Abstract
An important mechanism for gene regulation involves chromatin changes via histone modification. One such modification is histone H3 lysine 4 trimethylation (H3K4me3), which requires histone methyltranferase complexes (HMT) containing the trithorax-group (trxG) protein ASH2. Mutations in ash2 cause a variety of pattern formation defects in the Drosophila wing. We have identified genome-wide binding of ASH2 in wing imaginal discs using chromatin immunoprecipitation combined with sequencing (ChIP-Seq). Our results show that genes with functions in development and transcriptional regulation are activated by ASH2 via H3K4 trimethylation in nearby nucleosomes. We have characterized the occupancy of phosphorylated forms of RNA Polymerase II and histone marks associated with activation and repression of transcription. ASH2 occupancy correlates with phosphorylated forms of RNA Polymerase II and histone activating marks in expressed genes. Additionally, RNA Polymerase II phosphorylation on serine 5 and H3K4me3 are reduced in ash2 mutants in comparison to wild-type flies. Finally, we have identified specific motifs associated with ASH2 binding in genes that are differentially expressed in ash2 mutants. Our data suggest that recruitment of the ASH2-containing HMT complexes is context specific and points to a function of ASH2 and H3K4me3 in transcriptional pausing control.
INTRODUCTION
Establishment and propagation of gene-expression patterns involves covalent modification of histones (1–4). These modifications play an important role in such processes as cell fate determination, development and cancer (5). Proteins of the trithorax (trxG) and Polycomb (PcG) groups form a cellular memory system that functions to maintain a heritable transcriptional state. These proteins were first identified for their role in homeotic gene regulation in Drosophila, but are now understood to constitute a conserved mechanism (6,7). Both trxG and PcG proteins act in multimeric complexes; some members exhibit histone methyltransferase (HMT) activity, while others interpret these marks and translate them into changes in chromatin structure which ultimately leads to changes in gene expression (8). A common hallmark of activated genes is trimethylation of histone 3 on lysine 4 (H3K4me3) at promoter regions, but it remains unclear how this modification is linked to transcriptional activation. In Saccharomyces cerevisiae, a single HMT complex is recruited to genes by the ubiquitination of histone H2B, requiring prior recruitment of RNA Polymerase II and the PAF1 complex (9–11). In mammalian systems, instead, recent studies provide evidence that H3K4me3 is needed for enrolment of the basal transcription machinery and transcriptional initiation (12,13). A member of the trxG, ASH2 (absent, small or homeotic discs 2) is essential for the deposition of the H3K4me3, but does not have the SET domain [Su(var)3-9, E(Z) and Trx] characteristic of the HMTs (14,15). ASH2 is associated with several HMT complexes in various organisms (15–18) and interacts with transcription factors such as HCF-1, Menin or Myc (14,18–21).
The Drosophila wing imaginal disc has proven to be a useful model to study the role of ASH2. Mutants in ash2 show a variety of pattern formation defects in addition to homeotic transformations expected for a trxG protein (22–24). Expression profile analysis of ash2 mutant discs has revealed downregulation of wing development and patterning genes (14), supporting the view that trxG proteins are involved in maintaining the activated state of those genes. An important step towards understanding ASH2 function is the identification of its target genes and the association with the transcriptional machinery since it is not clear whether ASH2 acts globally on active genes or binds sites in the genome without directly regulating gene transcription. We investigated the relationship between ASH2 occupancy, histone modifications and the transcriptional machinery in the wing disc using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq). Here, we provide a comprehensive analysis of target genes in association with expression levels.
MATERIALS AND METHODS
Drosophila strains
All Drosophila strains and crosses were kept on standard media at 25°C. The strains used were: Canton S, ash2I1/TM6C and w;daughterless·GAL4;UAS·Ash2HA (14).
