Abstract
We used a genetic screen based on tRNA-mediated suppression (TMS) in a Schizosaccharomyces pombe La protein (Sla1p) mutant. Suppressor pre-tRNASerUCA-C47:6U with a debilitating substitution in its variable arm fails to produce tRNA in a sla1-rrm mutant deficient for RNA chaperone-like activity. The parent strain and spontaneous mutant were analyzed using Solexa sequencing. One synonymous single-nucleotide polymorphism (SNP), unrelated to the phenotype, was identified. Further sequence analyses found a duplication of the tRNASerUCA-C47:6U gene, which was shown to cause the phenotype. Ninety percent of 28 isolated mutants contain duplicated tRNASerUCA-C47:6U genes. The tRNA gene duplication led to a disproportionately large increase in tRNASerUCA-C47:6U levels in sla1-rrm but not sla1-null cells, consistent with non-specific low-affinity interactions contributing to the RNA chaperone-like activity of La, similar to other RNA chaperones. Our analysis also identified 24 SNPs between ours and S. pombe 972h- strain yFS101 that was recently sequenced using Solexa. By including mitochondrial (mt) DNA in our analysis, overall coverage increased from 52% to 96%. mtDNA from our strain and yFS101 shared 14 mtSNPs relative to a ‘reference’ mtDNA, providing the first identification of these S. pombe mtDNA discrepancies. Thus, strain-specific and spontaneous phenotypic mutations can be mapped in S. pombe by Solexa sequencing.
INTRODUCTION
tRNA gene number and arrangement are of interest to genome biologists (1–7) because tRNA abundance is matched to codon usage (3) and tRNA genes affect nuclear and genome organization [(8,9), reviewed in ref. 10, and see ref. 11]. Biased codon usage in functionally related mRNAs suggests that the relative levels of tRNA isoacceptors may reflect a means of genetic control (12–15). Most eukaryotes contain a variable number of tRNA genes, from ∼200 to several thousand (4). Yet, in humans as in species from yeast to dog, about 12 codons have no tRNA with direct Watson:Crick basepairing (Supplementary Table and ref. 4), and must rely on wobble decoding. Further, the ratio of different isoacceptor tRNA genes can vary significantly between species (16) (Supplementary Table S1). Thus, eukaryotic evolution has been accompanied by highly variable proliferation of certain tRNA genes while many others are absent or extremely underrepresented.
Promoters for eukaryotic tRNA genes are all quite similar and provide similar transcription output. As such, differences in tRNA levels usually occur by varying tRNA gene copy number (17) which appears to coevolve with codon usage (3,5). Sequence identity suggests that multicopy tRNA genes arise by duplication and are maintained by gene conversion [(4), reviewed in refs 10,18 and 19]. Chromatin at eukaryotic tRNA loci contains RNA polymerase III (pol III) and other factors including the RNA-binding, molecular chaperone protein, La (20,21). Transcription termination by pol III generates UUU-3′OH ends on its nascent RNA products (22,23) which is a binding site for La (24–26) and thus directs early RNA processing events (27).
Maturation of a nascent pre-tRNA to a mature functional molecule requires many enzymatic and carrier-mediated processes (28). Inherent variability in the sequence and structure of different tRNAs suggests that they will differ in their activity as substrate for a particular enzyme, binding protein, or process (27). Indeed, significant variability in the order of processing and modification steps exists for different tRNAs (27). In the collective ‘pathway’ of tRNA maturation some events differ in the extent to which they are concentration-driven, kinetics-limited, spatially ordered and amenable to alternate mechanisms (27). Processes that degrade pre-tRNAs, such as nuclear surveillance, compete with some of the productive steps (27). Accordingly, pre-tRNAs that are not efficiently engaged by productive processing become subjected to surveillance systems that actively degrade hypomodified, improperly processed or structurally impaired pre-tRNAs (28–31). As a chaperone for pol III transcripts, La protein can offset this type of decay (27,32). Thus, the course for a pre-tRNA can vary depending on if it is engaged by La and/or other factors (27,28). Moreover, recent findings that unprocessed pre-tRNAs can activate stress signals (33,34) that can impact the small interfering RNA (siRNA) (35) or other pathways (31,36–40) suggest that our understanding of tRNA metabolism is incomplete.
