Abstract
The ecologically important cyanobacterium Prochlorococcus possesses the smallest genome among oxyphototrophs, with a reduced suite of protein regulators and a disproportionately high number of regulatory RNAs. Many of these are asRNAs, raising the question whether they modulate gene expression through the protection of mRNA from RNase E degradation. To address this question, we produced recombinant RNase E from Prochlorococcus sp. MED4, which functions optimally at 12 mM Mg2+, pH 9 and 35°C. RNase E cleavage assays were performed with this recombinant protein to assess enzyme activity in the presence of single- or double-stranded RNA substrates. We found that extraordinarily long asRNAs of 3.5 and 7 kb protect a set of mRNAs from RNase E degradation that accumulate during phage infection. These asRNA–mRNA duplex formations mask single-stranded recognition sites of RNase E, leading to increased stability of the mRNAs. Such interactions directly modulate RNA stability and provide an explanation for enhanced transcript abundance of certain mRNAs during phage infection. Protection from RNase E-triggered RNA decay may constitute a hitherto unknown regulatory function of bacterial cis-asRNAs, impacting gene expression.
INTRODUCTION
Endoribonuclease E (RNase E) initiates the decay of most, if not all, mRNAs in prokaryotes by endonucleolytic cleavage followed by polyadenylation and/or exonucleolytic degradation (1). Therefore, RNase E is a crucial regulator of the global rate of RNA decay. The half-life of RNA is an important factor in the regulation of gene expression, directly influencing the translation rates of their encoded proteins and hence impacting the rapidity of the response of an organism to changing environmental conditions. Several studies have shown that stress conditions, such as the presence of antibiotics, nutritional stress and transitions in growth phase, cause a dramatic change in the rate of RNA turnover for a subset of genes (2–4).
The presumably best characterized RNase E homologue is that of Escherichia coli, which is organized in a multimer enzyme complex known as the degradosome (5). The degradosome consists of polynucleotide phosphorylase, RNA helicase B and the glycolytic enzyme enolase (5–8). Degradosome structures have not been identified in cyanobacteria or spinach chloroplasts, though all components necessary for the assembly of the degradosome are present in cyanobacteria, including Synechocystis PCC6803 and Prochlorococcus sp. (9–11). However, cyanobacterial RNase E (rne) genes of the species Synechocystis PCC6803 and Prochlorococcus sp. lack the carboxy-terminal sequence necessary for the assembly of the degradosome in E. coli (12). Nevertheless, similar and important functions of RNase E in both E. coli and Synechocystis PCC6803 can be assumed, as the disruption of rne is lethal in both organisms (11). The role of degradosome structures is still under debate because the deletion of the carboxy terminus in E. coli (associated with a loss of the degradosome structure) does not impact cell viability (11). According to the lack of the C-terminal extension, a feature of RNase G (Caf A family), and sequence homology to RNase E, cyanobacterial homologues have been grouped into the RNase E/G family (13).
Several recent studies have been published on whole-transcriptome expression analyses, revealing that a significant portion of the transcriptome is expressed on the antisense strand not only in Eukaryotes (14–17) but also in Bacteria and Archaea, where data indicate that between 3% to more than half of all genes are associated with asRNAs (18–24). In light of RNase E enzyme specificity—recognizing single-stranded A/U-rich sequences (25,26)—one wonders if the potential formation of asRNA–mRNA duplexes affects RNase E activity. In this context, in a recent microarray study on whole-genome expression of the podovirus P-SSP7 and its cyanobacterial host Prochlorococcus MED4, we found that transcript abundance of 41 host genes, including RNase E, increased during lytic infection, while the vast majority of host mRNAs declined in abundance (27). We suggested that the induction of host RNase E transcript may help the phage by generating a source of nucleotides that (after nucleotide reduction by the phage-encoded ribonucleotide reductase) are utilized by the phage for its own replication (27). Those results raised the question of how these 41 host mRNAs are protected from degradation in light of the enhanced transcription of RNase E.
Here, we set out to test the hypothesis that in the ecologically important marine cyanobacterium Prochlorococcus, asRNA–mRNA duplex formation masks single-stranded recognition sites of RNase E, leading to increased stability of mRNAs and their asRNAs. Therefore, using purified recombinant Prochlorococcus RNase E from E. coli, we assessed RNase E activity in the presence of single- or double-stranded RNA substrates and showed that asRNA–mRNA duplexes protected mRNA from RNase E degradation, enhancing mRNA stability of transcripts accumulated during phage infection.
MATERIALS AND METHODS
Construction of rne expression plasmid pQE70PMM1501
Oligonucleotides used for PMM1501 cloning were PMM1501_C_K12 fw and PMM1501_C_K12 rev. Lowercase letters indicate PaeI or BamHI site (Supplementary Table S1). Both oligonucleotides were optimized for E. coli codon usage (Kazusa Codon Usage Database) without impairing the PMM1501 wild-type amino acid composition. The resulting coding sequence of PMM1501 was cloned in pQE70 (QIAGEN, Germany).
