Abstract
The Eco29kI restriction-modification (R-M) system consists of two partially overlapping genes, eco29kIR, encoding a restriction endonuclease and eco29kIM, encoding methyltransferase. The two genes are thought to form an operon with the eco29kIR gene preceding the eco29kIM gene. Such an organization is expected to complicate establishment of plasmids containing this R-M system in naive hosts, since common logic dictates that methyltransferase should be synthesized first to protect the DNA from cleavage by the endonuclease. Here, we characterize the Eco29kI gene transcription. We show that a separate promoter located within the eco29kIR gene is sufficient to synthesize enough methyltransferase to completely modify host DNA. We further show that transcription from two intragenic antisense promoters strongly decreases the levels of eco29kIR gene transcripts. The antisense transcripts act by preventing translation initiation from the bicistronic eco29kIR–eco29kIM mRNA and causing its degradation. Both eco29kIM and antisense promoters are necessary for Eco29kI genes establishment and/or stable maintenance, indicating that they jointly contribute to coordinated expression of Eco29kI genes.
INTRODUCTION
Type II restriction-modification (R-M) systems are comprised of (i) a restriction endonuclease that recognizes a specific DNA sequence and introduces double-stranded breaks at or around the recognition site and (ii) a methyltransferase (methylase) that recognizes the same DNA sequence and methylates it. Methylation prevents site recognition by the endonuclease and thus protects DNA from cleavage.
Bacterial cells carrying R-M genes become resistant to infection by bacteriophages whose genomes contain unmethylated (unmodified) recognition sites. The genomes of certain phages are devoid of recognition sites of enzymes encoded by R-M systems common in host bacteria (1), indicating that R-M gene products limit the spread of bacteriophages and other mobile genetic elements parasitizing on host bacteria. This beneficial property may explain the wide dissemination of R-M systems in the eubacterial kingdom.
R-M systems genes are themselves often carried on mobile genetic elements capable of horizontal spread between different bacterial species. Premature appearance of endonuclease activity upon entry of a genetic element carrying R-M system genes into a naïve host can lead to host DNA degradation. To minimize the possibility of such an outcome, R-M systems evolved mechanisms helping to ensure that endonuclease expression is activated only after the host DNA is completely modified by the methylase. Several distinct mechanisms that ensure delayed appearance of the restriction endonuclease activity have been described (2). Numerous R-M systems encode an additional protein, the product of a C (control) gene, which orchestrates cooperative time-delayed switching from methylase gene transcription to endonuclease gene transcription (3–6). Other systems ‘monitor’ the activity and/or the amount of methylase synthesized, and activate the endonuclease gene transcription only after a certain threshold level of methylase is reached (7–10). This is achieved by either incorporation of a recognition site into the M gene promoter (with modification of the site leading to decreased promoter activity) or through the binding of methylase to an operator sequence that is unrelated to the recognition site and overlaps with the M promoter. In the latter case, the methylase contains an additional DNA binding domain that recognizes the operator. When methylase binds the operator, it prevents the binding of RNA polymerase to the M gene promoter. In both cases, decreased transcription from the M promoter leads to increased transcription from the overlapping R gene promoter. In yet another class of R-M systems, the genetic organization of the structural genes (a single operon with the R gene following the M gene) passively ensures preemptive methylase synthesis (11).
A group of R-M systems, including systems such as SalI, EcoRI and Eco29kI, are organized in what appears to be a counterintuitive way (12,13). They consist of two (R and M) genes organized in an operon with the R gene preceding the M gene. Inspection of DNA sequences upstream of the 5′-end of the operon does not reveal recognition sites that could be modified by the methylase or palindromic sequences (operators) that methylase can bind to. Thus, a question arises how can these systems establish themselves in a naïve host and be stably maintained once established. Recently, studies of the EcoRI system showed that a promoter located inside the R gene can drive expression of the downstream M gene (14). In addition, antisense promoters internally located within the R gene have also been identified (15). These antisense promoters appear to negatively regulate expression of the EcoRI endonuclease gene. The generality of these observations, as well as a mechanism(s) by which antisense promoters regulate R gene expression are unknown. In this article, we describe the regulation of gene expression in the Eco29kI system. We identify Eco29kI promoters, including antisense promoters located within the eco29kI.R gene and define a mechanism through which the antisense Eco29kI transcripts attenuate the synthesis of the Eco29kI restriction endonuclease.
MATERIALS AND METHODS
Bacteria strains, phages and plasmids
Escherichia coli Z85 and HB101 (4), and XL10-Gold (Stratagene) were used as host strains in experiments to study gene expression and degree of the λvir phage resistance. Phage λvir was propagated as described (16). All bacterial strains were grown in standard Luria-Bertani (LB) media at 37°C with appropriated antibiotics (17).
All plasmids used in this work are listed in Supplementary Table S1. The pUC128 plasmid containing the Eco29kI gene cassette (GenBank Accession No. AJ001708.1) is here referred to as p29. Plasmid p29-ResPmut was obtained from p29 by introduction of a down-mutation in −10 elements of both promoters upstream of the eco29kIR gene. Plasmid pECO29Cm was derived from the original pECO29 plasmid carrying the Eco29kI R-M system (18) by digestion with SnaBI and insertion of the cat gene. Plasmid pECO29Cm-49mut contains an additional mutation leading to a Tyr to Ala substitution at R.Eco29kI position 49. Plasmids pR-galK, pM-galK, pM + UTR-galK, pUTR-galK, pAS1-galK, pAS2-galK, pAS3-galK, pAS4-galK and pRAS-galK containing transcriptional fusions with the galK gene were constructed by amplifying various Eco29kI fragments from the p29 plasmid using appropriate gene-specific primers (listed in Supplementary Table S2), treating amplified fragments with BamH1 and HindIII, and subcloning them upstream of promoterless galK gene of BamH1 and HindIII treated plasmid pFD51 (19). Plasmids pR-lacZ, pRAS-lacZ, pRASM-lacZ, pRASMext-lacZ, pM-lacZ, pM-UTR-lacZ and pEco49mut-lacZ containing translational fusions with the lacZ gene were constructed by amplifying Eco29kI DNA fragments with appropriate gene-specific primers and subcloning, after digestion with BamHI and KpnI, into the pLacZ vector (this study). In the case of pRAS_SD-lacZ plasmid, the amplified fragment was digested with BamHI and HindIII and inserted into the pLacZ_SD vector (this study).