While La serves to facilitate pre-tRNA processing, it is nonessential in yeasts (24). La contains two conserved RNA binding motifs, the La motif (LAM) and an adjacent RRM1 that act differently during tRNA maturation (41). Two types of chaperone activity have been attributed to La proteins, molecular chaperone activity and RNA chaperone activity (27). Its principal RNA-binding activity, UUU-3′-OH binding, is mediated primarily by the LAM to protect against 3′ exonucleases and promote 3′ end processing and RNP maturation. Mutations in human La (hLa) or Schizosaccharomyces pombe La, Sla1p, that disrupt RNA 3′ end binding or intracellular trafficking cause tRNA processing defects consistent with dysfunctional molecular chaperone activity (42,43).
The RNA chaperone-like activity noted for human and yeast La proteins (44–47) is an enigmatic aspect of La function that is discernible from UUU-3′OH binding. La promotes the maturation of pre-tRNAs with structural impairments that otherwise subject them to misfolding and degradation (32,46). The typical β-sheet surface and associated loop-3 of La RRM1 constitute an RNA-binding site that confers RNA chaperone-like activity (32,47). Two classes of mutations in La RRM1 have been distinguished. Mutation of basic residues in RRM1 loop-3 do not impair UUU-3′OH binding but significantly decrease binding to other parts of the pre-tRNA, decrease RNA chaperone activity and are detrimental to tRNA maturation (47). Mutation of the two invariant aromatic residues on the RRM1 β-sheet surface debilitates hLa and Sla1p for maturation of structurally impaired pre-tRNAs in vivo, but with too subtle a defect in RNA binding for detection in vitro (32). Nonetheless, these β-sheet surface aromatic mutations inactivate hLa and Sla1p for maturation of structurally impaired pre-tRNASer even in cells that lack Rrp6p (32), the 3′ exonuclease component of nuclear surveillance that degrades one class of structurally impaired pre-tRNAs (30,32,48). This observation suggested a novel, rrp6-independent degradation pathway for structurally impaired pre-tRNASer in the sla1-rrm1 mutants (32).
To try to better understand this, we developed a genetic screen in S. pombe that relies on accumulation of a functional suppressor tRNASerUCA-C47:6U despite a debilitating substitution of a G:C to a G:U basepair in its variable arm (Figure 1A). In cells carrying wild-type Sla1p, the pre-tRNASerUCA-C47:6U from this allele is matured and functionally suppresses a nonsense codon in ade6-704, alleviating accumulation of red pigment. However, this allele does not produce tRNA in the sla1-rrm1 mutant carrying the point mutations in the RRM1 β-sheet surface at levels sufficient to mediate TMS (32). We isolated spontaneous revertants that restore tRNASer-mediated suppression in sla1-rrm cells and mapped the phenotypic mutation by whole genome sequencing. We establish that this approach is applicable to S. pombe and report that the phenotypic mutation is duplication of the suppressor tRNASer gene. We show that a surprising disproportionate increase in suppressor-tRNA levels that occurs with tRNA gene duplication requires sla1-rrm, provoking a model in which activation by La of tRNA maturation is dependent on the concentration of the pre-tRNA.
Figure 1.
