Abstract
The hexameric Escherichia coli RNA chaperone Hfq (HfqEc) is involved in riboregulation of target mRNAs by small trans-encoded RNAs. Hfq proteins of different bacteria comprise an evolutionarily conserved core, whereas the C-terminus is variable in length. Although the structure of the conserved core has been elucidated for several Hfq proteins, no structural information has yet been obtained for the C-terminus. Using bioinformatics, nuclear magnetic resonance spectroscopy, synchrotron radiation circular dichroism (SRCD) spectroscopy and small angle X-ray scattering we provide for the first time insights into the conformation and dynamic properties of the C-terminal extension of HfqEc. These studies indicate that the C-termini are flexible and extend laterally away from the hexameric core, displaying in this way features typical of intrinsically disordered proteins that facilitate intermolecular interactions. We identified a minimal, intrinsically disordered region of the C-terminus supporting the interactions with longer RNA fragments. This minimal region together with rest of the C-terminal extension provides a flexible moiety capable of tethering long and structurally diverse RNA molecules. Furthermore, SRCD spectroscopy supported the hypothesis that RNA fragments exceeding a certain length interact with the C-termini of HfqEc.
INTRODUCTION
In bacteria small trans-encoded regulatory RNAs (sRNAs) can modulate target gene expression in response to various stress conditions, a mechanism known as riboregulation. The majority of functionally characterized Escherichia coli sRNAs act as negative regulators by preventing ribosome loading onto the target mRNA through base-pairing with or in the vicinity of the ribosome binding site. As a result, the respective mRNA is prone to rapid decay [for review see refs (1–3)]. Positive regulation by sRNAs has been reported less frequently. In this case the sRNAs can act by an ‘anti-antisense’ mechanism and melt intramolecular secondary structures, which impede ribosome binding (4–6). Alternatively, an mRNA can act as a decoy for a sRNA leading to translation of another mRNA that is negatively controlled by this sRNA (7). Similarly, a sRNA can protect a homologous sRNA from degradation, which in turn results in activation of a target mRNA by the latter (8). A number of bacterial sRNAs associate with the global regulator Hfq (9) and often require this protein for function (10,11). Part of the Hfq requirement may be ascribed to Hfq-mediated stabilization of sRNAs (12–15). However, as many sRNAs display imperfect and non-contiguous target complementarity, the requirement for Hfq in riboregulation has mainly been attributed to its RNA chaperone function, which is believed to facilitate the interaction between the sRNA and the cognate mRNA (13,16–19).
Hfq belongs to the eukaryotic and archaeal family of Sm- and Sm-like proteins (13,19,20). Hfq orthologues have been found in a number of prokaryotes. The Hfq proteins of different organisms display an evolutionarily conserved core consisting of amino acid residues 7–66, whereas there is considerable variation at the C-terminal extension (20). The first X-ray structure of the Staphylococcus aureus Hfq protein (HfqSa) in complex with a short RNA oligonucleotide, was reported by Schumacher et al. (21). Since then 13 unique structures of Hfq-proteins from seven different organisms have been deposited in the Protein Data Bank [S. aureus (HfqSa; amino acids 1–77) 1KQ1, 1KQ2 (21); E. coli (HfqEc; amino acids 1–72) 1HK9 (22), 3GIB (amino acids 2–69) (23); Pseudomonas aeruginosa (HfqPae; amino acids 1–82) 1U1T, 1U1S (24), 3M4G, 3INZ (25); Methanococcus jannaschii (HfqMj; amino acids 1–71) 2QTX (26); Synechocystis sp. (HfqCSyn; amino acids 1–70) 3HFO; Anabaena sp. (HfqCAna; amino acids 1–72) 3HFN (27); Bacillus subtilis (HfqBs; amino acids 1–78) 3HSB (28), and 3HSA (to be released)].
The study by Schumacher et al. (21) showed that HfqSa forms homo-hexamers with a central pore and that a short poly(U) oligoribonucleotide was bound in a circular conformation along the inner basic rim. A mutational analysis performed with E. coli Hfq (HfqEc) has later provided evidence that sRNAs bind to the same positively charged proximal face of HfqEc (29). Unlike uridine-containing sequences, a recent structural study revealed binding of a poly(A) tract to the distal face of HfqEc using six tripartite binding motifs (23). Interestingly, a poly(A) tract in the 5′UTR of rpoS mRNA was shown to be relevant for Hfq-mediated riboregulation by the sRNA DsrA (30), and a mutational analysis by Mickulecky et al. (29), suggested that rpoS mRNA contacts the distal site. Nevertheless, a truncated HfqEc protein containing the conserved common core (amino acids 1–65) was deficient in binding to the full-length 5′-untranslated region (5′-UTR) of rpoS, suggesting that mRNA contacts to the distal site are not sufficient for stable binding of this ligand (18). Further studies provided genetic and biochemical evidence that the C-terminus of HfqEc is additionally required for binding of rpoS and other mRNAs, i.e. it constitutes a RNA interaction surface with specificity for longer RNAs (18).
Only the Hfq proteins of γ- and β-proteobacteria have an extended C-terminus, whereas some Gram-positive bacteria including S. aureus have Hfq proteins with short C-terminal extensions (20). Interestingly, Hfq-mediated riboregulation of mRNAs by sRNAs has been mainly demonstrated in bacterial species bearing Hfq proteins with C-terminal extensions (31). As yet, structural data on the C-terminus of HfqEc are lacking. Previous analyses predicted that the C-terminus of HfqEc is structurally disordered (18,32), which is a hallmark of RNA chaperones (33).
Here we have used an integrated approach employing bioinformatics, biophysical and structural studies to analyze the dynamic properties, and to determine the conformation and shape of the C-terminal extension of HfqEc. Collectively, these studies revealed that the C-terminus is intrinsically disordered and flexible, which appears to contribute to the interactions of Hfq with RNA substrates.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The E. coli strains BL21 DE3 (Novagen), AM111 (MC4100 hfq1::Ω) (34) and the corresponding F′ (lacIq) variant (35) have been described. They were grown at 37°C in Luria–Bertani (LB) medium or in M9 medium supplemented with 0.2% glucose, 2 mM MgSO4, 0.1 mM CaCl2 and 10 µg/ml thiamine. For RpoS synthesis the cells were grown in LB medium at 22°C. Where appropriate, ampicillin (100 µg/ml), kanamycin (25 µg/ml), tetracycline (30 µg/ml) or chloramphenicol (15 µg/ml) was added to the medium.
Construction of plasmids and expression of hfq variants
The plasmids pUH5Hfq and pUH65 used for synthesis of full-length HfqEc and HfqEc65, respectively, have previously been described (18,36). In these plasmids, the corresponding hfq gene was placed under transcriptional control of the lac promoter in plasmid pUC19 (New England Biolabs). To avoid negative translational autoregulation (37), the 5′-UTR of the hfq gene was replaced by the ribosome binding site of phage T7 gene 10 using a PCR approach.
The plasmids pUH85 and pUH75 used for synthesis of HfqEc85 and HfqEc75 were constructed as follows. The forward primer (5′-GCTCTAGAAATATAATAGTTTAACTTTAAGAAGGAGATATACATATGGCTAAGGGGCAATCTTTACAAGATCCGTTCCT-3′) contained the sequence derived from plasmid pET22b (underlined; Novagen) including the XbaI site and the Shine and Dalgarno sequence of gene 10 abutted with the first 35 nucleotides of the hfq gene (given in bold). The reverse primers (5′-TTGAATTCATTAATGATGGTAGTTACTGCTGGTACCGCCACC-3′) and (5′-TTGGATCCCTGCAGTTACTAGGCGTTGTTACTGTGATGAG-3′) contain two stop codons after the triplet encoding the Hfq-specific amino acids His85 and Ala75 as well as an EcoRI and BamHI site, respectively. The resulting PCR products were cleaved with XbaI and EcoRI/BamHI and then cloned into the same sites of plasmid pUC19 yielding plasmids pUH85 and pUH75, respectively.
The plasmids pProSA and pProBS used for purification of the S. aureus and B. subtilis Hfq proteins, respectively, have been described (18). For construction of plasmids pAH75 and pAH85, the corresponding hfq segments were sub-cloned as PvuII fragments from pUH75 and pUH85 and then inserted into the EcoRV and NruI sites of plasmid pACYC184. Construction of plasmids pAHfq and pAH65 is described (18).