Site-directed mutants were obtained using QuickChangeR Site-Direct mutagenesis kit (Stratagene).
RNA extraction and qRT–PCR
Escherichia coli Z85 and HB101 cells harboring various plasmids were grown until OD600 = 0.4 and total RNA was extracted using RNeasy Mini Kit (QIAGEN). RNA samples were next treated with DNase I (Fermentas). Total RNA (2 μg) was reverse-transcribed with 100 U of SuperScript III enzyme from First-Strand Synthesis Kit for RT-PCR (Invitrogen) in the presence of appropriate gene-specific primers (cDNAres_r for eco29kIR, cDNAmeth_r for eco29kIM, cDNAas_f for eco29kI antisense promoter transcripts, cDNAgal_r for galK, cDNAlac_r for lacZ, cDNAbla_r for bla and cDNAcat_r for cat). One microliter of completed reverse transcription reactions was used as a template for real-time PCR. Several sets of primers were used to assay transcript levels of plasmid-borne eco29kIR (res + asPCR_f and res + asPCR_r), eco29kIM (methPCR_f and methPCR_r), eco29kI antisense transcripts (res + asPCR_f and res + asPCR_r), galK (galPCR_f and galPCR_r) and lacZ (lacPCR_f and lacPCR_r). Expression levels of bla and cat genes (revealed, respectively, with blaPCR_f and blaPCR_r, and catPCR_f and catPCR_r primers) were used for normalization. Each real-time PCR mixture (25 μl) contained 10 μl SYBR Green I PCR Master Mix (2.5 × PCR buffer, Taq DNA-polymerase, dNTP, glycerol, Tween 20, SYBR Green I), 12 μl H2O, 1 μl of 20 μM forward and reverse primer, and 1 μl of cDNA template. Amplifications were carried out using ANK32 CyclerSystem (Syntol). Reaction products were analyzed using 2% agarose electrophoresis to confirm that signals detected originated from products of expected lengths. Each qRT–PCR reaction was performed at least in triplicate and average data are reported.
Primer extension and manual DNA-sequencing
For primer extension reactions, 2 μg of total RNA was reverse-transcribed with 100 U of SuperScript III enzyme from First-Strand Synthesis Kit for RT-PCR (Invitrogen) in the presence of 1 pmol of [γ-32P] end-labeled gene-specific primers. Reaction products were treated with RNase H, precipitated with ethanol, dissolved in 7 M urea–formamide loading buffer, and resolved on 7% sequencing gels. The products of sequencing reactions, performed with the same end-labeled primers and appropriate plasmid as a template using fmol DNA Cycle Sequencing System (Promega), were run alongside primer extension reactions. Reaction products were revealed using PhosphoImager.
β-Galactosidase assay
The activity of β-galactosidase was determined according to the protocol described by Miller (17) with modifications. Fifty microliter of overnight culture was inoculated into 5 ml of LB containing appropriate antibiotics and grown at 37°C until OD600 of 0.5–0.7. 1 ml culture aliquots were withdrawn, cells were collected by centrifugation and resuspended in 640 μl of Z-buffer pH 7.0 (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 µM β-mercaptoethanol). One hundred and sixty microliter of lysozyme (2.5 mg/ml) was added and samples were incubated for 5 min at 37°C followed by the addition of 8 μl of 10% Triton X-100. To start the reaction, 200 μl of 4 mg/ml ONPG was added to each probe. When the color of reactions became light yellow, reactions were terminated by the addition of 400 μl of 1M Na2CO3. The amount of β-galactosidase activity was determined using the following equation: Units = (1000 × A420)/(t × OD600), where t is the time of the reaction in minutes.
Footprinting and in vitro transcription
Transcription reactions were set in 14 µl and contained 50 ng of transcription templates and various amounts of E. coli RNAP σ70 holoenzyme in a buffer containing 20 mM Tris–HCl pH 8.0, 100 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 50 µg BSA, 5% glycerol. After 5–10 min incubation at 37°C, reactions were supplemented with 2 µl of nucleotide hot mix [2 mM АTP, GTP and CTP, 500 µМ UTP and 0.5 μCi [α-32P]-UTP (3000 Ci/mmol) and 20 µg heparin] and incubated for additional 10 min. Reactions were terminated by the addition of 15 µl of formamide-containing loading buffer and loaded on 7% sequencing gels.
KMnO4 footprinting was conducted under conditions used in transcription assays. Complexes were probed for 30 s at 37°C with 1 mM KMnO4 and modification reactions were stopped by the addition of β-mercaptoethanol and ammonium acetate to final concentration of 1 and 0.3 M, respectively. Samples were precipitated with ethanol, dried and resuspended in 90 μl H2O. After addition 10 μl piperidine, samples were incubated at 90°C for 20 min, ethanol-precipitated, dried and resuspended in 8 μl 7 M urea-formamide loading dye buffer. Samples were applied on 7% sequencing gel and revealed by Phosphoimager.
