Abstract
It has long been known that type II topoisomerases require divalent metal ions in order to cleave DNA. Kinetic, mutagenesis and structural studies indicate that the eukaryotic enzymes utilize a novel variant of the canonical two-metal-ion mechanism to promote DNA scission. However, the role of metal ions in the cleavage reaction mediated by bacterial type II enzymes has been controversial. Therefore, to resolve this critical issue, this study characterized the DNA cleavage reaction of Escherichia coli topoisomerase IV. We utilized a series of divalent metal ions with varying thiophilicities in conjunction with oligonucleotides that replaced bridging and non-bridging oxygen atoms at (and near) the scissile bond with sulfur atoms. DNA scission was enhanced when thiophilic metal ions were used with substrates that contained bridging sulfur atoms. In addition, the metal-ion dependence of DNA cleavage was sigmoidal in nature, and rates and levels of DNA cleavage increased when metal ion mixtures were used in reactions. Based on these findings, we propose that topoisomerase IV cleaves DNA using a two-metal-ion mechanism in which one of the metal ions makes a critical interaction with the 3′-bridging atom of the scissile phosphate and facilitates DNA scission by the bacterial type II enzyme.
INTRODUCTION
Virtually every eubacteria encodes two type II topoisomerases, gyrase and topoisomerase IV (1–6). These enzymes help to regulate the superhelical density of the bacterial chromosome and remove knots and tangles from the double helix. Gyrase and topoisomerase IV alter DNA topology by generating transient double-stranded breaks in the genetic material and passing an intact segment of DNA through the break (1–6). However, the two enzymes appear to play different roles in the cell (1–6). Gyrase is the only known type II enzyme that is able to actively underwind the double helix. It is involved primarily in regulating the superhelical density of chromosomal DNA and alleviating torsional stress that accumulates ahead of DNA tracking systems. In contrast, topoisomerase IV efficiently decatenates DNA and is the major enzyme that is responsible for unknotting and untangling the bacterial genome (1–6).
Beyond their critical physiological functions, gyrase and topoisomerase IV are targets for quinolone-based drugs (1,6–9). Members of this drug class, such as levofloxacin and ciprofloxacin, are among the most active and broad-spectrum antibacterial agents currently in clinical use. Quinolones kill bacterial cells by stabilizing covalent enzyme-cleaved DNA complexes, which are requisite intermediates in the DNA strand passage reactions of gyrase and topoisomerase IV (1,7,8,10–12). These ‘cleavage complexes’ are converted to permanent strand breaks by collisions with helicases and other DNA tracking enzymes, triggering an SOS response in the bacterial cell (1,7,8,10–12). Gyrase and topoisomerase IV both contribute to the cytotoxicity of quinolones (1,7,13–15). However, the enzyme primarily responsible for cell death appears to be species- and drug-dependent.
The ability of type II topoisomerases to cleave and ligate DNA is central to their catalytic functions (1–6). Topoisomerase IV is composed of two different subunits (ParC and ParE in Gram-negative species), each of which is present in two copies in the active enzyme (1–6). Each ParC subunit contains an active site tyrosine (position 120 in Escherichia coli) (1,4,16). This residue cuts one strand of the double helix and forms a covalent bond with the 5′-terminal phosphate that results from scission (1–6). A 3′-terminal –OH moiety also is generated during this process. The two scissile bonds are located across the major groove, four bases apart from one another, such that the DNA cleavage event results in 4-base, 5′-overhanging cohesive ends.
Despite the importance of gyrase and topoisomerase IV in the regulation of DNA topology in bacteria and the treatment of infectious diseases, many aspects of the DNA cleavage reaction are poorly understood. To this point, an area of emerging interest is the use of divalent metal ions in catalyzing the DNA scission event (17,18).
Based on the ability of mutant gyrase proteins to utilize metal ions, Noble and Maxwell (19) proposed that the bacterial enzyme utilizes a two-metal-ion phosphotransferase/hydrolase mechanism similar to that of DNA primases and polymerases (20,21). In this mechanism, one divalent cation activates a catalytic water or ribose hydroxyl for nucleophilic attack, the other coordinates a leaving group and both stabilize the pentavalent transition state. While the mutagenesis approach employed in this study provided evidence for the use of two divalent metal ions, it was not able to provide information on the specific roles of the metal ions in the DNA cleavage/ligation reaction of gyrase.
A greater understanding of metal ion usage by type II topoisomerases was provided by enzymological and mutagenesis studies with human topoisomerase IIα and topoisomerase IIβ (18,22–26). These studies indicated that the human type II enzymes also employ a two-metal-ion mechanism. Furthermore, they suggested that one of the metal ions has an important interaction with the bridging oxygen of the scissile bond that greatly stimulates DNA cleavage. An interaction with the non-bridging oxygen of the scissile bond also was noted, but the ability of this interaction to stimulate DNA cleavage varied between the two human isoforms (18,22–26).
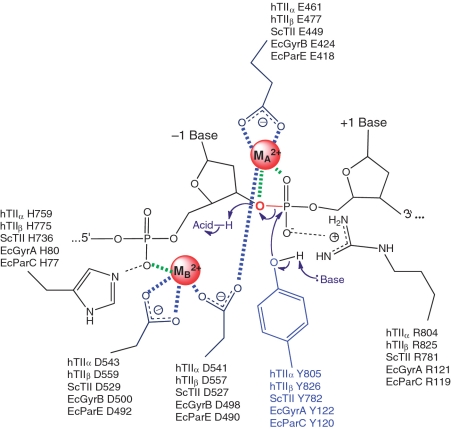
A recent crystal structure of the covalent yeast topoisomerase II-cleaved DNA complex contained both metal ions and led to the proposal that eukaryotic topoisomerase II employs a novel variation of the canonical two-metal-ion mechanism for DNA cleavage (Figure 1) (27). In this model, one metal ion (metal ion A) plays a direct role in the transition chemistry, most likely by promoting the leaving of the ribose 3′-OH. The other metal ion (metal ion B) appears to play a structural role in anchoring the DNA during cleavage.
Figure 1.
