Abstract
Much insight into the interactions of DNA and enzymes has been obtained using a number of single-molecule techniques. However, recent results generated using two of these techniques—atomic force microscopy (AFM) and magnetic tweezers (MT)—have produced apparently contradictory results when applied to the action of the ATP-dependent type III restriction endonucleases on DNA. The AFM images show extensive looping of the DNA brought about by the existence of multiple DNA binding sites on each enzyme and enzyme dimerisation. The MT experiments show no evidence for looping being a requirement for DNA cleavage, but instead support a diffusive sliding of the enzyme on the DNA until an enzyme–enzyme collision occurs, leading to cleavage. Not only do these two methods appear to disagree, but also the models derived from them have difficulty explaining some ensemble biochemical results on DNA cleavage. In this ‘Survey and Summary’, we describe several different models put forward for the action of type III restriction enzymes and their inadequacies. We also attempt to reconcile the different models and indicate areas for further experimentation to elucidate the mechanism of these enzymes.
INTRODUCTION
Recent years have seen the introduction and development of sophisticated single-molecule techniques that make possible the study of molecular interactions that could previously only be investigated using bulk measurements. Such techniques are increasing in popularity and are reputed to provide greater biochemical insight into complex molecular systems, for example molecular motors (1,2). While these single-molecule techniques would at first appear to be preferable to the less specific bulk methods, the data produced can lead to inconsistencies. This might be expected if the differences were only between findings made with the bulk and single-molecule experiments, but there can also be inconsistencies between data obtained using the different newer techniques. Here, we will address the mechanism of action of type III DNA restriction/modification (R/M) enzymes (3–6). Recent study of this enzyme system has indeed led to inconsistencies in data and interpretation. These enzymes operate as endonucleases on DNA in vivo and in vitro and they have been developed as tools for superSAGE (7,8) and for counting the number CAG repeats in repetitive DNA (9). Recent results using two different single-molecule techniques, atomic force microscopy (AFM) (10–12) and magnetic tweezers (MT) (13,14) lead to conclusions that are in disagreement. In this article we hope to address this disagreement.
Type III R/M enzymes, in common with many restriction enzymes (15,16), work best when two copies of their target sequence are present on the same DNA molecule. Type III R/M enzymes have asymmetric target sites; for example EcoP15I recognizes 5′-CAGCAG-3′. In contrast to most other restriction enzymes, the type III R/M enzymes generally prefer their target sites to be oriented in a head-to-head fashion (17,18) and large site separations of up to 3.5 kb between the two sites can still lead to cleavage. Two enzyme molecules are needed per double strand cut; in other words one enzyme per target sequence is the minimum for cleavage. Cleavage occurs at a defined location next to only one of the sites, and which one of the two sites is chosen for cleavage is random, although this is influenced by the base composition of the DNA. The EcoP15I R/M enzyme, for example cuts the sequence CAGCAG(N)25/27 as long as the underlined adenine is unmethylated. The preference for a two-site substrate gives some indication of the complexity of the system and indicates that some interaction needs to take place between the two copies of the enzyme bound to the same DNA molecule for cleavage to take place. Further, ATP hydrolysis is essential for the overall restriction process.
The type III R/M enzymes comprise two modification (Mod) subunits, each containing a target recognition domain (TRD) to bind to the target sequence and a methyltransferase catalytic (CAT) domain to monitor the methylation status of an adenine in the target (the second A in the sequence above for EcoP15I), and two restriction (Res) subunits each containing a DNA helicase and ATP-hydrolysing (ATP) domain and an endonuclease DNA cleavage (NUC) domain (6,19,20). They share structural features with other complex R/M enzymes such as the type I and type IIB R/M enzymes (3,6,21). Several models for the mode of action of type III R/M enzymes have been proposed. In this survey we describe each model and its faults in turn and then conclude with a possible model for the operation of these enzymes and suggestions for future experiments.