RESULTS
Mapping of Eco29kI promoters
R gene promoters
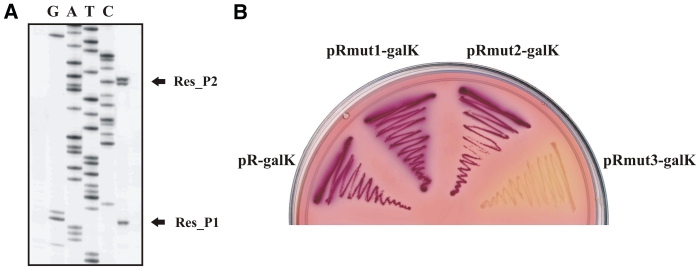
The p29 plasmid, which contains the entire Eco29kI R-M system, is stably maintained in E. coli cells and provides resistance to phages. Eco29kI promoters located upstream of the eco29kir gene were mapped by primer extension using total RNA prepared from cells harboring p29. Two primer extension products were observed (Figure 2A). Inspection of sequences upstream of primer extension end points revealed plausible promoter elements (Figure 1). In vitro transcription (Supplementary Figure S3) and KMnO4 probing (Supplementary Figure S1A) experiments confirmed that both primer extension products observed in vivo were produced from distinct E. coli RNAP σ70 holoenzyme promoters. We refer to upstream promoter as Res_P2. Approximately 70% of eco29kIR mRNA molecules are initiated from this promoter. The downstream promoter is referred to as Res_P1; it accounts for the remaining ∼30% of eco29kIR transcripts. Cells carrying a plasmid with transcriptional fusion of Eco29kI DNA upstream of the eco29kIR gene with promoterless galK contained high levels of GalK activity as judged by deep red color of cell colonies on McConkey plates (Figure 2B). This activity was unchanged when p29, a compatible plasmid carrying the entire Eco29kI R-M system, was introduced in the cells (Supplementary Figure S2). We therefore conclude that transcription from eco29kIR promoters is not affected by the presence of the Eco29kI gene products.
Figure 2.
Mapping eco29kIR gene promoters. (A) RNA was purified from E. coli cells harboring a plasmid with the beginning as well as upstream eco29kIR sequence fused to promoterless galK and subjected to primer extension reaction with a galK-specific primer. (B) Overnight growth of E. coli cells harboring plasmids with eco29kIR promoters fused to promoterless galK on a McConkey agar plate. The following plasmids were used: pR-galK—a DNA fragment (from −72 to +34) of eco29kIR fused to galK; pRmut1-galK—pR-galK with inactive Res_P1; pRmut2-galK—pR-galK with inactive Res_P2; pRmut3-galK—pR-galK with inactive Res_P1 and Res_P2.
Figure 1.
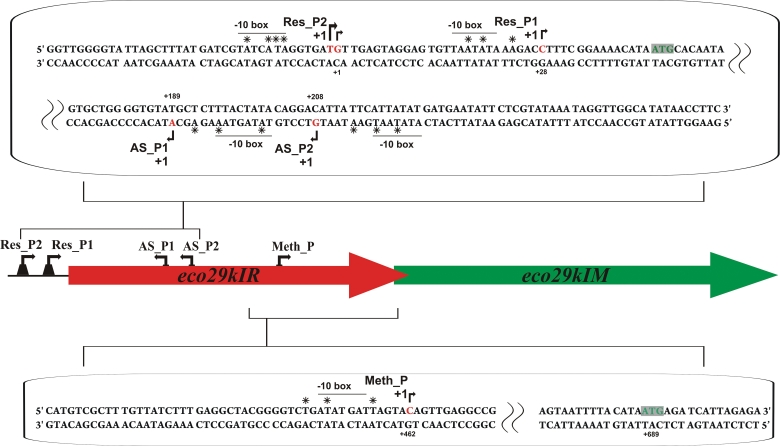
Genetic organization of restriction-modification system Eco29kI. The eco29kI genes are schematically shown by colored arrows, with arrow direction matching the direction of transcription. The DNA sequences of parts of the eco29kIR gene that include eco29kIR and AS promoters, and the eco29kIM promoter are expanded, respectively, above and below the eco29kI genes schematics (both DNA strands are shown). The initiating codons of both eco29kI genes are highlighted. The Res_P1, Res_P2 and Meth_P start sites are indicated by rightward arrows above the sequence. The AS_P1 and AS_P2 start sites are indicated by leftward arrows below the sequence. Experimentally defined −10-boxes are labeled and over- or underlined. Nucleotide positions substituted in engineered down-mutations discussed in the text are marked with ‘asterisks’.
Substitutions in −10 elements of either Res_P1 or Res_P2 that decreased the similarity to consensus −10 element sequence had no effect on galK expression as judged by colony color on McConkey agar (Figure 2B). However, simultaneous substitutions in −10 elements of both promoters abolished GalK synthesis as judged by the white color of colonies formed by cells harboring the mutant fusion plasmid (Figure 2B). Analysis of translational fusions between wild-type and mutant eco29kIR promoters and the lacZ gene positioned in-frame with the R.Eco29kI reading frame indicated that transcripts from both promoters are translated with comparable efficiency (data not shown).
M gene promoter
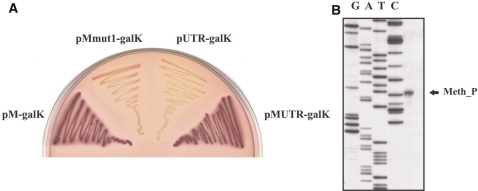
We next explored a possibility that an intragenic (i.e. located within eco29kIR) promoter may drive transcription of eco29kIM independently of eco29kIR promoters. Fragments of the eco29kIR gene located downstream of Res_P1/Res_P2 were cloned, in appropriate orientation, upstream of promoterless plasmid-borne galK gene. The resulting plasmids were transformed into galK− cells, and production of GalK was monitored. The results indicated that a DNA fragment whose endpoints (with respect to the Res_P2 transcription start point) corresponded to positions from +408 and +472 allowed robust expression of GalK activity (Figure 3A). Other eco29kIR fragments lacked rightward (Figure 1) promoter activity.
Figure 3.