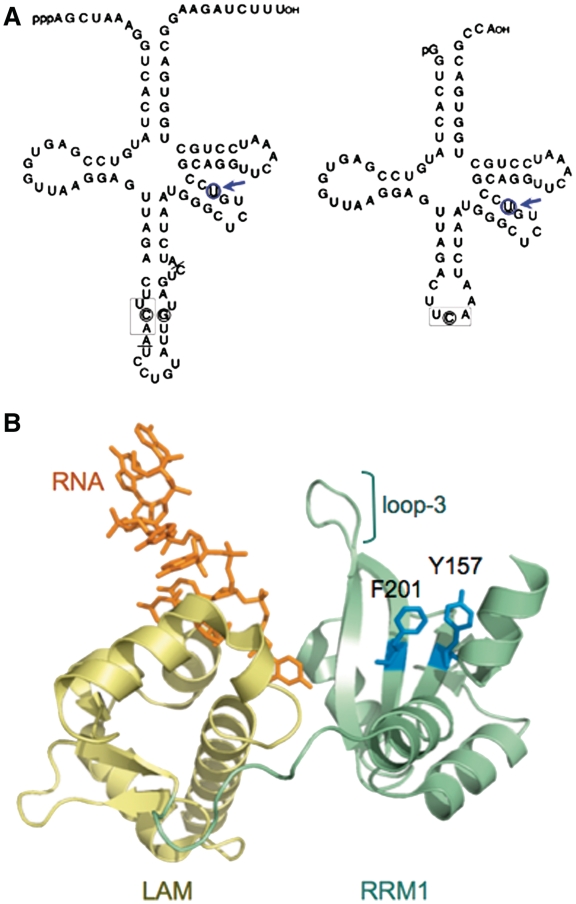
The Sla1p and suppressor-tRNASerUCA-C47:6U components of the yMWB3-15 Strain. (A) Precursor (left) and mature (right) suppressor tRNASerUCA-C47:6U harbors a C47:6U mutation on the variable arm indicated by a blue circle and arrow. The three anticodon residues are boxed. The intron in the precursor is demarcated by the solid lines at the intron junctions. (B) Structure of the LAM (gold) and RRM1 (green) of La protein bound to a 10-mer RNA ending in UUU-3′OH (orange) modified from PDB 2VON (71). The highly conserved β-sheet surface residues that extend from the RRM1 binding surface of Sla1p, Y157 and F201 mutated for this study, are highlighted in blue. Loop-3 which comprises part of the canonical RNA-binding site of RRM1 (47) is also indicated.
In the process of genome analysis, we also uncovered 24 strain-specific single-nucleotide polymorphism (SNPs) in the genomes of our laboratory strain and the reference strain 972h- (yFS101) which were separated ∼30 years ago and passed through multiple laboratories.
MATERIALS AND METHODS
Strains
Strains are listed in Table 3. sla1-Y157A or sla1-Y157A/F201A RRM1 mutations were introduced into ySH18 which harbors a copy of tRNASerUCA-C47:6U and associated pJK148 DNA at the leu1-32 locus as described (49) to generate yMWB2-4 (test strain) and yMWB3-15 (parent), respectively. yMWB3-15 was plated on media containing limiting (10 mg/l) adenine, and spontaneously arising white colonies (e.g. yDA317) were isolated after 3 days of growth and characterized as described.
Table 3.
Strains used in this study
| Strain | Genotype | Phenotype | Details |
|---|---|---|---|
| ySH18 | h- ade6-704, sla1-Δ, ura4-D, leu1-32::tRNA U47:6-leu1+ | Red | Progenitor |
| yMWB2-4 | h- ade6-704, sla1+::sla1Y157A-kanMX6, ura4-D, leu1-32::tRNA U47:6-leu1+, | Pink | Test strain |
| yMWB3-15 | h- ade6-704, sla1+::sla1Y157A/F201A-kanMX6, ura4-D, leu1-32::tRNA U47:6-leu1+ | Red | Parent strain |
| yMWB1-1 | h- ade6-704, sla1-Δ::kanMX6, ura4-D, leu1-32::tRNA U47:6-leu1+ | Red | Isogenic sla1-Δ test strain |
| yDA317 | h- ade6-704, sla1+::sla1Y157A/F201A-kanMX6, ura4-D, leu1-32::tRNA U47:6-leu1+ | White | Mutant strain |
| yFS101 | h- | White | Ref strain |
Construction of a customized yMWB3-15 reference sequence
The S. pombe genome used in this study originated from the sequence obtained at Sanger Institute, dated June 2008 (50). Modifications were introduced to better reflect the strains used in this study following a first pass of read alignment. SNP and insertion/deletion modifications noted by the Broad Institute (51) were incorporated after identifying identical mutations in the parent strain. pJK148 vector sequence including the suppressor tRNA insertion (49) was incorporated at the leu1-32 locus as described (52).
