Abstract
Type 2 internal ribosomal entry sites (IRESs) of encephalomyocarditis virus (EMCV), foot-and-mouth disease virus (FMDV) and other picornaviruses comprise five major domains H-L. Initiation of translation on these IRESs begins with specific binding of the central domain of initiation factor, eIF4G to the J-K domains, which is stimulated by eIF4A. eIF4G/eIF4A then restructure the region of ribosomal attachment on the IRES and promote recruitment of ribosomal 43S pre-initiation complexes. In addition to canonical translation factors, type 2 IRESs also require IRES trans-acting factors (ITAFs) that are hypothesized to stabilize the optimal IRES conformation that supports efficient ribosomal recruitment: the EMCV IRES is stimulated by pyrimidine tract binding protein (PTB), whereas the FMDV IRES requires PTB and ITAF45. To test this hypothesis, we assessed the effect of ITAFs on the conformations of EMCV and FMDV IRESs by comparing their influence on hydroxyl radical cleavage of these IRESs from the central domain of eIF4G. The observed changes in cleavage patterns suggest that cognate ITAFs promote similar conformational changes that are consistent with adoption by the IRESs of comparable, more compact structures, in which domain J undergoes local conformational changes and is brought into closer proximity to the base of domain I.
INTRODUCTION
Initiation of translation on most eukaryotic mRNAs involves binding of 43S pre-initiation complexes (comprising a 40S ribosomal subunit, initiator tRNA and eukaryotic initiation factors eIF2, eIF3, eIF1 and eIF1A) to the 5′-end of mRNA, followed by their scanning to the initiation codon and joining with a 60S ribosomal subunit to form an 80S ribosome (1). Initiation on a few viral mRNAs is 5′-end independent, and is instead mediated by an internal ribosomal entry site (IRES) in the 5′-untranslated region, which promotes recruitment of ribosomes to the mRNA either upstream of or directly to the initiation codon. Viral IRESs are classified into four major structural groups, and although each uses a different mechanism for initiation, the key characteristic that differentiates them all from the canonical end-dependent initiation mechanism is that ribosomal recruitment is based on specific non-canonical interactions of the IRES with canonical components of the translation apparatus, and because of this, needs only a subset of the initiation factors required by the canonical mechanism (1).
Type 2 IRESs, such as those of the picornaviruses encephalomyocarditis virus (EMCV), foot-and-mouth disease virus (FMDV) and Theiler's murine encephalomyelitis virus (TMEV) are about 450-nt long, and consist of five major domains, designated H-L (Figure 2). Sequence variation between these IRESs can reach 50%, but a similar overall structure is maintained by compensatory base changes in helical elements. Biochemical reconstitution experiments have shown that 48S complex formation on type 2 IRESs requires eIF2, eIF3 and a 40S ribosomal subunit (which together form the core of the 43S complex), the RNA helicase eIF4A and the central domain of eIF4G, but is only modestly stimulated by eIF4B, eIF1 and eIF1A, and does not require the cap binding protein eIF4E or the N-terminal domain of eIF4G to which it binds (2–5). The key interaction of type 2 IRESs with the translational apparatus is the specific binding of the central HEAT-1 domain of eIF4G to the IRES's J-K domains, which is enhanced by eIF4A (3,5–8). The eIF4G/eIF4A complex induces Adenosine-5′-triphosphate (ATP)-dependent conformational changes at the 3′-border of the IRES (9) that likely prepare this region for binding of the 43S complex in a process that presumably involves the eIF4G/eIF3 interaction. However, the exact mechanism by which eIF4G/eIF4A promotes attachment of 43S complexes to the IRESs has not been determined.
Figure 2.
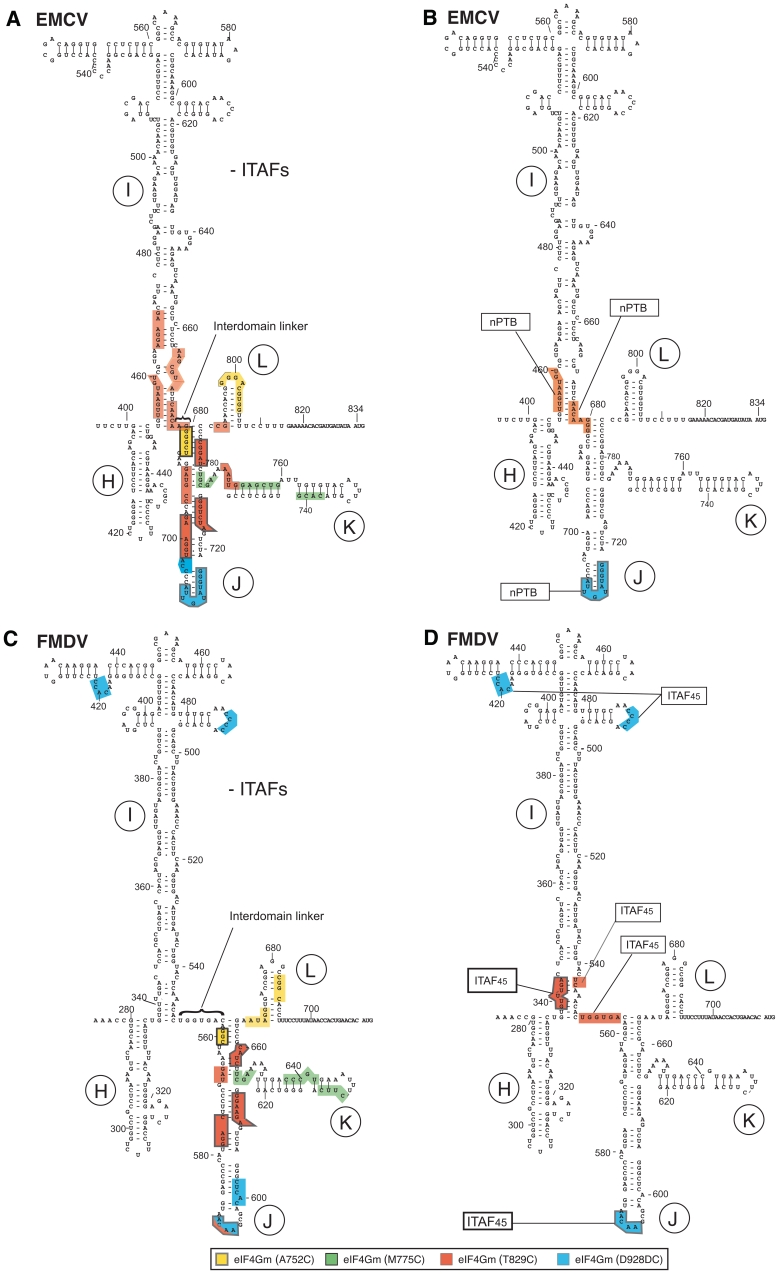
Sites of hydroxyl radical cleavage in EMCV and FMDV IRESs from Fe(II)-tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes influenced by cognate ITAFs mapped onto the secondary structure of IRESs. Sites of hydroxyl radical cleavage from positions on eIF4Gm mapped onto the secondary structures of (A and B) the EMCV IRES and (C and D) the FMDV IRES (GenBank accession no. X00871). Models in panels B and D show only those sites at which cleavage was enhanced by nPTB or ITAF45. Nucleotide numbering of the EMCV IRES and nomenclature of IRES domains are as in (ref. 37). Cleavage sites are show in colors that match the colors of corresponding spheres in Figure 1A. Sites of strong cleavage are indicated by thick edging.