Overexpression and purification of recombinant RNase E from Prochlorococcus MED4
Escherichia coli TOP10 F′ served as the host strain for construction and propagation of pQE70-PMM1501. Expression of recombinant PMM1501 protein was carried out in E. coli ArcticExpress (DE3) RIL (Stratagene, USA). Strains were grown in Luria broth, supplemented with ampicillin (100 µg/ml), streptomycin (75 µg/ml) and gentamicin (20 µg/ml), if required. An overnight culture was diluted in 2 l of Luria broth and grown at 37°C to an OD600 of 0.5–0.8. Subsequently, heterologous expression was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) (0.1 mM final concentration), and the culture was transferred to 10°C and grown for an additional 48 h. Thereafter, cells were harvested (4°C, 6000 rpm, 10 min) and resuspended in buffer A [50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, (pH adjusted to 8.0 using NaOH)], containing protease inhibitor PMSF (0.2 mM). Disruption of cells was carried out on ice by sonication. Cell debris was removed by centrifugation at 13 000 rpm for 30 min at 4°C, and the supernatant was collected. C-terminally His-tagged MED4 RNase E was purified under native conditions by Ni2+ nitrilotriacetic acid affinity chromatography (QIAGEN, Germany) according to the manufacturer’s instructions (QIAGEN handbook, fifth edition). All steps were performed at 4°C, but the pH of all buffers was adjusted at room temperature. The Ni-NTA matrix size was adjusted to 500 µl. Following equilibration with 10 volumes of buffer A, the column was charged with supernatant and subsequently washed with 60 volumes of buffer B [50 mM NaH2PO4, 500 mM NaCl, 50 mM imidazole, 0.1% (v/v) Triton X-100, 8% (v/v) glycerol (pH adjusted to 8.0 using NaOH)]. Adsorbed peptides were eluted by ligand exchange employing a step gradient of 250 and 500 mM imidazole in buffer containing 50 mM NaH2PO4, and 300 mM NaCl, pH 8.0 (adjusted with NaOH). Fractions containing specific endoribonuclease activity were identified by in vitro assays as described below (in vitro cleavage assay) and visualized by Coomassie-stained 10% SDS–PAA gels.
Size exclusion chromatography
Combined elution fractions of His-tag-purified MED4 RNase E were size fractionated on a superdex 200 10/300 GL column (GE Healthcare, Germany). The column was equilibrated with buffer A (see purification of recombinant protein) and calibrated using a gel filtration standard, ranging from 1.35 to 670 kDa (Bio-Rad, Germany). Fractionation was analysed and documented with Unicorn 4.12 software (Amersham Pharmacia, Germany).
In vitro synthesis of RNA
In vitro transcription was carried out with T7 polymerase and PCR fragments containing a T7 recognition site as templates using MEGAshortscriptTM High Yield Transcription KIT (Ambion, USA), according to the manufacturer’s instructions. Residual DNA was removed with 2 U TURBO DNase (Ambion, USA) for 15 min at 37°C. RNA was purified and concentrated by phenol–chloroform extraction and ethanol precipitation and finally quantified using a NanoDrop ND-1000 spectrophotometer (PEQLAB biotechnology, Germany). Ten micrograms of purified in vitro transcript was subjected to electrophoretic separation on a 7 M urea–6% polyacrylamide gel. The RNA fragment, corresponding to full-length transcript, was excised and recovered from the gel slice by electro-elution for 45 min at 11 mA per glass tube (Model 422 Electro-Eluter; BIO-RAD, Germany). Enrichment of gel-purified RNA was conducted by subsequent gel chromatography using RNA Clean & Concentrator™-25 according to the manufacturer’s instructions (Zymo Research, USA). If required, RNA was treated with tobacco acid pyrophosphatase or 5′-polyphosphatase (Epicentre, USA) according to the manufacturer’s instructions and subjected to an additional purification by phenol–chloroform extraction and ethanol precipitation (prior to gel purification). The following mRNAs and asRNAs were synthesized: PMM0684 RNA, PMM0685 RNA, PMED4_07431 RNA, PMED4_07411 RNA, 3 kb polycistronic mRNA and 3.5 kb asRNA. Oligonucleotides used for template generation are listed in Supplementary Table S1.
In vitro cleavage assay
For ribonuclease activity tests of duplex RNA, both mRNA and asRNA were diluted in H2O in the same reaction vial, heated to 95°C for 3–5 min to break intrinsic secondary and tertiary structures, and then briefly chilled on ice. The reaction mixture was complemented with 25 mM Tris–HCl (pH 8.0), 60 mM KCl, 5 mM MgCl2, 100 mM NH4Cl and 0.1 mM DTT. Hybridization of complementary sequence regions was carried out at room temperature for 20 min.
RNase E
In vitro cleavage was carried out for 15 min at 30°C in a 5-µl reaction mixture containing 25 mM Tris–HCl (pH 8.0), 60 mM KCl, 5 mM MgCl2, 100 mM NH4Cl, 0.1 mM DTT, 5% (w/v) glycerol, 0.2–1.3 pmol ssRNA or 0.4–2.6 pmol dsRNA, and 7 pmol affinity-purified RNase E. Endoribonuclease activity was abolished by addition of 1 µl 0.5 M EDTA and 1 volume loading buffer or by direct addition of 1 volume loading buffer. Finally, samples were heated to 95°C for 3–5 min and briefly chilled on ice before the cleavage reactions were analysed by employing electrophoretic separation on 7 M urea–6% polyacrylamide gels for 2 h at 6 mA.
RNase III and RNase T1
Assay conditions were identical to those of the RNase E approach, with the following modifications. RNA digestion was carried out at 37°C for 15 min using 0.1 U RNase III (Epicentre, USA; from E. coli) and 0.5 U RNase T1 (Sigma, Germany; from Aspergillus oryzae), respectively, or according to experimental requirements.
Fluorogenic cleavage assay
A fluorogenic RNA oligonucleotide with a FAM tag and a BHQ-1 quenching tag containing the RNase E recognition site of Synechocystis PCC6803 in the psbA gene leader sequence, psbA2-5′UTR [5′-Phos-A-A(2′-O-Methyl)–C(2′-O-Methyl)–A(2′-O-Methyl)–U(FAM)–C(2′-O-Methyl)–G(2′-O-Methyl)–A(2′-O-Methyl)–A–A–A–U–A–C–A–U–A–C(BHQ-1)-OH], was synthesized and PAGE purified by Purimex (Grebenstein, Germany). Digestion of the fluorogenic oligonucleotide results in a marked increase in fluorescence that can be monitored in real time under defined reaction conditions. Cleavage assays were carried out at 30°C in a 50-µl reaction mixture containing 25 mM Tris–HCl (pH 8.0), 60 mM KCl, 5 mM MgCl2, 100 mM NH4Cl, 0.1 mM DTT, 5% (w/v) glycerol, 1 µl affinity-purified RNase E (not quantified) and 0.1 µM (in combination with MED4 RNase E) or 0.06 µM (in combination with PCC6803 RNase E) fluorogenic RNA, or adjusted according to experimental requirements. To avoid evaporation during the course of procedure, samples were covered with 20 µl RNase-free immersion oil. Each cleavage experiment was assayed in technical triplicates. The cleavage reaction was monitored continuously by fluorometry using a VictorTM X3 multilabel plate reader (PerkinElmer; excitation at 480 nm, emission at 520 nm).