The four poly-N gaps in the Sanger reference were probed as follows. Gap-flanking sequences were used in a seeded search of unaligned reads. For the gap at Chr 2 position 80202 this led to isolation of continuous sequence tracts on either side. A BLAST search identified the previously resolved ∼18-kb gap-spanning sequence (53) which was inserted to replace the poly-N gap, and predicted genes in this ∼18 kb were incorporated into the sequence annotations. Introduction of this sequence provided continuous coverage at near average depth across the region. As a result, some Chr 2 gene positions in our reference are offset by ∼18 000 relative to the Sanger positions (Tables 1 and 4). Similar attempts to resolve the remaining three contig gaps (Chr 1: 29662; Chr 2: 1616568; and Chr 2: 1083349) were unsuccessful with this read data set.
Table 1.
Single-nucleotide polymorphisms obtained by Solexa sequencing of parent, mutant and test strains
| Classification | Position | SangerPos | Ref | Test | Parent | Mut | Locus | Verified |
|---|---|---|---|---|---|---|---|---|
| Test strain –known | I 1381336 | I 1381336 | T | C | T | T | nhp6 | Yes |
| Test strain –known | I 1381510 | I 1381510 | A | A | G | G | nhp6 | Yes |
| Test strain –known | I 1381719 | I 1381719 | G | A | G | G | Yes | |
| Test strain –known | I 1382042 | I 1382042 | T | T | C | C | Yes | |
| Test strain –known | I 1382159 | I 1382159 | A | A | G | G | Yes | |
| Test strain –known | I 1382225 | I 1382225 | C | T | C | C | Yes | |
| Test strain –known | I 1382753 | I 1382753 | A | A | G | G | sla1 | Yes |
| Test strain –known | I 1382837 | I 1382837 | A | A | G | G | sla1 | Yes |
| Test strain –known | I 1382838 | I 1382838 | A | A | C | C | sla1 | Yes |
| Test strain –known | I 1383602 | I 1383602 | A | G | A | A | Yes | |
| Test strain –known | I 1384146 | I 1384146 | T | C | T | T | tim40 | Yes |
| Test strain –unknown | I 877349 | I 877349 | C | C | T | T | nop58 | Yes |
| Test strain –unknown | I 2277237 | I 2277235 | C | C | A | A | Yes | |
| Test strain –unknown | I 1177692 | I 1177692 | A | G | A | A | dak1 | Yes |
| Parent # mutant | I 933736 | I 933736 | C | C | C | T | erg3 | Yes |
| Base deletion | II 621061 | II 603280 | T | miscRNA | Yes |
Position refers to our parent strain with roman numeral reflecting chromosome. ‘SangerPos’ refers to the position as found in the Sanger Institute reference genome. The corresponding gene locus is noted, if any. Nucleotide identities at a given position are shown for four strains, reference yFS101 (Ref), Test, Parent (WT, yMWB3-15) and mutant (Mut, yDA317). Multiple known mutations in our test strain at the nhp6-sla1-tim40 locus region on chromosome I, spanning positions 1381336–1384146 served as a control. Each mutation was verified by conventional PCR-mediated sequencing.
Table 4.