Importantly, initiation on type 2 IRESs also requires IRES trans-acting factors (ITAFs), which are RNA binding proteins that enhance IRES function but are not involved in the canonical initiation process. Although, they all need the same set of canonical initiation factors, different type 2 IRESs require distinct sets of ITAFs, likely due to minor sequence and structural differences between them, and these different requirements may influence viral tissue tropism. The ITAF for the EMCV IRES is the pyrimidine tract binding protein (PTB). Interestingly, initiation on the wild-type (wt) IRES is only modestly enhanced by PTB (2) whereas specific substitutions in the IRES or variation in the downstream reporter led to a strong PTB dependence (10). The TMEV IRES's requirement for PTB also varies significantly, depending on the viral strain used (5,11). The highly homologous neuronal paralog of PTB, nPTB, binds type 2 IRESs with the same specificity as PTB and promotes initiation on them equally well (12). In contrast to the EMCV IRES, efficient initiation on the FMDV IRES requires a second ITAF, ITAF45 (also known as Ebp1), as well as PTB (5,13). The FMDV IRES's requirement for ITAF45 has been confirmed in gene silencing experiments (14). It is expressed at very low levels in non-proliferating differentiated cells such as murine and human brain cells (15,16) and consistently, substitution of the IRES in the neurovirulent TMEV GDVII strain by that of FMDV (O1K) attenuated it, without compromising its translational activity in rabbit reticulocyte lysate or transfected BHK-21 cells (5).
A generally accepted hypothesis for the mechanism by which ITAFs promote internal initiation is that their specific binding to an IRES induces it to adopt the active conformation. PTB has four RNA binding domains (RBDs). RBD1 and RBD2 are independent domains and are connected by flexible linkers whereas RBD3 and RBD4 have a fixed relative orientation and can therefore bind two separated elements in one RNA, bring them to within ∼30 Å and thus induce RNA looping (17,18). Binding of PTB to multiple sites on type 2 IRES was first observed by foot printing (5,12,19). More recently, directed hydroxyl radical cleavage was used to map the interactions of individual domains of PTB with the EMCV IRES (20), showing that RBDs 1 and 2 bound to Domain K of the IRES, near its 3′-end, whereas RBDs 3 and 4 interacted with domain H and the base of domains I and L. These interactions are consistent with the proposed ability of PTB to induce structural changes in type 2 IRESs. ITAF45 has a structure that is characteristic of type II methionine aminopeptidases (MAPs) with additional insertions and a disordered positively charged C-terminal extension (residues 338–394) that is required for its binding to type 2 IRESs (14,21). ITAF45 modulates the accessibility of the FMDV IRES to chemical and enzymatic probes at the apex of domain I and at the junction of I, J and L domains (5). Importantly, even though it has been established that PTB and ITAF45 interact specifically with type 2 IRESs, there is as yet no proof that PTB or ITAF45 induce structural changes in these IRESs that promote their adoption of an active conformation, and the nature of these potential changes is not known.
Here, we investigated the influence of ITAFs on the conformations of EMCV and FMDV IRESs using the directed hydroxyl radical cleavage technique and now report that cognate ITAFs promote similar conformational changes in the type 2 EMCV and FMDV IRESs that are consistent with adoption by them of a comparable tertiary structure in which domain J and the base of domain I are brought into greater proximity, resulting in compaction of these IRESs.
MATERIALS AND METHODS
Plasmids
Expression vectors for His6-tagged eIF1 (4), eIF1A (4), eIF4A (wt) and eIF4B (2,3), nPTB (12), ITAF45 (5), Escherichia coli methionyl tRNA synthetase (22) and eIF4GI736–1115 (eIF4Gm) (9) and eIF4A cysteine mutants (23). The previously described variants of eIF4GI736–1115 (9) have been renamed to take into account a recent revision to the sequence of the largest eIF4GI isoform (GenBank accession no. NM182917). tRNAMeti and mRNAs containing EMCV and FMDV IRESs were transcribed using T7 polymerase and previously described transcription vectors (2,5,24).
Purification of initiation factors, ribosomal subunits and aminoacylation of initiator tRNA
Native eIF2, eIF3 and 40S ribosomal subunits were purified from rabbit reticulocyte lysate, and recombinant eIFs 1, 1A, 4A (wt), 4B, 4Gm (wt) and E. coli methionyl tRNA synthetase were expressed and purified from E. coli (25). Recombinant ITAF45 and nPTB were expressed and purified as described (26). The expression protocol was modified slightly to increase the yield of mutant forms of eIF4A and eIF4Gm. After overnight growth on LB/kanamycin (kan) plates, cells were transferred to 4 L LB/kan medium and incubated with shaking at 30°C until 0.3 < OD600 <0.4. 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce expression of recombinant proteins and incubation was continued for 2 h under the same conditions. Purification of mutant proteins was done as for the corresponding ‘wt’ proteins (25,26). In vitro transcribed tRNAiMet was aminoacylated using recombinant E. coli methionyl tRNA synthetase as described (25).
Fe(II)-BABE modification of eIF4A and eIF4Gm mutants
eIF4Gm and eIF4A cysteine mutants were derivatized with Fe(II)-bromoacetyl-amidobenzyl-EDTA (BABE) as described (9) by incubating 3000 pmol of a protein with 1 mM Fe(II)-BABE in 100 µl H300 buffer [80 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) (pH 7.5), 300 mM KCl, 2.5 mM MgCl2, 10% glycerol] for 30 min at 37°C. Derivatized proteins were separated from unincorporated reagent by buffer exchange on Microcon YM-30 filter units and stored at −80°C.