Experimental models for the operation of type III R/M enzymes
Translocation, loop extrusion and collision model
Initially it was believed that ATP hydrolysis by a type III R/M enzyme provided energy to allow translocation of DNA so that the enzymes bound at each target site would eventually collide (18,19). This is found with other DNA translocases such as the type I R/M enzymes (3–5). These type I R/M enzymes hydrolyse one ATP for roughly each base pair of the many thousands translocated during the restriction process (22). They do not turn over in the endonuclease reaction and loops of DNA are extruded by translocation as the enzyme apparently remains bound to its target site. In this model, two enzymes eventually collide with each other, with DNA cleavage resulting at the collision point. Since they also hydrolyse ATP, it was assumed that the type III R/M enzymes, in common with type I R/M enzymes, would also remain bound at their target sites during translocation and that loops of DNA would be extruded, Figure 1. Eventually all of the DNA between two target sites would be extruded into loops and the enzymes would collide with each other and cleave the DNA, but in this case at a defined site close to one of the initial recognition sequences (18,19). The translocation proceeds only from the 3′-side of the asymmetric target site and only the NUC domain in the Res subunit on the 3′-side of the translocating enzyme is active, to agree with the observed head-to-head orientation preference. We term this the translocation, looping and collision (TLC) model, Figure 1. Curiously, the type III R/M enzymes use only ∼1% of the amount of ATP consumed by a type I R/M enzyme (13,14,23,24), but they achieve the same biological effect, namely the cleavage of DNA containing two copies of the target sequence and a restriction of propagation of foreign invading DNA through a bacterial population. This ATP hydrolysis, driving DNA translocation and bringing about collision of two type III R/M enzymes has been presumed for many years to be the fundamental requirement for restriction by type III R/M enzymes. However, the way in which translocation might be achieved with so little ATP hydrolysis is difficult to understand and is a major barrier in transposing the TLC model from the type I R/M enzymes to the type III R/M enzymes.
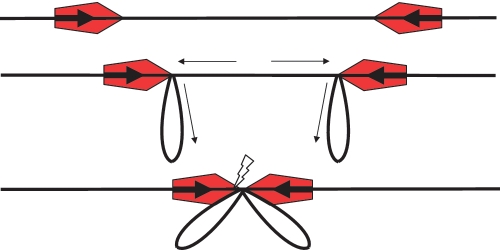
Figure 1.
The TLC model on a head-to-head substrate showing, from top to bottom: binding of enzymes (red pentagons with the active NUC domain of the Res subunit located at the leading point of the pentagon) to target sites (bold arrows), initiation of translocation to extrude loops, and, lastly, collision of the enzymes leading to cleavage at one or other target site (flash). Small arrows indicate the movement of the DNA.
DNA end effects and single site substrates pose mechanistic problems
Cleavage of various substrates other than a two-site head-to-head circular DNA has been observed. The TLC model has difficulty in accounting for this cleavage. Effective cleavage of circular single-site substrates (25), of linear single-site substrates (26–28), of linear substrates with two sites in a tail-to-tail orientation (14,28) and of substrates where the two sites actually contact each other (28) have all been observed. Furthermore, cleavage of linear versions of the head-to-head substrates was enhanced to the levels seen with circular substrates if the free ends of the DNA were blocked by a streptavidin molecule (14,29).
Considering only the substrates containing two target sites for the EcoP15I enzyme, the DNA sequence leading to cleavage is actually CAGCAG(N)∼−3200 to ∼+3500 bpGTCGTC. The limits of the spacing between the two copies of the target sequence are those observed experimentally and the negative number of non-specific bp is for a tail-to-tail orientation on a linear substrate. Sites oriented in a head-to-tail manner are considered to be poor substrates, but Raghavendra and Rao (27) note that even with this substrate some very limited cleavage can be seen to occur. The TLC model does not explain any of these observations. Thus new models for the operation of type III R/M enzymes became necessary.