Summary of polymorphisms found in our parent strain, yMWB3-15, by Solexa sequencing and the S. pombe reference genome, yFS101 (972h-)
| Chr | Position | SangerPos | Ref | WT | Cover | Gene | Name | Mutation | FS101 | MWB315 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 733352 | 733352 | G | A | 66 | SPAC22G7.04 | pan2 | e | G | A |
| 1 | 853328 | 853328 | T | G | 80 | SPAC23G3.01 | rpb2 | L872R | T | G |
| 1 | 877349 | 877349 | C | T | 71 | SPAC23G3.06 | nop58 | N98a | C | T |
| 1 | 943745 | 943745 | T | C | 48 | SPAC1687.22c | puf3 | I709M | T | C |
| 1 | 1E + 06 | 1263527 | A | G | 58 | * | A | G | ||
| 1 | 2E + 06 | 1664907 | T | A | 42 | * | T | A | ||
| 1 | 2E + 06 | 2207525 | G | T | 51 | SPAC1327.01c | (txn fctr) | L191I | G | T |
| 1 | 2E + 06 | 2277237 | C | A | 63 | SPAC4G9.12 | (glco kinase) | Y13stop | C | A |
| 1 | 2E + 06 | 2283617 | T | C | 42 | * | T | C | ||
| 1 | 3E + 06 | 2643596 | C | A | 67 | * | C | A | ||
| 1 | 3E + 06 | 2747078 | A | G | 49 | SPAC6F6.08c | cdc16 | c | A | G |
| 1 | 4E + 06 | 3548913 | T | A | 32 | * | T | A | ||
| 1 | 6E + 06 | 5543973 | T | C | 46 | * | T | C | ||
| 2 | 535941 | 518160 | G | A | 43 | * | G | A | ||
| 2 | 683208 | 665427 | T | C | 49 | * | T | C | ||
| 2 | 998064 | 980283 | T | C | 46 | SPBP35G2.08c | air1 | S81a | T | C |
| 2 | 1E + 06 | 990639 | C | T | 87 | SPBP35G2.13c | swc2 | S243a | C | T |
| 2 | 1E + 06 | 1422317 | T | A | 42 | SPBC691.02c | tip20 | L531a | T | A |
| 2 | 2E + 06 | 1553976 | A | T | 23 | * | A | T | ||
| 2 | 2E + 06 | 2007715 | A | G | 40 | * | A | G | ||
| 2 | 3E + 06 | 2806053 | G | A | 42 | * | G | A | ||
| 2 | 3E + 06 | 2901509 | G | T | 47 | miscRNA | T | T | ||
| 3 | 1E + 06 | 1316977 | T | A | 80 | SPCC1322.13 | ade6d | C215stop | NA | A |
| 3 | 2E + 06 | 1647827 | G | T | 33 | * | G | T | ||
| 3 | 2E + 06 | 1862803 | A | G | 48 | * | A | G | ||
| Mt | 1788 | 1788 | C | T | 9483 | rRNA | T | T | ||
| Mt | 2091 | 2091 | A | G | 7144 | rRNA | G | G | ||
| Mt | 4102 | 4102 | C | G | 7556 | rRNA | G | G | ||
| Mt | 4463 | 4463 | C | A | 7275 | rRNA | A | A | ||
| Mt | 6906 | 6906 | A | T | 6332 | CDS | S21C | T | T | |
| Mt | 8529 | 8529 | G | T | 7585 | cox1 | cox1 | E451D | T | T |
| Mt | 10309 | 10309 | T | C | 5261 | cob1 | cob1 | I46T | C | C |
| Mt | 11218 | 11218 | T | A | 9344 | aic ORF | I121N | A | A | |
| Mt | 11644 | 11644 | A | C | 6613 | aic ORF | H263N | C | C | |
| Mt | 13196 | 13196 | C | T | 7841 | aic ORF | a | T | T | |
| Mt | 13751 | 13751 | C | T | 6789 | cob1 | cob1 | a | T | T |
| Mt | 15082 | 15082 | A | – | 6204 | atp6 | atp6 | L109Ff | – | – |
| Mt | [15088] | [15088] | – | A | 5950 | atp6 | atp6 | R111Gf | A | A |
| Mt | 15200 | 15200 | G | T | 6428 | atp6 | atp6 | V149F | T | T |
| Mt | 18648 | 18648 | G | A | 5716 | cox2 | cox2 | V301I | A | A |
| Mt | 19152 | 19152 | A | G | 5906 | cox2 | cox2 | S198G | G | G |
Chr = chromosome, 1,2,3 or mitochondria (Mt). Cover = coverage.