Growth conditions of Prochlorococcus MED4
Cells were grown at 22°C in AMP1 or Pro99 medium (28) under 30 µmol quanta m−2 s−1 continuous white cool light. For phage infection experiments, the podovirus P-SSP7 was added at a ratio of three infective phage per cell (MOI = 3) such that the cell and phage concentrations were 108 cells/ml and 3 × 108 phage/ml, respectively. Prior to phage addition, the cells were growing exponentially with a generation time of 1.5 days as were the uninfected control cells.
RNA isolation
Total RNA was extracted from cultures using the hot phenol method (29,30). Briefly, cells were harvested by centrifugation at 12 000 rpm for 10 min at 20°C. The pellet was resuspended in RNA resuspension buffer [10 mM sodium acetate (pH 5.2), 200 mM sucrose, 5 mM EDTA]. Following the addition of half a volume of phenol, the suspension was incubated in a 60°C water bath for 15 min and subsequently incubated at room temperature for another 30 min. Half a volume of chloroform/isoamyl alcohol (24:1) was added, and the homogenate was thoroughly mixed, incubated for another 1–10 min at room temperature and centrifuged at 6000 rpm for 5 min at 20°C. The upper aqueous phase was extracted once more with one volume phenol/chloroform/isoamyl alcohol (25:24:1) and centrifuged for 5 min at 6000 rpm (20°C). The aqueous phase was precipitated with 1/10 volume 3 M sodium acetate, 3 volumes 100% ethanol and 0.2 µl glycogen at −20°C overnight. Subsequently, RNA was concentrated by centrifugation at 13 000 rpm for 30 min (4°C) and resuspended in RNase-free H2O.
In vitro synthesis and labelling of RNA
Oligonucleotide probes
Ten picomoles of oligodeoxynucleotide were radiolabelled at the 5′-end with 10 U T4 polynucleotide kinase (Fermentas, Germany), 3 nmol [γ-32P]ATP (Hartmann Analytic, Germany), 5 U RiboLock RNase inhibitor and 1 × reaction buffer A (Fermentas, Germany) in a total volume of 20 µl. Following incubation at 37°C for 30 min, the reaction was stopped by the addition of 25 mM EDTA and denaturation at 95°C for 5 min. The reaction mixture was diluted in H2O to a total volume of 40 µl and subjected to separation on pre-packed sephadexTM G-25 spin columns (GE Healthcare, Germany) to remove free [γ-32P]ATP. For probing PMM0685 mRNA or PMM0685, asRNA oligonucleotides PMM0685 rev and PMM0685-5′-end mRNA, respectively, were used (Supplementary Table S1).
Transcript probes
PCR fragments containing a T7 recognition site were synthesized for in vitro transcription from the T7 promoter in the presence of [α-32P]UTP (Hartmann Analytic, Germany) using T7 MAXIscript (Ambion, USA). Sense and antisense probes were prepared for the following genes: PMM0684, PMM0685, PMED4_07411, PMED4_07431. Oligonucleotides used for template PCR amplification are listed in Supplementary Table S1.
Northern blot analysis
RNA samples (1.5 µg total RNA or 1.3 or 2.6 pmol in vitro transcripts) were supplemented with loading buffer, denatured at 65°C for 5 min, and then briefly chilled on ice before electrophoretic separation on 7 M urea–6% polyacrylamide gels for 2 h at 6 mA or 1.5% formaldehyde–agarose or native gels for 2 h at 80 V, respectively. Polyacrylamide gels were transferred to Hybond-N nylon membranes (Amersham, Germany) by electroblotting for 1 h at 400 mA. Agarose gels were transferred overnight by capillary blotting using 20 × SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.0) as transfer solution. Membranes were hybridized overnight at 42 or 57°C with labelled oligonucleotide probes or labelled transcript probes, respectively, in 50% deionized formamide, 7% SDS, 250 mM NaCl and 120 mM Na(PO4) pH 7.2. Membranes were washed in 2× SSC/1% SDS for 10 min, in 1 × SSC/0.5% SDS for another 10 min and, according to signal intensity, 1–10 min in 0.1 × SSC/0.1% SDS. All wash steps were carried out at 5°C below the hybridization temperature with pre-warmed buffers. Membranes were exposed to imaging plates, and the resulting signals were visualized using a Personal Molecular Imager FX system with Quantity One software (Bio-Rad, Germany).
Reverse transcription–PCR
DNA was removed from total RNA using TURBOTM DNase (Ambion, USA). Treatment of RNA samples (3–15 µg) was performed in three consecutive incubation steps for 20 min each at 37°C using 6 U of DNase in total. The reaction was stopped with inactivation reagent (Ambion, USA), and the supernatant was purified and enriched by gel chromatography using RNA Clean & Concentrator™-25 according to the manufacturer’s instructions (Zymo Research, USA).