The end reversal model
The cleavage of single-site substrates, particularly of linear substrates led to the observation of ‘end’ effects and the idea that the translocation direction could be reversed once an end was reached (27). The enzyme would initially bind and recognize its target sequence, commence ATP hydrolysis and vacate the target site by tracking along the DNA in the 3′-direction, driven by the ATP hydrolysis, Figure 2. Only the NUC domain on the 3′-side of the target is active. If the translocating enzyme collides with another enzyme bound to a second target oriented to face the first target, cleavage will result. On reaching a free end on a single-site substrate, the enzyme would either fall off or, if the end was blocked, reverse its direction and track back along the DNA until it collides with another enzyme bound at the target site, subsequently producing DNA cleavage. Reversal of the translocation direction at the end of a linear DNA molecule includes a reversal in orientation of the active NUC domain to allow DNA cleavage. If both enzymes have translocated away from their target sites, the collision will be ineffective and the process will have to be repeated. We will refer to this as the ‘reversal’ model, Figure 2.
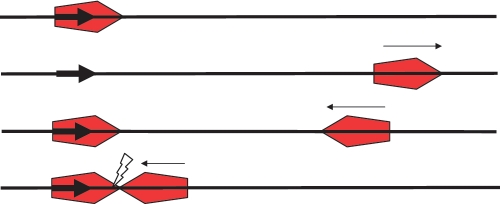
Figure 2.
The reversal model on a single-site substrate. Small arrows indicate the movement of the enzyme. Top: enzyme (red pentagon with the active NUC domain of the Res subunit located at the leading point of the pentagon) binds to target site (bold arrow). Second from top: site-specific binding triggers ATP-dependent translocation off the site in the 3′-direction. Third from top: collision of the enzyme with an end causes the enzyme to turn around. A second enzyme is shown bound at the target site. Bottom: collision of this reversed enzyme with an enzyme at the original target site initiates cleavage (flash).
At first sight, the reversal model can explain head-to-head, tail-to-tail and single-site linear cleavage effectively. Unfortunately, it cannot explain the cleavage of single-site circles, as no end exists to cause reversal. Instead, the translocating enzyme would simply run into the back of the second enzyme bound at the target site and the active NUC domains would not be juxtaposed to cleave. Furthermore, the model allows the enzyme to reverse when it hits an end and this reversal includes the requirement for the active NUC domain to reverse its orientation of DNA. Unfortunately, this would lead to effective cleavage of head-to-tail substrates, which would now be as good a substrate as head-to-head DNA. Since cleavage of head-to-tail substrates is very much poorer than cleavage of head-to-head substrates, the model provides only a partial explanation of the experimental observations.
MT experiments: the random walk sliding model
A similar model has been proposed on the basis of MT assays, tethered particle motion (TPM) assays and biochemical data (13,14,29,30). Cutting was observed on both head-to-head and tail-to-tail linear DNA, particularly when the ends were blocked by streptavidin. This cutting occurred even when the DNA was under sufficient tension to preclude any looping, even if coupled to ATP hydrolysis. Due to the low time resolution of the measurements, at low tension, transient looping could not be ruled out. The low ATP consumption was proposed to originate from an ATP-induced activating ‘conformational change’ when the enzyme was bound at a target site. This activation then allowed the enzyme to track along the DNA in a one-dimensional random walk without further energy consumption, Figure 3. The walk would go in either direction and would leave the original target site vacant for further enzymes to bind. The random walking enzyme could then collide with an enzyme bound at a second site and, if the orientation were correct, cleavage would occur. Head-to-head sites would allow cleavage to occur as the collision would be ‘head-on’ while tail-to-tail sites would require the first enzyme to walk past the second site prior to binding of the second enzyme. A subsequent collision would then be head-on as required for cleavage. Cleavage would occur as long as the second enzyme to bind had not commenced its random walk and was still at a target site as also proposed in the reversal model. We will refer to this model as the MT model, Figure 3.
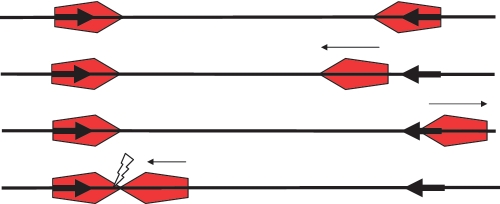
Figure 3.