Mapping the eco29kIM gene promoter. (A) Overnight growth of E. coli cells harboring plasmids with internal eco29kIR fragments fused to promoterless galK on a McConkey agar plate. The following plasmids were used: pM-galK—a fragment (from +408 to +472) of eco29kIR fused to galK; pMmut1-galK—pM-galK with inactive Met_P; pUTR-galK—a fragment (from +469 to +682) of eco29kIR fused to galK; pMUTR-galK—a fragment (from +408 to +682) of eco29kIR fused to galK. (B) Results of primer extension analysis carried out with RNA purified from cells harboring a plasmid with eco29kIM promoter fused to promoterless galK are presented.
Primer extension reactions with RNA prepared from cells harboring a plasmid with a fusion of the +408 to +472 eco29kIR fragment to galK revealed a single primer extension product (Figure 3B). Analysis of upstream sequence revealed a plausible −10 element located at correct distance from the transcription start point but no recognizable −35 promoter element (Figure 1). In vitro transcription (Supplementary Figure S3) and KMnO4 probing (Supplementary Figure S1B) confirmed that the RNAP σ70 holoenzyme recognizes the eco29kIM promoter (Met_P) and transcribes from it, initiating from the in vivo start site.
The Met_P transcription initiation start point is located 227 bp upstream of the initiating codon of the eco29kIM gene. To determine if the 227 bp-long untranslated leader region (UTR) contributes to control of eco29kIM gene expression, Met_P::galK fusions with or without UTR were prepared. Cells carrying plasmid with either fusion formed red-color colonies on McConkey plates (Figure 3A). Moreover, primer extension experiments with RNA prepared from these cells showed that the amount of Met_P-specific primer extension product was the same in both types of cells (data not shown), indicating that the presence of UTR does not influence the steady-state levels of Met_P transcripts. Cells carrying plasmids with a point mutation in the −10 promoter element of Met_P or a plasmid containing a UTR::galK fusion without Met_P formed white-color colonies (Figure 3A), indicating that UTR lacks promoter activity.
Though a down mutation abolishing Met_P activity did not change the protein sequence of the Eco29kI nuclease, we were unable to obtain a plasmid carrying the entire Eco29kI R-M system harboring a down-mutation in Met_P. Thus, the activity of this promoter seems to be required for Eco29kI establishment and/or maintenance.
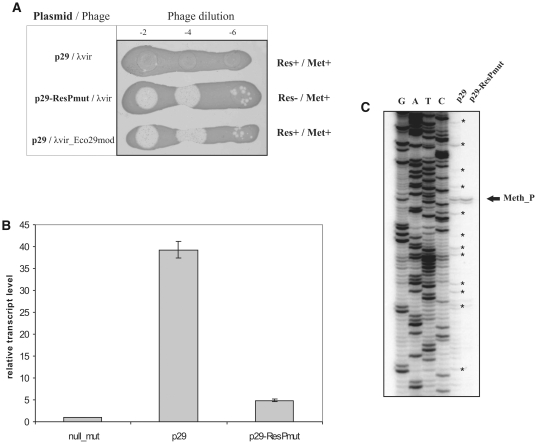
To show that eco29kIR promoter-independent eco29kIM transcription is biologically relevant, we determined the ability of cells harboring a plasmid expressing the complete Eco29kI system or derivative plasmids to restrict and/or modify the λvir phage (Figure 4A). As expected, the infection of cells harboring the entire Eco29kI system by unmodified phage was inefficient. Unmodified phage effectively infected cells carrying a plasmid with mutated eco29kIR promoters, also as expected. Importantly, phage progeny from the latter infections was not restricted by cells harboring complete R-M system, indicating that sufficient amounts of methylase are synthesized even in the absence of transcription from Res_P1/Res_P2.
Figure 4.
Production of M.Eco29kI from internal promoter Meth_P. (A) Horizontal lines show the overnight growth of E. coli Z85 strain harboring a plasmid carrying the entire Eco29kI R-M system (p29) or a mutant plasmid with inactivated eco29kIR promoters (p29-ResPmut) on LB plates. Cells were spotted with indicated dilutions of λvir phage lysate. Phages collected from infections of cells carrying the mutant plasmid (second row) were used in infections shown at the bottom of the panel (this phage is labeled λvir-Eco29mod). The phenotypes of cells with respect to Eco29kI Res and Met activities are indicated at the right. (B) Determining the relative abundance of bicistronic and monocistronic Eco29kI RNA by qRT–PCR. Reverse transcription reactions were carried out with a primer annealing at the end of the eco29kIM gene using RNA samples purified from cells harboring indicated plasmids and qRT–PCR was performed using eco29kIM specific primers. qRT–PCR reactions run with bla-specific primers were used for normalization. (C) Primer extension analysis carried out with RNA purified from cells harboring a plasmid with entire Eco29kI system (p29) or cells harboring the same plasmid with inactive eco29kIR promoters (p29-ResPmut) using a primer designed to reveal transcripts initiated from Meth_P. Reverse transcription reaction products that correspond to the bicistronic mRNA degradation intermediates are marked with asterisks.
Using plasmids carrying the entire Eco29kI system or a derivative plasmid with disrupted Res_P1/Res_P2 promoters, we showed by qRT–PCR with eco29kIM specific primer pairs that the steady-state levels of Met_P-initiated transcripts constituted ∼10% of the steady-state level of transcripts initiated from Res_P1/Res_P2 (Figure 4B). β-Galactosidase activity measurements indicated that Met_P-initiated transcripts were translated about as efficiently as Res_P1/Res_P2 initiated transcripts (see below).
Primer extension reactions with primers designed to reveal Met_P-initiated transcripts revealed a single primer extension product corresponding to Met_P-initiated transcript in RNA purified from cells carrying a mutant plasmid with inactivated eco29kIR promoters. In contrast, analysis of RNA prepared from cells carrying wild-type Eco29kI plasmid revealed, in addition to primer extension product corresponding to the Met_P transcript, a large number of products corresponding to RNAs whose 5′-ends were located both upstream and downstream of Met_P (Figure 4C). Since Met_P is the only rightward intragenic promoter in eco29kIR, primer extension products whose 5′-ends map upstream of Met_P must result from degradation of Res_P1/Res_P2-initiated transcripts (see also below).