Reverse transcription (RT) was performed with 0.75–1.5 µg total RNA using Superscript III (Invitrogen, Germany). Briefly, after denaturation at 95°C for 3–5 min, a mixture containing RNA and 0.5 µM of target- and strand-specific oligonucleotide ‘RT–mRNA’ (for amplification of mRNA) or ‘RT–asRNA’ (for amplification of asRNA) was incubated at 42°C for 20 min to permit annealing. The mixture was complemented with 4 U RiboLock RNase inhibitor (Fermentas, Germany), 1 mM dNTPs, 10 mM DTT, 5 mM MgCl2, 1 × reaction buffer (Invitrogen, Germany) and 200 U reverse transcriptase in a total volume of 20 µl. For first-strand synthesis, the reaction was incubated at 42°C for 2 h and the enzyme was inactivated for 5 min at 95°C. cDNA was amplified by PCR in a 50-µl reaction mixture comprising 0.2 µM oligonucleotides, 1 × HF reaction buffer (Finnzymes, USA), 1 U Phusion polymerase (Finnzymes, USA) and 0.2 mM dNTPs. The following thermo profile was applied: 98°C for 30 s; 32 cycles of 98°C for 10 s, 54°C for 30 s, and 72°C for 3 min; and 72°C for 5 min. To amplify cDNA derived from mRNA, the primers RT–mRNA (reverse primer) and GI-II-mRNA (forward primer) were used. To amplify cDNA derived from asRNA, the primers RT–asRNA (reverse primer) and RT–mRNA (forward primer) were used. Oligonucleotides used for RT–PCR are listed in Supplementary Table S1. The lack of contamination with genomic DNA was verified by omitting the enzyme in an additional reverse transcription control reaction.
RESULTS
Purification of Prochlorococcus RNase E
Expression of recombinant Prochlorococcus MED4 RNase E (PMM1501) protein required the adjustment of the codon usage of the first 63 nt and the last 42 nt of PMM1501 for sufficient expression in E. coli. C-terminally His-tagged pQE70-PMM1501 was overexpressed in E. coli strain Arctic Express (DE3) RIL and purified under native conditions by affinity column chromatography. Recombinant PMM1501 protein migrated according to its theoretical molecular weight of 70 kDa in SDS–polyacrylamide gels (Figure 1 and Supplementary Figure S1). Mass spectrometry analysis of the purified protein fraction did not reveal any contamination with E. coli-specific RNase E protein or other host ribonucleases (data not shown). Furthermore, western blots of the purified protein fraction were immunodecorated with antibodies targeting E. coli-specific RNase E, RNase R or PNPase. None of these antibodies detected measurable amounts of the respective target protein in the elution fraction used for cleavage assays (Supplementary Figure S1A). To test for the presence of other sources of E. coli-specific exonuclease activity in the purified protein fraction, we in vitro transcribed a poly-A RNA, a substrate for exonucleases in general (RNase R, PNPase, RNase II) but not for RNase E, and incubated it with RNase R or recombinant Prochlorococcus RNase E. Cleavage activity was only detected with RNase R but not with recombinant PMM1501 (Supplementary Figure S1B) verifying that the recombinant Prochlorococcus RNase E protein was not contaminated with nucleases from E. coli.
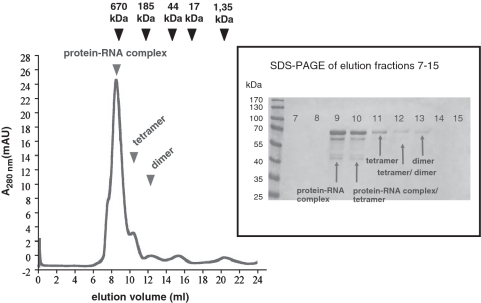
Figure 1.
Gel filtration profile of affinity-purified pQE70-PMM1501 protein. Sizes of RNase E conformers were estimated using Bio-Rad’s gel filtration standard (black arrows) containing thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.35 kDa). Elution peaks of RNase E conformers are indicated by black arrows. Peaks at elution fraction 15 and 21 correspond to buffer components. The inset on the right shows a 10% SDS–PAGE gel that was loaded with elution fractions of the gel filtration.
To assess the quaternary structure of affinity-purified pQE70-PMM1501 protein, we analysed the oligomerization state by gel filtration. According to the calibration profile, the main part of purified recombinant MED4 RNase E was eluted probably as a tetramer (Figure 1), which would suggest a similar conformer organization as that present in E. coli and Synechocystis PCC6803. These results are in good agreement with the presence of the highly conserved Zn-link motif in the Prochlorococcus RNase E sequence. Callaghan et al. (31) demonstrated that the Zn-link is involved in the organization of a functional active quaternary structure of RNase E/G in E. coli by mediating its dimerization and tetramerization.
Determination of the physicochemical properties of Prochlorococcus RNase E
To establish optimal RNA cleavage conditions and to determine the physicochemical properties of MED4 RNase E, we assessed cleavage of a fluorescently labelled synthetic oligonucleotide substrate containing the RNase E recognition site of Synechocystis PCC6803 psbA leader (32) (see ‘Material and Methods’ section). We found that monovalent cations are not required whereas divalent cations are essential for MED4 RNase E enzyme activity with an optimum at 12 mM Mg2+ (Supplementary Figure S2). The enzyme was active between pH 5 and 11, with optimal activity at pH 9 (Figure S2D). The optimal temperature for enzyme activity was 35°C (Supplementary Figure S2E), 10° above the optimal growth temperature for Prochlorococcus MED4 (33). Interestingly, the temperature optima for RNase E activity from Synechocystis PCC6803 [courtesy of M. Asayama (32)] was at 25°C (data not shown), which grows optimally at 30°C (34,35). Similar growth-deviant enzyme optima have been reported for purified glutamine synthetases of Prochlorococcus PCC9511 (a clonal culture of MED4) and Synechocystis PCC6803 with maximal enzyme activities at 55 and 35°C, respectively (36).The MED4 RNase E enzyme was quite stable up to 52°C, whereas a nearly complete inhibition of the enzyme was observed at 56°C (Supplementary Figure S2F).