The MT model on a head-to-head substrate. Small arrows indicate the movement of the enzyme. Top: enzymes (red pentagons with the active NUC domain of the Res subunit located at the leading point of the pentagon) are bound at each target site (bold arrows). Second and third from top: the right-hand enzyme diffuses off its target site after ATP activation and moves randomly backwards and forwards along the DNA. Note that the orientation of the moving enzyme on the DNA is maintained. Low levels of ATP hydrolysis keep the enzyme on the DNA. The left hand enzyme remains at its target. Bottom: the enzyme eventually collides with the left-hand enzyme initiating cleavage (flash). If the ends of the DNA are blocked with an obstacle (such as streptavidin), the moving enzyme generally does not fall off the DNA and cleavage probability is enhanced.
Although the model ignores structural information on the type III R/M enzymes, it implicitly assumes that only the NUC domain on the 3′-side of the target is active. The diffusion brings the active NUC domain into contact with another enzyme still bound at a second target site and cleavage will occur if the orientation correctly brings the active NUC domains in the two enzymes into contact. If this second enzyme has moved off the target site, then the whole process is ineffective and has to be repeated, as in the reversal model. The MT model explains cleavage of head-to-head and tail-to-tail substrates very effectively, even if the sites are adjacent, as it does not allow the enzyme to change the orientation of the active NUC domain on the DNA when an end is hit. On the other hand, because of this requirement, the model is unable to explain the good cleavage of single-site substrates, as two active NUC domains would never come into contact: one enzyme would run into the back of the other and the active NUC domains would not contact each other. This is true for both linear and circular single-site DNA molecules, as one would always obtain only head-to-tail collisions. Hence this model is also incomplete.
The transient looping and translocation model from AFM
AFM showed that in the presence of ATP, EcoP15I could cause DNA to form loops held together at their apex by one or two copies of the enzyme, depending upon whether one or two copies of the target sequence were present (10–12). On a linear molecule with a single target site, a single copy of the enzyme bound to the target site could capture another segment of the DNA molecule, resulting in one or two DNA loops. The lengths of the loops were in excellent agreement with well characterized theories of DNA flexibility and diffusive looping, rather than having sizes dictated by DNA translocation driven by ATP hydrolysis (12). The apex of the loop usually included the target sequence, but occasionally the specific target sequence was not at the apex, implying that the enzyme could move off the target sequence after the formation of the loop. It might also be proposed to bind two non-specific sites and form a loop, but since no loops were observed on substrates completely lacking target sites, this possibility can be discounted. On two-site substrates, multiple loops were observed emanating from a single apex. The apex apparently had two copies of the enzyme bound, as also observed for type I R/M enzymes (31), and the loops were again of the size expected from DNA flexibility. On neither substrate were additional single enzyme molecules observed, contrary to what one would expect from the MT model. Fast-scan AFM further showed the real-time formation and breaking of loops and the introduction of supercoiling due to translocation past the enzyme, as in the TLC model, once DNA was stably trapped in a loop (11). The observation of supercoiling correlates with the bulk observation that supercoiling influences cleavage efficiency (19). A model was proposed showing how the initial formation of loops via diffusion and segment capture, which would shorten the distance between two target sites, could subsequently reduce the amount of translocation required for inter-site collision of the NUC domains and the amount of ATP required (12), Figure 4. Since two-site substrates showed an enzyme dimer at the apex of multiple loops, the model also incorporated a specific dimerisation of the R/M enzyme and a probable enzyme structure which would account not only for the number of loops but also detect the orientation of the two target sites and whether they were on the same DNA molecule. We will refer to this as the AFM model, Figure 4.
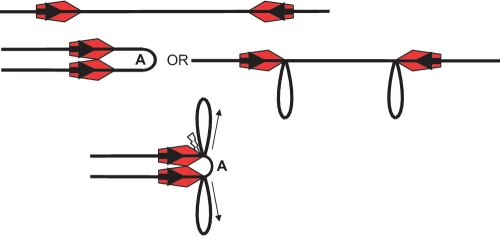
Figure 4.