Directed hydroxyl radical cleavage
To investigate hydroxyl radical cleavage of EMCV and FMDV IRESs from the surface of eIF4Gm in IRES/eIF4Gm/eIF4A ternary complexes (Figures 1, 3 and 4), 5 pmol EMCV RNA (nucleotides 280–974) or FMDV RNA (nucleotides 280–740) were incubated at 37°C for 10 min in a total reaction volume of 50 µl containing buffer A [20 mM HEPES (pH 7.6), 100 mM KCl, 2.5 mM MgCl2 and 5% glycerol] with 10 pmol [Fe(II)-BABE]-eIF4Gm and 10 pmol eIF4A in the presence/absence of 10 pmol nPTB and ITAF45, as indicated. To investigate hydroxyl radical cleavage of EMCV from the surface of eIF4Gm in 48S complexes (Figure 4), 5 pmol EMCV RNA (nucleotides 280–974) were incubated with 10 pmol [Fe(II)-BABE]-eIF4Gm, 20 pmol eIF4A, 10 pmol 40S subunits, 15 pmol eIF2, 10 pmol eIF3, 50 pml eIF1, 50 pmol eIF1A, 20 pmol eIF4B and 15 pmol Met-tRNAiMet in the presence/absence of 10 pmol nPTB in 50 µl buffer A supplemented with 1 mM ATP, 0.2 mM Guanosine 5′-[β,γ-imido] triphosphate (GMPPNP) and 0.25 mM spermidine for 10 min at 37°C. To investigate hydroxyl radical cleavage of EMCV and FMDV IRESs from the surface of eIF4A in IRES/eIF4Gm/eIF4A ternary complexes (Figure 5), 5 pmol EMCV RNA (nucleotides 280–974) or FMDV RNA (nucleotides 280–740) were incubated at 37°C for 10 min in 50 µl buffer A with 10 pmol [Fe(II)-BABE]-eIF4A and 20 pmol eIF4G in the presence/absence of 10 pmol eIF4B, nPTB and ITAF45, as indicated. To investigate hydroxyl radical cleavage of 18S rRNA from the surface of eIF4Gm (Figure 6), 43S pre-initiation complexes containing [Fe(II)-BABE]-eIF4Gm were formed by incubating 10 pmol 40S subunits, 15 pmol eIF2, 10 pmol eIF3, 50 pmol eIF1, 50 pmol eIF1A, 15 pmol Met-tRNAiMeti, 20 pmol eIF4A and 10 pmol [Fe(II)-BABE]-eIF4Gm in 50 ml buffer A supplemented with 1 mM adenosine-5′-triphosphate (ATP), 0.2 mM Guanosine 5′-[β,γ-imido] triphosphate (GMPPNP) and 0.25 mM spermidine for 10 min at 37°C. To generate hydroxyl radicals, reaction mixtures were supplemented with 0.05% H2O2 and 5 mM ascorbic acid and incubated on ice for 10 min. Reactions were quenched by adding 20 mM thiourea. mRNAs and 18S rRNA were phenol extracted, ethanol precipitated and analyzed by primer extension using AMV reverse transcriptase and appropriate [32P]-labeled primers. cDNA products were resolved in a 6% sequencing gel.
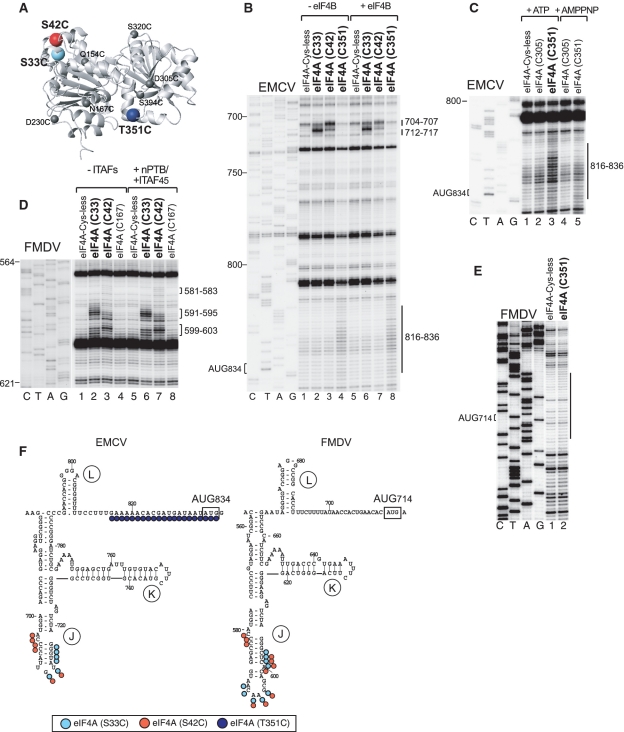
Figure 1.
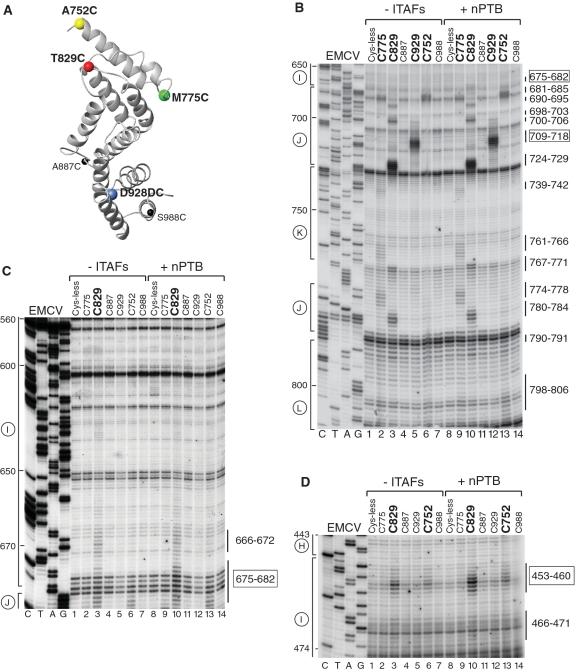
Influence of nPTB on hydroxyl radical cleavage of the EMCV IRES in IRES/eIF4Gm/eIF4A complexes from Fe(II)-tethered eIF4Gm. (A) Ribbon diagram of the HEAT-1 domain of eIF4G (PDB: 1hu3), with spheres indicating newly introduced cysteines. (B–D) Primer extension analysis of hydroxyl radical cleavage of the IRES from Fe(II) tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes in the presence/absence of nPTB. Lanes C, T, A, G depict the corresponding EMCV sequence. IRES domains and nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right; boxed numbers indicate sites at which nPTB enhanced cleavage.
Figure 3.
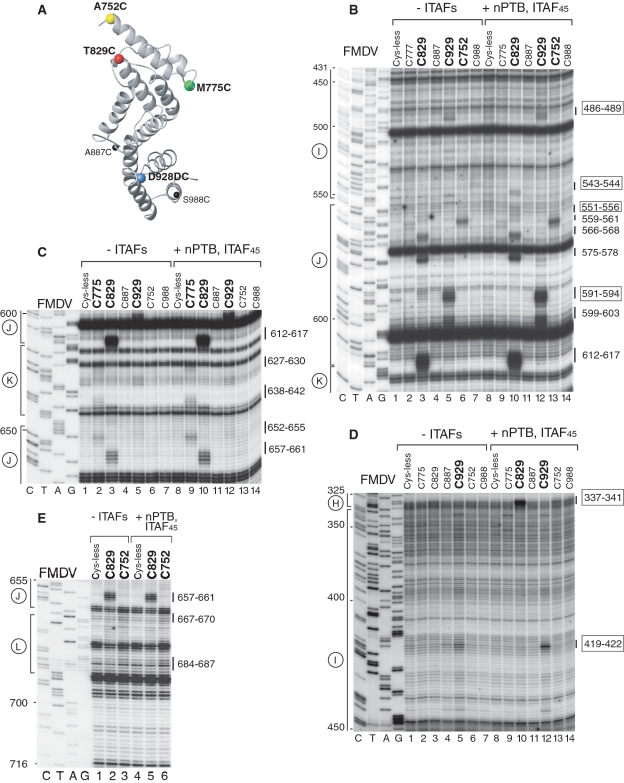
Influence of nPTB and ITAF45 on hydroxyl radical cleavage of the FMDV IRES in IRES/eIF4Gm/eIF4A complexes from Fe(II)-tethered eIF4Gm. (A) Ribbon diagram of the HEAT-1 domain of eIF4Gm, with spheres indicating newly introduced cysteines. (B–E) Primer extension analysis of hydroxyl radical cleavage of the IRES from Fe(II)-tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes in the presence/absence of nPTB and ITAF45. Lanes C, T, A, G depict the corresponding FMDV sequence. IRES domains and nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right; boxed numbers indicate sites at which cleavage was enhanced by nPTB and ITAF45.
Figure 4.