Transcript organization and expression of phage infection-induced transcripts located in genomic island II and in the ribosomal gene cluster
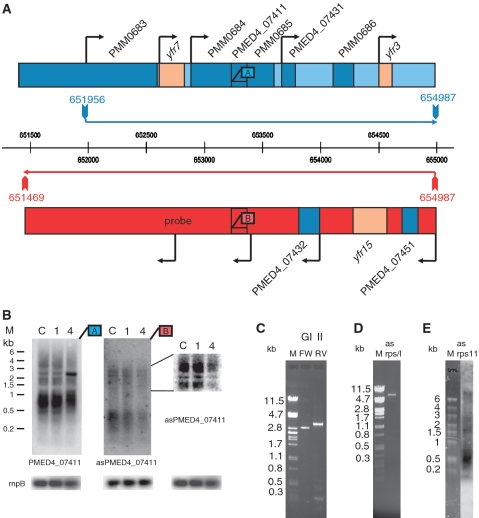
A previous study on global expression profiles of Prochlorococcus MED4 transcripts during phage infection revealed the induction of a 1.5-kb region in genomic island II consisting of three small ORFs, from PMM0684 to PMM0686. Only PMM0686 has a proposed function (clpS-like protease) (27). Here, we investigated the genetic organization of this region in more detail. RT–PCRs and northern blot analysis revealed that this region is organized as a polycistron extending into two upstream genes, ssrS (6S RNA, Yfr7) and PMM0683 (purK), and one downstream gene, yfr3 (Figure 2A and B). In total, the polycistronic region encompasses 3 kb from nucleotide position 651 956–654 987, as determined from RT–PCR data and verified by northern blot analysis (Figure 2B and C). In another global microarray analysis, asRNAs partially covering the complementary strand of the same polycistronic region were identified (37). According to RT–PCR data and northern blot analysis, these asRNAs originate from one large precursor of 3.5 kb at nucleotide 651 469–654 987 on the complementary strand (Figure 2B and C), leading to complete coverage of the polycistronic region in genomic island II. Both RNAs on the sense and antisense strand are induced or stabilized during phage infection (Figure 2B and Supplementary Figure S3) (27,37) supporting our hypothesis that mRNA–asRNA duplex formation can protect RNA from RNase E degradation. Similar cleavage patterns were observed during exponential growth and phage infection despite changes in relative accumulation of the different bands (Figure 2B). Another region that is induced during phage infection is that of the ribosomal protein gene cluster. An increase of gene expression has been measured for rps11 (PMM1536), rpl6 (PMM1544) through rpl5 (PMM1546) and rpl16 (PMM1551) (27). These five genes belong to two adjacent operons ranging from PMM1533 to PMM1558 (38). Using specific primers in RT–PCR, we detected a 7-kb asRNA from nucleotide 1 476 365–1 483 393 that covers the complete phage-induced region (Figure 2D and E). Notably, the detection of a 7-kb asRNA is the first report of a prokaryotic asRNA with such an extraordinary length. However, asRNAs covering more than one gene have been reported before in the bacterium Listeria monocytogenes (39).
Figure 2.
Organization of phage-induced transcripts from genomic island II. (A) Genome arrangement and transcript organization of the section of genomic island II starting with yfr7 (46). Protein-coding genes and non-coding RNA genes (yfr) are shown in dark-blue and light-brown boxes, respectively. Transcript lengths as determined by RT–PCR are marked with horizontal arrows, blue for the forward strand and red for the reverse strand. The vertical arrows correspond to the PCR primers. Black arrows indicate primary transcript starts that were determined by 454 deep sequencing (unpublished data). Binding sites of probes used for northern hybridization shown in (B) are indicated by boxes and labelled with letter A in blue for the forward strand or B in red for the reverse strand. (B) Expression changes of PMED4_07411 in genomic island II and its asRNA during phage infection as determined from northern blot analysis. Lane C—RNA from uninfected control cells. Lanes 1 and 4—RNA from cells during phage infection 1 or 4 h after phage addition. Total RNA (1.5 µg) was loaded per lane. A blot with higher resolution of a section of asPMED4_07411 is presented to the right of the asRNA hybridization. As a loading control rnpB was hybridized. Transcript lengths were estimated from Fermentas RNA high-range marker (M). Probes used for northern hybridizations are indicated by boxes labelled with letter A in blue for the forward strand or B in red for the reverse strand. (C) RT–PCRs of genomic island II (GI II) region on the forward (fw) and reverse (rv) strand revealing an asRNA of ∼3.5 kb. (D) RT–PCR of the asRNA transcript of ribosomal protein cluster (asrps/l) revealing an asRNA of approximately 7 kb. PCR fragment lengths were estimated from lambda PstI marker (M). The identity of cDNAs was verified by Sanger sequencing of PCR products. (E) Northern blot hybridized with a probe targeting the antisense region of rps11. The asrps11 probe was synthesized using oligonucleotides asrps11_FW and asrps11_RV (Supplementary Table S1). M—fermentas RNA high-range marker.