Proposed two-metal-ion mechanism for DNA cleavage and ligation mediated by type II topoisomerases. A novel variant of the canonical two-metal-ion mechanism employed by primases and polymerases is shown. Amino acids in the active site that are postulated to function in catalysis by human topoisomerase IIα (hTIIα) and topoisomerase IIβ (hTIIβ) Saccharomyces cerevisiae topoisomerase II (ScTII), and the corresponding amino acids in the GyrA (EcGyrA) and GyrB (EcGyrB) subunits of E. coli gyrase and the ParC (EcParC) and ParE (EcParE) subunits of E. coli topoisomerase IV are indicated. Metal ions (MA2+ and MB2+) are highlighted in red. The scissile bond (between the −1 and +1 bases) also is shown in red. Interactions between metal ions and nucleic acids or acidic amino acid residues are denoted in green or blue, respectively. In the proposed model, MA2+ makes critical contacts with the 3′-bridging atom (red) and a non-bridging atom of the scissile phosphate and plays a role in the transition chemistry and in stabilizing the leaving 3′-terminal oxygen. MB2+ contacts the non-bridging oxygen of the phosphate that connects the −2 and −1 bases upstream from the scissile bond and plays a structural role in anchoring the DNA during scission. Figure is adapted from Schmidt et al. (27).
Structures for non-covalent and covalent complexes formed between bacterial type II topoisomerases and DNA have been reported recently (28–31). In contrast to the work with yeast topoisomerase II, some of these structures contained divalent metal ions at site A or B, but none contained ions at both sites. These results led to an alternative proposal for the use of divalent cations by bacterial type II enzymes, suggesting that they use a ‘moving metal ion mechanism’ in which only one of the two metal ion sites is filled at any given time (30–33).
In order to resolve this fundamental issue regarding the use of divalent metal ions by the bacterial type II enzymes, we carried out enzymological studies with E. coli topoisomerase IV. These studies utilized a series of divalent metal ions with varying thiophilicities in conjunction with DNA cleavage substrates that replaced specific bridging and non-bridging oxygen molecules with sulfur. Results strongly suggest that topoisomerase IV mediates DNA scission via a two-metal-ion mechanism and that one of the required divalent cations makes critical interactions with the bridging atom of the scissile bond during the cleavage event. Thus, it is proposed that the bacterial and eukaryotic type II topoisomerases utilize a common metal ion-dependent mechanism for cleaving DNA.
MATERIALS AND METHODS
Enzymes and materials
E. coli topoisomerase IV was expressed and purified as described by Peng and Marians or by a minor modification of Corbett et al. (34,35). In the latter case, the cell pellet was resuspended in 20 mM HEPES (pH 7.5), 400 mM NaCl, 10% glycerol and 2 mM β-mercaptoethanol, lysed by sonication and centrifuged. The supernatant was loaded onto a HisTrap HP column (GE Healthcare) and the protein was eluted with a linear gradient of 0–200 mM imidazole. The protein containing fractions were pooled and desalted on a HiPrep desalting column (GE Healthcare). The His-tag was removed by overnight incubation with AcTEV at 4°C using an OD280nm ratio 1:100 AcTEV:protein, and the cleaved protein was filtered through a HisTrap HP column. The protein was concentrated and further purified on a Superdex 200 10/300 GL column (GE Healthcare). The concentrations of ParC and ParE were quantified by UV absorption. Negatively supercoiled pBR322 DNA was prepared from E. coli using a Plasmid Mega Kit (Qiagen) as described by the manufacturer.
A 50-bp oligonucleotide duplex was designed using a previously identified topoisomerase II cleavage site from pBR322. This site corresponds to the sequence designated as site 2 by Fortune et al. (36). Wild-type oligonucleotide sequences were generated using an Applied Biosystems DNA synthesizer. The 50-mer top and bottom sequences were 5′-TTGGTATCTGCGCTCTGCTGAAGCC↓AGTTACCTTCGGAAAAAGAGTTGGT-3′ and 5′-ACCAACTCTTTTTCCGAAGGT↓AACTGGCTTCAGCAGAGCGCAGATACCAA-3′, respectively (arrow denotes site of cleavage).
DNA containing a single 3′-bridging phosphorothiolate linkage at the scissile bond (−1/+1 position) was synthesized as described previously (37). Substrates containing a racemic phosphorothioate in place of a non-bridging oxygen at the scissile bond or at the −2/−1 position of the bottom strand were synthesized by Operon.
[γ-32P]ATP (∼5000 Ci/mmol) was obtained from ICN. Single-stranded oligonucleotides were labeled on their 5′-termini using T4 polynucleotide kinase (New England Biolabs). Following labeling and gel purification, complementary oligonucleotides were annealed by incubation at 70°C for 10 min and cooling to 25°C.
Topoisomerase IV-mediated cleavage of negatively supercoiled plasmid substrates
DNA cleavage assays were carried out using the procedure of Fortune and Osheroff (38). Unless otherwise stated, assay mixtures contained 35 nM topoisomerase IV (1:1 ratio of ParC and ParE subunits) and 10 nM negatively supercoiled pBR322 DNA in a total of 20 µl of 40 mM Tris–HCl (pH 7.9), 2.5 mM MgCl2, MnCl2, or CaCl2 and 2.5% glycerol. DNA cleavage mixtures were incubated for 10 min at 37°C, and enzyme-DNA cleavage intermediates were trapped by adding 2 µl of 5% SDS and 1 µl of 375 mM EDTA, pH 8.0. Proteinase K was added (2 µl of a 0.8 mg/ml solution), and reaction mixtures were incubated for 30 min at 45°C to digest the topoisomerase IV. Samples were mixed with 2 µl of 60% sucrose in 10 mM Tris–HCl (pH 7.9), 0.5% bromophenol blue and 0.5% xylene cyanol FF, heated for 2 min at 45°C, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris–acetate, (pH 8.3) and 2 mM EDTA containing 0.5 µg/ml ethidium bromide. DNA cleavage was monitored by the conversion of negatively supercoiled plasmid DNA to linear molecules. DNA bands were visualized by ultraviolet light and quantified using an Alpha Innotech digital imaging system.
