Abstract
The antizyme protein, Oaz1, regulates synthesis of the polyamines putrescine, spermidine and spermine by controlling stability of the polyamine biosynthetic enzyme, ornithine decarboxylase. Antizyme mRNA translation depends upon a polyamine-stimulated +1 ribosomal frameshift, forming a complex negative feedback system in which the translational frameshifting event may be viewed in engineering terms as a feedback controller for intracellular polyamine concentrations. In this article, we present the first systems level study of the characteristics of this feedback controller, using an integrated experimental and modeling approach. Quantitative analysis of mutant yeast strains in which polyamine synthesis and interconversion were blocked revealed marked variations in frameshift responses to the different polyamines. Putrescine and spermine, but not spermidine, showed evidence of co-operative stimulation of frameshifting and the existence of multiple ribosome binding sites. Combinatorial polyamine treatments showed polyamines compete for binding to common ribosome sites. Using concepts from enzyme kinetics and control engineering, a mathematical model of the translational controller was developed to describe these complex ribosomal responses to combinatorial polyamine effects. Each one of a range of model predictions was successfully validated against experimental frameshift frequencies measured in S-adenosylmethionine-decarboxylase and antizyme mutants, as well as in the wild-type genetic background.
INTRODUCTION
Polyamines are essential, ubiquitous polycations found in all eukaryotic and most prokaryotic cells. They are utilised in a wide range of core cellular processes such as binding and stabilising RNA and DNA, mRNA translation, ribosome biogenesis, cell proliferation and programmed cell death (1). Polyamine depletion results in growth arrest (2), whereas their over-abundance is cytotoxic (3,4). Thus, homeostatically regulating polyamine content within a non-toxic range is a significant regulatory challenge for the cell.
Ornithine decarboxylase (ODC) is the first and rate limiting enzyme in the biosynthesis of the polyamines (Saccharomyces cerevisiae pathway shown in Figure 1). The key regulator of ODC in a wide range of eukaryotes is the protein antizyme (5). There is a single antizyme isoform in S. cerevisiae, Oaz1 (6). Antizyme binds to and inhibits ODC, and targets it for ubiquitin-independent proteolysis by the 26S proteasome (7,8). Antizyme synthesis is in turn dependent upon a polyamine-stimulated +1 ribosomal frameshift event during translation of its mRNA. Antizyme degradation by the ubiquitin pathway is inhibited by polyamines (6). Polyamines thus regulate their homeostasis via a negative feedback system, by controlling Oaz1 synthesis and inhibiting its proteolysis.
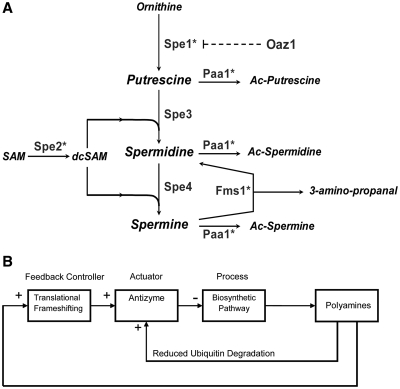
Figure 1.
The polyamine biosynthetic pathway in S. cerevisiae. (A) Antizyme (Oaz1), the main regulator is indicated; it targets ODC (Spe1) for degradation by the 26S proteasome. Metabolites are shown in italics and proteins in Roman type. Asterisks denote genes deleted in the spe1 spe2 paa1 fms1 deletant strain. See text for further details. (B) Block diagram representing the regulation of polyamine biosynthesis by antizyme as a modular feedback control system.
To understand how this negative feedback contributes to the complexity of polyamine homeostasis will require a robust quantitative characterisation of the underlying control mechanisms (9). One partial mathematical model of polyamine metabolism has been developed previously (10). However, this model lacks a description of antizyme regulation through frameshifting, or of polyamine regulation of antizyme degradation, and is therefore of limited use in understanding the consequences of perturbing a pathway structured with complex homeostatic controls. In control engineering terms, polyamine regulation consists of a process (the biosynthetic pathway), which is affected by an actuator (the protein antizyme) under the control of multiple negative feedback loops (Figure 1B). In this representation, the translational frameshift event plays the role of a feedback controller, and it is clear that a complete system model requires a quantitative characterisation of this controller.
In S. cerevisiae, polyamines stimulate antizyme frameshifting at an in-frame UGA codon that terminates antizyme ORF1 (6). Frameshifting is enhanced by the immediately preceding GCG codon, for which there is no tRNA whose anticodon matches the codon by Watson–Crick or Wobble base pairing in yeast (11,12), and by the poor context of the UGA stop codon which causes inefficient termination (13,14). Ribosomal frameshifting thus occurs in direct competition with an inefficient translation termination event and potentially, stop codon readthrough by a cellular tRNA. Indeed, polyamines have been reported to stimulate stop codon readthrough in mammalian systems and Escherichia coli (15,16). Which polyamines govern the ratio of frameshifting, termination and readthrough at the antizyme recoding site, and the concentrations that affect the ratio, is however far from clear. Although a number of reports indicate that exogenously supplied putrescine, spermidine and spermine are all stimulators of antizyme frameshifting, these studies were conducted in wild-type cells with active polyamine metabolism, making it impossible to define which polyamine is the active reagent (17).
The relatively recent discovery of Oaz1 in S. cerevisiae, and the conserved nature of polyamine metabolism, allows this genetically tractable organism to be used as a test-bed system to quantify polyamine-induced ribosomal frameshifting, and using that analysis, to develop and validate a mathematical model of the ribosomal frameshift feedback controller. Using a yeast with multiple mutations, incapable of polyamine synthesis or metabolism, the in vivo effects of exogenously supplied polyamines on antizyme frameshifting, readthrough and termination could be quantified. These measurements were in turn used to generate a mathematical model describing polyamine effects on ribosome fate at the OAZ1 translational recoding site. Successful validation of this model in different mutant and gene overexpression backgrounds marks a breakthrough in the process of developing a complete predictive model of the ribosome as a polyamine feedback controller.