The AFM model on a head-to-head substrate. Small arrows indicate the movement of the DNA. Top: enzymes (red pentagons with the active NUC domain of the Res subunit located at the leading point of the pentagon) are bound at each target site (bold arrows). Middle: ATP activates both enzyme:enzyme dimerisation (left hand side) and the trapping of loops via the flexibility of the DNA (right hand side). Loop A results from the dimerisation of the enzymes while the other loops result from trapping of DNA segments between the Mod subunit and the Res subunit. By this mechanism the inter-site distance is reduced prior to further translocation. Bottom: translocation, driven by ATP hydrolysis, leading to contraction of loop A in the head-to-head complex, pulls the loop taut between the two translocating Res subunits. It is postulated that this is the trigger for cleavage (flash) by the two NUC domains.
For this model it was suggested that the translocation movement would be directional and driven by ATP hydrolysis just as in the TLC model (12). However, we note that a one-dimensional random walk diffusion, activated by ATP hydrolysis as in the MT model, could be an alternative way of changing the size of the loops (30), although this would not account for the observed supercoiling (11,19).
The AFM model works for two-site substrates in explaining the cleavage of head-to-head and tail-to-tail substrates rather than head-to-tail substrates. It also allows discrimination of sites in trans as the protein–protein interaction is responsible for detecting the site orientation. The observation of looping on single-site substrates, which can be cleaved, can also be explained if the enzyme is allowed to become activated and undergo inter-segment transfer on the DNA, thereby clearing the target site to bind a second enzyme. However, in common with the other models there are weaknesses. In this case dimers were not observed by AFM on single-site DNA so this model cannot explain cleavage of such substrates. The AFM model also does not work for situations where the two target sites are adjoining, as there is insufficient space on the DNA. Hence, once again the model cannot be complete.
Perhaps supporting the AFM model as a plausible mechanism is the fact that looping to bring two target sequences in cis together is a feature of a substantial number of restriction enzymes (15,16) and has often been observed using AFM [e.g. (31,32)] and electron microscopy [e.g. (33)]. The loops often require enzyme ‘dimers’. Single-molecule methods such as MT and TPM assays have also led to the observation of looping (32,34–36). In some cases this looping is very obvious, but sometimes the forces generated on DNA by the technique reduce looping to very low levels so that it is hard to observe without special care, especially if the stability of the loop is low. For example the loops formed by Ecl18kI type II restriction enzyme were obvious via AFM, but formed in only a few percent of events in a TPM assay (32). Biochemical evidence is also derived from measurements of the enzyme molecular weight in the presence and absence of DNA (enzyme ‘dimers’ may be stable in the absence of DNA or require DNA binding to be stable), and from a dependence of cleavage of a long piece of DNA on the presence of a second short duplex, itself containing a target site [e.g. (37)]. Such an enhancement of cleavage has been observed for the type III R/M enzyme EcoPI, although in this case the cleavage occurred on a long piece of DNA lacking any target sites and was apparently an example of a ‘secondary’ cleavage activity of the enzyme (29).
A potential reconciliation of the models and conclusions
It should now be clear that none of the models, TLC, Reversal, MT or AFM, fit all current data. So is there some way to incorporate all the data in a model of operation encompassing the biochemical data but also resolving the discrepancies revealed by AFM, a predominantly ‘structural’ method and MT, a predominantly ‘kinetic’ method, bearing in mind that DNA cleavage under high applied force indicates that a requirement for looping can be circumvented?
The MT model has been put forward most forcefully by Szczelkun et al. (30), but the model disregards the loops observed by AFM and other biochemical evidence for looping, such as the obvious cleavage of single-site circular DNA (25,29) and the observation that cleavage is hindered by supercoiling of the DNA (19). The absence of apparent looping in the TPM assay when applied to the type III R/M enzymes is also cited as support for the MT model (13,14), but it is apparent that the data are not of sufficient resolution, when compared with the data obtained for the Ecl18kI type II restriction enzyme, to rule out looping (32). It is important to note that DNA looping and one-dimensional diffusion along a DNA contour are both fast events occurring on the millisecond timescale, much faster than the time resolution of the assays currently employed, be they single molecule or ensemble assays (1,38,39).