Resulting coding change, if any is noted in the ‘Mutation' column, *-intergenic, a-synonymous mutation in coding region, c-intronic, d-ade6-704 allele mutation confirming previous sequencing (65), e-intronic based on revised splice prediction (72), f-atp6 frameshift mutations result in two amino acid mutations leaving 110 unchanged.
Each nuclear SNP was verified by conventional PCR-mediated sequencing and the identities are noted in the rightmost columns.
For some applications, we incorporated the complete mitochondrial DNA sequence (54,55) Schizosaccharomyces pombe ad7-50h- (Project ID: 20755).
Illumina Solexa sequencing and identification of mutations
Sequencing of samples was according to standard methods. Unpaired 36-bp reads were collected and pooled under designation WT (yMWB3-15), MUT (yDA317) or TEST (yMWB2-4) for each. Our updated S. pombe reference genome was used in alignments performed using MAQ (Mapping and Assembly with Qualities) software (56) without any trimming. SNPs were called from sequence alignments. Deletions and insertions were identified and called by alignment of flanking sequence seeds to regions of zero coverage using the total read database, followed by verification by polymerase chain reaction (PCR) amplification of genomic DNA and conventional sequencing.
Software and sequence analysis tools
Reads were obtained and adjusted to Sanger FASTQ format and pooled for each strain dataset (57). MAQ software was utilized in alignment of reads and calling of SNPs (56). Alignments were manipulated with SAMtools (58). Programs used in our analysis were written primarily in Perl with some use of BioPerl modules (59) and are described and available for free download as the MATCH-G toolset at http://science.nichd.nih.gov/confluence/display/smcb/MATCH-G+Program or through the Maraia laboratory website http://science.nichd.nih.gov/confluence/display/smcb/Home. Briefly, these programs performed the following tasks: assignment of SNPs to the reference genome and determination of resulting mutations with corresponding confidence scores; determination of regions corresponding to zero read coverage; seeded search and alignment of zero coverage flanking sequence regions for determination of small deletions and insertions; comparison of annotated feature average mapped read depth between data sets; and a sliding averaging window comparison of mapped read depth between data sets.
Verification of mutations
Mutations were verified from DNA prepared from acid-washed bead-lysed S. pombe cells. Following basic phenol–chloroform extraction, lysate was treated with DNase-free RNase A and DNA was ethanol precipitated and quantitated by nanodrop. PCR amplification of regions of interest was performed and purified using a QIAGEN QIAquick PCR cleanup kit. PCR products were sequenced by ACGT, Inc. (Glenview, Ill) to verify mutations identified by Illumina sequencing.
Determination of duplications
Identification of regions of duplication was performed using mapped coverage depth by two approaches. In the first approach, for each annotated feature ‘context’ of the genome, the number of mapped reads at each position was summed and averaged over the full length of the annotated region. These averages were compared for yMWB3-15 and yDA317 to obtain a ratio corrected by a factor of 1.12 to reflect the difference in average mapped read depth for these strains. The ratios were compared at each feature to establish statistical distribution for all annotated features, filtering out only those <50 nt long, and copy number exceeding 1000 reads, to limit noise.