Chromatin immunoprecipitation
Canton S flies were used for histones and RNA Polymerase II ChIP-Seq experiments, and w;daughterless·GAL4;UAS·Ash2HA (Ash2-Hemagglutinin) immunoprecipitated with anti-HA antibody for ASH2. Polytene chromosomes stained with a newly generated anti-ASH2 antibody showed no differences between overexpressed and endogenous ASH2 binding (data not shown). Third instar larva wing imaginal discs isolated from the above flies were fixed as previously described (25) and used as a source of chromatin for ChIP-Seq experiments. The discs were pooled in 700 µl of sonication buffer and sonicated in a Branson sonifier. Conditions were established to obtain chromatin fragments, 200–1000 bp in length. Chromatin was centrifuged for 10 min at top speed at 4°C and the supernatant was recovered. As input sample, 10 µl of fixed and sonified chromatin were decrosslinked and purified. For histone modifications, three immunoprecipitations of 100 µl, corresponding to 100 discs each, were carried out in RIPA buffer (140 mM NaCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% Na deoxycholate, protease inhibitors). For non-histone proteins, six immunoprecipitations (IPs) of 100 discs were performed either in RIPA buffer for PolIIS2P and PolIIS5P or in IP buffer (0.5% NP40, 150 mM NaCl, 200 mM Tris–HCl pH8.0, 20 mM EDTA, protease inhibitor) for HA. As a pre-clearing step, 35 µl of 50% (v/v) protein A—Sepharose CL4B was added to the IPs and incubated for 1.5 h at 4°C in a rotating wheel. Protein A was removed by centrifugation at 3000 rpm for 2 min. A suitable amount of antibody (1–2 µg) was added to each chromatin aliquot and incubated on a rotating wheel overnight at 4°C. As a negative control, an aliquot was immunoprecipitated without antibody. Immunocomplexes were recovered by adding 35 µl of 50% (v/v) protein A-Sepharose (previously blocked in RIPA or IP/1% BSA for 2 h at 4°C) to the sample and incubating with rocking for 3 h at 4°C. Protein A was washed five times for 10 min each in 1 ml of RIPA buffer or IP buffer, once in 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM Na–EDTA, 10 mM Tris–HCl, pH 8.0, and twice in TE (10 mM Tris–HCl, pH 8.0, 1 mM Na–EDTA). Protein A was resuspended in 100 µl of TE and DNase-free RNase at 50 µg/ml was added and incubated for 30 min at 37°C. To purify the immunoprecipitated DNA, samples were adjusted to 0.5% SDS, 500 µg/ml Proteinase K and incubated overnight at 65°C. IP chromatin was purified with Qiagen PCR purification columns, following the manufacturer’s instructions. Two independent replicates were performed per ChIP-Seq.
For qPCR ChIPs, 40 wild-type (Canton S) and ash2I1 homozygous third instar larva were disrupted, fixed and processed as above. Only anterior-half larva were used. For total PolII ChIPs, we used from 5 to 10 µg of antibody and chromatin was immunoprecipitated in IP buffer. Immunocomplexes were recovered with a mixture of protein A/G. Real-time PCRs were normalized against the input sample and depicted as percentage of the input (see Supplementary Data S1 for selected primers).
The antibodies used for chromatin immunoprecipitation were: H3K4me3 (Abcam/ab8580) (Millipore-Upstate/07-473); H3K27me3 (Millipore-Upstate/07-449); H3K36me3 (Abcam/ab9050); PolIIS2P (Abcam/ab5095); PolIIS5P (Abcam/ab5131); HA tag (Abcam/ab9110) and PolII clone 8WG16 (Abcam/ab817).
Solexa/Illumina sequencing
All protocols for Solexa/Illumina ChIP-Seq analysis (sample preparation and sequencing) were carried out following the manufacturer’s protocol. For a detailed protocol, see Supplementary Data S1.
Bioinformatics analysis
We ran PeakSeq (26) to identify the regions significantly enriched on ChIP-Seq reads from each sample in comparison to the normalized input control (READLENGTH = 325, MAXGAP = 40, MINFDR = 0.05 and PVALTHRESH = 0.05). The optimal read length selected for PeakSeq (26) maximizes the overlap between reads in both forward and reverse strands on each sample. The resulting read maps and target lists were visualized as custom tracks in the University of California Santa Cruz (UCSC) Genome Browser (27). ChIP-Seq profiles and target regions were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository as wiggle (WIG) and Browser Extensible Data (BED) files, respectively, under the accession number GSE24115. Correlation between replicates was performed at three levels (coordinates of reads, number of targets and number of genes associated to the targets). Using RefSeq (28), we determined the genes overlapping at least one nucleotide to each target on each sample. We considered the Gene Ontology (GO) enrichments identified by DAVID (29) in Level 3 of biological process, molecular function and cellular component categories and in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. To identify probable novel genes, we combined the H3K4me3-enriched areas with the collection of full-length mRNAs from GenBank (30), selecting those regions that do not contain any RefSeq gene (28). The same procedure was used to detect putative alternative initial exons. We next determined the genome fragments in which ASH2, the H3K4me3 mark and RNA Polymerase II modifications were present (no gene was annotated within) and selected those elements overlapping with H3K4me3 targets presenting the characteristic occupancy pattern from the FlyBase (31) catalogue of non-coding RNAs. To produce the reads’ graphical distribution for each sample around the transcriptional start site (TSS), we calculated the weighted number of reads on each position from 2000 bp upstream to 2000 bp downstream of the TSS of all genes (according to RefSeq). For the graphical representation of the idealized gene, we normalized the location of the reads within the genes using a window of 100 units, calculating the mean at each point. We integrated this representation into the neighbouring genomic region corresponding to 1000 bp upstream and downstream of the idealized gene. To measure the background levels of ASH2 read counts in intergenic regions, we computationally searched the set of 10 kb regions that do not contain gene annotations (1900 regions according to RefSeq). Similar results were obtained by searching intergenic regions of multiple sizes (25, 50 and 100 kb). We found 12% of ASH2 reads in these intergenic regions, while 65% of ASH2 ChIP-Seq reads are found within RefSeq gene regions (this represents 5-fold enrichment on gene regions).