Antisense promoters
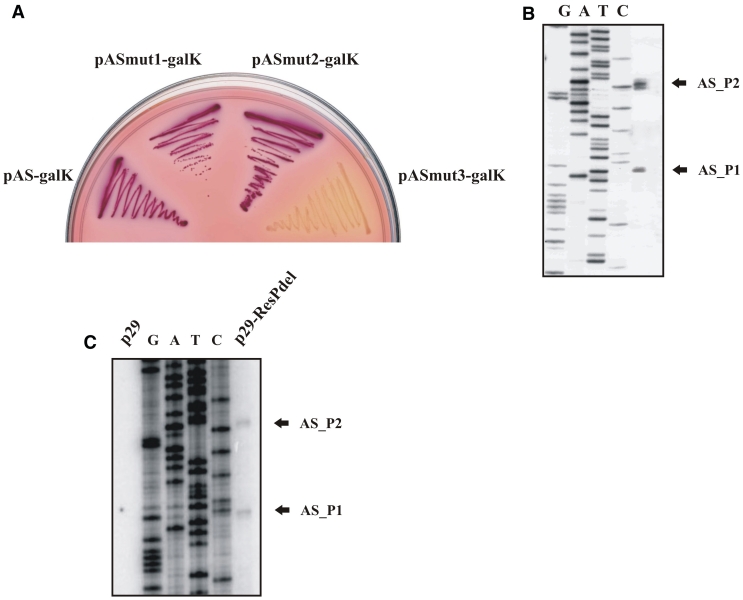
While constructing galK fusions with eco29kIR gene fragments we noticed that a fragment corresponding to eco29kIR positions from +178 to +252 with respect to the Res_P2 start point resulted in a strong GalK synthesis when cloned in an orientation opposite to the direction of eco29kIR transcription (Figure 5A). Primer extension analysis of RNA prepared from cells harboring this plasmid revealed two closely spaced primer extension products (Figure 5B) and in vitro analysis confirmed that σ70 RNAP holoenzyme forms promoter complexes and initiates transcription from both sites in vitro (Supplementary Figures S1C and S3). We call the corresponding promoters antisense promoters AS_P1 and AS_P2. AS_P2 (located further away from eco29kIR promoters) accounts for ∼60% of AS transcription. In the context of galK fusions, AS promoters appear to be as active as the eco29kIR promoters, as judged by qRT–PCR with galK specific primers (data not shown). Derivatives of AS::galK promoter fusion plasmids with point mutations decreasing the similarity of each (or both) AS promoters to consensus were constructed. Cells harboring a plasmid with mutations in both promoters formed white colonies on McConkey plates (Figure 5A). Attempts to incorporate these down-mutations in a plasmid carrying entire Eco29kI R-M system were unsuccessful. Since down mutations in AS promoters did not lead to changes in the R.Eco29kI protein sequence, the result indicates that activity of AS promoters is necessary for establishment and/or stable maintenance of the Eco29kI R-M system.
Figure 5.
Mapping AS promoters. (A) Overnight growth of E. coli cells harboring plasmids with AS promoters fused to promoterless galK gene on a McConkey agar plate. The following plasmids were used: pAS-galK—a DNA fragment (from +252 to +178) of Eco29kI system fused to promoterless galK; pASmut1-galK—pAS-galK with inactive AS_P1; pASmut2-galK—pAS-galK with inactive AS_P2; pASmut3-galK—pAS-galK with inactive AS_P1 and AS_P2. (B) The results of primer extension reaction carried out with RNA purified from cells harboring a plasmid with AS promoters fused to promoterless galK gene are shown. (C) Primer extension analysis of RNA samples purified from cells harboring a plasmid with intact Eco29kI system (p29), left lane, and the same plasmid with inactive eco29kIR promoters (p29-ResPmut, right lane) with primers designed to reveal AS promoters initiated transcripts.
Analysis of RNA purified from cells containing wild-type Eco29kI plasmid failed to reveal primer extension products corresponding to AS promoters. However, the expected primer extension products were readily detectable when a plasmid containing inactivated eco29kIR promoters was used (Figure 5C).
Functional interactions between the Eco29kI promoters
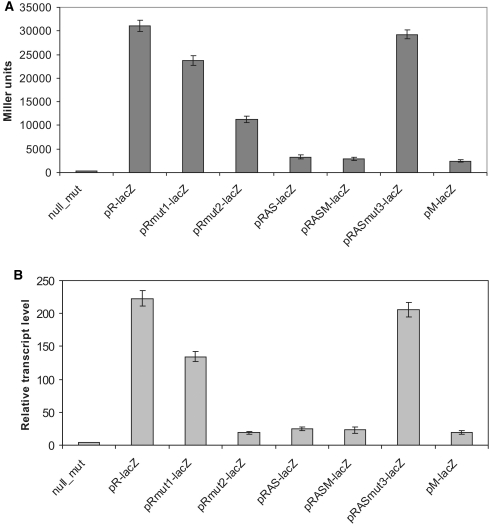
A series of translational fusions of eco29kIR promoters to the lacZ gene with intervening Eco29kI DNA of various lengths was prepared. The resulting plasmids were introduced in lacZ E. coli cells and the levels of LacZ activity and the amounts of lacZ RNA were determined. The results are summarized in Figure 6. As can be seen, inclusion of Eco29kI DNA fragments containing AS promoters had a strong negative effect on LacZ activity levels (Figure 6A) and the steady-state levels of lacZ transcripts (Figure 6B) synthesized from eco29kIR promoters. The inclusion of additional Eco29kI sequences between eco29kIR promoters and lacZ—up to and downstream of Met_P—had no further negative effect on LacZ activity or lacZ RNA abundance. AS promoters activity was responsible for decrease in LacZ synthesis and steady-state levels of eco29kIR promoter-initiated transcripts since inactivation of both AS promoters increased the levels of LacZ activity and lacZ transcripts to those seen in cells that contained plasmids with the shortest eco29kIR::lacZ fusion from which both AS promoters were absent (note that down mutations in AS promoters did not affect the sequence of the R.Eco29kI-LacZ fusion protein). Analysis of cells harboring plasmids with individually inactivated AS promoters indicated that the more active AS_P2 was primarily responsible for inhibition of lacZ expression from eco29kIR promoters (data not shown).