RNA–RNA duplex formation masks recognition sites of RNase E
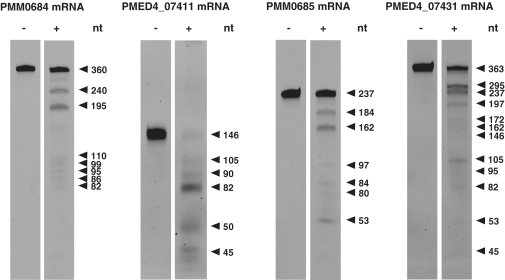
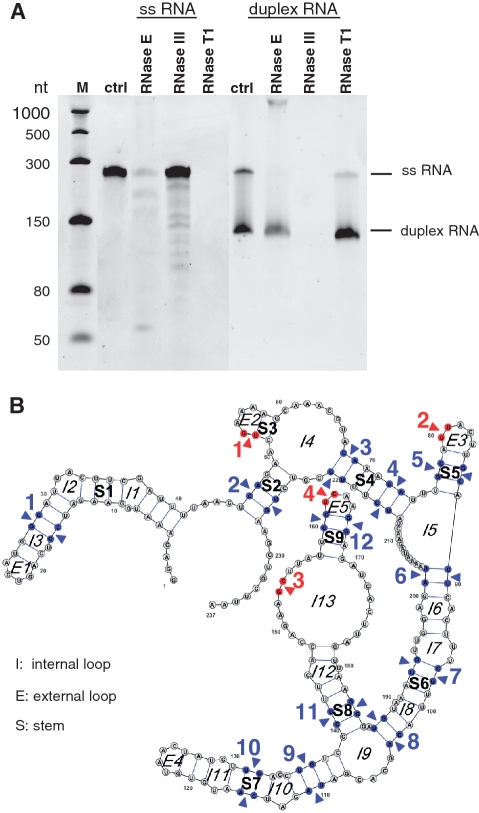
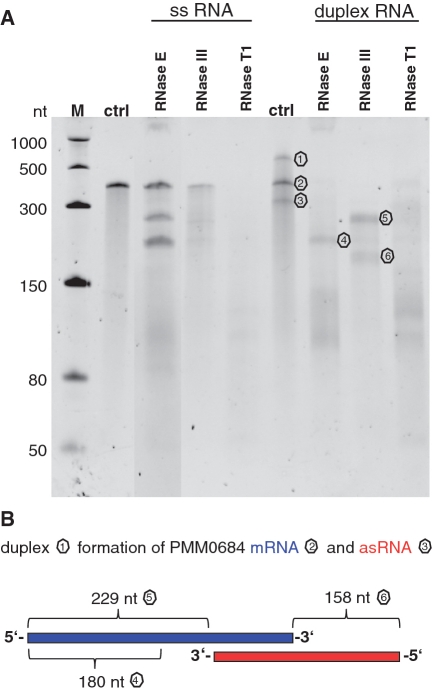
We established an in vitro RNase E cleavage assay to follow up our initial questions about how the activation of genes in genomic island II is regulated during phage infection and how these RNAs are protected from degradation in light of the enhanced transcription of RNase E. The mRNAs of four small ORFs of genomic island II (PMM0684, PMM0685, PMED4_07411, PMED4_7431), which were induced during phage infection (Figure 2B and Supplementary Figure S3), were synthesized in vitro and incubated with purified recombinant MED4 RNase E. All four transcripts were recognized by RNase E and were susceptible to specific cleavage (Figure 3). RNase E and RNase III cleavage sites of PMM0685 were investigated in more detail by northern hybridization (Supplementary Figure S4) using specific oligonucleotide probes targeting the 5′- and 3′-ends, respectively, of the ribonuclease-treated transcripts. The RNase E cleavage sites identified are listed in Table 1. All four cleavage sites were located in loop regions, indicating that the secondary structure is more relevant than sequence specificity for RNase E recognition (Figure 4B). RNase III cleavage sites were located in double-stranded RNA regions (Figure 4B) and are specified in Table 2. Figure 4A shows the results of RNase E, RNase III and RNase T1 digestions for single-stranded and duplex RNAs of PMM0685 in vitro-synthesized RNA. Both sense and antisense RNAs of duplex RNAs had identical sizes and an exact overlap. Incubation of single-stranded PMM0685 RNA with RNase E or RNase III yielded specific cleavage products, whereas RNase T1, which cleaves single-stranded RNA after guanine residues, led to complete digestion of PMM0685 in vitro-synthesized RNA. In contrast, the a priori duplex formation of sense and antisense RNA before RNase E treatment prevented RNase E digestion. RNase III treatment of duplex RNA resulted in complete digestion of the duplex, whereas treatment with RNase T1 did not show any cleavage products. These findings suggest that the secondary and tertiary structures of both RNA molecules were altered and hybridized to each other over their full-length. Clearly, the resultant structural changes hinder substrate accessibility of ribonucleases that recognize single-stranded regions (RNase E and RNase T1) and promote enzyme activity of RNA double strand-dependent enzymes like RNase III.
Figure 3.
RNase E cleavage assay of four in vitro transcripts of genomic island II with (+) and without (−) recombinant Prochlorococcus RNase E. Cleavage fragments were separated on a 7 M urea–6% PAA gel and stained with ethidium bromide. Fragment sizes were estimated from an NEB ssRNA marker.
Table 1.
RNase E cleavage sites of PMM0685 in vitro transcript
| Position within RNA backbone (5′–3′) | Cleavage site (5′–3′) |
|---|---|
| 53, 54 | U_GAAAA |
| 80, 81 | AU_UACU |
| 153, 154 | AAG_CUU |
| 162, 163 | CU_CAAG |
Figure 4.
Ribonuclease cleavage assay of 1.3 pmol single-stranded and 2.6 pmol duplex PMM0685 in vitro RNA. (A) Ethidium bromide-stained 7 M urea–6%PAA gel of PMM0685 single-stranded (ss) and duplex in vitro-synthesized RNA (ctrl) treated with RNase E, RNase III or RNase T1. RNA was incubated with RNases for 15 min. M denotes ssRNA marker from NEB. Though gel electrophoresis was performed under stringent denaturing conditions, only small amounts of duplex RNA were denatured and migrated according to the single-stranded RNA population. (B) The secondary-structure of PMM0685 was folded with RNAfold and drawn using VARNA. RNase E and RNase III cleavage sites, which were deduced from northern blot hybridizations with two oligonucleotide probes targeting the 5′- or 3′-end of PMM0685 in vitro-synthesized RNA, are marked with red arrows or blue arrows, respectively.
Table 2.