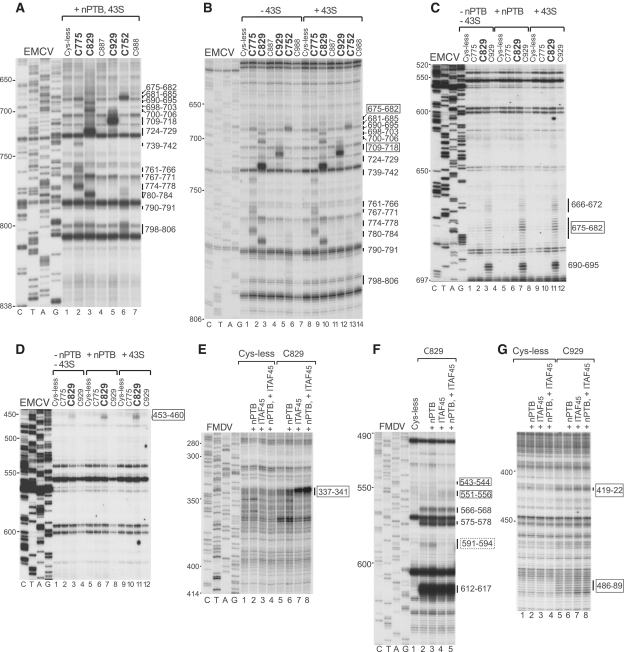
Comparison of changes induced by ITAFs and by 43S complexes in the pattern of hydroxyl radical cleavage of EMCV and FMDV IRESs from Fe(II) tethered eIF4Gm. Primer extension analysis of hydroxyl radical cleavage of (A–D) the EMCV IRES and (E–G) the FMDV IRES from Fe(II)-tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes in the presence of nPTB, ITAF45 or 43S complexes, as indicated. Lanes C, T, A, G depict corresponding EMCV and FMDV sequences. IRES nucleotides are indicated on the left of each panel; positions of cleaved nucleotides on the right are annotated to indicate sites of enhanced cleavage (solid boxes) and reduced cleavage (dashed box).
Figure 5.
Hydroxyl radical cleavage of EMCV and FMDV IRESs from eIF4A in IRES/eIF4Gm/eIF4A ternary complexes. (A) Ribbon diagram of eIF4A in the closed ATP/RNA-bound conformation (PDB: 3EX7), with spheres indicating newly introduced cysteines. Primer extension analysis of hydroxyl radical cleavage of (B and C) the EMCV IRES and (D and E) the FMDV IRES from Fe(II)-tethered eIF4A in IRES/eIF4Gm/eIF4A complexes in the presence/absence of eIF4B, nPTB/ITAF45, ATP and adenosine 5′-[β,γ-imido] triphosphate (AMPPNP), as indicated. Lanes C, T, A, G depict corresponding EMCV and FMDV sequences. IRES nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right. (F) Sites of hydroxyl radical cleavage from positions on eIF4A mapped onto the secondary structure of EMCV and FMDV IRESs. Cleavage sites are shown in colors that match the colors of corresponding spheres in (A).
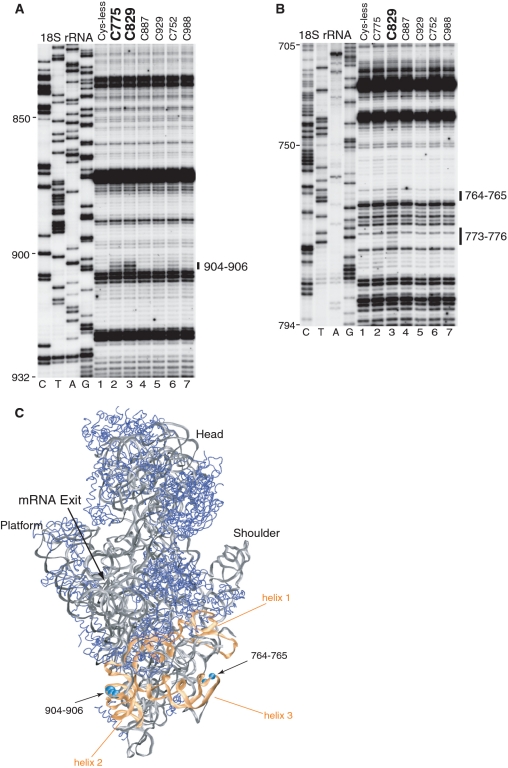
Figure 6.
Hydroxyl radical cleavage of 18S rRNA from eIF4Gm in eIF4Gm-associated 43S complexes. (A and B) Primer extension analysis of hydroxyl radical cleavage of 18S rRNA from Fe(II)-tethered eIF4Gm in eIF4Gm-bound 43S complexes. Lanes C, T, A, G depict the corresponding 18S rRNA sequence. 18S rRNA nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right. (C) Cleavages in 18S rRNA from eIF4Gm (cyan spheres) mapped onto the crystal structure of the yeast 80S ribosome (31). 18S rRNA and ribosomal proteins are shown as grey and blue ribbons. ES6 is colored orange and helices within it are numbered. The radius of the spheres is proportional to the efficiency of cleavage.
RESULTS
Conformational changes induced by ITAFs in type 2 IRESs
To test the hypothesis that ITAFs promote conformational changes in type 2 IRESs, we assayed hydroxyl radical cleavage of EMCV and FMDV IRESs from the surface of eIF4G in eIF4G/eIF4A/IRES complexes assembled with and without nPTB and ITAF45, with the rationale that differences in the intensity or pattern of cleavage caused by the presence of ITAFs would indicate their influence on the conformation of the final complex and/or eIF4G's affinity to the IRES. In this approach, locally generated hydroxyl radicals cleave the IRES in the vicinity of Fe(II) tethered to a unique cysteine residue on the surface of eIF4G via the linker 1-(p-bromo-acetamidobenzyl)-EDTA (BABE), after which cleavage sites are mapped by primer extension. For this, we employed six eIF4G736–1115 mutants (numbering of residues in eIF4G is based on its revised sequence); (GenBank accession no. NM182917) containing single surface-exposed cysteines, and a cysteine-less (Cys-less) variant, which we have previously used to determine the orientation of eIF4G on the EMCV IRES (9); (Figure 1A). The cysteine residue in the D928→D928C insertion mutant is designated Cys929 in the text below. eIF4G736–1115 (termed eIF4Gm) corresponds to the first HEAT domain of eIF4G and comprises five α-helical repeats (7). It binds to eIF4A and eIF3 and is sufficient for specific interaction of eIF4G with the J-K domain of EMCV and FMDV IRESs and to promote efficient 48S complex formation on them (5,6). All eIF4Gm cysteine mutants bound specifically to the J-K domain and supported 48S complex formation on EMCV and FMDV IRESs at levels comparable to that of the wt protein [(9); (data not shown)].
Interaction with eIF4Gm and orientation of domains in the EMCV IRES
Hydroxyl radicals generated from four positions on the surface of eIF4Gm (C752, C775, C829 and C929; shown as colored spheres in Figure 1A) cleaved the EMCV IRES in eIF4G/eIF4A/IRES complexes (Figures 1B–D; summarized in Figure 2A and Table 1). The pattern of cleavage was very similar to that observed previously (9). However, improved protocols for purification and conjugation of eIF4G mutants that allowed higher yields of active proteins to be obtained resulted in an increase in the sensitivity of the hydroxyl radical cleavage assay and allowed us to observe additional medium to weak cleavages, particularly in IRES domains I and K.