We reanalysed previously published data of wild-type and ash2 mutant transcriptomes (14), where two Affymetrix GeneChip Drosophila Genome 2.0 arrays (Affymetrix Inc.) were hybridized per sample. For wild-type flies, we defined three gene classes according to the expression level: highly expressed (5000 or more), expressed (50–5000) and silenced (50 or less). To calculate the Spearman’s rank correlation coefficient between gene expression and the number of targets of each ChIP-Seq experiment, we previously computed the target gene density of the microarray (using a window of 100 genes). To build the list of upregulated and downregulated genes in the mutant microarray, we selected the genes for which the ratio between the expression value in the mutant array and the wild-type wing disc transcriptome was either above 2.0 or below 0.5 (upregulated and downregulated in ash2 mutants, respectively) with an absolute difference between the values of at least 100 units. Both lists were intersected with ASH2 and H3K4me3 target genes to build the final set of 196 downregulated and 137 upregulated genes in ash2 mutants. To evaluate the statistical significance of the differences observed in gene size, number of exons and number of isoforms between upregulated and downregulated genes, we performed on each gene feature a two sample t-test that discriminates whether two distributions of means can be assumed to be equal (null hypothesis) or not. MEME (32) was run on the preferred ASH2 binding region of these genes in our ChIP-Seq experiments, which is (–370, +560) around the TSS, using TomTom (33) to scan the collections of known transcription factor binding sites (34,35). In addition, TRANSFAC and JASPAR catalogues were used to complement the motif search in the ASH2 binding regions (similarity threshold 85%) (36,37). We filtered out the predicted motifs that were not conserved at least in Drosophila pseudoobscura and four additional Drosophilids in the genome alignments of 12 Drosophila species (38). We implemented multiple scripts written in Perl and R in order to perform most of these tasks (format conversion among different tools, comparison of lists of targets, association of genes with lists of targets and graphical representations of reads around the TSS of genes and within the genes). This software is available upon request to the authors.
Other methods
Details of other procedures are provided in Supplementary Data S1.
RESULTS
ASH2 correlates with H3K4me3 and binds upstream this activating mark
ASH2 occupancy was mapped using chromatin isolated from third instar larva wing imaginal discs, obtaining 8009 target genes (Figure 1 and Supplementary Table S1). GO analysis revealed a significant enrichment in development and morphogenesis categories (Supplementary Figure S1). We also determined the genomic distribution of H3K4me3 and H3K36me3, as specific marks of positive transcriptional regulation and H3K27me3 as a negative mark (Supplementary Table S1). We identified 5730 target genes for H3K4me3, 4919 for H3K36me3 and 2999 for H3K27me3. Figure 1A shows an example of ASH2 binding between two peaks of H3K4me3 associated with katanin-60 and Mms19 genes, which display opposite transcriptional orientation. H3K36me3 extends over the gene region of both expressed genes. By contrast, in an example of a gene silenced in the wing disc, H3K27me3 is spread throughout the Deformed (Dfd) locus. As anticipated, there was extensive overlap between ASH2 occupancy and the H3K4me3 and H3K36me3 marks, but not with H3K27me3 (Figure 1B). A subset of 441 genes has both activating and silencing marks, and among those, 423 contain ASH2 binding sites. Genes from several pathways known to be expressed differentially in the wing disc are included in this group (Supplementary Figure S1), suggesting that the two marks are present in various cell types according to their transcriptional state. Thus, we can predict that uncharacterized genes of this group display heterogeneous expression patterns in the wing tissue.
Figure 1.
Landscape of ASH2 binding and histone methylation profiles in the wing imaginal disc. (A) UCSC Genome Browser overview of the ChIP-Seq reads across two regions of chromosome 3R: from the top, the input sample, ASH2, H3K4me3, H3K27me3, H3K36me3 and RefSeq genes. The height of the peaks represents the number of reads obtained for each mark in each region by ChIP-Seq. The enriched regions (targets) are shown in brown boxes below the corresponding sample. (B) Venn diagrams showing the intersection between ASH2, H3K4me3 and H3K27me3 (left) or H3K36me3 (right). (C) Projection of ASH2, H3K4me3, H3K36me3 and H3K27me3 over the TSS and the idealized gene.