Topoisomerase IV-mediated cleavage of oligonucleotide substrates
DNA cleavage assays were carried out by a modification of the procedure of Deweese et al. (37). Oligonucleotide substrates were always 5′-end-labeled. Unless otherwise stated, cleavage reactions contained 70 nM topoisomerase IV (1:1 ratio of ParC and ParE subunits) and 100 nM double-stranded oligonucleotide in a total of 10 µl of 40 mM Tris–HCl (pH 7.9), 10 mM MgCl2, MnCl2 or CaCl2, and 2.5% glycerol. In some cases, the concentration of divalent cation was varied or combinations of the cations were used. Reactions were initiated by the addition of enzyme and were incubated for up to 10 min at 37°C. DNA cleavage products were trapped by the addition of 2 µl of 10% SDS. DNA products were precipitated in 100% ethanol, washed with 70% ethanol, dried and resuspended in 5 µl of sample loading buffer (40% formamide, 10 mM NaOH, 0.02% xylene cyanol FF and 0.02% bromophenol blue). Cleavage products were resolved by electrophoresis in 14% denaturing polyacrylamide gels. To inhibit oxidation of cleaved oligonucleotides containing 3′-terminal –SH moieties and the formation of multimers in the gel, 100 mM DTT was added to the sample loading buffer (37). DNA cleavage products were visualized and quantified using a Bio-Rad Molecular Imager.
Topoisomerase IV-mediated cleavage of linear plasmid substrates
DNA cleavage sites were mapped using a modification of the procedure of O’Reilly and Kreuzer (39). A linear 4330 bp fragment (HindIII/EcoRI) of pBR322 plasmid DNA singly labeled with 32P on the 5′-terminus of the HindIII site was used as the cleavage substrate. The plasmid was linearized by treatment with HindIII. Terminal 5′-phosphates were removed by treatment with calf intestinal alkaline phosphatase and replaced with [32P]phosphate using T4 polynucleotide kinase and [γ-32P]ATP. The DNA was treated with EcoRI, and the 4330 bp singly end-labeled fragment was purified from the small EcoRI-HindIII fragment by passage through a CHROMA SPIN+TE-100 column (Clontech).
Reaction mixtures contained 1.4 nM labeled linear DNA and 35 nM topoisomerase IV (1:1 ratio of ParC and ParE subunits) in 50 µl of 40 mM Tris–HCl (pH 7.9), 0–2.5 mM MgCl2, MnCl2 or CaCl2 and 2.5% glycerol. Reactions were initiated by the addition of enzyme and were incubated for 10 min at 37°C. Cleavage intermediates were trapped by adding 5 µl of 5% SDS followed by 3 µl of 250 mM EDTA (pH 8.0). Topoisomerase IV was digested with proteinase K (5 µl of a 0.8 mg/ml solution) for 30 min at 45°C. DNA products were precipitated twice in 100% ethanol, washed in 70% ethanol, dried and resuspended in 5 µl of sample loading buffer. Samples were subjected to electrophoresis in 6% denaturing polyacrylamide gels. DNA cleavage products were visualized and quantified as above.
RESULTS AND DISCUSSION
DNA cleavage mediated by E. coli topoisomerase IV is promoted by an interaction between a divalent metal ion and the bridging atom of the scissile phosphate
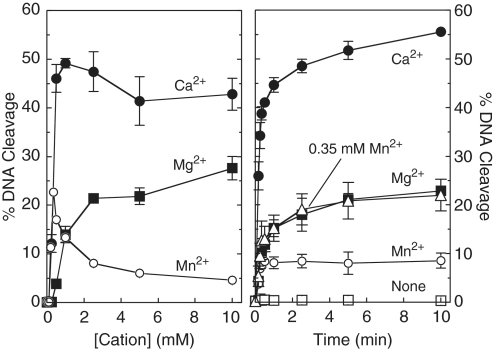
Escherichia coli topoisomerase IV, like other type II topoisomerases, requires a divalent metal ion to support DNA cleavage and overall catalytic activity (Figure 2) (40). Although the physiological metal ion appears to be magnesium, the bacterial enzyme can utilize a variety of divalent cations for the DNA cleavage reaction in vitro (40,41). Figure 2 shows the effects of metal ion concentration on topoisomerase IV-mediated DNA cleavage (left panel) as well as time courses for cleavage at constant metal ion concentrations (right panel). When negatively supercoiled plasmid was used as the substrate, Ca2+ supported the highest levels of DNA scission over a broad range of metal ion concentrations (left panel). Mg2+ and Mn2+ both supported DNA cleavage to a lesser extent (2- to 4-fold lower than Ca2+), but the optimal concentration of Mn2+ (∼0.35 mM) was nearly an order of magnitude lower than that of Mg2+ (∼2.5 mM).
Figure 2.
Cleavage of plasmid pBR322 by E. coli topoisomerase IV in the presence of different divalent metal ions. The effects of cation concentration on DNA cleavage (10-min reactions) are shown in the left panel. Mg2+ (closed squares), Mn2+ (open circles) or Ca2+ (closed circles) were titrated from 0.1 to 10 mM. Time courses for DNA cleavage in the presence of 2.5 mM divalent metal ions are shown in the right panel (Mg2+, closed squares; Mn2+, open circles; Ca2+, closed circles). Time courses in the absence of metal ion (open squares) and at the optimal concentration for Mn2+ (0.35 mM Mn2+, open triangles) also are shown. Error bars represent the standard deviation of three independent experiments.
Biochemical and structural studies with human and yeast type II topoisomerases suggest that active site metal ions in the eukaryotic enzyme interact with DNA at the bridging oxygen of the scissile bond (−1/+1 position), and non-bridging oxygen atoms at the scissile bond and the −2/−1 position (Figure 1) (18,24–27). While the interactions at the scissile bond are believed to help mediate the chemistry of DNA cleavage, the interaction with the −2/−1 non-bridging oxygen is thought to play a structural role in the reaction (27). In order to characterize the role of the metal ion in the DNA cleavage reaction of topoisomerase IV, a series of oligonucleotide substrates was employed that substituted sulfur for the bridging (i.e. phosphorothiolate) or non-bridging (i.e. phosphorothioate) oxygen atoms described above.
It should be noted that the 3′-terminal –SH moiety that is generated following cleavage of the 3′-bridging phosphorothiolate is a poor nucleophile for phosphorous (42,43) (data not shown). Because the phosphorothiolate substrate supports ligation poorly (if at all), topoisomerase IV-DNA cleavage complexes accumulate over time. This is in contrast to wild-type oligonucleotides or substrates with substituted non-bridging atoms, which establish rapid DNA cleavage-ligation equilibria and maintain lower levels of cleavage complexes.