MATERIALS AND METHODS
Microbial strains and growth conditions
Yeast strains (Table 1) were grown under standard conditions on synthetic defined minimal medium [SD; 0.67% w/v yeast defined minimal medium without amino acids (Formedium), 2% w/v glucose] supplemented with the appropriate amino acids or nucleotides at 2 mg/ml (except for leucine, at 6 mg/ml). Spermidine (10−5 M) was added to support growth of polyamine auxotrophs on SD medium plates. Escherichia coli strain XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’ proAB lacIq ZΔM15 Tn10 (Tetr)]; Stratagene) was used throughout for cloning experiments.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATahis3-1 leu2-0 met15-0 ura3-0 | EUROSCARF |
| spe1 (Y05034) | MATahis3-1 leu2-0 met15-0 ura3-0 spe1::kanMX4 (BY4741 genetic background) | EUROSCARF |
| spe2 (Y11743) | MATα his3-1 leu2-0 lys2-0 ura3-0 spe2::kanMX4 (BY4742 genetic background) | EUROSCARF |
| fms1 (Y10595) | MATα his3-1 leu2-0 lys2-0 ura3-0 fms1::kanMX4 (BY4742 genetic background) | EUROSCARF |
| oaz1 (Y02776) | MATahis3-1 leu2-0 met15-0 ura3-0 oaz1::kanMX4 (BY4741 genetic background) | EUROSCARF |
| spe1 spe2 paa1 (IS527/1a) | MATahis3-1 leu2-0 met15-0 ura3-0 spe1::kanMX4 spe2::natMX4 paa1::hphMX4 (BY4741 genetic background) | This study |
| spe1 spe2 paa1 fms1 (IS532/44d) | MATα his3-1 leu2-0 ura3-0 spe1::kanMX4 spe2::natMX4 paa1::hphMX4 fms1::kanMX4 | This study |
Generation of a spe1 spe2 paa1 fms1 multiple deletant yeast strain
A spe1 spe2 paa1 triple deletant strain was first generated by sequentially deleting the entire SPE2 and PAA1 coding sequences in a spe1::kanMX4 background (Y05034; Table 1), using short-flanking homology PCR-targeting of NatMX4 and HphMX4 markers, respectively (18). The spe1 spe2 paa1 deletant (IS527; Table 1) was then mated with the fms1 deletant strain (Y10595; Table 1), forming a diploid that was then sporulated using pre-sporulation and sporulation media (19) containing 50 μM spermidine and 100 μM β-alanine. Tetrad analysis identified multiply deleted spe1 spe2 paa1 fms1 strains (IS532; Table 1).
Determination of in vivo polyamine concentrations
Polyamines were assayed essentially using the method described with the following modifications (20). All glassware used in polyamine assays was washed with concentrated HCl and rinsed in distilled water to remove adherent polyamines. Colonies grown on SD medium agar plates supplemented with 10−5 M spermidine were inoculated into 3 ml SD overnight cultures, and sub-cultured overnight to an OD600 of 0.65–0.85 in 20 ml SD medium supplemented with different concentrations of polyamines. Cells were harvested, washed twice in ice-cold phosphate buffered saline (PBS) and resuspended in 400 µl ice-cold PBS (suspension A). To carry out high-pressure liquid chromatography (HPLC) analysis, 200 µl of suspension A was centrifuged (15 000g, 30 s) and the cell pellet resuspended for 1 h at 4°C in 200 µl of 1.5 M perchloric acid, resulting in a volume of 215 μl. Cells (10 µl) were enumerated. Cells were then harvested (12 000g, 10 min, 4°C) and 50 µl of the supernatant diluted in sterile distilled water to a final volume of 200 µl. In parallel, polyamine standards (Sigma Aldrich) were prepared using putrescine, spermidine and spermine over a range of 0.125–4 nmol in 200 µl of 0.2 M perchloric acid. The commercial preparation of putrescine and spermine used contained low-level spermidine contamination (<3%), although model fitting and data interpretation in every case took account of this low-level contamination. Samples were subjected to dansylation by alkalinizing with 50 µl of 1 g/ml sodium carbonate decahydrate, followed by the addition of 200 µl of dansyl chloride (15 mg/ml in acetone). The samples were incubated overnight in the dark at 30°C with shaking. Then 125 µl of 10 mg/ml proline was added and samples incubated at 37°C for 15 min. Polyamines were extracted using 500 µl of toluene by vortexing for 30 s, and centrifugation (15 000g, 3 min). The organic phase was recovered and evaporated to dryness under nitrogen. Finally, the samples were reconstituted in 200 µl of methanol and 10 µl of sample subjected to HPLC analysis using a Gilson 234 Injector to load samples onto a Hichrom HIRBP-2922 column (C8/C18, 5 μm; 150 × 4.6 mm). Samples were chromatographed using a methanol gradient (72–90%; 2 min held at 72%, increased to 90% >14 min, held for 4 min, before returning to 72% and re-equilibrated for 5 min) with a flow of 1 ml/min. At least three independent transformants of any given type were quantified by HPLC. Polyamine intracellular concentrations assumed a typical yeast cell volume of 40 µm3 (21); careful measurements showed the quadruple yeast mutant size did not differ from that of the wild type, and that addition of polyamines to the growth medium left cell size unaffected.
Plasmid vectors
To assay ribosomal frameshifting and readthrough at the antizyme frameshift site, pairs of complimentary oligonucleotides corresponding to the core OAZ1 frameshift sequence plus 17 and 10 nt of the flanking 5′ and 3′ sequences, respectively, were cloned into the unique NotI site of vector pAC98T using standard methods (22) (Supplementary Figure S1). pAC98T directs the expression of a β-galactosidase-luciferase fusion protein and is identical to pAC98 vector (23), differing only in the promoter that drives expression of the reporter fusion (24). Cloning the pair-wise combination of oligonucleotides, 5′-GGCCG GGA TTT AAG GAT TGG TGC GCG T GAC ATC CCT CTA G-3′, 5′-GGCC G GGA TTT AAG GAT TGG TGC GCG TGA CAT CCC TCT A-3′ and 5′-GGCCG GGA TTT AAG GAT TGG TGC GCG TGG CAT CCC TCT A-3′, into pAC98T produced plasmids: pAC98T-OAZ1-FS (measuring frameshift frequency), pAC98T-OAZ1-RT (measuring readthrough frequency) and pAC98T-OAZ1-Cont, respectively. In pAC98T-OAZ1-FS, the OAZ1 frameshift site is cloned such that luciferase is in the +1 frame relative to lacZ. By placing the luciferase ORF in the 0 frame (pAC98T-OAZ1-RT), downstream of the antizyme UGA stop codon, luciferase activity reports stop codon readthrough. The remaining plasmid, pAC98T-OAZ1-Cont, consists of the luciferase ORF in the 0 frame, preceded by the OAZ1 frameshift context, in which the UGA stop codon was replaced by a TGG tryptophan codon. This construct reports 100% production of the β-galactosidase–luciferase fusion. Plasmids were transformed into yeast using standard methods (25). High-intracellular concentrations of putrescine were engineered using a plasmid expressing a GAL1 promoter-controlled SPE1 gene (Open Biosystems). SD culture media containing a range of galactose concentrations controlled the expression of SPE1 in pGAL1-SPE1 transformants. Polyamine effects on stop codon readthrough in different contexts were assayed as described (Supplementary Figure S2, legend) using variants of pAC98 in which the lacZ and luc reading frames are separated by stop codons in different contexts (26).