We observed extensive overlap between the targets obtained by ChIP-Seq in wing discs and by ChIP-on-chip in embryos (39): up to 95% of H3K4me3 targets match any ChIP-on-chip region. The enriched regions detected by high-throughput sequencing were, though, more precise (average target length: 327.2 nt in ChIP-Seq and 2048.3 nt in ChIP-on-chip), confirming that this technique results in higher resolution (Supplementary Figure S2). These results suggest that there are few differences in the chromatin state between these two time points, although a subset of genes is cell type and developmental stage specific. Additionally, through the combination of our data with FlyBase/RefSeq gene collections, we refined the annotation of 21 genes (Supplementary Table S2) and uncovered 55 new regions (Supplementary Table S3) in the fly genome that show significant enrichment of activating marks in the wing discs. Using a similar approach, we identified 21 non-coding RNAs that display transcription-activating marks in the wing disc (Supplementary Table S4).
The projection of the mean reads over the TSS of the full set of genes or over an idealized gene (Figure 1C) reveals a single ASH2 peak from the promoter to the gene region (5-fold enrichment of ASH2 read counts in gene regions relative to typical intergenic regions, see ‘Materials and Methods’ section). The H3K27me3 distribution was found scattered throughout silenced regions. In contrast, H3K4me3 exhibits a main peak at the first 500 bp downstream of the TSS and a secondary one upstream, presumably caused by the presence of genes transcribed in the opposite direction (Figure 1C). This hypothesis is supported by the fact that one single peak is detected downstream the TSS when only plotting genes for which there are no other annotated genes in their vicinity (data not shown). Finally, we observed that the ASH2 peak localizes upstream of the main H3K4me3 peak in ∼80% of the genes, suggesting it contributes to this methylation in nearby nucleosomes.
ASH2 binding correlates with RNA Polymerase II and histone activating marks in expressed genes
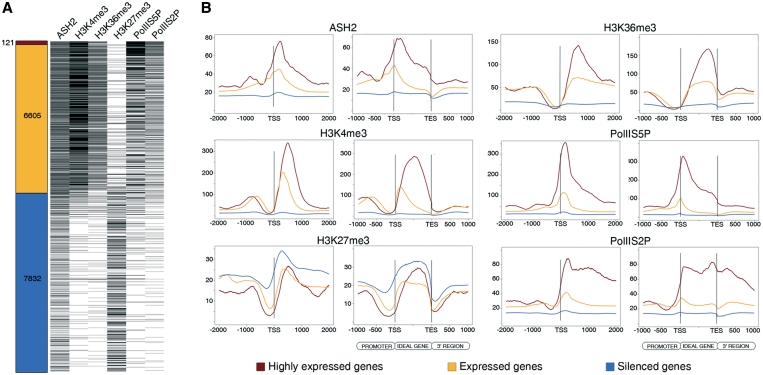
To uncover the relationship between ASH2 and transcription, we took advantage of previously published data on the wing disc transcriptome (14). We classified the genes into three categories according to their expression level: silenced, expressed and highly expressed genes (Figure 2A). We also performed ChIP-Seq analysis using specific antibodies against two modified forms of RNA Polymerase II: serine 5 phosphorylated (PolIIS5P, as a mark of the stalled polymerase at the TSS) and serine 2 phosphorylated (PolIIS2P, as the elongating mark) (Supplementary Table S1). We found 1080 genes containing only PolIIS5P mark (putatively stalled genes), 1452 genes showing only the elongating PolIIS2P mark and 1817 genes with both. As expected, PolIIS5P, like ASH2, peaks around the TSS, and the elongating polymerase (PolIIS2P) is present in actively transcribed regions, coinciding with H3K36me3 (Figure 2B). We uncovered a positive association between gene expression and number of ChIP-Seq reads as previously reported (40). The correlation between the expression level and the ChIP-Seq data (Spearman’s rank correlation coefficient, see ‘Material and Methods’ section) confirmed that the set of expressed genes is clearly enriched in ASH2 (correlation coefficient 0.88), H3K4me3 (0.93), PolIIS5P (0.95), PolIIS2P (0.95) and H3K36me3 (0.93) targets. In contrast, H3K27me3 (–0.84) is primarily associated with silenced genes (Figure 2A).
Figure 2.
Correlation between gene expression levels, ASH2, histone marks and RNA Polymerase II occupancy. (A) Ranking of genes according to their expression levels in the wing imaginal disc. Genes on the Affymetrix array (left) are classified as highly expressed (dark red), expressed (orange) or silenced (blue); target genes of ASH2, histone modifications, PolIIS5P and PolIIS2P are distributed relative to their expression levels (right). (B) Distribution of ChIP-Seq reads of genes belonging to each expression category in the Affymetrix array across the TSS and the idealized gene. Highly expressed genes are shown in red, expressed genes in orange and silenced genes in blue.