Metal ion–DNA interactions were determined by comparing the ability of topoisomerase IV to cleave substrates containing oxygen or sulfur atoms in the presence of metal ions of varying ‘hardness’ (i.e. thiophilicity) (42,43). Experiments took advantage of the fact that ‘soft’ metal ions such as Mn2+ (which are larger and more polarizable) often prefer sulfur over oxygen as an inner-sphere ligand, whereas ‘hard’ metals such as Mg2+ and Ca2+ (which have a smaller ionic radius and are non-polarizable) usually coordinate more readily with oxygen (44–50).
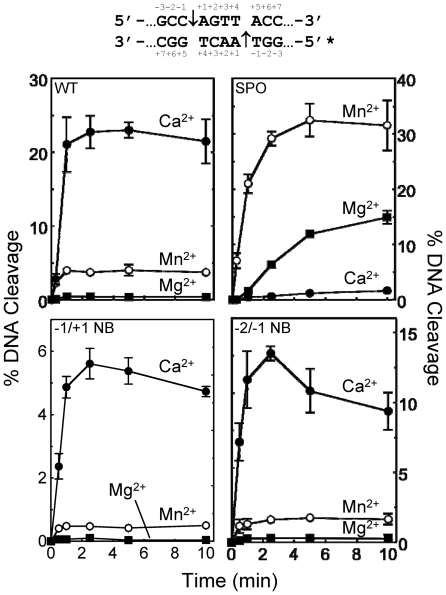
Results with the wild-type oligonucleotide (Figure 3, top left panel) were similar to those obtained with plasmid substrate, except that higher concentrations of metal ions were required to yield optimal levels of DNA scission (data not shown; see Figure 6). Once again, Ca2+ supported (by far) the highest levels of topoisomerase IV-mediated DNA cleavage. The only difference between results with the wild-type oligonucleotide and plasmid substrates was the fact that Mn2+ consistently supported higher levels of cleavage with the oligonucleotide than did Mg2+.
Figure 3.
Cleavage of oligonucleotide substrates by topoisomerase IV in the presence of different divalent metal ions. The central portion of the DNA cleavage substrate is depicted above the graphs. The asterisk denotes the 5′-end-labeled strand and the scissile bonds are indicated by arrows. All modifications are on the bottom strand. Results for the wild-type oligonucleotide (WT, top left panel), and substrates containing a 3′-bridging phosphorothiolate at the scissile (−1/+1) bond (SPO, top right), a non-bridging phosphorothioate at the scissile bond (−1/+1 NB, bottom left) and a non-bridging phosphorothioate at the −2/−1 position (−2/−1 NB, bottom right) are shown. DNA cleavage was carried out in the presence of 10 mM Mg2+ (closed squares), Mn2+ (open circles) or Ca2+ (closed circles). Error bars represent the standard deviation of three independent experiments.
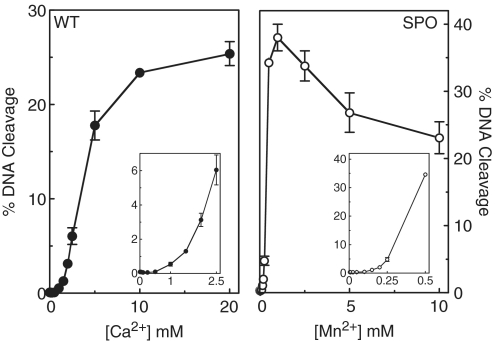
Figure 6.
Metal ion concentration dependence for DNA cleavage by topoisomerase IV with wild-type (WT) and phosphorothiolate (SPO) oligonucleotide substrates. Left panel, cleavage of a wild-type oligonucleotide in the presence of 5 µM to 20 mM Ca2+. The inset shows concentrations up to 2.5 mM Ca2+. Right panel, cleavage of an oligonucleotide containing a 3′-bridging phosphorothiolate in the presence of 5 µM to 10 mM Mn2+. The inset shows concentrations up to 0.5 mM Mn2+. All DNA cleavage reactions were incubated for 10 min. Error bars represent the standard deviation of three independent experiments.
A substrate containing a 3′-bridging sulfur was employed to determine whether metal ion interactions with the bridging/leaving group facilitate catalysis. If this were the case, relative levels (or rates) of scission with substrates that contain a 3′-bridging phosphorothiolate should increase in the presence of soft metals and decrease in reactions that contain hard metals.
As seen in Figure 3 (top right panel), metal ions produced a dramatically different cleavage pattern with the 3′-bridging phosphorothiolate as compared with the wild-type substrate. Cleavage rates were significantly higher and the levels of topoisomerase IV-mediated DNA scission were nearly 10-fold higher with the sulfur-containing substrate in the presence of the soft metal Mn2+. Conversely, the hard metal Ca2+, which supported the highest levels of cleavage with the wild-type substrate, generated the slowest rates of cleavage and the lowest levels of scission with the sulfur-containing oligonucleotide. Despite the fact that cleavage complexes accumulate over time with the 3′-bridging phosphorothiolate, levels of cleavage seen with Ca2+ fell >10-fold as compared with wild-type reactions. The precipitous drop in Ca2+-supported scission of the sulfur-containing substrate, coupled with the increased scission observed with Mn2+, demonstrates that there is an important interaction between the metal ion and the 3′-bridging atom of the scissile bond in the active site of E. coli topoisomerase IV that promotes the DNA cleavage event. Similar conclusions have been drawn for human topoisomerase IIα and topoisomerase IIβ (24,26).
Divalent metal ion interactions with non-bridging phosphate atoms
Interactions between metal ion A and the non-bridging oxygen atom of the scissile phosphate have been observed in the crystal structure of the covalent yeast topoisomerase II-cleaved DNA complex (27). While similar interactions in the active site of human topoisomerase IIα and topoisomerase IIβ have been proposed on the basis of kinetic experiments, their importance in the DNA cleavage reaction of these enzymes differs considerably (18,24,26). The relative ability of topoisomerase IIα to utilize different divalent metal ions with substrates that contain a racemic phosphorothioate at the non-bridging position of the scissile phosphate (−1/+1) was similar to that observed with the wild-type substrate (24). In contrast, the interaction between the metal ion and the non-bridging oxygen appears to play a more substantial role in the DNA scission reaction of topoisomerase IIβ (26). The relative ability of Ca2+ to support cleavage of the racemic non-bridging phosphorothioate decreased and fell below that of Mg2+ (26).