Ribosomal frameshifting and readthrough assay methods
Dicistronic assays for frameshifting and stop codon readthrough were performed essentially as described (24) with the following modifications. Yeast transformants in suspension A (200 µl; see ‘Determination of in vivo polyamine concentrations’ section above) were harvested and resuspended in 200 µl lysis buffer (0.1 M potassium phosphate buffer pH 7.8, 0.2% Triton X-100) and broken using glass beads (5 mm) in a FastPrep machine (MP Biomedicals) over three cycles of 20 s with cooling. The lysate was centrifuged (15 000g, 10 min, 4°C) and the supernatant diluted 250-fold before assay for β-galactosidase and firefly luciferase in 96 well plates using chemiluminescent reporter gene assays, respectively Galacto-Light Plus System (Applied Biosystems) and the Bright-Glo Luciferase Assay System (Promega) and a Glomax 96 luminometer (Promega; 1 s measurement after a 2-s delay following injection of 100-µl assay reagents). Frameshift frequencies were determined by first normalizing measured pAC98T-OAZ1-FS luciferase activities using the corresponding β-galactosidase activity. This value was expressed as a percentage of the corresponding normalized luciferase activity measured using pAC98T-OAZ1-Cont.
Development of a mathematical model and curve fitting to experimental data
The eight kinetic parameters used in the eventual frameshifting function (equation 5) were derived using the curve fitting procedure implemented in NLREG software (Nonlinear Regression Analysis Program Version 6.5, Phillip H. Sherrod, 1991-2010). NLREG uses a model/trust-region technique along with an adaptive choice of the model Hessian. The algorithm is a combination of Gauss-Newton and Levenberg-Marquardt methods.
The foundation of the minimization technique used by NLREG is to compute the sum of the squared residuals (SSR) for one set of parameter values and then slightly change each parameter value and recalculate the SSR to analyse how it is affected by the parameter value change. By dividing the difference between the initial and updated SSR values by the amount the parameter was changed, NLREG is able to provide the approximate partial derivative with respect to the parameter. This partial derivative is employed by NLREG to decide how to change the value of the parameter for the next iteration.
If the initial guess for the parameter is not far from the optimum value, the procedure will provide the best estimate for the parameter. Such a procedure was carried out simultaneously for all parameters representing a minimization problem in eight-dimensional space as there are eight kinetic parameters in the final frameshifting function (equation 5). In this way, the algorithm was used to estimate the kinetic parameters that deliver the best fit for the frameshifting function and experimental measurements. As an important characteristic of non linear regression analysis we specifically considered the SSR between the experimental frameshifting (yi; equation 1) and predicted frameshifting (yip; equation 2) values at each measured polyamine concentration.
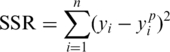 |
(1) |
In addition we also emphasize the regression characteristic that is the standard error of the estimated kinetic parameters. These standard errors can be associated with the variance in the experimental dataset and allowed us to compute the uncertainty range associated with prediction by the frameshifting function (equation 5). This uncertainty range is non-linear with respect to the magnitudes of polyamine kinetic parameters.
To perform optimization of the polyamine kinetic parameters in the frameshifting function (equation 5) by means of non linear regression analysis (NLREG software) we divided all available data into training and testing datasets, the latter to be used in model validation. The training data set consisted of a series of single, double and triple polyamine effects on frameshift levels in the quadruple yeast mutant spe1 spe2 paa1 fms1. To begin with we trained the function to predict frameshifting from single polyamine data sets. Optimized kinetic parameters from the single effect served as an initial guess for NLREG analysis of double polyamine effects on frameshifting. Two experimental training data sets were used for the double polyamine effect on frameshifting: the combined spermine and spermidine (with no putrescine) data set; the combined spermidine and putrescine (with no spermine) data set. The optimised parameters from both these data sets are used as two initial guesses for the triple polyamine effect on frameshifting. At the end stage, both these initial guesses deliver a unique optimized polyamine kinetic parameters solution expressed by the best fit of the frameshifting function (equation 5) to all available frameshift and polyamine concentrations in the quadruple deletant yeast. The testing data set (the model validation data set) consisted of frameshift in oaz1, wild-type (WT) BY4741, spe2[pGAL1-SPE1] and spe2[pRS426] yeast strains. We used the frameshifting function with final optimized kinetic parameters to predict frameshifting levels in the testing data set.
The single experimental effect of spermidine on frameshifting was analysed by non-linear regression analysis (NLREG) with the Michaelis-Menten function (equation 2) which is a simplified form of the frameshift function (equation 5) with zero spermine and putrescine,
| (2) |
where Kmd designates the Michaelis constant for spermidine and Vmaxd the maximal velocity (% frameshift) for spermidine. Spermidine concentration is designated by [Spd]. BFS indicates the basal level of frameshifting frequency, added as a constant. The best curve fit (equation 2) to experimental data has SSR = 5.89 and holds the optimal set of kinetic parameters estimated for single spermidine effect on frameshifting.
A similar analysis was performed for the single putrescine effect on frameshifting with NLREG model of function (equation 3), which is a simplification of equation 5 in the absence of spermine and spermidine.
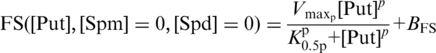 |
(3) |
Here, K0.5p, designates the affinity constant for putrescine and Vmaxp, the maximal velocity (% frameshift) for putrescine. The putrescine concentration is designated by [Put]. The Hill coefficient is designated p. BFS indicates the basal level frameshifting frequency, added as a constant. The best curve fit (equation 3) to experimental data holds sum of squared residuals magnitude 44.55 and corresponds to the optimal set of kinetic parameters estimated for a single putrescine effect on frameshifting. The Hill function (equation 4) represents the single spermine effect on frameshifting, and its kinetic parameters were optimized using NLREG, producing a best-fit curve for the single spermine treatment with sum of squared residuals of magnitude 2.89.
| (4) |
Here, K0.5m, designates the affinity constant for spermine and Vmaxm, the maximal velocity (% frameshift) for spermine. The spermine concentration is designated by [Spm]. The Hill coefficient is designated m. BFS indicates the basal level frameshifting frequency, added as a constant.
We then analysed the frameshift effect of pairwise polyamine treatments on the quadruple yeast deletant to further optimise values of the frameshift function kinetic parameters. For the pairwise experimental treatments, the third polyamine was excluded from the medium. Using such data, a search was performed for the best fit of the function (equation 5) to predict experimentally observed frameshift frequencies. By these means we obtain the optimised polyamine kinetic parameters to use as the initial guess for triple polyamine effect on frameshifting training data set. The NLREG final results for sum of squared residues for double spermine and spermidine (with no added putrescine) treatment was SSR = 78.18 and for spermidine and putrescine (with no added spermine) was SSR = 118.49.