Figure 6.
Expression of eco29kIR is negatively affected by the activity of antisense-promoters. (A) β-Galactosidase activity in cells harboring plasmids with translational fusions of the lacZ reporter with the following fragments of eco29kIR or their mutational derivatives: pR-lacZ—a DNA fragment (from −44 to +168) of Eco29kI system fused to lacZ; pRmut1—pR-lacZ with inactive Res_P1 promoter; pRmut2—pR-lacZ with inactive Res_P2 promoter; pRAS-lacZ—a DNA fragment (from −44 to +282) of Eco29kI system fused to lacZ; pRASM-lacZ—a DNA fragment (from −44 to +464) of Eco29kI system fused to lacZ; pRASmut3-lacZ—pRAS-lacZ with inactive AS promoters; pM-lacZ—a DNA fragment (from +364 to +806) of Eco29kI system fused to lacZ. (B) Relative lacZ transcript abundance measured with qRT–PCR. The amount of cat transcript revealed with appropriate primers was used to normalize lacZ transcript abundance in each reaction mix.
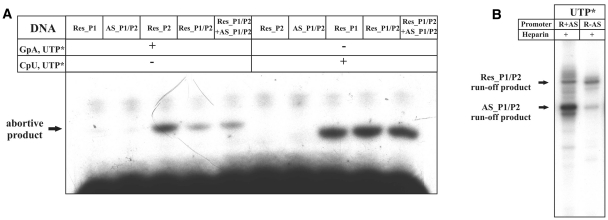
The data presented above indicate that steady-state levels of eco29kIR promoters-initiated transcripts are negatively affected by AS promoters activity, either directly or indirectly. The presence of AS promoters had no effect on in vitro abortive RNA synthesis from eco29kIR promoters by purified σ70 RNAP holoenzyme (Figure 7A), indicating that promoter complex formation on eco29kIR and AS promoters proceeds independently. This is an expected result, considering the ∼160 bp distance between transcription start points of Res_P1 and AS_P1.
Figure 7.
In vitro transcription and promoter complex formation on eco29kIR and AS promoters. (A) Escherichia coli RNAP σ70 holoenzyme was combined with Eco29kI DNA fragments carrying indicated promoters and abortive transcription initiation reactions with dinucleotide monophosphate primers specific for Res_P1 (GpA) or Res_P2 (CpU) and 32P UTP were performed. Reaction products (GpApU and CpUpU for Res_P1 and Res_P2, respectively, bold-type face denoting 32P phosphate) were resolved by denaturing PAGE and revealed using a Phosporimager. (B) Single round transcription using wild-type template Eco29kI DNA (R + AS) and a template DNA with inactive AS promoters (R − AS).
In principle, transcription from AS promoters could decrease transcription from eco29kIR promoters by dislodging eco29kIR promoter complexes. Such a ‘sitting duck’ model of transcription interference requires that the rate of RNAP clearance from the interfering (AS) promoter is much faster that the clearance rate of promoter, whose activity is interfered with (eco29kIR promoters). Examples of such regulation were reported for several lambdoid phages (20) as well as, more recently, for an R-M system Ecl18kI (9). We failed to observe significant differences in RNAP clearance rates from eco29kIR and AS promoters in vitro (Supplementary Figure S3). Most importantly, single-round in vitro run-off transcription assays revealed that the same amount of eco29kIR promoter-initiated transcripts is produced from templates containing wild-type or mutant, inactivated, AS promoters (Figure 7B). We therefore conclude that interference from AS promoters is an unlikely cause of decreased eco29kIR promoter in vivo activity.
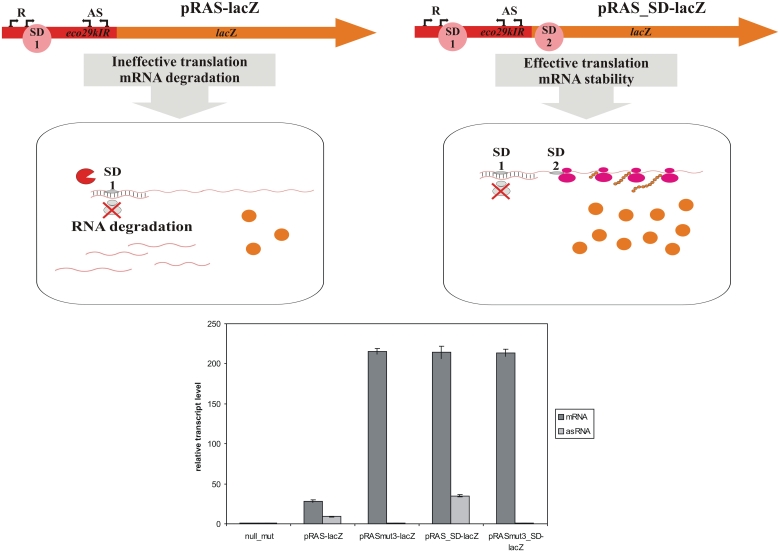
Because of our inability to detect a direct negative effect of AS promoters on eco29kIR promoters, we wondered if an indirect (i.e. unrelated to AS-initiated transcription per se) effect could be responsible for observed strong repression of LacZ synthesis and decrease in the abundance of eco29kIR promoters transcripts in the presence of AS promoters. The most likely possibility would be a mechanism that destabilizes eco29kIR promoters-initiated messages in vivo due to complementary interactions with AS promoters-initiated transcripts (21). Such a mechanism would be consistent with apparent degradation of eco29kIR transcripts revealed during primer extension analysis of RNA prepared from cells carrying complete Eco29kIR system (Figure 4C). Since we were unable to detect AS transcripts when eco29kIR promoters are active (Figure 5C), we envision that both AS transcripts and eco29kIR transcripts are degraded once the RNA duplex is formed. Degradation could be a consequence of poor translation of the eco29kIR mRNA containing a double-stranded region at its 5′-end (21). To check this possibility, we created a modified eco29kIR::lacZ translational fusion plasmid by inserting a Shine–Dalgarno sequence in front of the first codon of the lacZ gene. As a result, the continuous eco29kIR::lacZ reading frame was disrupted, but the lacZ ORF could be translated separately. A corresponding construct with down mutations in AS promoters was also created. Cells harboring such plasmids produced high levels of β-galactosidase. Importantly, β-galactosidase levels were not dependent on the presence of active AS promoters. Real-time PCR analysis (Figure 8) and primer extension analysis (Supplementary Figure S4) revealed that the level of eco29kIR transcripts was the same in cells carrying both plasmids. In cells carrying wild-type AS promoters, AS transcripts were also detected (data not shown; note that these transcripts were undetectable in the context of wild-type Eco29kIR or eco29kIR::lacZ fusions). We therefore conclude that downstream translation initiation abolishes the inhibitory effect of AS promoters on the steady-state levels of eco29kIR promoters transcripts while simultaneously increasing the steady-state level of AS transcripts. It therefore follows that the inhibitory effect of AS transcripts on eco29kIR transcript abundance is unrelated to RNA duplex formation per se. Rather, inhibition of translation initiation that normally results from duplex formation leads to destabilization of both eco29kIR sense and antisense messages.