Given these differences, we wanted to determine whether the use of metal ions by topoisomerase IV was more similar to that of topoisomerase IIα or topoisomerase IIβ. Therefore, we examined the ability of different metal ions to support topoisomerase IV-mediated cleavage of the non-bridging phosphorothioate (−1/+1) substrate. As seen in Figure 3 (bottom left panel), the relative use of metal ions approximated that seen with the wild-type substrate. Once again, the order of metal ion utilization was Ca2+ > Mn2+ > Mg2+. On the basis of these data, E. coli topoisomerase IV appears to utilize metal ion A (at least with respect to interactions at the non-bridging oxygen of the scissile phosphate) in a manner similar to that of human topoisomerase IIα.
As discussed above, the crystal structure of the covalent yeast topoisomerase II-DNA cleavage complex revealed a novel interaction between metal ion B and the non-bridging oxygen of the phosphate at the −2/−1 position (27). It is believed that this contact plays a structural role in anchoring the double helix during the cleavage/ligation reaction. Therefore, the effects of this interaction on rates of DNA cleavage were determined. Once again, the use of metal ions was similar to those seen with the wild-type substrate (Figure 3, bottom right panel). These findings cannot rule out an interaction between metal ion A or B and the −2/−1 non-bridging oxygen atoms. However, if these interactions exist in the active site of topoisomerase IV, their effects on the overall rate of DNA cleavage appear to be equivocal.
A two-metal-ion mechanism for DNA cleavage mediated by topoisomerase IV
Biochemical, kinetic and mutagenesis studies with human topoisomerase IIα and topoisomerase IIβ indicate that both DNA cleavage/ligation active sites contain two divalent metal ions (22–26). Furthermore, the eukaryotic enzymes cleave DNA using a two-metal-ion mechanism in which metal ion site A and site B must both be occupied (18,24–26). This model for eukaryotic topoisomerase II is strongly supported by a recent structural study of the covalent yeast topoisomerase II-cleaved DNA complex (27). In this structure, both metal ion sites on each enzyme protomer are filled simultaneously.
Noble and Maxwell (19) found that mutant gyrase proteins with substitutions at residues believed to complex with metal ions displayed greater DNA cleavage in the presence of Mg2+ and Ca2+ than did either divalent cation alone. Based on these findings, they proposed that the prokaryotic type II enzyme utilized a two-metal-ion mechanism.
This conclusion was challenged by a recent crystallographic study that examined non-covalent complexes between Staphylococcus aureus gyrase, DNA and inhibitors (31). Although structures were generated that contained a metal ion at site A or B, none was observed that included metal ions at both sites. Based on these findings with gyrase, the authors proposed that type II topoisomerases utilize a moving metal ion mechanism in which only one of the two metal ion sites in each protomer is filled at any given time (31–33). In this model, metal ion binding to site B promotes DNA binding, albeit in a conformation that does not support efficient DNA cleavage. Once this conformation has been achieved, the first metal ion dissociates and is replaced by a second metal ion bound to site A, which promotes efficient DNA cleavage.
Recently, two structures of the covalent Streptococcus pneumoniae topoisomerase IV-cleaved DNA complex were reported. One of the structures contained no metal ions (28), while the other contained a single Mg2 (consistent with metal ion A) at each active site (30). Although the authors ‘[could not] exclude the participation of a more weakly bound Mg2+’, they also suggested the possibility that topoisomerase IV might utilize a single dynamic metal ion for DNA cleavage (30).
An important issue should be noted regarding the proposed moving (or dynamic) metal ion mechanism: the affinity of metal ion B, which would be the first to bind the enzyme in this model, is ∼5- to 10-fold lower than that of metal ion A (Figures 4–6), which is the cation that participates directly in the chemistry of the cleavage reaction (24–26).
Figure 4.
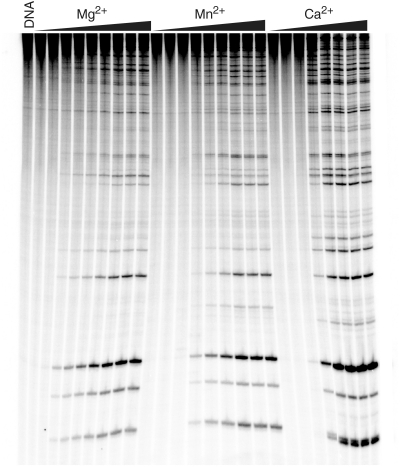
Metal ion concentration dependence for topoisomerase IV-mediated DNA cleavage of linearized pBR322 plasmid. A 4330-bp fragment of pBR322 was singly end-labeled and employed as the cleavage substrate. Reaction products were resolved by acrylamide gel electrophoresis. DNA cleavage in the presence of varying concentrations of Mg2+ (0, 0.25, 0.5, 0.65, 0.85, 1.0, 1.5, 2.0 and 2.5 mM), Mn2+ (0, 0.025, 0.050, 0.075, 0.100, 0.125, 0.150, 0.175 and 0.200 mM) or Ca2+ (0, 0.05, 0.15, 0.25, 0.40, 0.50, 0.75 and 1.00 mM) (left, center or right, respectively) is shown. A DNA standard (DNA) also is shown. The gel is representative of three independent experiments.
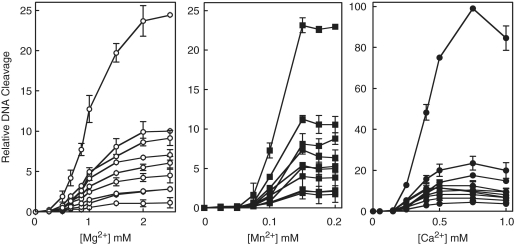
Figure 5.
Metal ion concentration dependence for topoisomerase IV-mediated DNA cleavage of linearized pBR322 plasmid. Data from Figure 4 and additional experiments were quantified. Results for Mg2+(left), Mn2+(middle) and Ca2+ (right) are shown. All values were quantified relative to the most abundant cleavage product (generated at 0.75 mM Ca2+), which was set to 100. Error bars represent the standard deviation of three independent experiments.
Two independent approaches were used to determine whether bacterial type II topoisomerases utilize a two-metal-ion or a moving metal ion mechanism for DNA cleavage. In the first, the concentration dependence of divalent metal ions for DNA cleavage mediated by E. coli topoisomerase IV was assessed. Initial studies utilized end-labeled linear pBR322 as the cleavage substrate. Several striking features were seen (Figure 4). The most obvious is that no DNA scission was observed at low concentrations of Mg2+, Mn2+ or Ca2+. For each of the three metal ions, cleavage products were generated only after a threshold concentration had been reached (∼0.05–0.25 mM, depending on the metal ion) and began to accumulate coordinately for all observable DNA bands. Furthermore, when quantified, the metal ion dependence of DNA cleavage appeared to be sigmoidal in shape (Figure 5).