RESULTS
A spe1 spe2 paa1 fms1 deletant yeast strain disabled for polyamine metabolism
Addition of any polyamine to the growth medium stimulates in vivo antizyme frameshifting in cell cultures (17). However, these studies were conducted in systems where metabolic interconversion of the added polyamines was possible, preventing the rigorous analysis of the effects of individual polyamines on antizyme frameshifting and readthrough. It was therefore essential to define how each polyamine governs frameshift and readthrough efficiency in a system in which polyamine conversion is prevented.
To generate such a metabolically isolated system, the SPE1, SPE2, PAA1 and FMS1 genes were deleted to generate a mutant yeast strain in which de novo synthesis and in vivo metabolic interconversion of polyamines taken up from the medium is prevented (Figure 1). The lack of Spe1 and Spe2 prevents putrescine synthesis and its conversion to spermidine and spermine (27,28,29,30), while FMS1 deletion prevents the reverse spermine to spermidine reaction (31,32,33). The absence of Paa1 prevents polyamines from being acetylated (34) (Figure 1). Polyamine analysis of the now polyamine auxotrophic quadruple deletant confirmed that de novo synthesis had been eliminated (Table 2). A block in metabolic interconversion of polyamines was confirmed by separately supplying each of the three polyamines exogenously to the deletant (10−2 mM), and subsequently detecting only that single polyamine in cell extracts (Supplementary Table S1).
Table 2.
Intracellular polyamine concentrations wild-type BY4741 and spe1 spe2 paa1 fms1 deletant strainsa
| Polyamine | Strains |
|
|---|---|---|
| BY4741 | spe1 spe2 paa1 fms1 | |
| Putrescine (mM) | 0.17 ± 0.01 | NDb |
| Spermidine (mM) | 2.88 ± 0.53 | NDb |
| Spermine (mM) | 0.74 ± 0.15 | NDb |
aAnalyzed by high-pressure liquid chromatography (HPLC).
bND, not detectable by HPLC (detection limit was 0.02 mM).
Putrescine, spermidine and spermine stimulate antizyme frameshifting in qualitatively and quantitatively different ways
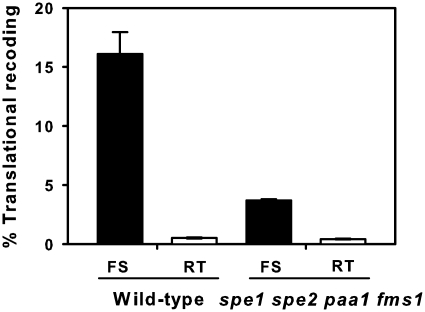
In order to understand how polyamines govern yeast antizyme synthesis, we analysed in vivo how each of the different polyamines affect the three mutually exclusive processes of frameshifting, readthrough and termination at the antizyme frameshift site in the quadruple deletant yeast. Frameshift and readthrough were measured in actively growing wild-type and spe1 spe2 paa1 fms1 cultures using dicistronic reporter plasmids containing the antizyme frameshift site, along with readthrough controls (‘Materials and Methods’ section). In the absence of added polyamines, ∼16.1% of the translating ribosomes shifted to the +1 frame in the wild-type genetic background, and only 3.7% in the quadruple deletant (Figure 2). The readthrough frequencies for both wild-type and quadruple deletant strains were similar and low (0.52 and 0.43%, respectively; Figure 2). Thus frameshifting, but not stop codon readthrough, appears to be polyamine sensitive.
Figure 2.
Antizyme ribosomal frameshifting and readthrough in wild-type BY4741 and spe1 spe2 paa1 fms1 deletant strains. Average percent frameshifting (FS) and readthrough (RT) for wild-type and quadruple deletant strains grown in polyamine-free media. Filled bars represent average frameshift and open bars represent readthrough. Standard deviations are presented for all bars (typically <15% of mean value; n = 3).
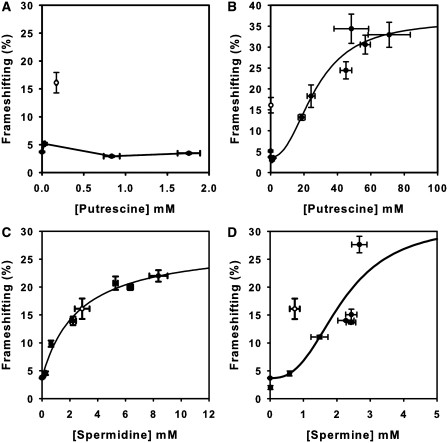
The effect of polyamines on frameshifting at the OAZ1 frameshift site was next examined in the quadruple deletant strain grown in media with varying extracellular polyamine concentrations. The addition of increasing extracellular concentrations of putrescine (10−1, 1, and 10 mM) produced either no effect, or a very minor 1.7-fold stimulatory effect on frameshift frequencies (Figure 3A). However, other studies have shown that very high concentrations of putrescine can stimulate frameshifting at the yeast Ty1 retrotransposon frameshift site, specifically under conditions where very low concentrations of spermidine and spermine were present (35). We therefore overexpressed the SPE1 gene under the control of the GAL1 promoter to engineer higher intracellular concentrations of putrescine. Using galactose in the culture medium to induce SPE1, extremely high levels of putrescine (18.6–71 mM) were attained. At these very high-intracellular concentrations, frameshifting was indeed stimulated, to a maximum of 34% (Figure 3B). Strikingly, the putrescine frameshift response curve was sigmoidal, indicating that the binding of putrescine to the ribosome to stimulate frameshifting was in some way cooperative and may involve multiple binding sites (Figure 3B).
Figure 3.
Single treatment effects of putrescine, spermidine and spermine on frameshifting at the antizyme frameshift site. Average percent frameshifting (filled circles) was measured using a dicistronic assay and plotted versus intracellular polyamine intracellular concentrations in the spe1 spe2 paa1 fms1 deletant strain. (A) Putrescine effects on frameshifting. The highest putrescine concentration used contained 0.16 mM contaminating spermidine, and was therefore not used in the curve fitting process. (B) Putrescine-stimulated frameshift frequencies were measured in the quadruple deletant strain transformed with pGAL1-SPE1 grown on a range of galactose concentrations to regulate SPE1 expression. (C) The effect of intracellular spermidine on frameshift frequency. (D) The effect of intracellular spermine on frameshift frequency. The highest concentration contained 0.2 mM contaminating spermidine. For reference, the wild-type strain BY4741 frameshift frequency (open circles) is represented on all graphs. Error bars (horizontal and vertical) indicate standard deviations for three independent transformants, analysed in triplicate.