The AFM model proposes that the observed loops are formed via the natural flexibility of DNA and that ATP hydrolysis then drives their expansion or contraction until they cannot change further, at which point cleavage is induced. If instead one proposes that after their initial formation, the loops expand or contract via randomly diffusing backwards and forwards through the enzyme in a process maintained via ATP hydrolysis then one could link the AFM and MT models together to explain the data on two-site substrates.
A key question is why do the MT experiments not reveal loops? It is worth highlighting that in all of the proposed models, cleavage is preceded by a specific ‘dimerisation’ of two enzyme molecules in such a way as to position two active NUC domains together. So the question is really when does this dimerisation occur? For two-site substrates, we suggest that the AFM surface enhances the ability of an EcoP15I bound to its target site to stick to another EcoP15I bound at a second site, noting that such collisions are inevitable and rapid on any long piece of DNA in three and two dimensions, as long as the DNA is not under tension (35,36,39). The inter-segmental collision is indeed one of the fastest ways to communicate between sites on DNA, especially when other DNA-binding proteins are bound along the DNA contour (38,40). If the kinetics of this collision interaction, including its sticking to the AFM surface, are faster than the kinetics of activating the sliding mechanism, which allows the enzyme to leave its site, then the AFM will predominantly see these looped complexes, whereas the MT experiments will predominantly see the sliding complexes. Thus the MT and AFM data may be seeing the same pathway but with different aspects being highlighted.
Thus, reconciliation between the AFM and MT models is possible for the cleavage of substrates with two target sites but there is clearly also a need for further experimentation on the cleavage of single-site substrates, as this is still unexplained, and on the structural domains and interfaces present in the enzyme. Further assays could include an examination of whether a type III enzyme bound to its target would interfere with cleavage by a type II restriction enzyme that had an overlapping target site. The MT model would suggest that little hindrance would occur as the type III enzyme would diffuse away to leave the site for the type II restriction enzyme clear of obstruction. The AFM ‘dimerisation via looping’ model could be tested by varying the spacing between two target sites, as it would predict a 10 bp periodicity for the ability of the two enzymes to dimerize. Future experiments on type III restriction enzymes might also include a direct visualisation of fluorescently-labelled enzymes diffusing on hydrodynamically stretched DNA (1,2) and more sophisticated dynamic looping experiments using higher time resolutions (here we note the potential of new CMOS-based single photon imagers (41) with microsecond resolution). Perhaps the most useful in vitro experiments to perform now are an analysis of the multiple DNA binding sites on each copy of the enzyme and further study of the three-dimensional structure, particularly the ‘dimerisation’ and subunit interfaces, of these complex enzymes to constrain speculations on their mode of operation.
In addition, any more complete model needs to give further consideration to the target situation in vivo and specifically whether the foreign DNA is naked, i.e. devoid of other non-sequence-specific proteins (40,42), stretched out or coiled up. All of these factors will influence the ability of the restriction enzymes, and not just type III enzymes, to find and bind their target sequences and ultimately to protect the cell from invasion. Furthermore, the model of action has to be fast enough to prevent replication of the invading foreign DNA because the R/M system will generally only have ∼1 min, if one considers the life cycle of, for example a lytic phage, in which to act on foreign DNA.
FUNDING
Biotechnology and Biological Sciences Research Council (BBS/B/1065X to AFM work). Funding for open access charge: Wellcome Trust grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
DTFD gratefully acknowledges the Master and Fellows of Emmanuel College, Cambridge for the award of a Derek Brewer Visiting Fellowship during which time this article was prepared.