Full-length HfqEc was produced in E. coli BL21 DE3 (Novagen) and the S. aureus, B. subtilis and truncated E. coli Hfq proteins were produced in the hfq deficient strain AM111F′ (lacIq). One liter of Luria–Bertani medium was supplemented with 100 µg/ml of ampicillin, 50 µg/ml kanamycin and 15 µg/ml tetracycline where appropriate. Protein for NMR-studies was obtained from cells grown in M9-medium, supplemented with 15NH4Cl (Isotec) and 13C-glucose (Isotec). The cells were grown to an OD600 of 0.6–0.8 and the expression of the respective hfq variants was induced by addition of 0.5 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sigma). After 4 h the cells were harvested by centrifugation (4,000 g/15 min).
In vitro synthesis of RNA
For hfq126 mRNA synthesis plasmid pUhfqwt (37) cleaved with AflIII was used as a template for in vitro transcription with the T7-MEGAshortscript kit (Ambion). The run-off transcripts were purified on 6% polyacrylamide-8M urea gels following standard procedures. The mRNA concentration was determined spectrophotometrically measuring absorption at 260 nm. The DsrA34 RNA, comprising the nucleotides 26–59 of the DsrA-sequence was purchased from Dharmacon, USA.
Purification of Hfq proteins
The Hfq proteins are heat-stable, and the purification therefore involved an initial fractionation by heating and subsequent processing by FPLC (Äkta, GE-Healtcare). The final purification-scheme for E. coli Hfq and variants thereof comprised four steps: (i) An initial washing step by Ni2+-affinity (HfqEc and HfqEc85) or hydrophobic interaction chromatography (HfqEc75, HfqEc65); (ii) a filtration over an anion-exchange column to remove nucleic acids; (iii) a concentrating step by respectively Ni2+-affinity or HIC; and (iv) a final step of size-exclusion chromatography. The detailed procedure is described in Supplementary Data. Purification of HfqBs and HfqSa was performed as previously described (18).
Western blot analysis
The RpoS, Hfq and L14 (loading control) protein levels were determined in strain AM111 harboring plasmids pACYC184 (control), pAHfq, pAH65, pAH75 and pAH85, respectively. The strains were grown at 22°C in LB medium to OD600 of 0.5, at which time 2 ml aliquots were withdrawn, pelleted and boiled in protein sample buffer. Equal amounts of total protein were separated on 12% SDS–polyacryamide gels and blotted to a nitrocellulose membrane. The blots were blocked with 5% dry milk in TBS buffer, and then probed with anti-RpoS (NeoClone, Madison), anti-Hfq (Pineda, Berlin) and anti-L 14 (provided by Dr I. Moll) antibodies to detect the RpoS, Hfq and L14 proteins, respectively. The antibody–antigen complex was visualized as described (38).
FRET assays
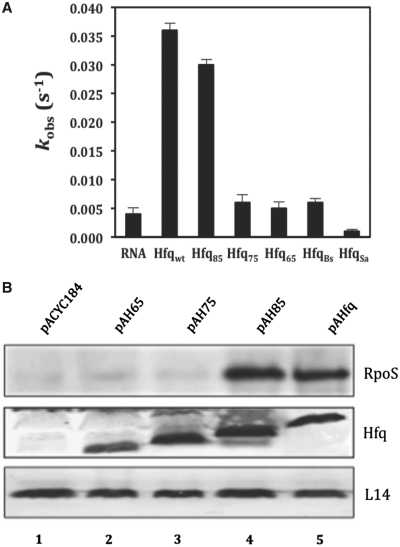
This method is described in more detail in refs (32,39). Two complementary, fluorophore-tagged RNA 21mers [Cy5-5′-AUGUGGAAAAUCUCUAGCAGU-3′ (Cy5-21R+) and Cy3-5′-ACUGCUAGAGAUUUUCCACAU-3′ (Cy3-21R−)] were used in the annealing experiment. The tagged RNA oligonucleotides were purchased from VBC-Biotech (Vienna, Austria). Using a Tecan GENios Pro microplate reader, the first oligoribonucleotide was injected into wells with or without Hfq protein (100 nM final Hfq-hexamer concentration), and the measurement was started with the injection of the second oligoribonucleotide. The reaction was performed in annealing buffer (50 mM Tris–HCl pH 7.5, 3 mM MgCl2 and 1 mM DTT) at 37°C. The final concentration of the RNAs was 5 nM, in a sample volume of 40 µl. The reaction was allowed to proceed for 180 s, and with Cy3 excited, donor and acceptor dye fluorescence emissions were measured once every second. The time-resolved ratio of the fluorescence emissions (FRET index FCy5/FCy3) was normalized to 1 at t180s and least-square fitted with Prism 4.03 (GraphPad Software Inc., San Diego, CA, USA) with the second-order reaction equation for equimolar initial reactant concentrations: y = A [1 − (kobs t + 1)−1], where y = fraction annealed, kobs = observed annealing reaction constant, A = maximum reaction amplitude. The observed reaction constant kobs shown in Figure 1A was calculated as the average of three individually fitted reactions.
Figure 1.
The region between amino acids 75 and 85 is instrumental for RNA binding and in vivo function of HfqEc. (A) RNA annealing activities of HfqBc, HfqSa, full-length HfqEc and C-terminally truncated variants thereof. Five nanomolar single-stranded, complementary 21-nt-long oligoribonucleotides with fluorophores at their 5′-end were annealed at 37°C in the absence or presence of 100 nM protein. Relative fluorescence resonance energy transfer was calculated as the ratio of acceptor to donor fluorescence (FCy5/FCy3) as described in ‘Materials and Methods’ section. The time-resolved curves were least-square fitted with the second-order reaction equation for equimolar initial reactant concentrations: y = A[1 − (kobs t + 1)−1]; y = fraction annealed, kobs = observed annealing reaction constant, A = maximum reaction amplitude. The reaction rate kobs was calculated from the average of three independent experiments. (B) RpoS protein synthesis in the E. coli hfq- strain AM111 harboring plasmids pACYC184 (control, lane 1), pAH65 (encoding HfqEc65; lane 2), pAH75 (encoding HfqEc75; lane 3), pAH85 (encoding HfqEc85; lane 4) and pAHfq (encoding full-length HfqEc; lane 5), respectively. The western-blot analysis was carried out with anti-RpoS, anti-Hfq antibodies and with antibodies against ribosomal protein L14 (loading control) using equal amounts of total cellular protein as described in ‘Materials and Methods section’. Only the relevant sections of the western-blot showing the RpoS-, Hfq- and L14-specific signals are shown.
SRCD Spectroscopy
SRCD spectra were collected on the CD1 beamline at the ISA facility, University of Aarhus, Denmark. For each protein, the extinction coefficient at 280 nm {HfqEc ∼0.34, HfqEc85 ∼0.41, HfqEc75 ∼0.30, HfqEc65 ∼0.35 [(mg/ml)−1 × cm−1]} was calculated from its amino acid sequence (http://www.scripps.edu/∼cdputnam/protcalc.html) and the concentration derived from the A280 measured on a Nanodrop 1000 spectrophotometer immediately prior to the SRCD spectrum being acquired. For the complexes with RNA, the protein concentration was assessed prior to RNA addition to allow for a 1:1 molar ratio of Hfq hexamer to RNA. The protein and RNA was mixed at elevated temperature (∼60°C), and incubated for 5 h at 65°C, and then allowed to slowly cool to room temperature. Subsequently the complexes were centrifuged for 1 h at 10 000 g, at room temperature.
Each protein sample (concentrations of ∼5 mg/ml) was loaded into a 0.015 mm path length quartz Suprasil demountable cell (Hellma Ltd) and three spectra were collected at 20°C over the wavelength range from 280 to 170 nm, with a 1 nm step size and a 2 s dwell time. Data processing was carried out using the CDTool software (40). Replicates scans were averaged, subtracted from the average of three baseline spectra (either buffer or buffer plus RNA), smoothed with a Savitsky–Golay filter, and calibrated against a spectrum of camphoursulphonic acid that was obtained at the beginning of the data collection (41). The spectra were converted to delta epsilon units using mean residue weight values [calculated as molecular mass/number of residue−1] of 110.6, 114.3, 113.7 and 112.5 for the HfqEc, HfqEc85, HfqEc75, and HfqEc65 constructs, respectively, based on molecular weights of 11166.3, 7317.5, 8417.7, 9415.7, and numbers of residues of 102, 64, 75, 85. The low wavelength cutoff of the data was determined as previously described (42). Data were analysed with the DichroWeb analysis server (43) and are reported as the averaged results (±1 SD) from the CONTINLL (44,45), SELCON3 (46) and CDDSTR (47) algorithms, using the SP175 reference dataset (48). The NRMSD is a goodness-of-fit parameter (49) calculated for the CONTINLL-calculated structure and the experimental data.