Table 1.
[Fe(II)-BABE]-eIF4G cleavage sites in the EMCV IRES
| EMCV IRES | Residue | Cleaved nucleotides | Cleavage intensity w/o ITAFs | Influence of ITAFs on cleavage |
|---|---|---|---|---|
| Domain I | C752 | 453–457 | Weak | Enhanced by nPTB (weak) |
| C829 | 453–460 | Medium | Enhanced by nPTB | |
| Enhanced in 48S complexes | ||||
| 466–471 | Very weak | |||
| 666–672 | Weak | |||
| 675–682 | Weak | Enhanced by nPTB | ||
| Enhanced in 48S complexes | ||||
| Domain J | C752 | 681–685 | Strong | |
| C829 | 690–695 | Medium | ||
| 698–703 | Strong | |||
| 724–729 | Very strong | |||
| 780–784 | Medium | |||
| C929 | 700–706 | Weak | ||
| 709–718 | Very strong | Enhanced by nPTB | ||
| Enhanced in 48S complexes | ||||
| Domain K | C775 | 739–742 | Weak | |
| 761–766 | Weak | |||
| J–K domain Junction | C775 | 774–778 | Medium | |
| C829 | 767–771 | Medium | ||
| Domain L | C752 | 798–806 | Very weak | |
| C829 | 790–791 | Very weak |
The strongest cleavage occurred from C929 and C829. Hydroxyl radicals generated from C929 strongly cleaved at nucleotides 709–718 in the apex of domain J (Figure 1B, lane 5). Strong cleavage from C829 occurred at nucleotides 724–729 and nucleotides 698–703 on opposite stands in the middle of domain J (Figure 1B, lane 3), which overlap a conserved sequence motif that comprises bulged AC and GA dinucleotides and two helical elements of two and 4 bp and that has been identified as an important determinant of eIF4G's interaction with type 2 IRESs (27,28). C829 also cleaved with medium intensity at nucleotides 767–771 in the A-rich loop at the J-K domain junction and at nucleotides 780–784 at the base of the domain J (Figure 1B, lane 3). Strong cleavage also occurred from C752 at the base of domain J (nucleotides 681–685) (Figure 1B, lane 6). This cleavage pattern is consistent with positioning of the N-terminus of eIF4G's HEAT-1 domain near the base and of its C-terminus near the apex of Domain J of the EMCV IRES.
Importantly, cleavage was also observed at the base of domain I. Thus, medium intensity cleavage occurred at nucleotides 453–460 at the very base of domain I from C829 and on the other side of the very base of this domain at nucleotides 675–682 and 666–672 (Figures 1C and D, lane 3). Weak cleavage at nucleotides 453–458 also occurred from C752 (Figure 1D, lane 6). The observation of cleavage from C829 in domains I and J suggests that at least the base of domain I is tilted toward domain J, resulting in compaction of the IRES.
Only weak cleavage from a single position, C775, was observed in domain K (nucleotides 761–766 and 739–742; Figure 1B, lane 2), indicating that domain K is most likely not a primary determinant of IRES/eIF4G interaction. Cleavage in domain L (nucleotides 798–806) was very weak and occurred only from C752 (Figure 1B, lane 6). No cleavages occurred downstream of domain L, upstream of domain I or in its apical half.
Interaction with eIF4Gm and orientation of domains in the FMDV IRES
The same probing analysis was undertaken using the FMDV IRES to investigate whether its interaction with eIF4G is similar to that of the EMCV IRES (summarized in Figure 2C and Table 2). As in the case of the EMCV IRES, the strongest cleavage occurred from C929 in the apex of domain J (nucleotides 591–596) and from C829 in the middle of domain J (nucleotides 612–617), and hydroxyl radicals generated from C752 induced medium intensity cleavage at the base of Domain J (nucleotides 559–561) (Figure 3B, lanes 3, 5 and 6). Thus the orientation of eIF4Gm on the FMDV IRES is similar to that on the EMCV IRES with its N- and C-termini positioned near the base and the apex of domain J, respectively. Cleavage in FMDV domain K from C775 (nucleotides 627–630, 638–642) was again weak, and nearly identical in location to that which occurred from this residue on the EMCV IRES (Figure 3C, lane 2). This indicates that, as for the EMCV IRES, domain K is likely not a primary determinant of eIF4Gm's interaction with the FMDV IRES. As for EMCV, cleavage in FMDV domain L was very weak (nucleotides 667–670, 684–687) and also occurred from C752 (Figure 3E, lane 3). No cleavages were observed downstream of domain L. Whereas, cleavage within domains J and K of EMCV and FMDV IRESs was identical, cleavage in domain I differed significantly. In contrast to the EMCV IRES, no cleavage from any position occurred in the base of FMDV domain I (Figure 3B and D). However, low-intensity cleavage occurred in the apex of domain I (nucleotides 419–422 and 486–489) from C929 (Figure 3B and D, lane 5). Thus the base of domain I in the FMDV IRES is not oriented as close to the base of domain J as in the EMCV IRES. Importantly, in contrast to the EMCV IRES (Figure 2A), these domains are separated in the FMDV IRES by an unstructured 6 nt-long spacer (Figure 2C). On the other hand, cleavage from C929 in apical regions of both domains I and J suggest the possibility of a kink in domain I that brings its apex in proximity to C929 positioned at the apex of domain J.
Table 2.
[Fe(II)-BABE]-eIF4G cleavage sites in the FMDV IRES
| FMDV IRES | Residue | Cleaved nucleotides | Cleavage intensity w/o ITAFs | Influence of ITAFs on cleavage |
|---|---|---|---|---|
| Domain I | C829 | 337–341 | Induced by ITAF45 (Very strong) | |
| 543–544 | Induced by ITAF45 (weak) | |||
| 551–556 | Induced by ITAF45 (weak) | |||
| C929 | 419–422 | Medium | Enhanced by ITAF45 | |
| 486–489 | Weak | Enhanced by ITAF45 | ||
| Domain J | C752 | 559–561 | Medium | |
| C775 | 559–561 | Weak | ||
| C829 | 566–568 | Medium | ||
| 575–578 | Strong | |||
| 591–594 | Weak | Eliminated by ITAF45 | ||
| 612–617 | Very strong | |||
| C929 | 575–578 | Weak | ||
| 591–594 | Very strong | |||
| 599–603 | Medium | |||
| 657–661 | Medium | |||
| Domain K | C775 | 627–630 | Weak | |
| 638–642 | Weak | |||
| J-K domain Junction | C775 | 652–655 | Medium | |
| Domain L | C752 | 667–670 | Very weak | |
| 684–687 | Very weak |
Influence of nPTB on the conformation of the EMCV IRES
Addition of nPTB to eIF4Gm/eIF4A/EMCV IRES complexes enhanced cleavage from C829 and to a much lesser extent from C752 in the basal helix of domain I (nucleotides 453–460) (Figure 1D, compare lanes 3, 6 and 10, 13) and from C829 in the opposite strand of this helix (nucleotides 675–682) (Figure 1C, compare lanes 3 and 10). It also enhanced the already strong cleavage from C929 at the apex of domain J (nucleotides 709–718) (Figure 1B, lanes 5 and 12). Results obtained using PTB1 (data not shown) and nPTB in this and other experiments were similar, consistent with a previous report (12). This enhancement likely reflects local conformational changes in domain J rather than an increase in affinity of eIF4Gm to it, because PTB enhanced cleavage only in this non-conserved apical region and not at nucleotides 698–703 or nucleotides 724–729, which overlap the adjacent conserved motif that contains determinants of eIF4G's binding to the IRES. Such changes in the pattern of cleavage (summarized in Figure 2B) would be consistent with conformational changes in the IRES that increase the proximity of the base of domain I to the N-terminal region of IRES-bound eIF4Gm, and of the apex of domain J to the C-terminal region of the factor, which in turn suggests adoption by the IRES of a more compact conformation in which the bases of domains I and J are brought into closer proximity.