Of the genes containing the PolIIS5P modification but not PolIIS2P, 193 are ASH2 target genes silenced in wing disc and belong to categories related to pupal and adult functions such as learning or memory, mating and circadian behaviour (Supplementary Figure S3). This number is likely to be an underestimate due to the stringent normalization protocol followed. On the other hand, most genes showing PolIIS2P alone or both modifications of the RNA Polymerase II are actively transcribed targets of ASH2 and H3K4me3 (Supplementary Figure S4). Finally, 35% of ASH2 target genes are silenced in the wing disc and include GO terms related to signal transduction and metabolism (Supplementary Figure S5). A subset of these silenced genes (1171) possess H3K27me3 but not H3K4me3 (Figure 1B) and is enriched in signal transduction and receptor activity categories (938 genes; Supplementary Figure S6). The identified functional categories suggest that many silenced ASH2 target genes are involved in dynamic biological processes and would thus require the ability to respond rapidly to signals. The phosphorylation state of PolII likely reflects this stalled state of the polymerase in the promoters of the above-mentioned ASH2 target genes.
ASH2 differentially modulates gene expression and may be recruited to active promoters by specific transcription factors
In light of our results, we reanalysed the expression data obtained previously in microarray analyses of ash2 mutant discs (14). By comparing wild-type with ash2I1 mutants, we identified 342 downregulated genes and 368 upregulated genes in wing imaginal discs (see ‘Materials and Methods’ section). A significant fraction of these differentially expressed genes are ASH2 target genes: 294 downregulated genes (85%) and 253 upregulated genes (69%). We next selected those genes that also present the H3K4me3 mark and found 196 ASH2 and H3K4me3 target genes among the downregulated genes and 137 among the upregulated ones. These genes display distinct features in terms of GO categories (Figure 3A). The downregulated set of genes is enriched in development and transcription categories, whereas the upregulated list is enriched in ribosomal and mitochondrial metabolism categories. Downregulated genes and upregulated genes also show significant differences (see ‘Materials and Methods’ section) in gene size (on average 14 068 and 6858 bp, respectively, P-value <10−5), number of exons (5.9 and 3.6 exons, P-value <10−13) and number of alternative forms as annotated in RefSeq (2.3 and 1.3 alternative transcripts, P-value <10−11). Moreover, genes showing a higher expression level in the mutant condition do not correspond to silenced genes in the wild-type disc. Instead, those genes were already expressed and only increased their values in the absence of ASH2. The projection of ASH2 and H3K4me3 reads over the TSS of downregulated and upregulated genes uncovers no differences in their occupancy. The difference in number of reads of H3K4me3 may reflect the number of cells presenting this activating mark in the wing disc (Figure 3B). Taken together, our data suggest that ASH2 action is dependent on interactions with other transcriptional regulators.
Figure 3.
ASH2 direct targets and the transcriptome of ash2 mutants. (A) Gene Ontology (GO) term enrichment of ASH2 and H3K4me3 target genes identified as downregulated (left) and upregulated (right) in ash2 mutant versus wild-type wing discs. The number of genes in each category is shown within the bars. (B) Distribution of ASH2 (left) and H3K4me3 (right) reads over the transcriptional start site (TSS) of upregulated and downregulated genes in the mutant array. In the ASH2 plot, the black box depicts the preferred position of ASH2 over the gene. This region (–370 to +560) was aligned to identify enriched motifs within each list of genes.
To address this possibility and to understand the sequence determinants of ASH2 binding, we proceeded to computationally characterize the regions around the TSS of upregulated and downregulated genes. Using motif discovery tools, we first identified multiple regulatory sites specifically present on each set of sequences. Complementary to this approach, we used TRANSFAC and JASPAR to scan these regions and enrich the initial collection of motifs. We next filtered out those predictions that were not confirmed by phylogenetic footprinting using the genomes of 12 Drosophilae by selecting only those sites conserved in D. pseudoobscura and at least four additional Drosophilids (enrichment calculated in comparison to the total number of conserved sites of each class in the D. melanogaster genome, see ‘Materials and Methods’ section). Roughly, 50% of ASH2-regulated genes presented at least one evolutionarily conserved motif (see Figure 4A). As anticipated, we found the GAGA motif, known to engage the GAGA transcription factor GAF, within the ASH2-binding regions of a significant subset of downregulated genes (58 genes, P-value <10−12). Recently published data from ChIP-on-chip analysis of GAF in Drosophila embryos (39) support our predictions, since 74% of GAGA predicted sites are located within GAF ChIP-on-chip regions. Interestingly, ASH2 binding regions are enriched in E2F-binding sites (42 genes, P-value <10−7) known to recruit E2F transcription factors. A different situation was observed in the set of upregulated genes, where we identified a non-canonical E-box (48 genes, P-value <10−10) and a DRE motif (39 genes, P-value <10−8), known to recruit the DNA replication-related element factor (DREF) (41). We also identified a common motif in both lists of genes (TGGTCACACTG) that is reportedly involved in the recruitment of Mnt/Max complexes (42). In fact, 18 putative Mnt/Max sites overlap with binding regions previously defined by DamID analysis (42) supporting our predictions. One novel motif was additionally identified in each group (Motifs 1 and 2 in Figure 4A). We believe that these novel sequences, together with the transcription factors, participate in ASH2 binding. Again, given the stringent protocol employed to identify these motifs, our results are likely to underestimate the actual number of binding sites. In order to decipher putative cis-regulatory modules underlying ASH2-binding regions, we depict the genes in both sets containing two or more motifs (Figure 4B). We next focused on those cases in which the binding motifs are located at a distance up to 100 bp, thus constituting a plausible regulatory unit. As shown in Figure 4C, we characterized several ASH2-binding regions that manifest specific preferences concerning local positioning and order between the components of each potential module.