RNase III cleavage sites of PMM0685 in vitro transcript
| Position within RNA backbone (5′–3′) | Cleavage site (5′–3′) |
|---|---|
| 16,17/27,28 | C_C/G_G |
| 47,48/225,226 | C_A/G_U |
| 68,69/220,221 | C_A/G_U |
| 72,73/216,217 | G_A/C_U |
| 77,78/86,87 | G_C/C_G |
| 89,90/201,202 | C_U/G_A |
| 97,98/193,194 | C_G/G_U |
| 102,103/187,188 | C_A/G_U |
| 110,111/136,137 | U_A/G_U |
| 115,116/131,132 | C_A/G_U |
| 141,142/183,184 | C_U/G_G |
| 160,161/166,167 | C_C/G_G |
RNA duplexes that overlapped only partially with overhangs at the 5′- or 3′-end lead to partial degradation of the duplex by RNase E (Figure 5). As expected, digestion patterns indicated that double-stranded regions were still protected, whereas single-stranded regions became susceptible to RNase E cleavage (Figure 5). When a 3-kb polysictronic mRNA in vitro transcript was annealed with a 3.5 kb in vitro asRNA (that mimic the in vivo transcripts) complete protection of the polycistron after RNase E treatment was observed (Supplementary Figure S5). The in vitro assay suggests that indeed the 3.5 kb asRNA located in genomic island II is important for the stabilization of mRNAs located opposite the asRNA. These results suggest that complete coverage of the mRNA by an asRNA is required to prevent RNase E digestion. It is conceivable that the same protective mechanism applies for the ribosomal protein gene cluster as the asRNA covers the complete phage induced region (Figure 2). Therefore, complete overlap of duplex RNAs may be a general requirement for mRNA protection.
Figure 5.
Ribonuclease cleavage assay of 1.3 pmol single-stranded and 2.6 pmol partially overlapping duplex PMM0684 in vitro-synthesized RNAs. (A) 7 M urea–6% PAA gel of PMM0684 single-stranded and duplex in vitro RNAs (ctrl) treated with RNase E, RNase III or RNase T1. M denotes ssRNA marker from NEB. (B) Scheme of duplex formation of PMM0684 in vitro-synthesized mRNA (blue) and asRNA (red). Regions of duplex RNA remaining after digestion with RNase E or RNase III in (A) are labelled with corresponding numbers in (B).
DISCUSSION
In this study, we describe a new function for asRNA, a class of regulatory RNA that has gained considerable attention due to its recently discovered ubiquitous occurrence in all kingdoms of life. Previously, we (27) suggested that the subset of host genes upregulated during phage infection might be regulated by the phage or might be part of a host-specific response. For either scenario, it remained unclear how asRNAs are regulated. A prolongation of mRNA half-life initially appeared unlikely because RNase E expression was stimulated as well. Here, we demonstrate the asRNA-mediated regulation of RNA stability, directly influencing the half-life of mRNAs that are protected from RNA decay by asRNAs that completely mask RNase E recognition sites. Whether the increase in transcript level is a final defensive step of the host during phage infection or whether increased mRNA levels are supportive of viral replication [in a similar fashion as has been suggested for phage-encoded D1 protein (40)] remains to be investigated in future studies. The asRNA-modulated regulation of gene expression during phage infection might be a common feature in both the host and the phage. Millard et al. (41) reported an asRNA that physically connects between the photosynthesis gene psbA encoding the D1 core protein of photosystem II with a homing endonuclease (F-Cphl) in the cyanophage S-PM2. Transcripts of the asRNA as well as of psbA and F-Cphl are induced during the lytic infection cycle of S-PM2 in its host Synechococcus sp. WH7803.
RNase E activity is known to impact the stability of both host and phage mRNA during infection of E. coli by bacteriophages T7 and T4 (42,43). In both cases, the C-terminal structural domain of RNase E is involved. For example, the T7 protein kinase phosphorylates the structural domain of host RNase E. However, this region is not present in the Prochlorococcus MED4 homologue. Therefore the indirect regulation of RNase E activity in Prochlorococcus by RNA duplex formation reported here reflects an alternative mechanism of regulation of enzyme activity that appears to be required for an organism lacking the region modified in other host-phage systems.
Our results clearly show that the duplex RNAs resulting from mRNA–asRNA hybridization are perfect substrates for RNase III in vitro (Figure 4). Therefore, an interesting question is how these duplex RNAs become protected from RNase III degradation in vivo. Nothing is known about RNase III protein stability in Prochlorococcus; however, the RNase III transcript, along with the majority of the transcriptome, is down-regulated during phage infection (27). Furthermore, ribosome activity on the mRNAs is likely to interfere with the full-length hybridization of both transcripts in vivo. In addition, other factors might exist in the cell that protect long duplex complexes from degradation.
The modulation of RNase E activity by RNA–RNA interaction has been reported before. However, base pairing of the small regulatory RNA RyhB with the 5′-UTR of sodB mRNA, changing the structure of the mRNA leader, results in an additional RNase E cleavage site, which in turn triggers another pathway for sodB RNA decay (44). Hence, in this example, RNA–RNA interaction does not hide but produces new RNase E recognition sites. The RNase E-dependent regulation of sodB turnover is very complex, and enzyme activity is also influenced by RNA–protein interaction between sodB and Hfq (an RNA chaperone that recognizes single-stranded AU-rich regions in the same manner as RNase E). Both RNase E and Hfq can bind mRNAs close to each other, resulting in protection of the RNase E substrate from nucleolytic degradation (44). Such a complex regulation cannot be expected for Prochlorococcus MED4 because an hfq homologue is lacking in the genome. Of the 12 completely sequenced Prochlorococcus strains, only MIT9313 and MIT9303, both belonging to the evolutionarily oldest Prochlorococcus clade, possess an hfq homologue.
Alternatively, RNase E activity can also be influenced by inhibitory RNase-binding proteins that dynamically regulate RNA decay and processing. In Synechococcus elangatus PCC 7942, the two heat shock proteins Dnak2 and DnaJ2 inhibit RNase E activity in an ATP-dependent manner, suggesting that both proteins are involved in RNA degradation through interaction with RNase E (45). Both proteins are present in Prochlorococcus and hence could provide an additional regulatory circuit for RNA half-lives.