Figure 8.
Downstream translation initiation abolished the inhibitory effect of AS transcripts. The scheme at the top shows two kinds of eco29kIR promoter lacZ fusions studied. Relative abundance of lacZ transcripts measured with qRT–PCR in cells harboring the following plasmids: pRAS-lacZ—an eco29kIR fragment (from −44 to +282) cloned into pLacZ; pRASmut3-lacZ—pRAS-lacZ with inactive AS promoters; pRAS_SD-lacZ—an eco29kIR fragment (from −44 to +282) cloned into pLacZ_SD; pRASmut3_SD-lacZ—pRAS_SD-lacZ with inactive AS promoters is shown below. The amount of cat transcript revealed with appropriate primers was used to normalize lacZ transcript abundance in each reaction mix.
DISCUSSION
The results of our work lead to the following view of gene expression regulation of the Eco29kI system. There are three groups of promoters in this system: the eco29kIR promoters, the AS promoters and the eco29kIM promoter. The eco29kIM promoter activity is sufficient to provide enough methylase for complete methylation of phage (and, presumably, host) DNA. The intrinsic strength of eco29kIR promoters is significantly higher than the eco29kIM promoter strength. However, transcription from AS promoters decreases the overall level of R.Eco29kI synthesis from eco29kIR promoter-initiated RNA to make it roughly equal to that of the M.Eco29kI synthesis level. In the absence of AS promoters, the amounts of eco29kIR promoters transcripts (and, therefore, of restriction endonuclease) would have been almost 10 times higher than the amount of Met_P-initiated transcripts (and of methyltransferase). Such high levels of restriction endonuclease apparently interfere with Eco29kI genes establishment, maintenance, or both. As a result, down mutations in AS promoters are not tolerated.
AS promoter-initiated transcripts decrease the level of eco29kIR promoters transcripts only if translation initiation of mRNA synthesized from eco29kIR promoters occurs in the area located between the eco29kIR and AS promoters. Apparently, AS transcripts associate with eco29kIR transcripts and prevent translation of the eco29kI open reading frame, which causes the degradation of untranslated eco29kIR transcripts. AS-initiated transcripts themselves also get degraded when they associate with untranslated eco29kIR transcripts. The lack of eco29kIR translation appears to be the primary signal for degradation of both transcripts. If translation initiation is allowed to occur downstream of the area where the AS/eco29kIR RNA duplex is formed, no eco29kIR or AS transcripts degradation is observed.
Genetic organization of Eco29kI is similar to SalI and EcoRI, where the R gene also preceeds the M gene. Comparison of our data with results obtained with these other systems earlier indicates that the common architecture leads to common transcription regulation. First, all three systems contain a promoter (two very closely spaced promoters in the case of EcoRI) that drives the expression of the downstream M gene independently of the R gene expression. The activity of this promoter alone is sufficient for production of enough methylase to modify both the host and phage DNA. In the case of EcoRI, upstream regulatory elements that, according to the authors (14), may control the activity of M promoter by decreasing its transcription initiation level were reported. Eco29kI appears to lack such elements.
In the EcoRI system, the presence of two antisense promoters that decrease the steady-state levels of transcripts initiated from R promoters has been reported and the mechanism of down-regulation of ecoRIR promoter activity by the EcoRI AS promoters has been investigated (15). Using RNase I protection assays, the authors showed that the steady-state level of ecoRIR transcripts was increased when AS promoters were inactivated. The effect was most pronounced when probes specific for portions of ecoRIR RNA located downstream of AS promoters were used to measure ecoRIR RNA abundance. The effect did not depend on ecoRIR transcript stability, as revealed by a Rifampicin challenge and northern blot analysis with ecoRIR RNA-specific probes. The authors therefore proposed that a truncated ecoRIR initiated transcript is produced as a result of AS promoter activity. After excluding several possibilities such as transcription termination/attenuation, the authors came to conclusion that the hypothetical truncated ecoRIR mRNA is generated by a pause or arrest of the transcription caused by a roadblock at AS promoters. According to them, ‘in the case of the ecoRIR gene, binding of RNA polymerase to the reverse promoters may change the DNA conformation and generate such a roadblock, influencing the transcription elongation complex of another RNA polymerase’ (15). The high degree of similarity of transcriptional organization of EcoRI and Eco29kI systems makes us to propose that the EcoRI AS promoters regulate the abundance of R transcripts through an indirect mechanism that is also operational in the case of Eco29kI, namely through post-transcriptional formation of a duplex RNA structure that prevents translation of the R message followed by degradation. The unaltered stability of full-sized ecoRIR message in the presence of functional AS promoters is likely caused by the fact that only those ecoRIR transcripts that anneal to AS transcripts are degraded; those that fail to anneal (and by design these would be the transcripts revealed in the Rifampicin challenge experiments by the Kobayashi group) would remain fully stable, as was indeed observed in their experiment.