Treating the quadruple deletant cultures with a range of extracellular spermidine concentrations (5 × 10−4, 10−3, 2.5 × 10−3, 10−2, 10−1 and 1 mM) produced a progressive increase in frameshifting, reaching a maximum of 22.0% with an intracellular concentration of spermidine of 8.36 mM (Figure 3C). Spermidine is thus a strong stimulator of antizyme frameshifting. Finally, treatment of the deletant with increasing extracellular concentrations of spermine (10−3, 10−2, 10−1, 0.8, 0.9 and 1 mM) produced similar increases in frameshifting. A maximal frameshift stimulation of 27.6% (a 7.5-fold increase) was observed at an intracellular concentration of 2.67 mM. Spermine is thus an even more potent stimulator of frameshifting than spermidine. The sigmoidal nature of the spermine response curve also suggests that binding of this polyamine to the ribosome is co-operative and/or involves multiple binding sites (Figure 3D).
Polyamines do not stimulate stop codon readthrough in yeast
In contrast to their marked effect on antizyme frameshifting, polyamines trigger only slight increases (1.5-fold) in readthrough of the antizyme premature UGA stop codon in yeast (Supplementary Figure S2A, B and C). These findings contrast with other studies reporting that polyamines can enhance stop codon readthrough in E. coli and in mammalian systems (15,16), and enhance readthrough between 2- and 9-fold at the antizyme 2 premature stop codon in COS7 cells (17). These observations call into question whether polyamines in yeast affect frameshifting specifically, or whether they more generally enhance mis-translation events. Accordingly, the effect of polyamines on translational readthrough of stop codons in a variety of different contexts was assessed using dicistronic reporters in the quadruple deletant yeast strain. Exogenous polyamine concentrations were chosen that significantly enhance translational frameshift frequencies (see legend, Supplementary Figure S3). The results show clearly that irrespective of the nature of the stop codon and its context, neither spermidine nor spermine significantly enhanced readthrough of any of the three stop codons (Supplementary Figure S3).
Frameshift stimulation by polyamine combinations reveals evidence of competition for common ribosomal binding sites
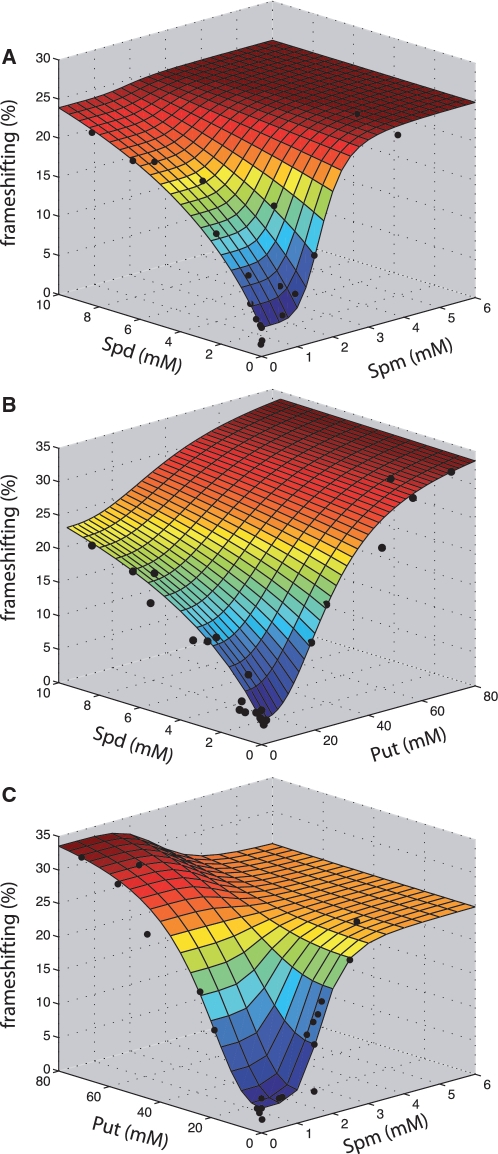
Having established that spermidine and spermine (and at high concentrations, putrescine), stimulate antizyme frameshifting, combination-specific polyamine effects on frameshifting and readthrough were tested. The spe1 spe2 paa1 fms1 deletant strain was therefore cultured in medium containing different combinations of extracellular polyamines (spermidine and spermine; putrescine and spermidine; putrescine and spermine). The results showed that together, spermidine and spermine (combined 0.01–1.04 mM) produced approximately median frameshift frequencies between the values that each polyamine stimulated individually. At higher combined concentrations of spermidine and spermine (>2.5 mM) frameshift frequencies were observed higher than those obtained when testing spermidine and spermine individually (Figure 4A).
Figure 4.
Combinatorial effects of polyamines on frameshifting at the antizyme frameshift site; experimental data and model prediction. Frameshifting frequencies were measured using a dicistronic reporter vector containing the OAZ1 recoding site. Intracellular polyamine concentrations were measured by HPLC. The combined effect of spermidine and spermine (A), putrescine and spermidine (B) and putrescine and spermine (C) on frameshifting are plotted as filled circles, and the model prediction (mesh surface) depicting the fitted frameshift function (Equation 5). Note that panel (C) data points are derived from combined putrescine/spermine treatments some of which contained trace spermidine contamination, but the mesh surface indicates predicted responses to dual spermine/putrescine treatment only. For clarity, experimental data standard deviations are not presented but were typically <15% of mean value (n = 3).
The effect of putrescine, which at low concentrations does not stimulate frameshifting (Figure 3A), was then assessed in combination with either spermidine or spermine, which individually were strong frameshift stimulators. Across a range of putrescine concentrations, the stimulatory effects of adding either 0.0025, 0.01 or 1 mM spermidine were somewhat suppressed in comparison to the expected levels of frameshifting generated by spermidine alone (Figure 4B). However, when the level of putrescine was maintained, but the concentration of spermidine was incrementally raised, this minor inhibitory effect of putrescine was overcome (Figure 4B). No such suppression of spermine-induced frameshifting by putrescine was observed (Figure 4C). At low concentrations, putrescine therefore acted as a weak suppressor of the stimulatory effect of spermidine on frameshifting. Polyamines thus do not significantly stimulate stop codon readthrough, but exert combinatorial and competitive effects on frameshifting consistent with their similar chemical nature.