An important question is whether these conformational changes induced in the IRES are related to its active conformation during initiation. We took advantage of the fact that 48S complex formation on the EMCV IRES occurs efficiently in the absence of PTB to compare the pattern of cleavage of the IRES in eIF4Gm/eIF4A complexes, in eIF4Gm/eIF4A/nPTB complexes and in 48S complexes assembled without nPTB. We first verified that all elements of the IRES in 48S complexes remain accessible to eIF4Gm-mediated cleavage. Cleavages in 48S complexes assembled in the presence of nPTB were the same as in eIF4Gm/eIF4A/nPTB/IRES complexes (Figure 4A) confirming that the IRES was not shielded by components of the 43S complex. Like addition of nPTB, inclusion of 43S complexes into reaction mixtures with the IRES and eIF4Gm/eIF4A resulted in enhanced cleavage from C929 at the apex of domain J (nucleotides 709–718) (Figure 4B, compare lanes 5 and 12), and enhanced cleavage from C829 at the base of domain I (nucleotides 453–460 and 675–682) (Figures 4C and D, compare lanes 3, 7 and 11). Thus the changes in the pattern of cleavage in the IRES in 48S complexes assembled without nPTB parallel the changes induced by nPTB in IRES/eIF4Gm/eIF4A complexes, indicating that the conformation of the IRES in 48S complexes is likely similar to that induced by nPTB in IRES/eIF4Gm/eIF4A complexes off the ribosome. This in turn suggests that nPTB induces adoption of the active conformation by the EMCV IRES.
Influence of ITAF45 and nPTB on the conformation of the FMDV IRES
Efficient 48S complex formation on the FMDV IRES requires PTB and ITAF45, and we therefore first investigated the influence of both ITAFs together on eIF4G-mediated cleavage of this IRES. nPTB and ITAF45 together moderately enhanced the strong cleavage from C929 in the apex of domain J (nucleotides 591–594) (Figure 3B, compare lanes 5 and 12) and slightly enhanced the weak cleavage in the apex of domain I (nucleotides 419–422 and 486–489) (Figures 3B and D, lanes 5 and 12). However, the strongest influence of these ITAFs was on the cleavage from C829. Thus, nPTB and ITAF45 induced very strong cleavage in the 5′-terminal strand of the basal helix of domain I (nucleotides 337–341) (Figure 3D, lanes 3 and 10) and weak cleavage in the opposite strand (nucleotides 543–544 and 551–556) from C829 (Figure 3B, lanes 3 and 10). nPTB and ITAF45 also eliminated the weak cleavage from C829 in the apex of domain J (nucleotides 591–594) (Figure 3B, lanes 3 and 10). These changes (summarized in Figure 2D) are qualitatively similar to those induced by nPTB in the pattern of eIF4G-mediated cleavage of the EMCV IRES (Figure 2B), in that they consisted of enhanced cleavage by C929 at the apex of domain J and either induction of cleavage (FMDV) or enhancement of cleavage (EMCV) by C829 at the base of domain I. Thus, although, in contrast to EMCV, no cleavage was observed in the FMDV IRES at the base of domain I in the absence of ITAFs, the global patterns of cleavage in both IRESs in the presence of ITAFs became very similar. Both IRESs thus likely adopt similarly compact conformations in the presence of cognate ITAFs in which the base of domain I and the base of domain J are brought into close proximity.
To determine which ITAF was primarily responsible for inducing these conformational changes in the FMDV IRES, cleavage from eIF4Gm was investigated in the presence of nPTB and ITAF45 individually. These experiments indicated that these changes were mostly attributable to the presence of ITAF45, which alone induced cleavage in the basal helix of domain I (nucleotides 337–341, 543–544, 551–556) (Figures 4E and F), eliminated cleavage in the apex of domain J from C829 (Figure 4F) and enhanced cleavage from C929 in the apex of domain I (Figure 4G). However, it is important to note that the strongest induction of cleavage in the 5′-terminal strand in the basal helix of domain I (nucleotides 337–341) was synergistically mediated by ITAF45 and nPTB (Figure 4E, compare lanes 7 and 8).
The position of eIF4A in IRES/eIF4G/eIF4A complexes assembled on EMCV and FMDV IRESs
eIF4A is essential for initiation on all type 2 IRESs. Apart from increasing the affinity of eIF4G for the IRES (6), it also induces ATP-dependent conformational changes in the IRES around the initiation codon that have been hypothesized to be required for attachment or proper positioning of 43S complexes (9,23). eIF4A consists of two domains joined by a flexible linker (Figure 5A). The eIF4A-NTD binds to the C-terminal helix of eIF4Gm whereas the eIF4A-CTD binds to its N-terminal two HEAT-repeats (29,30). To obtain insights into the molecular mechanism of eIF4A's action during initiation on EMCV and FMDV IRESs, we employed hydroxyl radical cleavage to determine its position in IRES/eIF4G/eIF4A complexes. For this, we used nine eIF4A mutants containing single surface-exposed cysteines at well-distributed positions (shown as spheres in Figure 5A), which were previously used to determine the position of eIF4A in IRES/eIF4G/eIF4A complexes assembled on type 1 IRESs (23).
Hydroxyl radicals from three positions (C33, C42 and C351) induced cleavage in the EMCV IRES (summarized in Figure 5F). Strong cleavage from C33 and C42 occurred in the apex of domain J (nucleotides 704–707 and 712–717; Figure 5B, lanes 2 and 3). Importantly, hydroxyl radicals generated from C351, which is close to eIF4A's RNA binding region, cleaved an extensive region of the IRES immediately upstream of and including the initiation codon (nucleotides 816–836) with weak to medium intensity (Figure 5B, lane 4). Cleavage from C351 was dependent on the presence of ATP (Figure 5C, lanes 3 and 5). None of the cleavages described above occurred in eIF4G's absence (data not shown), and neither their intensity nor location were influenced by eIF4B (Figure 5B, lanes 6–8).
Analogous cleavages from C33 and C42 were induced in the apex of domain J of the FMDV IRES (Figure 5D, lanes 1–4). Cleavage from these eIF4A residues in both IRESs overlapped with those induced from C929 of eIF4Gm (Figures 1B and 3B, lane 5), consistent with the proximity of these positions to each other in eIF4G/eIF4A complexes, as determined by NMR and X-ray crystallography (29,30). Although the position of these cleavages was not altered by ITAFs, the intensity of cleavage at the apex of domain J was slightly enhanced by them (Figure 5D, lanes 5–8). This is consistent with the enhancement by these ITAFs of cleavage by eIF4G-Cys929 in the same region, at nucleotides 591–594 (Figure 3B). However, in contrast to the EMCV IRES, cleavage around the initiation codon in the FMDV IRES from C351 of eIF4A was very weak, at the limit of significance (Figure 5E).