These data strongly suggest that each enzyme protomer contains two distinct sites for metal ion binding in the topoisomerase IV–DNA complex, and that both are critical for cleavage of each strand of the double helix. One metal ion binds to the complex with higher affinity (metal ion A) and saturates the site at levels that reflect the threshold concentration at which cleavage appears. The second (metal ion B) also is essential for DNA cleavage and associates with the topoisomerase IV–DNA complex with apparent KD values of ∼1.0, 0.12 and 0.40 mM, respectively, for Mg2+, Mn2+ and Ca2. It is not obvious how the concentration dependence of DNA cleavage seen in Figures 4 and 5 can be interpreted by a model in which both metal ion-binding sites are not filled simultaneously.
Results similar to those seen with the plasmid substrate also were observed using the oligonucleotide cleavage substrate described earlier. A biphasic concentration dependence for Ca2+-supported cleavage of the wild-type substrate and for Mn2+-supported cleavage of the phosphorothiolate substrate is shown (Figure 6). Once again, these findings suggest a model that requires two simultaneously bound metal ions to support DNA scission mediated by E. coli topoisomerase IV.
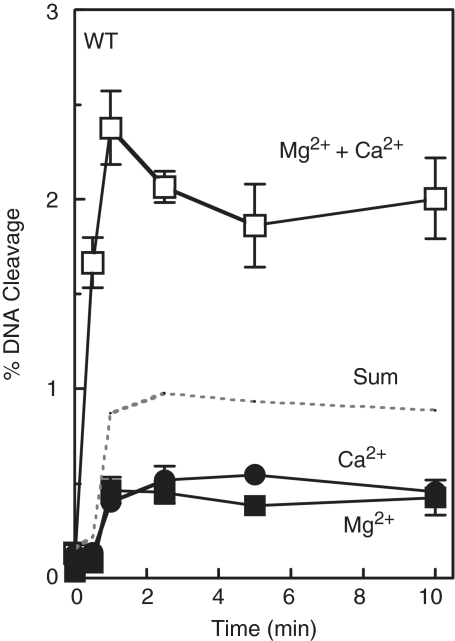
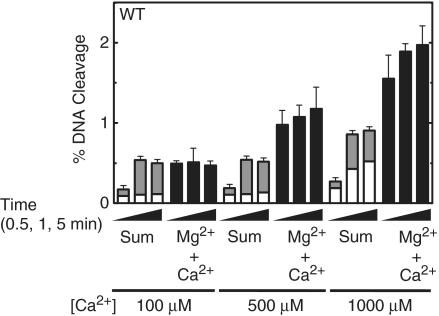
As a second approach to determine the number of metal ions required to support the DNA cleavage reaction of bacterial type II topoisomerases, metal ion mixing experiments were performed (19,24,26). Initial studies utilized the wild-type oligonucleotide substrate and mixtures of Mg2+ and Ca2+. This metal ion pair was used because Mg2+ supports DNA cleavage of the wild-type substrate poorly, even at high concentrations, while Ca2+ efficiently supports high levels of scission. The first time courses for DNA cleavage were generated in the presence of either 10 mM Mg2+, a saturating level, or 1 mM Ca2+, a concentration that saturates the site for metal ion A without appreciably filling the site for metal ion B (Figure 6). As seen in Figure 7, similar low levels of cleavage were generated when either one of the metal ions was included in assays. However, when both metal ions were present in reactions, rates of cleavage increased several-fold above levels predicted from the simple arithmetic sum of scission observed in the presence of the individual metal ions. Furthermore, a large enhancement in the rate and level of DNA scission (compared with calculated sums) was observed when 10 mM Mg2+ was combined with a range of Ca2+ concentrations (0.1–1 mM) that partially filled or saturated the high affinity metal ion site (Figure 8).
Figure 7.
Cleavage of the wild-type (WT) oligonucleotide substrate in the simultaneous presence of two different divalent metal ions. Time courses were carried out in the presence of 10 mM Mg2+ alone (closed squares), 1 mM Ca2+ alone (closed circles) or a mixture of 10 mM Mg2+ and 1 mM Ca2+ (open squares). The arithmetic sum of the cleavage with Mg2+ or Ca2+ alone is represented by the dotted line. Error bars represent the standard deviation of three independent experiments.
Figure 8.
Topoisomerase IV-mediated cleavage of the wild-type (WT) oligonucleotide in the presence of different divalent metal ions. Cleavage reactions were carried out for 0.5, 1 or 5 min in the presence of 10 mM Mg2+ alone (open bars), 100–1000 µM Ca2+ alone (gray bars) or a mixture of 10 mM Mg2+ and 100–1000 µM Ca2+ (black bars). The calculated sum of cleavage in the presence of Mg2+ or Ca2+ alone is shown (stacked open and gray bars). Error bars represent the standard deviation of three independent experiments.
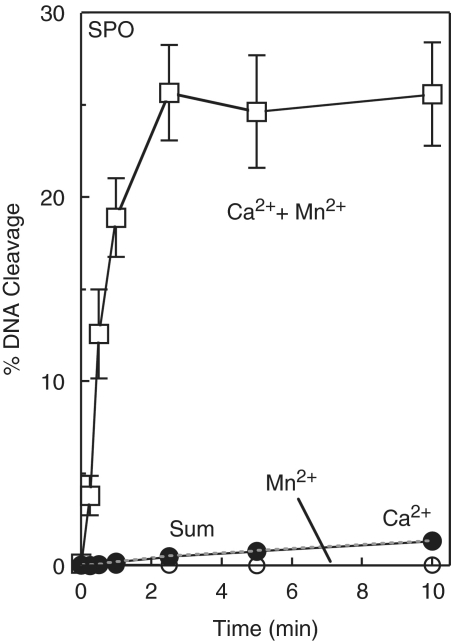
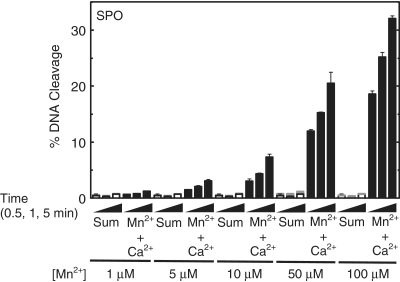
Even more dramatic results were observed when the oligonucleotide that contained a bridging phosphorothiolate was utilized as the substrate. These experiments employed Ca2+, which supports only low levels of cleavage of the sulfur-containing substrate, and Mn2+, which supports the highest levels of topoisomerase IV-mediated scission of this substrate. The first experiment used 10 mM Ca2+ and 100 µM Mn2+, which (as above) represents the concentration that fills only metal ion site A (Figure 6). As seen in Figure 9, the initial rate of cleavage was ∼200-fold faster and the final level of cleavage increased ∼20-fold in reactions that contained both metal ions as compared with the arithmetic sum of cleavage observed in reactions that contained only one of the two individual metal ions. Once again, similar results were observed in reactions that combined 10 mM Ca2+ and 1–100 µM Mn2+ (Figure 10).