REFERENCES
- 1.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 2.Hilario J, Kowalczykowski SC. Visualizing protein-DNA interactions at the single-molecule level. Curr. Opin. Chem. Biol. 2010;14:15–22. doi: 10.1016/j.cbpa.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao DN, Saha S, Krishnamurthy V. ATP-dependent restriction enzymes. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:1–63. doi: 10.1016/s0079-6603(00)64001-1. [DOI] [PubMed] [Google Scholar]
- 4.Dryden DTF, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 6.Madhusoodanan UK, Rao DN. Diversity of DNA methyltransferases that recognize asymmetric target sequences. Crit. Rev. Biochem. Mol. Biol. 2010;45:125–145. doi: 10.3109/10409231003628007. [DOI] [PubMed] [Google Scholar]
- 7.Raghavendra NK, Rao DN. Exogenous AdoMet and its analogue sinefungin differentially influence DNA cleavage by R.EcoP15I–usefulness in SAGE. Biochem. Biophys. Res. Commun. 2005;334:803–811. doi: 10.1016/j.bbrc.2005.06.171. [DOI] [PubMed] [Google Scholar]
- 8.Matsumura H, Yoshida K, Luo S, Kimura E, Fujibe T, Albertyn Z, Barrero RA, Krüger DH, Kahl G, Schroth GP, et al. High-throughput SuperSAGE for digital gene expression analysis of multiple samples using next generation sequencing. PLoS ONE. 2010;5:e12010. doi: 10.1371/journal.pone.0012010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Möncke-Buchner E, Reich S, Mücke M, Reuter M, Messer W, Wanker EE, Krüger DH. Counting CAG repeats in the Huntington’s disease gene by restriction endonuclease EcoP15I cleavage. Nucleic Acids Res. 2002;30:e83. doi: 10.1093/nar/gnf082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reich S, Gössl I, Reuter M, Rabe JP, Krüger DH. Scanning force microscopy of DNA translocation by the Type III restriction enzyme EcoP15I. J. Mol. Biol. 2004;341:337–343. doi: 10.1016/j.jmb.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Crampton N, Yokokawa M, Dryden DTF, Edwardson JM, Rao DN, Takeyasu K, Yoshimura SH, Henderson RM. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl Acad. Sci. USA. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crampton N, Roes S, Dryden DTF, Rao DN, Edwardson JM, Henderson RM. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramanathan SP, van Aelst K, Sears A, Peakman LJ, Diffin FM, Szczelkun MD, Seidel R. Type III restriction enzymes communicate in one-dimensional without looping between their target sites. Proc. Natl Acad. Sci. USA. 2009;106:1748–1753. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Aelst K, Tóth J, Ramanathan SP, Schwarz FW, Seidel R, Szczelkun MD. Type III restriction enzymes cleave DNA by long-range interaction between sites in both head-to-head and tail-to-tail inverted repeat. Proc. Natl Acad. Sci. USA. 2010;107:9123–9128. doi: 10.1073/pnas.1001637107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halford SE, Bilcock DT, Stanford NP, Williams SA, Milsom SE, Gormley NA, Watson MA, Bath AJ, Embleton ML, Gowers DM, et al. Restriction endonuclease reactions requiring two recognition sites. Biochem. Soc. Trans. 1999;27:696–699. doi: 10.1042/bst0270696. [DOI] [PubMed] [Google Scholar]
- 16.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 17.Meisel A, Bickle TA, Krüger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 18.Meisel A, Mackeldanz P, Bickle TA, Krüger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J. Mol. Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 20.Wagenführ K, Pieper S, Mackeldanz P, Linscheid M, Krüger DH, Reuter M. Structural domains in the type III restriction endonuclease EcoP15I: characterization by limited proteolysis, mass spectrometry and insertional mutagenesis. J. Mol. Biol. 2007;366:93–102. doi: 10.1016/j.jmb.2006.10.087. [DOI] [PubMed] [Google Scholar]
- 21.Dryden DTF. S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. In: Cheng X, Blumenthal RM, editors. Bacterial DNA methyltransferases. Singapore: World Scientific Publishing; 1999. pp. 283–340. [Google Scholar]
- 22.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiser J, Yuan R. Purification and properties of the P15 specific restriction endonuclease from Escherichia coli. J. Biol. Chem. 1977;252:451–456. [PubMed] [Google Scholar]