Up to now, various strategies of genetic regulation at the transcriptional level that are used by Nature to ensure temporally coordinated expression of R-M systems genes were described. Our results, together with earlier data, suggest that in R-M systems where restriction endonuclease genes precede methyltransferase genes, a mixed type of regulation is operational. Independent synthesis of methyltransferase gene is accomplished through a separate promoter located within the upstream R gene. The level of the R gene transcript is negatively regulated by intragenic antisense promoters. The regulation occurs at the post-transcriptional level, through destabilization of R promoter-initiated transcripts. It remains to be seen how widespread is the post-translational regulation in other R-M systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Presidium of Russian Academy of Sciences, Molecular and Cellular Biology programme grant; National Institutes of Health grant (GM64530 to K.S.); federal program Scientific and Scientific-Pedagogical Personnel of Innovative Russia 2009–2013, State Contract 02.740.11.0771 (to K.S.) and P1045 to (M.Z.). Funding for open access charge: Russian Academy of Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagornykh MO, Bogdanova ES, Protsenko AS, Zakharova MV, Solonin AS, Severinov KV. Regulation of gene expression in type II restriction-modification system. Genetika. 2008;44:606–615. doi: 10.1134/s1022795408050037. [DOI] [PubMed] [Google Scholar]
- 3.Bogdanova E, Djordjevic M, Papapanagiotou I, Heyduk T, Kneale G, Severinov K. Transcription regulation of the type II restriction-modification system AhdI. Nucleic Acids Res. 2008;36:1429–1442. doi: 10.1093/nar/gkm1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenova E, Minakhin L, Bogdanova E, Nagornykh M, Vasilov A, Heyduk T, Solonin A, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction-modification system. Nucleic Acids Res. 2005;33:6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mruk I, Blumenthal RM. Real-time kinetics of restriction-modification gene expression after entry into a new host cell. Nucleic Acids Res. 2008;36:2581–2593. doi: 10.1093/nar/gkn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanova E, Zakharova M, Streeter S, Taylor J, Heyduk T, Kneale G, Severinov K. Transcription regulation of restriction-modification system Esp1396I. Nucleic Acids Res. 2009;37:3354–3366. doi: 10.1093/nar/gkp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakharova M, Minakhin L, Solonin A, Severinov K. Regulation of RNA polymerase promoter selectivity by covalent modification of DNA. J. Mol. Biol. 2004;335:103–111. doi: 10.1016/j.jmb.2003.09.081. [DOI] [PubMed] [Google Scholar]
- 8.Christensen LL, Josephsen J. The methyltransferase from the LlaDII restriction-modification system influences the level of expression of its own gene. J. Bacteriol. 2004;186:287–295. doi: 10.1128/JB.186.2.287-295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Protsenko A, Zakharova M, Nagornykh M, Solonin A, Severinov K. Transcription regulation of restriction-modification system Ecl18kI. Nucleic Acids Res. 2009;37:5322–5330. doi: 10.1093/nar/gkp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Som S, Friedman S. Regulation of EcoRII methyltransferase: effect of mutations on gene expression and in vitro binding to the promoter region. Nucleic Acids Res. 1994;22:5347–5353. doi: 10.1093/nar/22.24.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lubys A, Menkevicius S, Timinskas A, Butkus V, Janulaitis A. Cloning and analysis of translational control for genes encoding the Cfr9I restriction-modification system. Gene. 1994;141:85–89. doi: 10.1016/0378-1119(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez MA, Chater KF, Rodicio MR. Complex transcription of an operon encoding the SalI restriction-modification system of Streptomyces albus G. Mol. Microbiol. 1993;8:243–252. doi: 10.1111/j.1365-2958.1993.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor CD, Humphreys GO. Expression of the EcoRI restriction-modification system and the construction of positive-selection cloning vectors. Gene. 1982;20:219–229. doi: 10.1016/0378-1119(82)90041-5. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Ichige A, Kobayashi I. Regulation of the EcoRI restriction-modification system: identification of ecoRIM gene promoters and their upstream negative regulators in the ecoRIR gene. Gene. 2007;400:140–149. doi: 10.1016/j.gene.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Kobayashi I. Negative regulation of the EcoRI restriction enzyme gene is associated with intragenic reverse promoters. J. Bacteriol. 2007;189:6928–6935. doi: 10.1128/JB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Gold Spring Harbor, NY: Gold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Pertzev AV, Ruban NM, Zakharova MV, Beletzkaja IV, Petrov SI, Kravetz AN, Solonin AS. Eco29kI, a novel plasmid encoded restriction endonuclease from Escherichia coli. Nucleic Acids Res. 1991;20:1991. doi: 10.1093/nar/20.8.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rak B, von Reutern M. Insertion element IS5 contains a third gene. EMBO J. 1984;3:807–811. doi: 10.1002/j.1460-2075.1984.tb01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 20.Dodd IB, Kalionis B, Egan JB. Control of gene expression in the temperate coliphage 186. VIII. Control of lysis and lysogeny by a transcriptional switch involving face-to-face promoters. J. Mol. Biol. 1990;214:27–37. doi: 10.1016/0022-2836(90)90144-B. [DOI] [PubMed] [Google Scholar]
- 21.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.