Figure 9.
Cleavage of the phosphorothiolate (SPO) oligonucleotide substrate in the simultaneous presence of two different divalent metal ions. Time courses were carried out in the presence of 10 mM Ca2+ alone (closed circles), 100 µM Mn2+ alone (open circles) or a mixture of 10 mM Ca2+ and 100 µM Mn2+ (open squares). The arithmetic sum of the cleavage with Ca2+ or Mn2+ alone is represented by the dotted line. Error bars represent the standard deviation of three independent experiments.
Figure 10.
Topoisomerase IV-mediated cleavage of the phosphorothiolate (SPO) oligonucleotide in the presence of different divalent metal ions. Cleavage reactions were carried out for 0.5, 1 or 5 min in the presence of 10 mM Ca2+ alone (open bars), 1–100 µM Mn2+ alone (gray bars) or a mixture of 10 mM Ca2+ and 1–100 µM Mn2+ (black bars). The calculated sum of cleavage in the presence of Mg2+ or Ca2+ alone is shown (stacked open and gray bars). Error bars represent the standard deviation of three independent experiments.
In the experiments discussed above, rates and levels of DNA cleavage were greatly enhanced by mixing low concentrations of a metal ion that effectively fills site A with saturating levels (i.e. high enough to fill site B) of a second metal ion that cannot act effectively at site A. The most straightforward interpretation of the above findings is that the first (i.e. low concentration) metal ion acts primarily at site A and the second (i.e. saturating) metal ion acts primarily at site B. Taken together, these results argue for the simultaneous use of two metal ions during the DNA cleavage reaction mediated by bacterial topoisomerase IV and provide strong evidence for a two-metal-ion mechanism for DNA cleavage.
Although the importance of metal ion binding to sites A and B is clear, specific relationships between the two sites have yet to be delineated. An intriguing question is whether the metal ion in site B on one ParC subunit might work in concert with the metal ion in site A on the other ParC subunit to promote DNA scission (31). Indeed, there is precedent for trans cooperation between residues on opposite protomers of yeast topoisomerase II in the catalysis of DNA cleavage (51,52). At the present time, we cannot distinguish between a two-metal-ion mechanism in which site A and site B cooperate in cis or trans.
Although type II topoisomerases require divalent metal ions in order to cleave the double helix, the role of these cations in promoting DNA scission by the bacterial type II enzymes has been controversial. Based on the findings discussed above, we propose that topoisomerase IV cleaves DNA using a two-metal-ion mechanism similar to that shown in Figure 1. In this model, metal ion A makes a critical interaction with the 3′-bridging atom of the scissile phosphate and facilitates DNA cleavage mediated by the bacterial type II enzyme.
FUNDING
National Institutes of Health grant GM033944 (to N.O.); the NHLBI Intramural Research Program of the National Institutes of Health (to K.C.N.); Biotechnology and Biological Sciences Research Council (BBSRC), UK (to A.M.). Funding for open access charge: National Institutes of Health grant GM033944; Laboratory Development Funds (to N.O.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Amanda C. Gentry, Adam C. Ketron, Katie J. Aldred and Dr. Qasim A. Khan for critical reading of this article.
REFERENCES
- 1.Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Velez-Cruz R, Osheroff N. Encyclopedia of Biological Chemistry. San Diego: Elsevier Inc.; 2004. Topoisomerases: eukaryotic and prokaryotic type II; pp. 806–811. [Google Scholar]
- 4.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Deweese JE, Osheroff MA, Osheroff N. DNA topology and topoisomerases: teaching a “knotty” subject. Biochem. Mol. Biol. Educ. 2009;37:2–10. doi: 10.1002/bmb.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sissi C, Palumbo M. In front of and behind the replication fork: bacterial type IIA topoisomerases. Cell. Mol. Life Sci. 2010;67:2001–2024. doi: 10.1007/s00018-010-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson VE, Osheroff N. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr. Pharm. Des. 2001;7:337–353. doi: 10.2174/1381612013398013. [DOI] [PubMed] [Google Scholar]
- 8.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiles JA, Bradbury BJ, Pucci MJ. New quinolone antibiotics: a survey of the literature from 2005 to 2010. Expert Opin. Ther. Pat. 2010;20:1295–1319. doi: 10.1517/13543776.2010.505922. [DOI] [PubMed] [Google Scholar]
- 10.Anderson VE, Gootz TD, Osheroff N. Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 1998;273:17879–17885. doi: 10.1074/jbc.273.28.17879. [DOI] [PubMed] [Google Scholar]
- 11.Anderson VE, Zaniewski RP, Kaczmarek FS, Gootz TD, Osheroff N. Quinolones inhibit DNA religation mediated by Staphylococcus aureus topoisomerase IV: changes in drug mechanism across evolutionary boundaries. J. Biol. Chem. 1999;274:35927–35932. doi: 10.1074/jbc.274.50.35927. [DOI] [PubMed] [Google Scholar]
- 12.Anderson VE, Zaniewski RP, Kaczmarek FS, Gootz TD, Osheroff N. Action of quinolones against Staphylococcus aureus topoisomerase IV: basis for DNA cleavage enhancement. Biochemistry. 2000;39:2726–2732. doi: 10.1021/bi992302n. [DOI] [PubMed] [Google Scholar]
- 13.Willmott CJ, Critchlow SE, Eperon IC, Maxwell A. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J. Mol. Biol. 1994;242:351–363. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 14.Shea ME, Hiasa H. Distinct effects of the UvrD helicase on topoisomerase-quinolone-DNA ternary complexes. J. Biol. Chem. 2000;275:14649–14658. doi: 10.1074/jbc.275.19.14649. [DOI] [PubMed] [Google Scholar]
- 15.Wentzell LM, Maxwell A. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J. Mol. Biol. 2000;304:779–791. doi: 10.1006/jmbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 16.Marians KJ, Hiasa H. Mechanism of quinolone action. A drug-induced structural perturbation of the DNA precedes strand cleavage by topoisomerase IV. J. Biol. Chem. 1997;272:9401–9409. doi: 10.1074/jbc.272.14.9401. [DOI] [PubMed] [Google Scholar]