Modeling the ribosomal frameshift feedback controller
Using the experimental data above, a mathematical function was introduced to capture the complex individual and combinatorial effects of the three polyamines on ribosomal frame maintenance. We noted that the single treatment spermidine frameshift response curve (Figure 3C) bore a striking similarity to a Michaelis–Menten enzyme kinetic function. Frameshifting was thus represented as an enzyme-like activity of the ribosome, stimulated by spermidine ‘substrate’. After first subtracting the background frameshift frequency from all measurements, a curve-fitting analysis was conducted to determine the maximal frameshifting 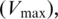 and affinity constant (Km) for spermidine interaction with the ribosome. Frameshift response curves for single putrescine and spermine treatments had indicated co-operative binding effects (Figures 3B and D), so Hill functions were used to represent their individual effects on frameshifting, and curve fitting (using non-linear regression methods; Materials and Methods) used to minimize residual sum of squares and define the optimal frameshifting
and affinity constant (Km) for spermidine interaction with the ribosome. Frameshift response curves for single putrescine and spermine treatments had indicated co-operative binding effects (Figures 3B and D), so Hill functions were used to represent their individual effects on frameshifting, and curve fitting (using non-linear regression methods; Materials and Methods) used to minimize residual sum of squares and define the optimal frameshifting 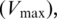 affinity constant (K0.5), and Hill coefficients for ribosome interaction (Table 3).
affinity constant (K0.5), and Hill coefficients for ribosome interaction (Table 3).
Table 3.
Polyamine frameshift model kinetic parameters
| Polyamine | Kinetic parameter; symbol | Value | Standard error (single polyamine) | Uncertainty range (combined treatment) |
|---|---|---|---|---|
| Putrescine | Maximum velocity 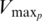 (percentage of frameshift) (percentage of frameshift) |
32% | ±7.6 | 24.4–39.6 |
| Affinity constant: K05p | 26.6 mM | ±7.4 | 19.2–33.9 | |
| Hill coefficient: p | 2.4 | ±1.4 | 1.1–3.8 | |
| Spermidine | Maximum velocity 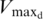 (percentage of frameshift) (percentage of frameshift) |
33.9% | ±12.5 | 21.4–46.3 |
| Michaelis constant: Kmd | 6.8 mM | ±4.2 | 2.6–10.9 | |
| Spermine | Maximum velocity 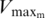 (percentage of frameshift) (percentage of frameshift) |
21.3% | ±2.7 | 18.6–24 |
| Affinity constant: K05m | 1.7 mM | ±0.2 | 1.5–1.8 | |
| Hill coefficient: m | 5.2 | ±2.3 | 2.9–7.5 |
To describe the combinatorial effects of the three polyamines, we recognized the chemical and structural similarities of the polyamines and proposed that they act as competing pseudo-substrates for the same active site(s) on the ribosome. Consistent with this, spermine and spermidine combinations can produce intermediate frameshift frequencies at certain concentrations (Figure 4), consistent with their structural similarity. We therefore generated an enzyme kinetic function ‘FS’ based on the kinetics of an enzyme with competing substrates (36) that described the action of the pseudo-substrates (Put, Spd, Spm) on a single enzyme activity (i.e. frameshift efficiency). The total frameshift frequency was, therefore, the sum of three individual frequencies contributed by each of the three polyamines. In the global frameshift function (equation 5), subsidiary frameshift functions describe the contribution made by each polyamine to the total frameshift frequency, moderated by competition from the other two polyamines through additional [concentration]/[affinity constant] terms in the denominator of each subsidiary term as described for an enzyme with competing substrates (36);
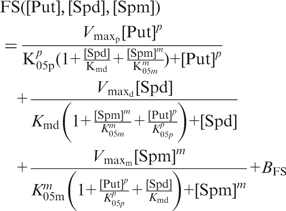 |
(5) |
In equation 5, ‘K’ parameters define affinity constants, ‘V’ parameters indicate velocities (frameshifting) (see Table 3 for details) and concentrations of the individual polyamines are indicated by [Spd], [Spm] and [Put]. The Hill coefficients are designated by m (spermine) and p (putrescine). BFS indicates the constant basal-level FS measured in the absence of polyamines.
The feedback-controller function describing the competing polyamine substrates accurately predicts the combinatorial effects of pair-wise polyamine treatments, which accurately map onto the 3D model mesh (Figure 4), validating our assumption that the three polyamines effectively compete for the same site(s) on the ribosomal ‘enzyme’. The biological principle underpinning the mathematical model is thus an accurate representation of the polyamine molecular interactions with the ribosome.
Validation of the frameshift feedback controller model using polyamine pathway mutants
To rigorously validate the model of the ribosomal frameshift feedback controller, other engineered yeast genetic backgrounds were used to generate hitherto unexplored combinations and concentrations of the three polyamines. Accordingly, frameshifting and intracellular polyamine concentrations were measured in a spe2 mutant with and without SPE1 gene overexpression, in an oaz1 deletant, and in the wild-type strain. The measured intracellular polyamine concentrations from each mutant strain were fed into the frameshift function, and the predicted in silico frameshift values compared with the corresponding in vivo measurements.
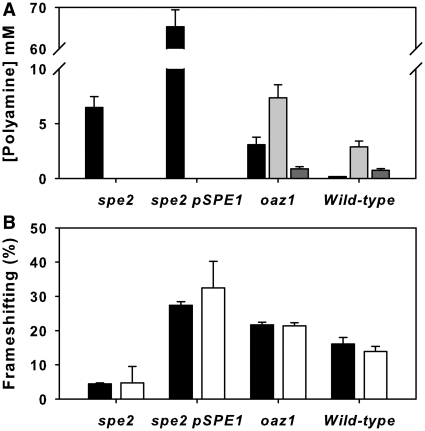
In the spe2 deletant strain, putrescine accumulated to moderately high levels (6.5 mM; Figure 5A). The measured 4.5% frameshifting was accurately predicted by the model (4.7% frameshift, Figure 5B). Overexpressing the SPE1 gene under the control of the GAL1 promoter in this strain (spe2 pSPE1), produced 65.3 mM intracellular putrescine and a frameshift frequency of 27.4%, again, well approximated by the model (32.4%; Figure 5B). To further validate the model, the oaz1 antizyme deletant was selected, which generates atypical concentrations of all three polyamines (high levels of putrescine and spermidine, normal spermine; Figure 5A), and high levels of frameshifting (21.7%), and which thus tested the ability of the model to integrate the competing polyamine effects. This level of frameshift was accurately predicted by the feedback controller function (21.4%; Figure 5). Finally, we verified that the wild-type frameshift frequency (16.1%) was accurately predicted by the model (13.9%; Figure 5B). The model thus demonstrated an ability to accurately predict the frameshift efficiencies generated by a wide range of different genetic conditions that were typified by markedly different polyamine steady states.