Figure 4.
Characterization of ASH2 binding regions. (A) Motifs identified in the region around the TSS of 196 downregulated (left) and 137 upregulated genes (right) in ash2 mutants. The following information is shown for each motif: transcription factor, motif logo, distribution of sites around the TSS, total number of predictions and number of conserved sites in at least five Drosophila species (in parenthesis). (B) Venn diagrams showing the intersection between genes harbouring the characteristic motifs in ASH2 binding regions of downregulated and upregulated sets of genes. (C) Identification of modules constituted of two different motifs in ASH2 binding regions (maximum distance to define a module is 100 bp). A selection of four classes out of the full set of combinations is shown here.
RNA Polymerase II phosphorylated in Ser5 is reduced in ash2 mutants
To clarify the role of ASH2 in transcriptional regulation we performed ChIP-qPCR analysis of individual genes in wild-type and ash2I1 mutant larva and analysed H3K4me3 and RNA Polymerase II modifications. The genomic regions were selected based on the following criteria: ASH2 targets with differential expression in ash2 mutants possessing at least one predicted motif around their TSS. We chose two downregulated genes: engrailed (en) and Cyclin A (CycA); two upregulated genes: mitochondrial Ribosomal protein L40 (mRpL40) and Ribosomal protein L36 (RpL36); and one gene whose expression does not change in the mutant condition used as a control: Sphingosine-1-phosphate lyase (Sply) (Figure 5A and Supplementary Figure S7). We observed that, consistent with the general function of ASH2, H3K4me3 is reduced in all genes in ash2 mutant flies, independently of their transcriptional state. Strikingly, we did not detect any change in PolIIS2P, but a decrease in PolIIS5P was observed in the TSS of the three classes of genes. A possible disengagement of PolIIS2P along the gene should be discounted since no change in its occupancy was observed when performing ChIP analysis on the 3′ gene region (Figure 5A). To confirm these observations we performed immunostaining on polytene chromosomes. As shown in Figure 5B, there was a general decrease of PolIIS5P on ash2 mutant larva in comparison to wild-type. In agreement with our ChIP experiments, PolIIS2P does not show clear differences between mutant and control larva.
Figure 5.
Phosphorylated forms of RNA Polymerase II in ash2 mutants. (A) ChIP analysis of wild-type (light grey) and ash2I1 (dark grey) larva with H3K4me3 (left), PolIIS5P (centre) and PolIIS2P (right) antibodies. For H3K4me3 and PolIIS5P, primers were designed across the TSS of two downregulated genes (en and Cyclin A), two upregulated genes (mRpL40 and RpL36) and one control gene (Sply) in ash2I1 flies in comparison to wild type. For PolIIS2P, primers from the TSS and the 3′ region of the same genes were used. Real-Time PCR results were normalized against the input sample and are depicted as percentage of the input. Error bars represent the Standard Error of the Mean. (B) Polytene chromosomes of wild-type (upper figure) and ash2I1 mutant (lower figure) third instar larva. Chromosomes were stained with PolIIS5P (left) and PolIIS2P (right) antibodies (in green and white). DNA was stained with DAPI (in red).
To further analyse RNA Polymerase II (PolII) occupancy over these genes, we performed ChIP-qPCR experiments with an antibody that recognizes total PolII and calculated the ratio between TSS and 3′ region (Supplementary Figure S8). Those genes that exhibit a clear enrichment of the polymerase at TSS in wild-type flies (en and RpL36; TSS/3′ ratio >1) display a decrease in PolII at the TSS relative to 3′ region in ash2 mutants. We detected a slight decrease of the TSS/3′ ratio in the case of CycA, mRpL40 and Sply (ratio ∼1 in wild-type flies). The uniform distribution of PolII along these genes might mask a reduction at the TSS in ash2 mutants. Furthermore, the presence of total PolII occupancy is likely to be underestimated, since the antibody used primarily recognizes the unphosphorylated form of the polymerase (43). Taken together, these results support the idea that the mechanism of action of ASH2 in terms of RNA Polymerase II modifications does not differ between developmentally regulated and housekeeping genes. Analysis of additional control mechanisms, such as RNA capping or splicing, require further experimentation.