- 24.Sears A, Peakman LJ, Wilson GG, Szczelkun MD. Characterization of the Type III restriction endonuclease PstII from Providencia stuartii. Nucleic Acids Res. 2005;33:4775–4787. doi: 10.1093/nar/gki787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Möncke-Buchner E, Rothenberg M, Reich S, Wagenführ K, Matsumura H, Terauchi R, Krüger DH, Reuter M. Functional characterization and modulation of the DNA cleavage efficiency of type III restriction endonuclease EcoP15I in its interaction with two sites in the DNA target. J. Mol. Biol. 2009;387:1309–1319. doi: 10.1016/j.jmb.2009.02.047. [DOI] [PubMed] [Google Scholar]
- 26.Raghavendra NK, Rao DN. Functional cooperation between exonucleases and endonucleases–basis for the evolution of restriction enzymes. Nucleic Acids Res. 2003;31:1888–1896. doi: 10.1093/nar/gkg275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghavendra NK, Rao DN. Unidirectional translocation from recognition site and a necessary interaction with DNA end for cleavage by Type III restriction enzyme. Nucleic Acids Res. 2004;32:5703–5711. doi: 10.1093/nar/gkh899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mücke M, Reich S, Möncke-Buchner E, Reuter M, Krüger DH. DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites. J. Mol. Biol. 2001;312:687–698. doi: 10.1006/jmbi.2001.4998. [DOI] [PubMed] [Google Scholar]
- 29.Peakman LJ, Szczelkun MD. S-adenosyl homocysteine and DNA ends stimulate promiscuous nuclease activities in the Type III restriction endonuclease EcoPI. Nucleic Acids Res. 2009;37:3934–3945. doi: 10.1093/nar/gkp267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szczelkun MD, Friedhoff P, Seidel R. Maintaining a sense of direction during long-range communication on DNA. Biochem. Soc. Trans. 2010;38:404–409. doi: 10.1042/BST0380404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neaves KJ, Cooper LP, White JH, Carnally SM, Dryden DTF, Edwardson JM, Henderson RM. Atomic force microscopy of the EcoKI Type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 2009;37:2053–2063. doi: 10.1093/nar/gkp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaremba M, Owsicka A, Tamulaitis G, Sasnauskas G, Shlyakhtenko LS, Lushnikov A, Lyubchenko YL, Laurens N, van den Broek B, Wuite GJL, et al. DNA synapsis through the transient tetramerization triggers the cleavage by Ecl18kI restriction enzyme. Nucleic Acids Res. 2010;38:7142–7154. doi: 10.1093/nar/gkq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Topal MD, Thresher RJ, Conrad M, Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991;30:2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- 34.van den Broek B, Noom MC, Wuite GJL. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gemmen GJ, Millin R, Smith DE. Tension-dependent DNA cleavage by restriction endonucleases: two-site enzymes are “switched off ” at low force. Proc. Natl Acad. Sci. USA. 2006;103:11555–11560. doi: 10.1073/pnas.0604463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gemmen GJ, Millin R, Smith DE. DNA looping by two-site restriction endonucleases: heterogeneous probability distributions for loop size and unbinding force. Nucleic Acids Res. 2006;34:2864–2877. doi: 10.1093/nar/gkl382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuter M, Kupper D, Meisel A, Schroeder C, Krüger DH. Cooperative binding properties of restriction endonuclease EcoRII with DNA recognition sites. J. Biol. Chem. 1998;273:8294–8300. doi: 10.1074/jbc.273.14.8294. [DOI] [PubMed] [Google Scholar]
- 38.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sankararaman S, Marko JF. Formation of loops in DNA under tension. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:021911. doi: 10.1103/PhysRevE.71.021911. [DOI] [PubMed] [Google Scholar]
- 40.Li G-W, Berg OG, Elf J. Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nat. Phys. 2009;5:294–297. [Google Scholar]
- 41.Li D-U, Arlt J, Richardson J, Walker R, Buts A, Stoppa D, Charbon E, Henderson R. Real-time fluorescence lifetime imaging system with a 32 × 32 0.13μm CMOS low dark-count single-photon avalanche diode array. Opt. Express. 2010;18:10257–10269. doi: 10.1364/OE.18.010257. [DOI] [PubMed] [Google Scholar]
- 42.Flyvbjerg H, Keatch SA, Dryden DTF. Strong physical constraints on sequence-specific target location by proteins on DNA molecules. Nucleic Acids Res. 2006;34:2550–2557. doi: 10.1093/nar/gkl271. [DOI] [PMC free article] [PubMed] [Google Scholar]