The reduced suite of protein regulators and the disproportionately high number of regulatory RNAs in Prochlorococcus has been linked to its adaptation to an extremely oligotrophic habitat, combined with a streamlined genome (37). This study provides the first insights into the functional role of asRNA regulators and emphasizes the importance of this class of regulatory molecules in the global network of gene regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Israeli Foundation (GIF) young investigator grant (2167-1743.9/2007 to C.S.); the German Science Foundation grant (GRK1305 to C.S. and D.S.); and an Israel Science Foundation (ISF) Morasha grant (1504/06 to D.L.); D.L. is a Shillman fellow. Funding for open access charge: University budget.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joke Lambrecht for performing RT–PCRs and northern hybridizations of rps11, Irena Pekarsky for preparing Prochlorococcus cultures and phage for the infection experiments and Wolfgang R. Hess for critical reading of the manuscript and helpful comments. We thank Agamemnon Carpousis, Murray Deutscher and Gianni Dehò for antibodies against E. coli RNase E, RNase R and PNPase respectively.
REFERENCES
- 1.Melefors O, Lundberg U, von Gabain A. RNA processing and degradation by RNase K and RNase E. In: Belasco J, Brawerman G, editors. Control of Messenger RNA Stability. San Diego, CA: Academic Press; 1993. pp. 53–70. [Google Scholar]
- 2.Bechhofer DH, Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1987;84:498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett TC, Bugrysheva JV, Scott JR. Role of mRNA stability in growth phase regulation of gene expression in the group A Streptococcus. J. Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 5.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 6.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Py B, Higgins C, Krisch H, Carpousis A. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 8.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaberdin VR, Miczak A, Jakobsen JS, LinChao S, McDowall KJ, von Gabain A. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl Acad. Sci. USA. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage RE, Shabanowitz J, Hunt DF, Schuster G, Gruissem W. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- 11.Rott R, Zipor G, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- 12.Schein A, Sheffy-Levin S, Glaser F, Schuster G. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA. 2008;14:10571068. doi: 10.1261/rna.907608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K, Cohen SN. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 2003;48:349–360. doi: 10.1046/j.1365-2958.2003.03435.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 16.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henz SR, Cumbie JS, Kasschau KD, Lohmann JU, Carrington JC, Weigel D, Schmid M. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 2007;144:1247–1255. doi: 10.1104/pp.107.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JM, Zhang S, Saha S, Santa Anna S, Jiang C, Perkins J. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 2001;183:7371–7380. doi: 10.1128/JB.183.24.7371-7380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 21.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 2009;305:1–17. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 25.Mackie GA. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J. Biol. Chem. 1992;267:1054–1061. [PubMed] [Google Scholar]
- 26.Cohen SN, McDowall KJ. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- 27.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, Kettler G, Sullivan MB, Steen R, Hess WR, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 28.Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, Frois-Moniz K, Waterbury J, Chisholm SW. Culturing the marine cyanobacterium Prochlorococcus. Limnol. Oceanogr. Methods. 2007;5:353–362. [Google Scholar]
- 29.Lindell D, Post AF. Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl. Environ. Microbiol. 2001;67:3340–3349. doi: 10.1128/AEM.67.8.3340-3349.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steglich C, Futschik M, Rector T, Steen R, Chisholm SW. Genome-wide analysis of light sensing in Prochlorococcus. J. Bacteriol. 2006;188:7796–7806. doi: 10.1128/JB.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callaghan AJ, Redko Y, Murphy LM, Grossmann JG, Yates D, Garman E, Ilag LL, Robinson CV, Symmons MF, McDowall KJ, et al. ‘Zn-link’: a metal-sharing interface that organizes the quaternary structure and catalytic site of the endoribonuclease, RNase E. Biochemistry. 2005;44:4667–4675. doi: 10.1021/bi0478244. [DOI] [PubMed] [Google Scholar]
- 32.Horie Y, Ito Y, Ono M, Moriwaki N, Kato H, Hamakubo Y, Amano T, Wachi M, Shirai M, Asayama M. Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol. Genet. Genomics. 2007;278:331–346. doi: 10.1007/s00438-007-0254-9. [DOI] [PubMed] [Google Scholar]
- 33.Zinser E, Johnson Z, Coe A, Karaca E, Veneziano D, Chisholm S. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol. Oceanogr. 2007;52:2205–2220. [Google Scholar]
- 34.Rippka R, Deruelles J, Waterbury JB, Herdmann M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. General Microbiol. 1979;111:1–61. [Google Scholar]
- 35.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 36.El Alaoui S, Diez J, Toribio F, Gomez-Baena G, Dufresne A, Garcia-Fernandez JM. Glutamine synthetase from the marine cyanobacteria Prochlorococcus spp: characterization, phylogeny and response to nutrient limitation. Environ. Microbiol. 2003;5:412–423. doi: 10.1046/j.1462-2920.2003.00433.x. [DOI] [PubMed] [Google Scholar]
- 37.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal phototroph: Non-coding RNAs in Prochlorococcus. PLoS Genetics. 2008;4:e10000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steglich C, Lindell D, Futschik M, Rector T, Steen R, Chisholm SW. Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol. 2010;11:R54. doi: 10.1186/gb-2010-11-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 40.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 41.Millard AD, Gierga G, Clokie MR, Evans DJ, Hess WR, Scanlan DJ. An antisense RNA in a lytic cyanophage links psbA to a gene encoding a homing endonuclease. ISME J. 2010;4:1121–1135. doi: 10.1038/ismej.2010.43. [DOI] [PubMed] [Google Scholar]
- 42.Marchand I, Nicholson AW, Dreyfus M. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol. Microbiol. 2001;42:767–776. doi: 10.1046/j.1365-2958.2001.02668.x. [DOI] [PubMed] [Google Scholar]
- 43.Ueno H, Yonesaki T. Phage-induced change in the stability of mRNAs. Virology. 2004;329:134–141. doi: 10.1016/j.virol.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe S, Sato M, Nimura-Matsune K, Chibazakura T, Yoshikawa H. Protection of psbAII transcript from ribonuclease degradation in vitro by DnaK2 and DnaJ2 chaperones of the cyanobacterium Synechococcus elongatus PCC 7942. Biosci. Biotechnol. Biochem. 2007;71:279–282. doi: 10.1271/bbb.60647. [DOI] [PubMed] [Google Scholar]
- 46.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, Chisholm SW. Genomic Islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.