- 17.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deweese JE, Osheroff N. The use of divalent metal ions by type II topoisomerases. Metallomics. 2010;2:450–459. doi: 10.1039/c003759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Yu P, Lin TC, Konigsberg WH, Steitz TA. Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry. 1996;35:8110–8119. doi: 10.1021/bi960178r. [DOI] [PubMed] [Google Scholar]
- 21.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West KL, Meczes EL, Thorn R, Turnbull RM, Marshall R, Austin CA. Mutagenesis of E477 or K505 in the B' domain of human topoisomerase IIβ increases the requirement for magnesium ions during strand passage. Biochemistry. 2000;39:1223–1233. doi: 10.1021/bi991328b. [DOI] [PubMed] [Google Scholar]
- 23.Leontiou C, Lakey JH, Lightowlers R, Turnbull RM, Austin CA. Mutation P732L in human DNA topoisomerase IIβ abolishes DNA cleavage in the presence of calcium and confers drug resistance. Mol. Pharmacol. 2006;69:130–139. doi: 10.1124/mol.105.015933. [DOI] [PubMed] [Google Scholar]
- 24.Deweese JE, Burgin AB, Osheroff N. Human topoisomerase IIα uses a two-metal-ion mechanism for DNA cleavage. Nucleic Acids Res. 2008;36:4883–4893. doi: 10.1093/nar/gkn466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deweese JE, Guengerich FP, Burgin AB, Osheroff N. Metal ion interactions in the DNA cleavage/ligation active site of human topoisomerase IIα. Biochemistry. 2009;48:8940–8947. doi: 10.1021/bi900875c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deweese JE, Burch AM, Burgin AB, Osheroff N. Use of divalent metal ions in the DNA cleavage reaction of human type II topoisomerases. Biochemistry. 2009;48:1862–1869. doi: 10.1021/bi8023256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 29.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 30.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 32.Oezguen N, Schein CH, Peddi SR, Power TD, Izumi T, Braun W. A “moving metal mechanism” for substrate cleavage by the DNA repair endonuclease APE-1. Proteins. 2007;68:313–323. doi: 10.1002/prot.21397. [DOI] [PubMed] [Google Scholar]
- 33.Dupureur CM. One is enough: insights into the two-metal ion nuclease mechanism from global analysis and computational studies. Metallomics. 2010;2:609–620. doi: 10.1039/c0mt00013b. [DOI] [PubMed] [Google Scholar]
- 34.Peng H, Marians KJ. Escherichia coli topoisomerase IV: purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 35.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 36.Fortune JM, Dickey JS, Lavrukhin OV, Van Etten JL, Lloyd RS, Osheroff N. Site-specific DNA cleavage by Chlorella virus topoisomerase II. Biochemistry. 2002;41:11761–11769. doi: 10.1021/bi025802g. [DOI] [PubMed] [Google Scholar]
- 37.Deweese JE, Burgin AB, Osheroff N. Using 3′-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIα. Biochemistry. 2008;47:4129–4140. doi: 10.1021/bi702194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortune JM, Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage. J. Biol. Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 39.O’Reilly EK, Kreuzer KN. A unique type II topoisomerase mutant that is hypersensitive to a broad range of cleavage-inducing antitumor agents. Biochemistry. 2002;41:7989–7997. doi: 10.1021/bi025897m. [DOI] [PubMed] [Google Scholar]
- 40.Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 41.Hiasa H. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry. 2002;41:11779–11785. doi: 10.1021/bi026352v. [DOI] [PubMed] [Google Scholar]
- 42.Burgin AB, Jr, Huizenga BN, Nash HA. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 1995;23:2973–2979. doi: 10.1093/nar/23.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henningfeld KA, Arsian T, Hecht SM. Alteration of DNA primary structure by DNA topoisomerase I. Isolation of the covalent topoisomerase I-DNA binary complex in enzymatically competent form. J. Am. Chem. Soc. 1996;118:11701–11714. [Google Scholar]
- 44.Basu S, Strobel SA. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4-P6 domain. RNA. 1999;5:1399–1407. doi: 10.1017/s135583829999115x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson RG. Acids and bases. Science. 1966;151:172–177. doi: 10.1126/science.151.3707.172. [DOI] [PubMed] [Google Scholar]
- 46.Pecoraro VL, Hermes JD, Cleland WW. Stability constants of Mg2+ and Cd2+ complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of MgATP and MgADP. Biochemistry. 1984;23:5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- 47.Sigel RKO, Song B, Sigel H. Stabilities and structures of metal ion complexes of adenosine 5′-O-thiomonophosphate (AMPS2-) in comparison with those of its parent nucleotide (AMP2-) in aqueous solution. J. Am. Chem. Soc. 1997;119:744–755. [Google Scholar]
- 48.Curley JF, Joyce CM, Piccirilli JA. Functional evidence that the 3′-5′ exonuclease domain of Escherichia coli DNA polymerase I employs a divalent metal ion in leaving group stabilization. J. Am. Chem. Soc. 1997;119:12691–12692. [Google Scholar]
- 49.Sontheimer EJ, Gordon PM, Piccirilli JA. Metal ion catalysis during group II intron self-splicing: parallels with the spliceosome. Genes Dev. 1999;13:1729–1741. doi: 10.1101/gad.13.13.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szewczak AA, Kosek AB, Piccirilli JA, Strobel SA. Identification of an active site ligand for a group I ribozyme catalytic metal ion. Biochemistry. 2002;41:2516–2525. doi: 10.1021/bi011973u. [DOI] [PubMed] [Google Scholar]
- 51.Liu Q, Wang JC. Identification of active site residues in the “GyrA” half of yeast DNA topoisomerase II. J. Biol. Chem. 1998;273:20252–20260. doi: 10.1074/jbc.273.32.20252. [DOI] [PubMed] [Google Scholar]
- 52.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]