NMR
All NMR experiments were performed at 310 K on a Varian Inova 600 MHz spectrometer equipped with 5 mm triple resonance cryo-probe and pulsed field gradients, and a Varian Inova 800 MHz spectrometer equipped with 5 mm conventional triple resonance probe equipped with pulsed field gradients. NMR spectra were processed with NMRPipe (50) and analyzed with Sparky software. The protein samples for NMR-experiments were all prepared by size-exclusion chromatography in 50 mM Na-PO4 pH ∼7.2, 200 mM NaCl and concentrated to ∼1 mM HfqEc65 (with respect to the monomer). Samples were supplemented with ∼10% (v/v) 2H2O to provide the deuterium signal for the field-frequency lock, as well as 0.1–0.2% (w/v) NaN3 to inhibit bacterial growth. Backbone signal assignment for the C-terminally truncated mutant HfqEc65 was obtained by a suite of standard (sensitivity enhanced) three-dimensional (3D) triple resonance experiments, acquired pairwise: HNCA (51)/HN(CO)CA (52), HNCACB (53), HNCO (54) were recorded for sequential backbone chemical shift assignment. 15N relaxation times (T1, T2) were measured using well-known gradient sensitivity-enhanced 2D methods with 1H detection (55,56).
SAXS
Small angle X-ray scattering experiments were acquired at beamline X33 (57,58) at the DORIS III synchrotron storage-ring (DESY, Hamburg, Germany), using a MAR345 image-plate detector and X-ray wavelength of λ = 1.5 Å, with a sample to detector distance of 2.7 m. This setup covers a range of momentum transfer 0.12 < s < 0.45 nm−1 (s = 4π sinθ/λ, where 2θ is the scattering angle). The data were processed in the ATSAS program-package (59). Data were acquired at 37°C from four concentrations of the full-length HfqEc (18.5, 9.3, 4.6, 2.3 mg/ml), and similar concentration of the C-terminally truncated core-construct HfqEc65 (19.0, 9.5, 4.8, 2.4 mg/ml). Protein concentration was measured with 2 µl samples on a Nanodrop 1000 UV/vis spectrophotometer at 280 nm, immediately prior to data-acquisition. The data were averaged and normalized to the intensity of the incident beam prior to subtraction of buffer scattering. The difference data curves were scaled, merged and initial analysis performed using PRIMUS (60). Invariants were derived by standard approaches for comparison (61). The intrinsic scattering (I0) and radius of gyration (Rg) was derived by Guinier analysis for the data range s < 1.3/Rg, approximating I(s) = I0 exp[−(s Rg)2/3]. The reverse transform program GNOM (62) was employed for calculation of the distance-distribution function, under the assumption of correct estimation of the particle maximum dimension (Dmax). The program also outputs a value for I0 and Rg. The particle excluded volume was calculated from the Porod Equation (63):
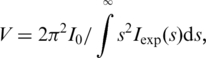 |
(1) |
where Iexp(s) is the experimental data.
In addition, the program Dammif (64) run without symmetry restraints outputs the particle excluded volume. The solute molecular mass was further evaluated by the standard approach of comparison of the intrinsic forward scattering (I0) with that recorded from a reference solution of known molecular mass and concentration. In the present study a single sample of 3.68 mg/ml bovine serum albumin (Mr = 66.4 kDa) was used.
Low-resolution shape-models of the SAXS data were generated ab initio using the program Dammif (64). The program represents the protein as a collection of densely packed dummy-residues inside a sphere with the diameter DMAX. Each dummy-residue belongs either to the particle or the solvent, and the shape is described by a binary string of length M. Starting from a random string, simulated annealing is employed to search for a compact model that fits the shape scattering curve I(s) to minimize discrepancy χ2:
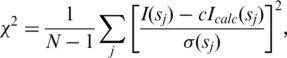 |
(2) |
where N is the number of experimental points, c is a scaling factor and Icalc(sj) and σ(sj) are the calculated intensity and the experimental error at the momentum transfer sj, respectively.
The program BUNCH (65) employs a combined rigid body and ab initio modeling approach to search for a spatial arrangement of domains with known high-resolution structure, and probable conformations of flexible domains represented as dummy-residues. Utilizing the known structure of a HfqEc65 monomer (amino acids 6–65), the program was run employing a 6-fold symmetry retaining the hexameric organization of the Hfq core. Theoretical scattering from hexameric Hfq core encompassing residues 6–65 (PDBid: 1HK9) was calculated using the program CRYSOL (66). The C-termini (6 × 37 residues) were represented as chains of dummy residues connected to Ser65 of Hfq. The program employs a simulated annealing procedure to manipulate the local conformation of the chains representing the termini, to minimize the χ2-discrepancy as described in Equation (2). The program allows for specifying restraints for the conformation of the flexible chains. In present study, the disordered nature of the C-terminus was represented by lowering of the restraint on the dihedral angle between adjacent dummy residues.
In order to test the packing of the C-terminus into the groove formed by amino acids 35–55 of one subunit and the β-sheet of the adjacent subunit as observed in the crystal structure of HfqEc65 (amino acids 6–65), we calculated rigid body models using the program BUNCH applying distance restraints on His70. When placing His70 in the groove, to force the C-termini into the groove we observe a decrease in the quality of the fit. When placing His70 on the outside of the N-terminal helix, to constrain the C-terminus in the vicinity of the conserved patch of charged residues present in Hfq (22), no significant change to the quality of the fit was observed. All graphical representations were made using MacPymol (67).
RESULTS
Annealing deficiency of Hfq proteins naturally devoid of a C-terminus and of C-terminally truncated E. coli Hfq variants
We have recently shown using fluorescence resonance energy transfer assays (FRET) that full-length HfqEc stimulated annealing of two complementary RNA-oligonucleotides with a rate constant that was ∼3-fold higher than that observed for HfqEc65, comprising only the evolutionarily conserved core region of Hfq (amino acids 1–65) (18). When two non-complementary RNA oligonucleotides were used, no significant increase in the FRET signal was discernable in the presence of HfqEc65, whereas an increase in FRET was observed for HfqEc. In contrast to HfqEc, the dual rate binding constant kdb for HfqEc65 was ∼10-fold lower, suggesting that HfqEc65 is severely impaired in annealing of two RNA oligonucleotides (18).
To test whether Hfq proteins naturally lacking a C-terminal extension are deficient or impaired in annealing activity, FRET assays were performed with the Hfq proteins of B. subtilis (HfqBs; 73 amino acids) and S. aureus (HfqSa; 77 amino acids). When compared to the absence of protein and similarly to HfqEc65, neither HfqBs nor HfqSa stimulated annealing of the complementary oligonucleotides (Figure 1A), again suggesting that the evolutionarily conserved core is not sufficient to support duplex formation of two RNAs.
With the aim to identify sub-sequences in the C-terminus of HfqEc that support RNA binding, we tested the ability of two C-terminally truncated HfqEc variants, HfqEc75 (amino acids 1–75) and HfqEc85 (amino acids 1–85) for their capacity to mediate low temperature (20°C) translational activation of rpoS mRNA by the sRNA DsrA during exponential growth (68). Under these conditions, HfqEc65, as shown before (18), and HfqEc75 were defective in stimulating DsrA-mediated RpoS synthesis, whereas the stimulation by HfqEc85 was indistinguishable from that of full-length HfqEc (Figure 1B). To verify these experiments the two C-terminally truncated HfqEc variants, HfqEc75 (amino acids residues 1–75) and HfqEc85 (amino acids residues 1–85) were analyzed in FRET assays with regard to their annealing activity. Interestingly, HfqEc85 was proficient in stimulating annealing of the two RNA ligands, whereas HfqEc75 exhibited the same low annealing activity as HfqEc65 (Figure 1A). As HfqEc75 was defective in both, the FRET and in vivo assay, these studies implicated amino acids residues 75–85 in RNA binding.