Ribosomal position of eIF4Gm in initiation complexes
To get insights into the orientation of type 2 IRESs in 48S complexes, we investigated the ribosomal position of eIF4Gm in eIF4G-bound 43S complexes and in 48S complexes formed on the EMCV IRES. The position of eIF4Gm was mapped by hydroxyl radical cleavage of 18S rRNA using the eIF4Gm cysteine mutants described above (Figure 1A). In eIF4Gm-bound 43S complexes, hydroxyl radicals generated from two positions in the N-terminal region of eIF4Gm cleaved 18S rRNA in the eukaryote-specific expansion segment (ES) 6, which emerges at the solvent side of the platform just below helix 26 and branches into three long irregular helices. The first runs perpendicularly to the body in the direction of the shoulder, the second is oriented parallel to the length of the body pointing toward its bottom, and the third is also situated perpendicular to the body but points more into solution away from the body than the first (31); (Figure 6C). In eIF4Gm-associated 43S complexes, medium-intensity cleavage occurred at the base of the second helix: hydroxyl radicals from C829 and C775 cleaved exposed nucleotides 904–906 with medium intensity (Figure 6A, lanes 2 and 3). In addition, hydroxyl radicals generated from C829 induced weak cleavage at the apex of the third helix (nucleotides 764–765 and 773–776) (Figure 6B, lane 3). Cleavage was identical in eIF4Gm-bound 43S complexes and in 48S complexes assembled on the EMCV IRES, but did not occur when eIF4Gm was incubated with 40S subunits alone (data not shown). No cleavage was observed in any other region of 18S rRNA, including the platform or the head of the 40S subunit.
It is difficult to reconcile cleavage from the same position (C829) at sites in different helices that are so far apart (Figure 6C) unless ES6 undergoes substantial conformational rearrangement e.g. upon formation of 43S pre-initiation complexes. Since the third helix protrudes away from the surface and does not contact other portions of the 40S subunit, it would be more likely to undergo major conformational changes than the base of the second helix. The model for the eIF3 position on the 40S subunit obtained by superimposing the cryo-EM eIF3/HCV IRES structure onto the cryo-EM structure of the 40S/HCV IRES complex (32) indicates that the left leg of eIF3 is located between the Platform and ES6. This places it near and just above the regions of 18S rRNA cleaved from eIF4G. If greater reliance is placed on the substantially more intense cleavage at nucleotides 904–906, then these data suggest that the HEAT-1 domain of eIF4G is located on the solvent side of the 40S subunit just below the platform, in which case eIF4Gm would likely interact with the left leg of eIF3 (32). However, the position of eIF4G that was deduced by fitting the EM structure of the eIF3/eIF4G complex onto the eIF3 density in a 40S/eIF3/HCV IRES model suggested that eIF4G interacts with the left arm of eIF3 and is instead located in close proximity to the head/neck of the 40S subunit (32), over 100 Å from the positions of cleavage in 18S rRNA that we observed. Although the apparent contradiction concerning the ribosomal position of eIF4G cannot currently be resolved, the possibility that the absence of cleavage in the neck/head area is due to the extensive shielding of this region of 18S rRNA by ribosomal proteins cannot be excluded.
DISCUSSION
Interaction of eIF4G with type 2 IRESs
Hydroxyl radical cleavage experiments reveal important similarities between the interaction of eIF4Gm with EMCV and FMDV IRESs. They confirmed that the interaction of eIF4Gm primarily involves domain J, since cleavage in domain K was weak and was observed from only a single position (Cys775). These observations are consistent with prior reports that in the absence of domain K, specific binding of eIF4G to domain J still occurs, albeit with reduced affinity (33). The pattern of cleavage of EMCV and FMDV IRESs indicates that in both cases, the HEAT1 domain of eIF4G binds with its C-terminus oriented toward the base of IRES domain J and its N-terminus in proximity to its apex. The sites of interaction of eIF4Gm with type 2 IRESs inferred from the cleavage data are consistent with chemical and enzymatic foot-printing analyses (5,8). Importantly, the sites of strongest cleavage, which were from Cys829 in the lower half of domain J (Figure 2), coincided with a conserved element that has been identified as a key determinant of eIF4G's binding to type 2 IRESs (27,28).
The influence of ITAFs on the conformation of type 2 IRESs
Although eIF4Gm induced similar patterns of cleavage in J-K domains of EMCV and FMDV IRESs, and no cleavages occurred upstream of domain I or downstream of domain L in both cases, cleavage in domain I in the absence of ITAFs differed between these IRESs. Thus cleavage at the base of domain I was observed only in the EMCV IRES, implying that tilting of the base of domain I toward domain J occurs in the EMCV but not in the FMDV IRESs, and cleavage at the apex of domain I was observed only in the FMDV IRES, suggesting that a kink in domain I brings its apical cloverleaf element in proximity to the region of eIF4Gm that is positioned at the apex of domain J. These distinct aspects of the patterns of cleavage indicate that in the absence of ITAFs, the orientation of domain I relative to the J-K domain differs in these IRESs. Importantly, the I and J-K domains are separated by only 1 nt in the EMCV IRES, but by a 6-nt long unstructured linker in the FMDV IRES (Figure 2).
The primary effects of inclusion of cognate ITAFs in probing reactions were (i) strong enhancement of cleavage from Cys829 of eIF4Gm at the base of domain I by nPTB in the case of the EMCV IRES and induction of strong cleavage from this eIF4Gm residue at an identical position in the FMDV IRES by ITAF45 and (ii) enhancement by nPTB and ITAF45 of cleavage from eIF4G-Cys929 at the apex of domain J in the EMCV IRES and the FMDV IRES, respectively. ITAF45 also eliminated cleavage at the apex of FMDV domain J from Cys829, which was not observed in the EMCV IRES. As a result of these changes, the patterns of cleavage from eIF4Gm in complexes of EMCV and of FMDV IRESs with cognate ITAFs became broadly similar. This would be consistent with adoption by these IRESs of a comparable tertiary structure in which domain J undergoes local conformational change and domain J and the base of domain I are brought into greater proximity, resulting in compaction of these IRESs. The nature of the changes induced by cognate ITAFs in type 2 IRESs suggests that the latter consist of independent domains connected by linkers and that the individual domains can pivot or rotate relative to each other and pack together in a manner that is promoted by ITAFs.
Although ITAF45 was primarily responsible for altering the pattern of eIF4Gm-mediated cleavage of the FMDV IRES, there was strong synergism between it and nPTB in promoting cleavage at the base of domain I, which would explain the positive influence of PTB on FMDV IRES function (34) and the synergistic activation of this process by both ITAFs (5,13). It should also be noted that the conformational rearrangement induced by nPTB and ITAF45 in these IRESs is likely more extensive than apparent here, because our assay identified only those changes that occurred in proximity to eIF4G.
A significant question is whether adoption of an altered conformation by the EMCV and FMDV IRESs in the presence of cognate ITAFs is in fact responsible for the translational ‘activation’ of IRESs by these factors. The observation that the changes in the pattern of cleavage from eIF4Gm in the EMCV IRES in 48S complexes assembled in the absence of nPTB were similar to those induced by nPTB suggests that nPTB does indeed induce the active conformation of the EMCV IRES.
