolding and aggregation. J Neuroimmune Pharmacol. 2009;4:10–16. doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Malkova S, Lyubchenko YL. alpha-Synuclein misfolding: single molecule AFM force spectroscopy study. J Mol Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Khandogin J, Brooks CL., 3rd. Linking folding with aggregation in Alzheimer's beta-amyloid peptides. Proc Natl Acad Sci U S A. 2007;104:16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valerio M, Porcelli F, Zbilut JP, Giuliani A, Manetti C, Conti F. pH effects on the conformational preferences of amyloid beta-peptide (1–40) in HFIP aqueous solution by NMR spectroscopy. ChemMedChem. 2008;3:833–843. doi: 10.1002/cmdc.200700324. [DOI] [PubMed] [Google Scholar]
- 23.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 24.Ryuta J, Morita E, Tanaka T, Shimanuki Y. Crystal-Originated Singularities on Si Wafer Surface after Sc1 Cleaning. Jpn J Appl Phys 2. 1990;29:L1947–L1949. [Google Scholar]
- 25.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Warnke J, Lyubchenko YL. Nanoprobing of alpha-synuclein misfolding and aggregation with atomic force microscopy. Nanomedicine. 2010 doi: 10.1016/j.nano.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Krasnoslobodtsev AV, Shlyakhtenko LS, Lyubchenko YL. Probing Interactions within the synaptic DNA-SfiI complex by AFM force spectroscopy. J Mol Biol. 2007;365:1407–1416. doi: 10.1016/j.jmb.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tinoco I, Jr., Bustamante C. The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys Chem. 2002;101–102:513–533. doi: 10.1016/s0301-4622(02)00177-1. [DOI] [PubMed] [Google Scholar]
- 29.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyubchenko YL, Kim BH, Krasnoslobodtsev AV, Yu J. Nanoimaging for protein misfolding diseases. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2:526–543. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 32.Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 33.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 34.Urbanc B, Betnel M, Cruz L, Bitan G, Teplow DB. Elucidation of amyloid beta-protein oligomerization mechanisms: discrete molecular dynamics study. J Am Chem Soc. 2010;132:4266–4280. doi: 10.1021/ja9096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei G, Jewett AI, Shea JE. Structural diversity of dimers of the Alzheimer amyloid-beta(25–35) peptide and polymorphism of the resulting fibrils. Phys Chem Chem Phys. 2010;12:3622–3629. doi: 10.1039/c000755m. [DOI] [PubMed] [Google Scholar]
- 36.Miller Y, Ma B, Nussinov R. Polymorphism of Alzheimer's Abeta17–42 (p3) oligomers: the importance of the turn location and its conformation. Biophys J. 2009;97:1168–1177. doi: 10.1016/j.bpj.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Udgaonkar JB. Conformational conversion may precede or follow aggregate elongation on alternative pathways of amyloid protofibril formation. J Mol Biol. 2009;385:1266–1276. doi: 10.1016/j.jmb.2008.11.033. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Takeda T, Klimov DK. Mapping conformational ensembles of abeta oligomers in molecular dynamics simulations. Biophys J. 2010;99:1949–1958. doi: 10.1016/j.bpj.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi T, Yagi H, Goto Y, Matsuzaki K, Hoshino M. A disulfide-linked amyloid-beta peptide dimer forms a protofibril-like oligomer through a distinct pathway from amyloid fibril formation. Biochemistry. 2010;49:7100–7107. doi: 10.1021/bi100583x. [DOI] [PubMed] [Google Scholar]
- 40.Sandberg A, Luheshi LM, Sollvander S, Pereira de Barros T, Macao B, Knowles TP, Biverstal H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, Lannfelt L, Dobson CM, Hard T. Stabilization of neurotoxic Alzheimer amyloid-beta oligomers by protein engineering. Proc Natl Acad Sci U S A. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ainavarapu SR, Brujic J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrion-Vazquez M, Li H, Fernandez JM. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda T, Klimov DK. Probing the effect of amino-terminal truncation for Abeta1–40 peptides. J Phys Chem B. 2009;113:6692–6702. doi: 10.1021/jp9016773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 44.Yang G, Cecconi C, Baase WA, Vetter IR, Breyer WA, Haack JA, Matthews BW, Dahlquist FW, Bustamante C. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc Natl Acad Sci U S A. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stine WB, Jr., Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.