Bioinformatics analyses predict an intrinsically disordered, solvent exposed C-terminus of HfqEc
The C-terminus of HfqEc (amino acids 66–102) contains a low portion (17%) of ‘order promoting’ residues (Asn, Cys, Ile, Leu, Phe, Trp, Tyr and Val), and a high fraction (64%) of ‘disorder promoting’ residues (Ala, Arg, Gln, Glu, Gly, Lys, Pro and Ser). As observed in several bioinformatic studies (69,70) this is a fingerprint of intrinsically disordered proteins. For comparison, in folded globular proteins, these values are 36% for ‘order’ and 47% for ‘disorder promoting’ residues, which gives a ratio of ∼0.76. Interestingly, in the first half of the HfqEc C-terminus (amino acids 68–85) this ratio is ∼0.63, whereas for the last half we calculate ∼0.08. Overall this indicated that the C-terminal extension, which was indispensable for function in the assays used in this study (Figure 1), is predicted to be disordered.
The metaPrDOS algorithm, which generates disorder predictions based on a consensus principle between the individual predictors PrDOS, DISOPRED2, DisEMBL, DISPROT-VSL2P, DISpro and IUPred (http://prdos.hgc.jp/meta) (71), predicts the C-terminal moiety comprising residues 69–102, to be intrinsically disordered, with high signals (above 0.70) for amino acids 73–102 (Supplementary Figure S1). Secondary-structure prediction servers Jpred (72) and PSIPRED (73) do not detect secondary-structure elements in the C-terminal segment (66–102) of HfqEc (data not shown).
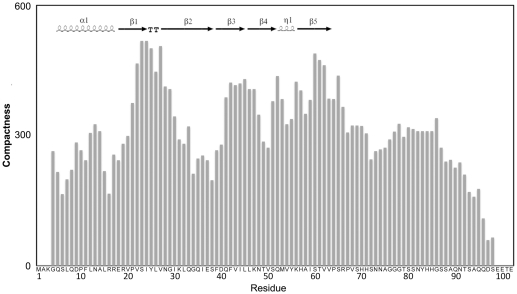
In addition, a protein meta-structural analysis was performed, which extracts two parameters for each residue on the basis of the protein primary sequence, one related to compactness and the other to secondary structure (74). The compactness parameter (Ci) provides a quantitative measure of the residue embedded in the protein tertiary structure. High values (>300) are found for residues in the buried core of globular domains, whereas low values correspond to surface exposed or conformationally flexible residues. The overall Ci values for the C-terminal segment (amino acids 70–102) are lower than for the core of Hfq, suggesting that this region is conformationally flexible and surface exposed. In addition, the compactness of the Hfq-core region and that of the C-terminus were calculated separately. The average compactness value of the Hfq core region increased (from 292 of the full-length protein) to about 337, whereas the corresponding value for the C-terminal extension alone decreased to 170, indicating that the C-terminus is less compactly folded when compared to the Hfq core. The conclusion that the C-terminus is largely disordered is further supported by the fact that a similar average compactness value (Cav) were found for a set of experimentally characterized intrinsically unstructured proteins [Myc C-terminal domain (75): Cav = 131; osteopontin (76): Cav = 181; α-synuclein (77): Cav = 255; TAU (78): Cav = 224; Sic1 (79): Cav = 229].
This bioinformatic analysis (Figure 2; Supplementary Figure S1) together with the 3D structure of Hfq (22), in which residues 65–71 are found in an extended conformation leaning on the body Hfq core, suggest that the segment encompassing amino acid residues 65 up to 75 displays propensity to be structured, while this tendency is much less pronounced for the rest of the C-terminal extension with high indications for being intrinsically disordered. These features of HfqEc resemble intrinsically disordered protein sequences, which show a propensity to form local elements and/or contain preformed elements of secondary structure or hydrophobic clusters (80,81).
Figure 2.
Meta-structure analysis of HfqEc. Compactness values (Ci) are plotted with respect to residue position. A high value indicates a buried residue (the average Ci of all residues in the Protein Data Bank is 300). Inserted in upper-left is the known secondary-structure content of Hfq (amino acids 6–65; PDBid: 1HK9) generated by ESPript (http://espript.ibcp.fr/ESPript/ESPript/), TT denotes β-turn and η1 the small 310-helix. Bottom, primary amino acid sequence of HfqEc.
Nuclear magnetic resonance and synchrotron radiation circular dichroism spectroscopy reveal the unstructured nature of the C-terminal extension of HfqEc
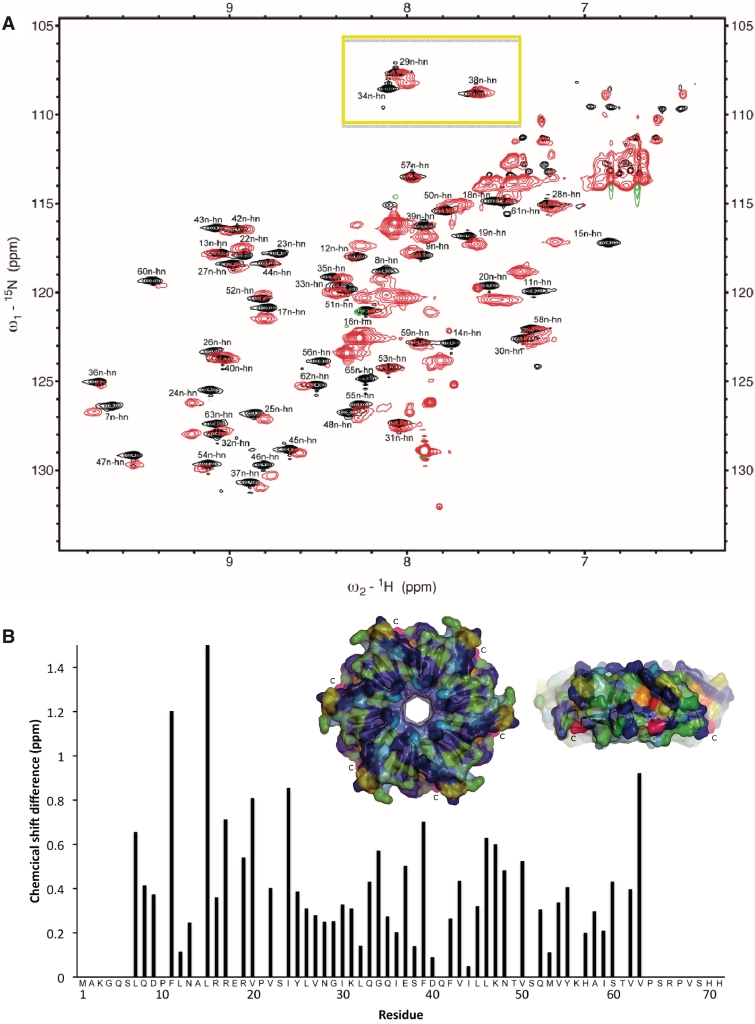
To obtain more information on the structural and dynamic properties of the C-terminus of HfqEc in solution we employed nuclear magnetic resonance spectroscopy (NMR). A series of NMR-spectra were recorded for full-length HfqEc and for HfqEc65. All NMR experiments for sequential signal assignments were conducted with HfqEc65, which—due to its lower molecular weight—was more amenable for NMR studies. Figure 3A shows the fingerprint region of the 15N-HSQC spectra of HfqEc65 superimposed on the 15N-HSQC spectra of full-length HfqEc. The spectrum of HfqEc65 is well resolved and sequential backbone signal assignment was possible for residues 6–65. Residues 1–5 were not observed possibly due to exchange broadening, suggesting some conformational flexibility of these residues. The spectra of HfqEc65 and full-length HfqEc overlapped largely, which allowed for assignment of the HfqEc spectra by simple peak comparison. However, many of the observed peaks corresponding to well-structured residues in the core of the protein show severe line broadening in the HfqEc spectrum due to fast relaxation. A plot of the 15N–1HN chemical shift differences between HfqEc65 and full-length HfqEc is shown in Figure 3B. Significant chemical shift differences were observed for residues in the region preceding the C-terminal segment (amino acids 45–55) and at the end of the N-terminal α-helix (residues Phe11, Leu15, Arg17, Val20), indicating that the presence of the C-terminus affects residues in these regions. In the 3D structure from X-ray studies performed by Sauter et al. (22), the segment encompassing amino acids 65–70 was found to pack in an extended conformation against the core of the Hfq hexamer, in a groove formed by the N-terminal α-helix of one domain and β-sheets formed by amino acids 34–55 of the adjacent subunit. A conserved patch of charged residues comprised by Arg16, Arg17, Glu18 and Arg19 are present in Hfq. Interestingly, amino acids substitutions of Arg17 in HfqEc compromised phage Qß replication (82), suggesting that this region is of functional importance.
Figure 3.