Figure 5.
Validation of the ribosomal controller frameshift model. (A) Polyamine concentrations were measured in a spe2 mutant with and without SPE1 gene overexpression, in an oaz1 deletant, and in the wild-type strain (putrescine; filled bar, spermidine; light grey, spermine; dark grey). Error bars represent standard deviations (n = 3). (B) Ribosomal frameshift frequencies were measured in the same mutant panel (filled bars) and the intracellular polyamine concentrations measured in the mutants were fed into the frameshift function to predict the frameshift frequency (open bars; error bars represent model uncertainty range originating with variation in the experimental data).
DISCUSSION
In this study, a novel quadruple yeast gene knockout strategy was devised in which de novo synthesis and metabolic interconversion of supplied polyamines is prevented. Using this tool, this study aimed to define for the first time how each of the polyamines individually stimulates the frameshifting, termination and readthrough events at the antizyme frameshift site in vivo. With this information, a predictive mathematical model of the ribosomal frameshift controller (Figure 1B) could then be generated which will form the cornerstone of future systems biological analysis of polyamine biosynthesis.
The observed polyamine effects on antizyme stop codon readthrough in the quadruple knockout yeast strain (Supplementary Figure S2) showed clear, system-specific differences in the way that polyamines influence translation termination. Previous studies have reported stimulation of stop codon readthrough in E. coli (16). Polyamines can also stimulate readthrough in in vitro translation systems (15). However, in this study, readthrough of the antizyme stop codon, and of other stop codons in a range of weak and strong contexts was refractory to polyamine stimulation (Supplementary Figures S2 and S3). This highlights potential differences between yeast and E. coli ribosome responses, and between in vivo measurements and those made in cell-free systems. However, the observations made simplify the development of a mathematical description of the yeast ribosomal feedback controller, since only a two-way competition between frameshifting and termination need be considered.
The analysis of polyamine frameshift responses revealed that although spermidine stimulates frameshifting with a hyperbolic (Michaelis–Menten) function, in contrast, putrescine and spermine each appear to bind to the ribosome in a co-operative manner (Figure 3B and D; Hill coefficients of 2.4 and 5.2, respectively). We speculate that this co-operativity could be generated by the existence of multiple polyamine binding sites on the ribosome, and that frameshifting stimulation requires gradual polyamine occupancy of these sites. This conclusion is consistent with photo-affinity labeling studies revealing spermine binding at several sites in the 23S rRNA of E. coli, including those regions involved in positioning and translocation of tRNA (37,38,39). Why spermidine responses are not co-operative is unclear (Figure 5B), but by extension, this may suggest the existence of a single spermidine binding site. This could in turn explain why at low concentrations putrescine slightly inhibits spermidine-induced frameshifting; putrescine occupancy of the putative, single spermidine-sensitive site may compete with spermidine binding, without putrescine itself triggering frameshifting (which requires multiple site occupancy; Figure 3B). However, the high-affinity constant for putrescine–ribosome interaction (10-fold higher than spermidine; Table 3) ensures that this is only a subtle effect.
Putrescine stimulation of antizyme frameshifting has been reported previously, although in cell culture systems where metabolic interconversion of putrescine into spermidine and spermine could occur (17,40). In our study, where interconversion was prevented by gene knockout, low concentrations of putrescine did not stimulate frameshifting (Figure 3A). This would be consistent with the fact that putrescine is a pathway intermediate with limited polycation character and function; as such, it is not desirable for low concentrations of putrescine to trigger frameshifting, antizyme production and pathway shut-down. Previously, reports indicated that a yeast spe2 deletion (exhibiting somewhat raised putrescine concentrations) triggered high levels of frameshifting at the yeast retrotransposon Ty1 frameshift site, but only following a period of spermidine deprivation (35,41). Experimentally, our study also confirmed that putrescine stimulates frameshifting at high concentrations (Figure 3B). Our experimental data, and the derived mathematical frameshift function, also indicates that spermidine would suppress the putrescine-induced frameshifting to some extent (Figure 4B). It is possible that any differences between our study and others (35,41) in the degree of spermidine suppression of putrescine frameshifting might stem from their use of a Ty1 frameshift site, rather than antizyme, to report polyamine responses.
Pair-wise treatments of polyamines (Figure 4) revealed that spermidine and spermine together generate frameshift frequencies intermediate between their individual effects. Consistent with their similar positively charged chemical structure, this indicated that polyamines might compete for the same binding sites on the ribosome. We used this discovery as the platform for the development of a mathematical model capable of describing frameshift responses to a full range of polyamine conditions in the cell. Importantly, although the model parameter fitting process primarily made use of single and pair-wise polyamine data sets, the complete model was able to accurately predict frameshift efficiencies measured in both the wild-type strain, and in an antizyme mutant (oaz1), each of which contain very different (triple) polyamine combinations.
The successful process of validating model predictions supports the underlying assumption that, in vivo, polyamines compete for common polyamine binding sites on the ribosome, and thus individually act as inhibitors of frameshift stimulation by either of the other two polyamines. The approach has also allowed the in vivo parameterisation of that competition. The distinct hyperbolic and co-operative responses of frameshifting to spermidine and spermine respectively determine non-linear frameshift responses to the polyamine milieu. This regulatory complexity justifies the need for a mathematical function to describe the four-dimensional relationship between frameshifting and the three polyamines. The model in addition provides a robust and quantitative tool to analyze the action of therapeutic polyamine analogues and other potential frameshift agonists on translational frameshifting. The validated ribosomal frameshift model will in addition act as a fulcrum for the development of complete mathematical models of the polyamine pathway and its regulation.