DISCUSSION
Work using various model organisms and cultured cells has provided high-resolution profiles of histone modifications and transcription factor binding across different genomes (40,44,45). In this study, we use direct sequencing of ChIP DNA from wing disc to analyse ASH2 function. Because the cell composition of isolated wing disc tissue is rather homogeneous, we have been able to set apart several attributes. First, ASH2 occupancy correlates with the presence of phosphorylated forms of RNA Polymerase II and activating histone marks in expressed genes. On the other hand, we cannot dismiss a direct role for ASH2 in gene repression as well, as ASH2 also targets silenced genes. In support of this, ASH2-interacting proteins HCF-1 and dMyc are involved in both transcriptional activation and repression (14,18,46). Alternatively, silenced ASH2 target genes could be arrested in an intermediate ready-to-go state of transcription, which may be activated by external signals. Second, our results agree with previous observations in Drosophila and Xenopus embryos, where dually marked domains do not seem to be a common feature (39,44). It has been reported that bivalently marked chromatin, containing both H3K4 and H3K27 trimethylation, is a hallmark of developmentally regulated silenced promoters in mammalian embryonic stem cells (47,48). In contrast, these marks can be coupled to the differential expression pattern of several genes throughout the wing disc, therefore indicating the presence of each individual mark in different cells. A recent report using a similar genome-wide approach in undifferentiated cell-enriched Drosophila testis reveals that differentiation-associated genes are also linked with monovalent modifications (49). Third, we use ASH2 binding together with activating marks of transcription as a powerful tool to identify previously unannotated genes.
The actively transcribed genes in the wing disc are occupied by nucleosomes with histone modifications that are hallmarks of both initiation and elongation, as described in human cells (50). We have uncovered a positive correlation between activating marks of transcription (both H3K4me3 and H3K36me3) and ASH2 occupancy. Our study has also determined that ASH2 contributes to H3K4me3 in nearby nucleosomes. H3K4me3 is associated with the TSS of active genes, whereas H3K27me3 spreads over large regions of chromatin to promote silencing and H3K36me3 is found in actively transcribed regions (40,51,52). Only genes containing H3K36me3 undergo further elongation and produce mature transcripts [reviewed in (53)].
Transcriptional regulation is a multistep process controlled by a large complex machinery at the level of recruitment, initiation, pausing and elongation of RNA Polymerase II (53,54). A series of recent genome-wide studies indicate that many developmental and inducible genes, prior to their expression, contain RNA Polymerase II bound predominantly in their promoter proximal regions in a stalled state (53,55,56). Nevertheless, not only silenced genes show an enrichment of the RNA Polymerase II density at their TSS as the stalled polymerase is also present at this region in active genes (57). The presence of ASH2 and H3K4me3 together with PolIIS5P at the TSS of expressed genes is consistent with previous reports proposing that promoter-proximal stalling serves not only to fully repress but also to attenuate transcription of active genes. As recently described, transient stalling of polymerase is a general feature of early elongation, even in highly active genes (58).
The analysis of ash2 mutant flies indicates that ASH2 is performing its canonical function promoting H3K4me3, regardless of the effect on the transcriptional state of its target genes and the context specificity of its recruitment to promoters. In light of the results obtained with RNA Polymerase II modifications in the mutants, we conclude that ASH2 influences different aspects of transcription. The specific binding motifs identified in differentially regulated genes, together with the co-occupancy of ASH2 and PolIIS5P at the TSS, suggests a role in transcription initiation. Nevertheless, the reduction of PolIIS5P in mutant flies points to a fast escape from stalling in the absence of ASH2.
Distinct sets of accessory factors are associated with polymerase stalling and its escape from this state, acting either by direct interaction with RNA Polymerase II, or by manipulating the chromatin environment (59). Among these factors, there are proteins associated with polymerase stalling, such as the DRB sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF), and others that contribute to escape from stalling, such as the positive transcription-elongation factor-b (P-TEFb) complex and the general transcription factors TFIIS and TFIIF [(53) and references herein]. It remains to be elucidated whether ASH2 interacts directly with some of these factors. However, NELF and GAF have been found linked to promoter-proximal pausing at many genes in Drosophila (60). A connection between ASH2 and polymerase stalling in developmental genes could, therefore, be envisioned through GAF, since it is known that GAF is a recruiter of PcG and trxG complexes to DNA (8). In fact, about half of the downregulated genes in ash2 mutants presenting GAGA sites are NELF targets (data not shown). Furthermore, it has been recently reported that c-Myc regulates RNA Polymerase II pause release by recruiting P-TEFb to its target genes (61), and it is known that ASH2 interacts with Myc in flies (19). The enrichment of Ebox and Mnt/Max motifs found in upregulated genes in ash2 mutants points to a function of ASH2 through Myc in their transcriptional regulation. A subset of these motifs was characterized in H3K4me3 regions by Schuettengruber et al. (39). We have been able to associate these motifs with downregulated and upregulated genes in ash2 mutants, suggesting differential transcriptional regulation.