Comparative NMR analysis of HfqEc65 and full-length HfqEc. (A) Overlay of 15N-HSQC spectra of HfqEc65 [black (positive) and blue (negative) contours] and full-length HfqEc [red (positive) and green (negative) contours] demonstrating the unstructured nature of the C-terminal extension, with multiple intense peaks occurring in the 7.5–8.5 ppm region of the 1H-dimension. The glycine region in the upper part of the HfqEc spectra (yellow box) is lacking the expected four peaks for the four glycine residues of the C-terminus. (B) Chemical 1H–15N shift differences calculated for assigned peaks plotted against residue positions. Chemical shift differences >0.5 ppm are considered significant. The insert shows a surface representation of the proximal face and side of E. coli Hfq (amino acids 6–65) hexamer. Chemical 1H–15N shift differences between HfqEc65 and full-length HfqEc are color-coded from blue (zero) to red (largest shift). Residues which could not be unambiguously assigned in the full-length HfqEc spectra are in blue. ‘C’ indicates the C-terminal Ser-65 of HfqEc65, noted in pink.
The spectrum of full-length HfqEc shows additional intense peaks, mainly in the region of 7.5–8.5 ppm (in the 1H dimension), corresponding to the partially disordered C-terminus. Interestingly, in the region of the 15N-HSQC-spectra expected for Gly residues in a random-coil conformation, we did not observe any of the four extra glycine residues present in the C-terminus, indicating that they are indeed not fully unstructured. One possible explanation for their disappearance is that they are subjected to an additional dynamic process, leading to further line broadening, which could result from conformational exchange between a partially structured and a fully unstructured state. Another cause for disappearance of correlation peaks is rapid intermolecular exchange of amide protons with bulk water. This exchange process is particularly facilitated in disordered and conformationally flexible parts of the protein lacking stabilizing backbone hydrogen bonds.
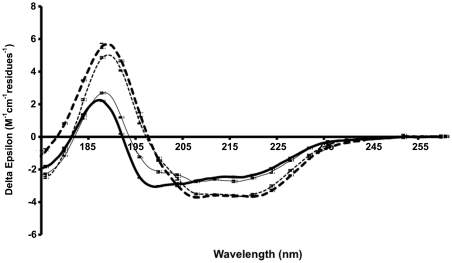
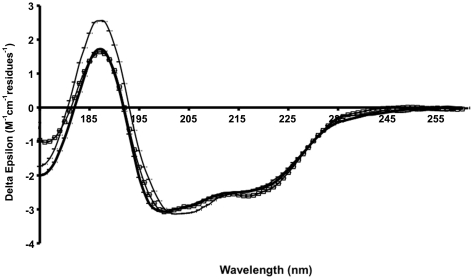
In order to study the secondary structure composition of the C-terminal extension of HfqEc in solution, we used synchrotron radiation circular dichroism (SRCD) spectroscopy. The additional data in the low-wavelength region accessible by this technique relative to that attainable in a conventional CD instrument is important for detecting natively-disordered types of secondary structure. The spectra of the protein constructs HfqEc65 (amino acids 1–65), HfqEc75 (amino acids 1–75), HfqEc85 (amino acids 1–85) and HfqEc (full-length, amino acids 1–102) collected under the same experimental conditions are shown in Figure 4.
Figure 4.
SRCD spectra of Hfq constructs. HfqEc65 (thin dotted line), HfqEc75 (thick dotted line), HfqEc85 (thin solid line), full-length HfqEc (thick solid line). The error bars represent one standard deviation between replicate scans.
The spectrum of the HfqEc65 is typical for a mixed α/β protein and the calculated secondary structure based on these data produced values close to those calculated from the crystal structure (Table 1). When the HfqEc65 protein is extended by 10 residues (HfqEc75) there is a slight increase in spectral magnitude at both 222 and 190 nm, generally indicative of an increase in helix content, which is reflected in the analysis (Table 1) as a slight increase (2%) in the calculated helical content. This may be explained by stabilization of the N-terminal α-helix by the segment 65–75 through a similar packing arrangement observed in the crystal structure of E. coli Hfq (amino acid residues 1–72 PDBid: 1HK9) (22), giving rise to increased ellipticity. This is in good agreement with the NMR observations showing that this C-terminal stretch of residues interacts with the N-terminal α-helix.
Table 1.
Secondary structure content of E. coli Hfq constructs and their complexes with RNA, evaluated from SRCD spectra using the DichroWeb analysis server (43)
| Protein | RNA | Theoretical %a |
Experimental % |
NRMSDb | ||||
|---|---|---|---|---|---|---|---|---|
| α-Helix | β-Strand | Other | α-Helix | β-Strand | Other | |||
| HfqEc | 11 | 21 | 68 | 11 ± 3 | 33 ± 2 | 56 ± 2 | 0.202 | |
| HfqEc85 | 13 | 25 | 62 | 12 ± 5 | 35 ± 3 | 55 ± 2 | 0.045 | |
| HfqEc75 | 15 | 28 | 57 | 22 ± 2 | 29 ± 1 | 49 ± 1 | 0.044 | |
| HfqEc65 | 17 | 32 | 51 | 20 ± 2 | 34 ± 2 | 46 ± 4 | 0.086 | |
| HfqEc | hfq126 | 12 ± 4 | 33 ± 2 | 54 ± 2 | 0.091 | |||
| HfqEc | sodB83 | 12 ± 5 | 37 ± 2 | 53 ± 2 | 0.135 | |||
| HfqEc | DsrA34 | 10 ± 4 | 35 ± 2 | 55 ± 3 | 0.056 | |||
| HfqEc75 | DsrA34 | 22 ± 3 | 29 ± 1 | 49 ± 2 | 0.077 | |||
aBold values listed under theoretical correspond to the crystal structure of HfqEc65; other ‘theoretical’ values are calculated based on the crystal structure and assuming the extension residues are disordered.
bNormalized root mean-square deviation between calculated and experimental spectra using the CONTINLL algorithm.
In general, with each additional extension to the C-terminus (i.e. HfqEc85 and full-length HfqEc) there is a reduction in magnitude of the spectrum combined with a blue shift of the peak located at ∼208 nm. This is indicative of an increase in the contribution of natively disordered-type structures to the spectrum, consistent with the assumption in the ‘theoretical’ values that the additional C-terminal residues are almost entirely unstructured; the percentage of ‘other’ increases as the number of residues are added.
The overall decrease in helix percentage as more residues are added—up to the full-length construct is caused by the original and constant number of helical residues representing a lower proportion of the total molecule in these constructs, not because the helical residues become unstructured. On the other hand, the percentage of ‘experimental’ β-structure in all examined variants is relatively constant at ∼33%, while the expected ‘theoretical’ values decrease, as this is calculated assuming the additional regions in the constructs to be disordered. This suggests that the additional C-terminal residues contribute to some extent to an increase in the number of β-structure residues in HfqEc, which is in agreement with results from FTIR spectroscopy studies (83) showing the C-terminus to increase the overall β-structure content of the protein.
Solution structure, hydrodynamic properties and shape of the C-terminus of full-length HfqEc
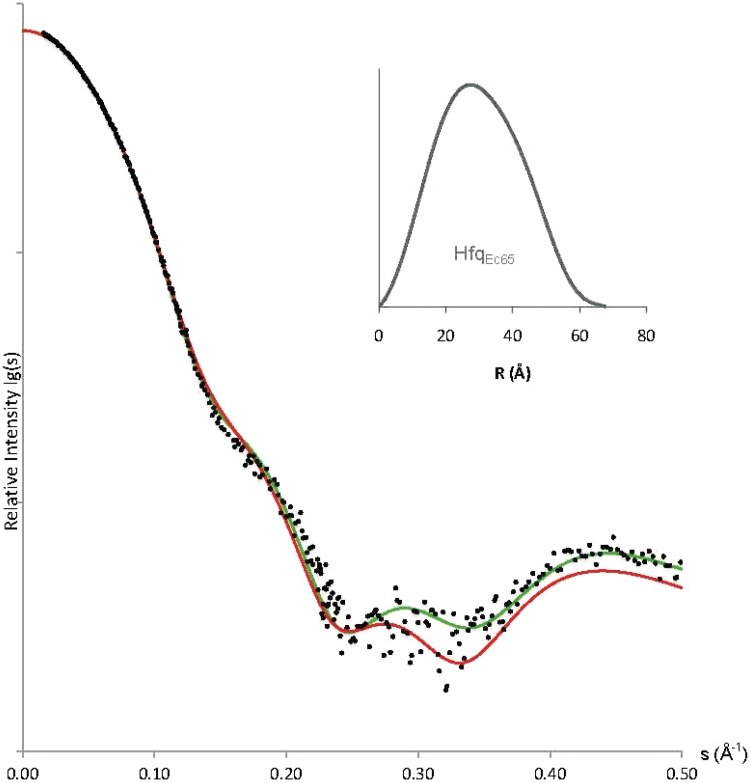
To further study the behavior of Hfq in solution, we employed a series of solution small angle X-ray scattering (SAXS) experiments in combination with NMR spectroscopy measurements of the transverse relaxation rate of Hfq. Figure 5 shows the fit between experimental X-ray scattering data and the calculated scattering curve (66) for residues 6–65 of Hfq (PDBid: 1HK9) (22), confirming the close agreement between crystal and solution structure of the hexameric Hfq core. By modeling the five missing N-terminal residues, we observe only a marginal improvement of the fit (χ2 ∼ 1.55 1.40), indicating a modest contribution to the overall scattering of these 6 × 5 = 30 unstructured residues in HfqEc65.