Beyond the field of polyamine biology, the ribosomal controller model we have developed is directly relevant to understanding other feedback translational regulation systems, for example the CPA1 arginine-regulated expression system (42). Translational feedback modules, with their inherently rapid response characteristics, could also represent useful building blocks in synthetic biology. In contrast to the well-understood example of transcriptional feedback, the current predictive mathematical understanding of translational control is very poor, representing a major stumbling block to its use in the design of synthetic circuits. The model presented in this paper therefore not only helps define the workings of a key metabolic pathway important in a number of human pathologies, but will also allow the introduction of novel translational-based feedback control modules to the synthetic biologist's toolkit.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BB/F019084/1 to I.S. and H.M.W., and BB/F019602/1 to D.G.B.). Funding for open access charge: Biotechnology and Biological Sciences Research Council.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem. J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace HM, Fraser AV. Inhibitors of polyamine metabolism. Amino Acids. 2004;26:353–365. doi: 10.1007/s00726-004-0092-6. [DOI] [PubMed] [Google Scholar]
- 3.Poulin R, Coward JK, Lakanen JR, Pegg AE. Enhancement of the spermidine uptake system and lethal effects of spermidine overaccumulation in ornithine decarboxylase-overproducing L1210 cells under hyposmotic stress. J. Biol. Chem. 1993;268:4690–4698. [PubMed] [Google Scholar]
- 4.Tobias KE, Kahana C. Exposure to ornithine results in excessive accumulation of putrescine and apoptotic cell death in ornithine decarboxylase overproducing mouse myeloma cells. Cell Growth Differ. 1995;6:1279–1285. [PubMed] [Google Scholar]
- 5.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palanimurugan R, Scheel H, Hofmann K, Dohmen RJ. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–4867. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol. Cell. Biol. 1992;12:3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Bates DG, Postlethwaite I, Ma L, Iglesias PA. Robustness analysis of biochemical network models. Syst. Biol. 2006;153:96–104. doi: 10.1049/ip-syb:20050024. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Caso C, Montanez R, Cascante M, Sanchez-Jimenez F, Medina MA. Mathematical modeling of polyamine metabolism in mammals. J. Biol. Chem. 2006;281:21799–21812. doi: 10.1074/jbc.M602756200. [DOI] [PubMed] [Google Scholar]
- 11.Vimaladithan A, Farabaugh PJ. Special peptidyl-tRNA molecules can promote translational frameshifting without slippage. Mol. Cell. Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundararajan A, Michaud WA, Qian Q, Stahl G, Farabaugh PJ. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell. 1999;4:1005–1015. doi: 10.1016/s1097-2765(00)80229-4. [DOI] [PubMed] [Google Scholar]
- 13.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 15.Hryniewicz MM, Vonder Haar RA. Polyamines enhance readthrough of the UGA termination codon in a mammalian messenger RNA. Mol. Gen. Genet. 1983;190:336–343. doi: 10.1007/BF00330661. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida M, Kashiwagi K, Kawai G, Ishihama A, Igarashi K. Polyamines enhance synthesis of the RNA polymerase sigma 38 subunit by suppression of an amber termination codon in the open reading frame. J. Biol. Chem. 2002;277:37139–37146. doi: 10.1074/jbc.M206668200. [DOI] [PubMed] [Google Scholar]
- 17.Petros LM, Howard MT, Gesteland RF, Atkins JF. Polyamine sensing during antizyme mRNA programmed frameshifting. Biochem. Biophys. Res. Commun. 2005;338:1478–1489. doi: 10.1016/j.bbrc.2005.10.115. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 19.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 20.Fontaniella B, Mateos JL, Vicente C, Legaz ME. Improvement of the analysis of dansylated derivatives of polyamines and their conjugates by high-performance liquid chromatography. J. Chromatogr. A. 2001;919:283–288. doi: 10.1016/s0021-9673(01)00841-x. [DOI] [PubMed] [Google Scholar]
- 21.Johnston GC, Ehrhardt CW, Lorincz A, Carter BL. Regulation of cell size in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, and Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes EM, Nieduszynska SR, Brunton FK, Gibson J, Glover LA, Stansfield I. Control of gag-pol gene expression in the Candida albicans retrotransposon Tca2. BMC Mol. Biol. 2007;8:94. doi: 10.1186/1471-2199-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 26.Williams I, Richardson J, Starkey A, Stansfield I. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:6605–6616. doi: 10.1093/nar/gkh1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohn MS, Tabor CW, Tabor H. Regulatory mutations affecting ornithine decarboxylase activity in Saccharomyces cerevisiae. J. Bacteriol. 1980;142:791–799. doi: 10.1128/jb.142.3.791-799.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitney PA, Morris DR. Polyamine auxotrophs of Saccharomyces cerevisiae. J. Bacteriol. 1978;134:214–220. doi: 10.1128/jb.134.1.214-220.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohn MS, Tabor CW, Tabor H. Isolation and characterization of Saccharomyces cerevisiae mutants deficient in S-adenosylmethionine decarboxylase, spermidine, and spermine. J. Bacteriol. 1978;134:208–213. doi: 10.1128/jb.134.1.208-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol. Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joets J, Pousset D, Marcireau C, Karst F. Characterization of the Saccharomyces cerevisiae FMS1 gene related to Candida albicans corticosteroid-binding protein 1. Curr. Genet. 1996;30:115–120. doi: 10.1007/s002940050109. [DOI] [PubMed] [Google Scholar]
- 32.White WH, Gunyuzlu PL, Toyn JH. Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J. Biol. Chem. 2001;276:10794–10800. doi: 10.1074/jbc.M009804200. [DOI] [PubMed] [Google Scholar]
- 33.Chattopadhyay MK, Tabor CW, Tabor H. Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase. Proc. Natl Acad. Sci. USA. 2003;100:13869–13874. doi: 10.1073/pnas.1835918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B, Sutton A, Sternglanz R. A yeast polyamine acetyltransferase. J. Biol. Chem. 2005;280:16659–16664. doi: 10.1074/jbc.M414008200. [DOI] [PubMed] [Google Scholar]
- 35.Balasundaram D, Dinman JD, Tabor CW, Tabor H. SPE1 and SPE2: Two essential genes in the biosynthesis of polyamines that modulate +1 ribosomal frameshifting in Saccharomyces cerevisiae. J. Bacteriol. 1994;176:7126–7128. doi: 10.1128/jb.176.22.7126-7128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornish-Bowden A. Fundamentals of enzyme kinetics. London, UK: Portland Press; 2004. [Google Scholar]
- 37.Amarantos I, Zarkadis IK, Kalpaxis DL. The identification of spermine binding sites in 16S rRNA allows interpretation of the spermine effect on ribosomal 30S subunit functions. Nucleic Acids Res. 2002;30:2832–2843. doi: 10.1093/nar/gkf404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL. Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nucleic Acids Res. 2005;33:2792–2805. doi: 10.1093/nar/gki557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amarantos I, Kalpaxis DL. Photoaffinity polyamines: Interactions with AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli ribosomes. Nucleic Acids Res. 2000;28:3733–3742. doi: 10.1093/nar/28.19.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard MT, Shirts BH, Zhou J, Carlson CL, Matsufuji S, Gesteland RF, Weeks RS, Atkins JF. Cell culture analysis of the regulatory frameshift event required for the expression of mammalian antizymes. Genes Cells. 2001;6:931–941. doi: 10.1046/j.1365-2443.2001.00477.x. [DOI] [PubMed] [Google Scholar]
- 41.Balasundaram D, Dinman JD, Wickner RB, Tabor CW, Tabor H. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delbecq P, Werner M, Feller A, Filipkowski RK, Messenguy F, Pierard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol. Cell. Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.