Several effector proteins that can bind to H3K4me3 determine the functional outcome of this histone modification. The activities of these binding proteins range from activation and repression of transcription, chromatin remodelling or splicing efficiency among others (62). An additional role for ASH2 during transcript elongation and maturation should not be excluded. Indeed, it has been suggested that methylated H3K4 serves to facilitate the competency of pre-mRNA maturation through the bridging of spliceosomal components (63). The fact that downregulated and upregulated genes in ash2 mutants display clear differences in size and genomic organization (gene size, alternative isoforms and number of exons) suggests they may be regulated in a different way during transcription and processing of RNA, as previously suggested [for review see (64)]. Finally, recent reports indicate an association of RNA Polymerases II and III at promoter regions of housekeeping genes (65–67) and a recruitment of RNA Polymerase III through Myc interacting with the cofactor BRF has also been described (68). However, preliminary experiments discard the implication of other polymerases in the transcription of these housekeeping genes in the absence of ASH2. Taken together, our results support a model in which an ASH2-containing complex would act at different levels of transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación (GEN2006-28564-E, BMC2006-07334, ACI2009-0903 and CSD2007-00008), Juan de la Cierva fellowship (to E.B.); Universitat de Barcelona, APIF fellowship (to A.C.); NIH (to M.S). Funding for open access charge: ACI2009-0903 and Universitat de Barcelona.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Guigó and J.F. Abril for kindly providing access to their computer facilities, A. Mazo, M. Buschbeck and C. Byars Baker for insightful suggestions and A. Mateo for technical support. We also thank the Ultrasequencing Unit of the CRG (Barcelona, Spain) and the Functional Genomics Core Facility of the IRB (Barcelona, Spain).
REFERENCES
- 1.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 6.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 10.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 11.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 12.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beltran S, Angulo M, Pignatelli M, Serras F, Corominas M. Functional dissection of the ash2 and ash1 transcriptomes provides insights into the transcriptional basis of wing phenotypes and reveals conserved protein interactions. Genome Biol. 2007;8:R67. doi: 10.1186/gb-2007-8-4-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 16.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl Acad. Sci. USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamson AL, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angulo M, Corominas M, Serras F. Activation and repression activities of ash2 in Drosophila wing imaginal discs. Development. 2004;131:4943–4953. doi: 10.1242/dev.01380. [DOI] [PubMed] [Google Scholar]
- 24.Beltran S, Blanco E, Serras F, Perez-Villamil B, Guigo R, Artavanis-Tsakonas S, Corominas M. Transcriptional network controlled by the trithorax-group gene ash2 in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2003;100:3293–3298. doi: 10.1073/pnas.0538075100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 33.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergman CM, Carlson JW, Celniker SE. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 35.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 38.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 39.Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Hyun J, Jasper H, Bohmann D. DREF is required for efficient growth and cell cycle progression in Drosophila imaginal discs. Mol. Cell. Biol. 2005;25:5590–5598. doi: 10.1128/MCB.25.13.5590-5598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009;10:1213–1219. doi: 10.1038/embor.2009.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev. Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu SG, Fire A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma. 2010;119:73–87. doi: 10.1007/s00412-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallant P. Drosophila Myc. Adv. Cancer Res. 2009;103:111–144. doi: 10.1016/S0065-230X(09)03005-X. [DOI] [PubMed] [Google Scholar]
- 47.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 48.Herz HM, Nakanishi S, Shilatifard A. The curious case of bivalent marks. Dev. Cell. 2009;17:301–303. doi: 10.1016/j.devcel.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 49.Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, Chen X. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11:R42. doi: 10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 52.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 53.Wu JQ, Snyder M. RNA polymerase II stalling: loading at the start prepares genes for a sprint. Genome Biol. 2008;9:220. doi: 10.1186/gb-2008-9-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- 56.Price DH. Poised polymerases: on your mark … get set … go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 60.Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vermeulen M, Timmers HT. Grasping trimethylation of histone H3 at lysine 4. Epigenomics. 2010;2:395–406. doi: 10.2217/epi.10.11. [DOI] [PubMed] [Google Scholar]
- 63.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeitlinger J, Stark A. Developmental gene regulation in the era of genomics. Dev. Biol. 2010;339:230–239. doi: 10.1016/j.ydbio.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 65.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc. Natl. Acad. Sci. USA. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steiger D, Furrer M, Schwinkendorf D, Gallant P. Max-independent functions of Myc in Drosophila melanogaster. Nat. Genet. 2008;40:1084–1091. doi: 10.1038/ng.178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.