1.40), indicating a modest contribution to the overall scattering of these 6 × 5 = 30 unstructured residues in HfqEc65.
Figure 5.
Overlay of experimental X-ray scattering data from HfqEc65 (black dots), on the calculated scattering curves for Hfq crystal structure (PDB: 1HK9, aa 6–65, red curve) and the complete model of HfqEc65 (aa 1-65, green curve) where N-terminal residues were restored using the Bunch program. Inserted is the pair-distribution function for HfqEc65 (grey).
For HfqEc65 we estimated a molecular mass of 44 ± 5 kDa and for full-length HfqEc 58 ± 7 kDa by normalization of the intrinsic scattering (I0) to a standard sample of bovine serum albumin (BSA) (84), which agrees with the theoretical values for hexamers of 43.2 and 67.2 kDa, respectively. We estimate the solute excluded volume (Porod’s volume) for HfqEc65 (70 ± 5) × 103 Å3 and for full-length HfqEc (110 ± 8) × 103 Å3, further indicating that both proteins adopt their native hexameric quaternary structure. In addition, Guinier analysis established for HfqEc65 an Rg of (24.2 ± 0.5) Å, in good agreement with Rg of 23.5 Å calculated for the crystal structure of HfqEc65. Comparatively, for full-length HfqEc Rg was (32.3 ± 0.5) Å (60). The estimated largest dimension (DMAX) of the particle was for HfqEc65 (68 ± 5) Å and for HfqEc (112 ± 8) Å (62).
By NMR 15N-relaxation measurements we measured for HfqEc65 (43.2 kDa) a T2 = 40.0 ± 2.1 ms (R2 = 25.0 ± 1.3 s−1). These homogeneous 15N-relaxation data indicate that the core domain of Hfq is uniformly rigid and does not undergo large amplitude fluctuations on timescales relevant for NMR.
For full-length HfqEc (67.2 kDa), we measured an extremely fast 15N relaxation of T2 = 5–10 ms for the signals of the core-region. Compared to HfqEc65, HfqEc exhibits considerably reduced 15N-transverse relaxation times (T2 = 5–10 ms). Consequently the precision of the T2 measurement is much lower due to the resulting lower S/N-ratio. Interestingly, there is a larger residue by residue variation of 15N T2 relaxation times in HfqEc than in HfqEc65, which is an indication of a more anisotropic behavior due to the extended C-termini. The observed relaxation rate is much faster than what could be expected for the increase in molecular mass, indicating an unusual increase in the hydrodynamic radius. For a perfect solid sphere, the radius of gyration (Rg) is only slightly smaller than the hydrodynamic radius (Rh), whereas increased discrepancy between the two reflects an anisometric structure of the particle. With a relation of 1.33 between the Rh3 of HfqEc65 and HfqEc from SAXS measurements, reflecting the expected increase in molecular weight, the relation of 4–8 between the relaxation rates measured by NMR (which are directly proportional to Rh3) indicates a pronounced anisometric structure of HfqEc. A possible explanation for these observations is that the partially unstructured C-termini are laterally extended into the surrounding solution. Thus the overall molecular shape is ‘puffed up’, causing the molecule to appear much larger than a comparable well-folded and tightly packed globular protein.
To validate this hypothesis, the observed 15N T2 relaxation properties were modeled through simulation of the hydrodynamic properties of HfqEc65 and HfqEc using their molecular coordinates as input. For HfqEc65 the coordinates from the X-ray crystal structure of Hfq of E. coli (amino acid residues 6–65, PDBid: 1HK9) (22) were used, onto which the N-termini had been added as unstructured. For full-length HfqEc a representative all-atom ensemble was created by restrained simulated annealing using the X-plor/CNS software system (85) constraining the backbone dihedral angles of the C-terminus to the β-region of the Ramachandran plot and applying the observed radius of gyration as overall restraint.
Using the HYDRONMR software (86) for simulation of relaxation times both protein systems were very well reproduced in the simulation and good agreement was obtained between experimental and simulated relaxation behavior. For HfqEc65 15N transverse relaxation times of T2 = 35.0 ± 2.0 ms (R2 = 28.6 ± 1.6 s−1) were predicted; for the ensemble of full-length HfqEc models an average value of 15N T2 = 9.7 ± 2.0 ms (R2 = 103.1 ± 21.3 s−1) was found in the simulation.
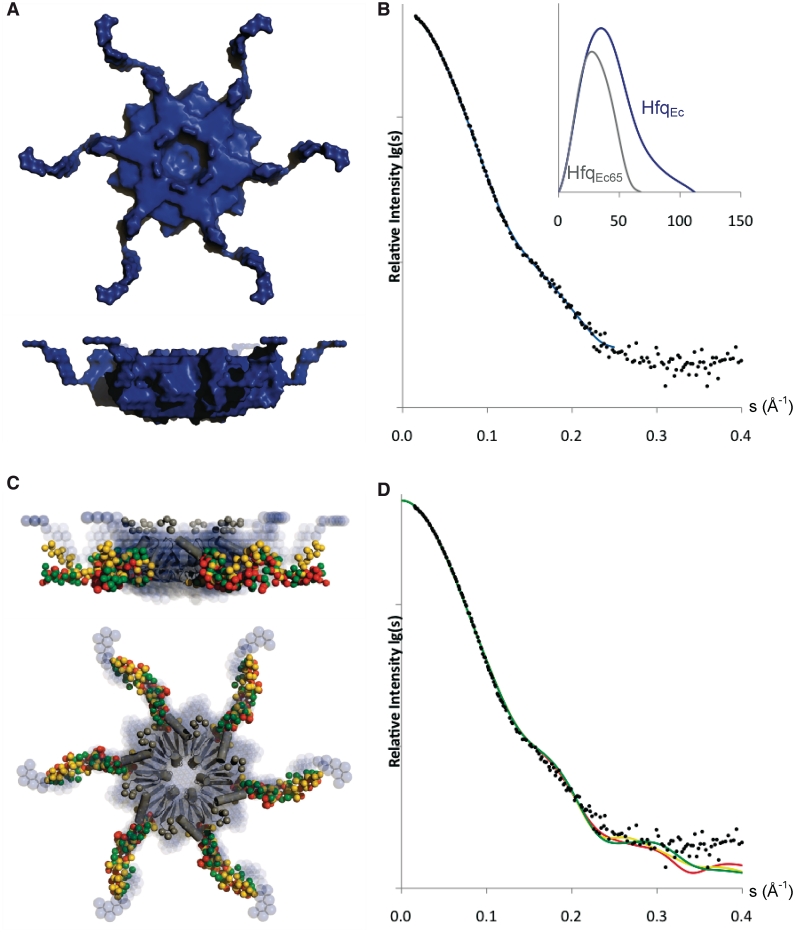
Implying 6-fold symmetry the low-resolution shape of HfqEc was constructed ab initio in the program Dammif (64). As shown in Figure 6A, the C-termini extend away from the core of the protein in a propeller like shape. The dimensions of the hexameric core of the ab initio model (diameter ∼72 Å, thickness ∼35 Å) are larger than those derived from the crystal structure (diameter ∼65 Å, thickness ∼28 Å) (22), which can be ascribed to the presence of the termini. The output Rg 32.6 Å by Dammif is in agreement with the Rg derived from the Guinier-analysis (32.3 Å), and the partial specific volume of the protein (HfqEc65 ∼74 × 103 Å3 and HfqEc ∼116 × 103 Å3; calculated by Dammif without implying symmetry) is within range of the estimated invariant values. Figure 6B shows the calculated scattering curve of the ab initio model, which agrees with the experimental data on full-length HfqEc, with a χ2 of 1.31, and the distance distribution function for HfqEc65 and for full-length HfqEc, with the difference clearly indicating the extended nature of the C-terminal domain of full-length HfqEc.