Directed hydroxyl radical cleavage experiments have shown that RBD1 and RBD2 of PTB interact with the apical loop of domain K of the EMCV IRES, whereas RBD3/4 bind to domain H and the base of domains I and L, which implies that PTB constrains and stabilizes the three-dimensional structure of the IRES (20). The similarity between PTB's footprint on the EMCV IRES and on other type 2 IRESs (5,12,19) and the sequence conservation between these IRESs suggests that individual domains of PTB interact with all type 2 IRESs in a similar manner. Foot-printing data (5) suggest that ITAF45 also interacts with multiple non-overlapping sites in the FMDV IRES. It is therefore likely that both PTB and ITAF45 interact simultaneously with several domains of type 2 IRESs, limiting their relative mobility and thus stabilizing the IRES in a specific conformation.
The mechanism of initiation of translation on type 2 IRESs
If type 2 IRESs consist of stable domains that can rotate relative to each other and that may require one or more ITAFs to promote their packing together for the IRES to adopt its active conformation, this raises the question of which stage in the process of IRES-mediated initiation is dependent on alteration of the conformation of the IRES.
The first stage in initiation on type 2 IRESs is the specific binding of eIF4G/eIF4A to the J-K domain, but this stage is not dependent on ITAFs (6). This binding induces conformational changes in the vicinity of the initiation codon in an ATP-dependent manner (9) that are likely to be required for productive binding of a 43S complex and proper positioning of mRNA in the mRNA binding cleft of the 40S subunit. Thus, one possibility is that changes in the IRES induced by ITAFs might be required for proper orientation of eIF4A so that it can efficiently induce the necessary conformational changes in the area of the IRES flanking the initiation codon. However, even in the absence of ITAFs, eIF4G/eIF4A were able to induce these conformational changes (9) and hydroxyl radicals generated from eIF4A-Cys351 (which is close to the mRNA binding surface of eIF4A in its closed ATP-bound form) (35), induced cleavage at and immediately upstream of the initiation codon (Figure 6B) in ternary IRES/eIF4G/eIF4A complexes. This indicates that even in the absence of ITAFs, eIF4A is appropriately oriented in IRES/eIF4G/eIF4A complexes.
Importantly, although specific interaction of the J-K domains of type 2 IRESs with eIF4G/eIF4A is crucial for initiation on them, the J-K domains alone do not have IRES activity (36,37) even though they retain the ability to bind specifically to eIF4G/eIF4A (9). In addition to the J-K domain, domain I is also essential for the activity of type 2 IRESs. Thus, deletion or mutation of elements at the apex of this domain abrogates the function of these IRESs without impairing their interaction with eIF4G (8,36,38–40), suggesting that this region plays an essential (but as yet uncharacterized) role in initiation on type 2 IRESs at a step other than recruitment of eIF4G/eIF4A.
The results of directed hydroxyl radical probing indicate that ITAFs promote relative reorientation of I and J-K domains. Reorientation of domain I might be required to overcome potential steric impediments to stable binding of 43S and IRES/eIF4G/eIF4A complexes, such as possible clashes between elements of the IRES and eIF3 and/or the 40S subunit. However, even if there are no such steric clashes, this conformational rearrangement may nevertheless contribute to the correct positioning of domain I, facilitating the establishment of specific interactions with eIF3 or the 40S subunit itself to anchor the IRES on the 43S complex, or even to induce conformational changes in 43S complexes that promote proper initial attachment or proper positioning of the coding region in the mRNA binding cleft, in a manner analogous to the activity of domain II of hepatitis C virus and related IRESs (41). Further mechanistic insights into this stage of initiation on type 2 IRESs will require determination of the structure of IRES-bound initiation complexes to establish the conformation and interactions of domain I in them.
FUNDING
National Institutes of Health (AI51340 to C.U.T.H.); (GM59660 to T.V.P.); Human Frontiers Science Program (RPG0055/2006-C to T.V.P.); National Cancer Institute (Howard Temin K01 CA119107 to A.M.); Boston University Peter Paul Career Development Professorship (to A.M.). Funding for open access charge: National Institutes of Health (AI51340 and GM59660).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are very grateful to M. Yusupov and A. Ben-Shem for sharing coordinates for the structure of the yeast 80S ribosome prior to publication.
REFERENCES
- 1.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestova TV, Hellen CUT, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pestova TV, Shatsky IN, Hellen CUT. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16:6870–6888. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 5.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CUT. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 6.Lomakin IB, Hellen CUT, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CUT, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 8.Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 9.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CUT. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaminski A, Jackson RJ. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 12.Pilipenko EV, Viktorova EG, Guest ST, Agol VI, Roos RP. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001;20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA. 2007;13:1366–1374. doi: 10.1261/rna.469707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38–2G4, which varies with the cell cycle. Exp. Cell. Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- 16.Yoo JY, Wang XW, Rishi AK, Lessor T, Xia XM, Gustafson TA, Hamburger AW. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer. 2000;82:683–690. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 18.Lamichhane R, Daubner GM, Thomas-Crusells J, Auweter SD, Manatschal C, Austin KS, Valniuk O, Alain FH-T, Rueda D. RNA looping by PTB: evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc. Natl Acad. Sci. USA. 2010;107:4105–4110. doi: 10.1073/pnas.0907072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolupaeva VG, Hellen CU, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 20.Kafasla P, Morgner N, Pöyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol. Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Kowalinski E, Bange G, Bradatsch B, Hurt E, Wild K, Sinning I. The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 2007;581:4450–4454. doi: 10.1016/j.febslet.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 22.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl Acad. Sci. USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pestova TV, Hellen CUT. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisarev AV, Unbehaun A, Hellen CUT, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 26.Kolupaeva VG, de Breyne S, Pestova TV, Hellen CUT. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007;430:409–439. doi: 10.1016/S0076-6879(07)30016-5. [DOI] [PubMed] [Google Scholar]
- 27.Clark AT, Robertson ME, Conn GL, Belsham GJ. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J. Virol. 2003;77:12441–12449. doi: 10.1128/JVI.77.23.12441-12449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassili G, Tzima E, Song Y, Saleh L, Ochs K, Niepmann M. Sequence and secondary structure requirements in a highly conserved element for foot-and-mouth disease virus internal ribosome entry site activity and eIF4G binding. J. Gen. Virol. 2004;85:2555–2565. doi: 10.1099/vir.0.80106-0. [DOI] [PubMed] [Google Scholar]
- 29.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl Acad. Sci. USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 32.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 33.Saleh L, Rust RC, Füllkrug R, Beck E, Bassili G, Ochs K, Niepmann M. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J. Gen. Virol. 2001;82:757–763. doi: 10.1099/0022-1317-82-4-757. [DOI] [PubMed] [Google Scholar]
- 34.Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 1997;71:8330–8339. doi: 10.1128/jvi.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 36.Evstafieva AG, Ugarova TY, Chernov BK, Shatsky IN. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 1991;19:665–671. doi: 10.1093/nar/19.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duke GM, Hoffman MA, Palmenberg AC. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kühn R, Luz N, Beck E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1990;64:4625–4631. doi: 10.1128/jvi.64.10.4625-4631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson ME, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hellen CUT. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]