Figure 6.
Shape models of HfqEc. (A) Surface representation of ab initio model. (B) Calculated scattering curve of ab initio model (blue) overlaid with experimental X-ray scattering data, inserted is the pair-distribution function for HfqEc65 (grey) and full-length HfqEc (blue). (C) Superimposition of the three best rigid-body HfqEc models from three iterations of modeling (red, gold and green). In the center, the crystal structure of the HfqEc (amino acids 6–65) hexameric core is represented as grey ribbons and the N-termini as grey spheres. The ab initio model is represented as blue spheres in the background. (D) Calculated scattering curves to a maximum range of momentum transfer (s) of 0.4 Å−1 for the three best rigid-body models (corresponding in red, gold and green).
Next, we took an approach of rigid-body modeling with the program Bunch (65). We first built the N-terminal residues onto the crystal structure of HfqEc65 (Figure 5), and proceeded to utilize the outcome model as a rigid-body in reconstruction of the C-terminal domains of full-length HfqEc, again implying 6-fold symmetry in the particle. Figure 6C shows the superposition of the model of HfqEc from three iterations of rigid-body modeling. Clear agreement was observed for the conserved hexameric core between the ab initio and rigid-body models, whereas some variability was observed for the C-termini. Iterative runs of Bunch (×1000) generated models with low variability in the shape of the C-termini. By adjusting restraints in the Bunch-program we could fit the experimental data to χ2 ∼ 1.65–1.75. We observed bad fits (χ2 > 2.3) for models with the C-termini completely extended towards either the proximal or distal face of the hexamer. The calculated Rg 31.8 Å (average of three best fits) for the models is in good agreement though slightly smaller than the corresponding value of ∼32.3 Å derived from the Guinier plot. Figure 6D shows the fit with the experimental data for the three best rigid-body models, as determined by χ2-values generated by the program Bunch. These models fit the experimental data to a range of momentum transfer of 0.35 Å−1, and indicate that the C-termini extend laterally away from the hexameric core. The overall trend appears to be a wave-like shape, with a characteristic loop in the beginning of the C-termini and some variability in the tip. Interestingly, this wave-like behavior is also seen in the meta-structure analysis, where a β-turn like structural element is found in the disordered C-terminus (Figure 2).
SRCD spectroscopy reveals an interaction of the C-terminus with RNA
In order to monitor changes in the structure of the C-terminal extension of HfqEc in the presence of RNA, we next used SRCD spectroscopy. Both HfqEc65 and full-length HfqEc bind the sRNA DsrA (82) and also the Hfq binding fragment of DsrA (DsrA34; data not shown) with comparable affinity, indicating that the C-terminus is not required for binding of the sRNA. As the SRCD spectra of HfqEc alone and in the presence of DsrA34 were indistinguishable (Figure 7), the known binding mode of DsrA to the inner core of the HfqEc hexamer (29), apparently does not affect the structure of the protein. In contrast to sRNAs, longer (m)RNA fragments did not bind to the HfqEc65 but depended on the presence of the C-terminus (18). Interestingly a 126-nt long fragment of hfq mRNA, which failed to bind to HfqEc65 but bound to full-length HfqEc (18), produced significant increases in the 185–200 nm region only upon binding to full-length HfqEc, reflecting an increase in the amount of ordered (β-strand or α-helical) conformations for this part of Hfq. The increase is consistent with a combination of decrease of disordered and increase in ordered residues. Altogether this suggests that RNA binding leads to an ordering of the C-terminal extension of Hfq. Many intrinsically disordered proteins undergo transitions to more ordered states or fold into stable secondary or tertiary structures upon binding to their targets. They undergo coupled folding and binding, forming complexes with high specificity and relatively low affinity, which is critical for processes in which not only specific association but also subsequent dissociation of binding partners is required (87).
Figure 7.
SRCD spectra of HfqEc (thick line), in complex with DsrA34 (open squares), and hfq126 RNA (thin line). The error bars shown represent one standard deviation between replicate scans.
DISCUSSION
In this integrative study on the dynamics and structure of the C-terminus of the Hfq protein from E. coli, we used bioinformatics and biophysical methods combined with molecular and structural biology. In brief, the different methods collectively revealed that the C-termini of HfqEc are intrinsically disordered and that they extend laterally away from the doughnut shaped core of the hexamer. Such flexible, intrinsically disordered regions are believed to provide conformational fluctuations, which can facilitate intermolecular interactions (33). Intrinsically disordered protein sequences are known to contain preformed, structured recognition motifs with functional implications in a hierarchical model of binding (88). Conversely, the unstructured nature of a binding motif is also thought to have a functional implication in compensating the gain in enthalpy upon binding by the loss of conformational entropy (89). This effect, termed isothermal enthalpy/entropy compensation, effectively uncouples binding-specificity from binding-strength, which enables specific binding partners to associate and dissociate reversibly. Recently this effect has been demonstrated in polyuridine-tract recognition of the splicing factor U2AF (90), and is known be valid for DNA-binding proteins (91). Interestingly, a recent study (92) revealed that Hfq65 and Hfq75 are defective in DNA binding, implicating the C-terminus as well as the distal surface in DNA interactions.
Binding of a poly(A) tract to six tripartite binding motifs at the distal face of HfqEc has been demonstrated in structural studies (23). However, as HfqEc65 was deficient in mediating low-temperature translational activation of rpoS mRNA by the sRNA DsrA (18) and HfqEc65 did not bind to rpoS mRNA containing the A-rich motifs implicated in distal site binding (30), these interactions are apparently not sufficient for the function of HfqEc in rpoS regulation. In addition, and in agreement with the observed annealing deficiency of HfqEc75, HfqBs and HfqSa (Figure 1A), the three proteins were defective in complementing full-length HfqEc in low temperature activation of rpoS [Figure 1B; (18)]. Furthermore, HfqBs and HfqSa did not complement HfqEc in RyhB-mediated translational repression of sodB mRNA (16). When compared to self-annealing HfqSa reduced the annealing rate of the two oligonucleotides. Although we have not further studied this behaviour of HfqSa it appears possible that this protein binds one or the other RNA too tightly, thereby reducing self-annealing. In contrast, HfqEc85 was functional in the annealing assay (Figure 1A) and stimulated rpoS activation by DsrA at low temperature. Thus, this study implicates the region between amino acids 75 and 85 as a minimal portion supporting the interactions with longer RNA fragments, whereas the core region, i.e. HfqEc65 is fully proficient in sRNA binding. Thus, the structurally flexible and disordered C-termini appear to provide a moiety involved in tethering of longer and structurally diverse RNA molecules, which could be followed by stable accommodation of the substrate at the distal site, i.e. at the polyA binding motifs as suggested for rpoS mRNA (23). Such a rearrangement would be in line with the concept of isothermal enthalpy/entropy compensation, as discussed above. While we see a certain spread in the conformations of the C-termini from our SAXS analysis, with the best fitting conformations extending laterally away from the hexameric core—the crystallization process apparently selects conformers with the C-termini on the proximal face, indicating this conformation to be one of the low energy states of the molecule. Using NMR, SAXS and small angle neutron scattering, we have obtained evidence that the DsrA34 fragment binds to the proximal region and extends into the solvent (M. B.-F., unpublished data). Thus, annealing of the sRNA with the mRNA may occur either during the accommodation of mRNA at the distal site or after the mRNA is bound at the distal site.
Although these biophysical studies predestine the C-termini of HfqEc for interactions with RNA, they are seemingly at variance with the observations that the Hfq orthologues of M. jannaschii (HfqMj; 71 amino acids) and Listeria monocytogenes (HfqLm; 77 amino acids) complemented HfqEc in rpoS translation (at least at 37°C), and RyhB-mediated decay of sodB mRNA (26,93). While HfqEc has relatively short predicted intrinsically disordered segments in the N-terminus (the first 5–6 amino acids), in HfqMj the first 13 N-terminal residues are predicted to be disordered, while the C-terminus is not (Supplementary Figure S2), which could indicate a role of the N-terminus in RNA binding. Nonetheless, HfqSa, being with 77 amino acids as long as HfqLm with also similar pattern of predicted unstructured regions, was in our hands defective in RyhB-mediated translational repression of sodB mRNA (18). Thus, the possibility exists that Hfq proteins of different origin interact differently with distinct substrates. In this regard it is notable that HfqEc65, although defective in the annealing assay and in supporting translation of rpoS (Figure 1) was fully functional in phage Qβ replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The work in the K.Dj.-C., U.B. and R.K laboratories was supported by grants F1722 (K.Dj.-C.), F1720 (to U.B) and F1719 (to R.K.) in the framework of the Special Research Program (SFB17) on ‘Modulators of RNA fate and function’ by the Austrian Science Fund. U.K. Biotechnology and Biological Research Council (BBSRC) project grant (to B.A.W.); SRCD beamtime grants (to B.A.W.). Funding for open access charge: Austrian National Science fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The group of D. Svergun, particularly Weifeng Shang, Adam Round, Alexei Kikhney and Manfred Roessle, at the X33 beamline, DESY, EMBL-Hamburg, is gratefully acknowledged for help with SAXS data-collection. Greg J. R. Ewing is acknowledged for help with shell scripting, and Euripedes de Almeida Ribeiro for helpful discussions. Access to the CD1 beamline at ISA (the Institute for Storage Ring Facilities, Aarhus University, Denmark) is acknowledged under the EU Integrated Infrastructure Initiative, Integrated Activity on Synchrotron and Free Electron Laser Science (IA-SFS), contract number RII3-CT-2004-506008.
REFERENCES
- 1.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 2.Kaberdin VR, Bläsi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 3.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 6.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandin P, Gottesman S. Regulating the regulator: an RNA decoy acts as an OFF switch for the regulation of an sRNA. Genes Dev. 2009;23:1981–1985. doi: 10.1101/gad.1846609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Masse E, Majdalani N, Gottesman S. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 2003;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 12.Moll I, Leitsch D, Steinhauser T, Bläsi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 14.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Bläsi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2006;59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 16.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 17.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Bläsi U, Moll I, Kachalova G, Yokoyama S, Vassylyev D, Garber M, et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 25.Moskaleva O, Melnik B, Gabdulkhakov A, Garber M, Nikonov S, Stolboushkina E, Nikulin A. The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:760–764. doi: 10.1107/S1744309110017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen JS, Boggild A, Andersen CB, Nielsen G, Boysen A, Brodersen DE, Valentin-Hansen P. An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA. 2007;13:2213–2223. doi: 10.1261/rna.689007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 2009;276:3904–3915. doi: 10.1111/j.1742-4658.2009.07104.x. [DOI] [PubMed] [Google Scholar]
- 28.Baba S, Someya T, Kawai G, Nakamura K, Kumasaka T. Expression, crystallization and preliminary crystallographic analysis of RNA-binding protein Hfq (YmaH) from Bacillus subtilis in complex with an RNA aptamer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:563–566. doi: 10.1107/S1744309110009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Rajkowitsch L, Schroeder R. Coupling RNA annealing and strand displacement: a FRET-based microplate reader assay for RNA chaperone activity. Biotechniques. 2007;43:304–310. doi: 10.2144/000112530. [DOI] [PubMed] [Google Scholar]
- 33.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 34.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 35.Sonnleitner E, Moll I, Bläsi U. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology. 2002;148:883–891. doi: 10.1099/00221287-148-3-883. [DOI] [PubMed] [Google Scholar]
- 36.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Bläsi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 37.Vecerek B, Moll I, Bläsi U. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vecerek B, Beich-Frandsen M, Resch A, Bläsi U. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 2010;38:1284–1293. doi: 10.1093/nar/gkp1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lees JG, Smith BR, Wien F, Miles AJ, Wallace BA. CDtool-an integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal. Biochem. 2004;332:285–289. doi: 10.1016/j.ab.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Miles AJ, Wien F, Lees JG, Rodger A, Janes RW, Wallace BA. Calibration and standardisation of synchrotron radiation circular dichroism and conventional circular dichroism spectrophotometers. Spectroscopy. 2003;17:653–661. [Google Scholar]
- 42.Miles AJ, Wallace BA. Synchrotron radiation circular dichroism spectroscopy of proteins and applications in structural and functional genomics. Chem. Soc. Rev. 2006;35:39–51. doi: 10.1039/b316168b. [DOI] [PubMed] [Google Scholar]
- 43.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 45.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 46.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 47.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins: Structure Function and Genetics. 1999;35:307–312. [PubMed] [Google Scholar]
- 48.Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 49.Mao D, Wachter E, Wallace BA. Folding of the mitochondrial proton adenosinetriphosphatase proteolipid channel in phospholipid vesicles. Biochemistry. 1982;21:4960–4968. doi: 10.1021/bi00263a020. [DOI] [PubMed] [Google Scholar]
- 50.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 52.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13 C/15 N-enriched proteins. J. Biomol. NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 53.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3 D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. B. 1993;101:201–205. [Google Scholar]
- 54.Kay LE, Ikura M, Bax A. Proton-proton correlation via carbon-carbon couplings: A three-dimensional NMR approach for the assignment of aliphatic resonances in proteins labeled with carbon-13. J. Am. Chem. Soc. 1990;112:888–889. [Google Scholar]
- 55.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 56.Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE. Spectral density function mapping using 15 N relaxation data exclusively. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 57.Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 2007;40:s190–s194. [Google Scholar]
- 58.Round AR, Franke D, Moritz S, Huchler R, Fritsche M, Malthan D, Klaering R, Svergun DI, Roessle M. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J Appl. Crystallogr. 2008;41:913–917. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1-towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 2007;40:S223–S228. [Google Scholar]
- 60.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 61.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 62.Semenyuk AV, Svergun DI. GNOM-a program package for small-angle scattering data processing. J. Appl. Crystallogr. 1991;24:537–540. [Google Scholar]
- 63.Porod G. General theory. In: Kratky OGaO., editor. Small-angle X-ray Scattering. Vol. 17. London: Academic Press; 1982. 51 pp. [Google Scholar]
- 64.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Svergun D, Barberato C, Koch MHJ. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 67.DeLano WL. MacPyMOL: A PyMOL-based Molecular Graphics Application for MacOS X. Palo Alto, CA: DeLano Scientific LLC; 2007. [Google Scholar]
- 68.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 69.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 70.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 72.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 74.Konrat R. The protein meta-structure: a novel concept for chemical and molecular biology. Cell Mol. Life Sci. 2009;66:3625–3639. doi: 10.1007/s00018-009-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fieber W, Schneider ML, Matt T, Krautler B, Konrat R, Bister K. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J. Mol. Biol. 2001;307:1395–1410. doi: 10.1006/jmbi.2001.4537. [DOI] [PubMed] [Google Scholar]
- 76.Schedlbauer A, Ozdowy P, Kontaxis G, Hartl M, Bister K, Konrat R. Backbone assignment of osteopontin, a cytokine and cell attachment protein implicated in tumorigenesis. Biomol. NMR Assign. 2008;2:29–31. doi: 10.1007/s12104-007-9076-2. [DOI] [PubMed] [Google Scholar]
- 77.Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc. Natl Acad. Sci. USA. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7:e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Demchenko AP. Recognition between flexible protein molecules: induced and assisted folding. J. Mol. Recognit. 2001;14:42–61. doi: 10.1002/1099-1352(200101/02)14:1<42::AID-JMR518>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 81.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 82.Sonnleitner E, Napetschnig J, Afonyushkin T, Ecker K, Vecerek B, Moll I, Kaberdin VR, Bläsi U. Functional effects of variants of the RNA chaperone Hfq. Biochem. Biophys. Res. Commun. 2004;323:1017–1023. doi: 10.1016/j.bbrc.2004.08.190. [DOI] [PubMed] [Google Scholar]
- 83.Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, Hajnsdorf E, Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 84.Akiyama S. Quality control of protein standards for molecular mass determinations by small-angle X-ray scattering. J. Appl. Crystallogr. 2010;43:237–243. [Google Scholar]
- 85.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 86.Garcia de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 87.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 88.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc. Natl Acad. Sci. USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 90.Jenkins JL, Shen H, Green MR, Kielkopf CL. Solution conformation and thermodynamic characteristics of RNA binding by the splicing factor U2AF65. J. Biol. Chem. 2008;283:33641–33649. doi: 10.1074/jbc.M806297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 92.Updegrove TB, Correia JJ, Galletto R, Bujalowski W, Wartell RM. E. coli DNA associated with isolated Hfq interacts with Hfq's distal surface and C-terminal domain. Biochim. Biophys. Acta. 2010;1799:588–596. doi: 10.1016/j.bbagrm.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.