In the second one, context-independent approach, we used a sliding averaging window of 400 bases and average mapped read depth was compared for yMWB3-15 and yDA317. Positions where the average depth difference exceeded 1.8-fold were identified as indicative of a duplication or loss of copy event and combined into continuous units.
Intentional introduction of additional suppressor tRNASerUCA-C47:6U gene
The ∼400-bp suppressor tRNASerUCA-C47:6U PstI–SacI gene fragment (49) and ∼150 bp of flanking DNA on either side was PCR amplified from the pJK148 vector used to create yWMB3-15, and ligated into pFA6A at the SalI and BamHI sites. pFA6A with and without the suppressor tRNA insert were linearized with NdeI and transformed into yMWB3-15 and yMWB1-1 in parallel. Transformed cells were plated onto YES media then replica plated onto YES + G418 to screen for integrants.
Three G418-resistant transformed colonies from the empty pFA6A and five from the suppressor tRNA-containing pFA6A were isolated and streak purified. These were then spotted onto media with limiting adenine (10 mg/l) alongside yMWB3-15 and yDA317 for comparison to examine suppression phenotype.
Northern blot and genomic DNA PCR verification of duplication
Total RNA was prepared from cells and northern blotted as described (49) using 10% TBE-urea gels and iBlot transfer apparatus (InVitrogen). The RNA was transferred to a Genescreen-Plus membrane which was UV-cross-linked and vacuum baked and hybridized with a 32P-5′-GACAGAGCCCATTAGATTTGAAG DNA complementary to spliced mature suppressor tRNASerUCA-C47:6U in the presence of an equal amount of unlabelled oligo GGCAGAGCCCATTAGATTTGAAG to block hybridization of the 32P-5′-probe with the highly related endogenous tRNASerUGA, a method known as homolog exclusion probing (60). After washing, the membrane was sealed, exposed and quantitated by PhosphorImager. The membrane was stripped, re-hybridized with a U5 snRNA probe to standardize quantitation.
Genomic copy number of the suppressor tRNA gene was verified by semi-quantitative duplex PCR utilizing a single copy region of the genome (sla1 gene) for comparison. Purified genomic DNA was incubated with two primer pairs (sla1F: CCGAATATTGTTACGATTTAAGCATT, sla1R: AACATTGTCTTCATGAGTAGG AAA, SUPF: GCGGGCCTCTTCGCTATTA, SUPR: GGCTCCTATGTTGTGTGG AATT) and amplified as follows (95°C 5 min; 25 cycles at 95°C 45 s, 55°C 45 s, 72°C 90 s; 72°C 3 min, 4°C). Twenty-five cycles had been demonstrated to be within the linear range of amplification for both primer sets (data not shown). Bands were stained with ethidium bromide, visualized by UV and quantitated using VisionWorksLS software. A ∼700-bp band density corresponding to the suppressor tRNA was divided by the ∼1000-bp band density corresponding to sla1. The ratios for each of four independent PCR data sets were normalized using yMWB3-15 ratio as representing a 1:1 copy number.
Tandem duplication was detected by a PCR assay strategy (61) using four primers: (i) ATAAGGAAGCCTTGGGAGGA, (ii) TATCCGCTCACAATTCCACA, (iii) CCAATTCGCCCTATAGTGAGTC and (iv) TGGATGAACATTGTAAATGGTAGG. The positions of primers 1–4 are indicated in Figure 4A. PCR was performed on purified genomic DNA in 25 mM Tricine pH 8.7, 85 mM KOAc, 8% glycerol, 1% DMSO and 1.2 mM Mg(OAc)2 using a mixture of polymerases 2U Tth (Promega) and 0.1U Vent (NEB) in 50 ul. Cycling was 95°C 5 min; 30 cycles at 95°C 45 s, 55°C 45 s, 68°C 7 min; 72°C 3 min, 4°C. Five microliters of each reaction was resolved on 1% agarose gel.
Figure 4.
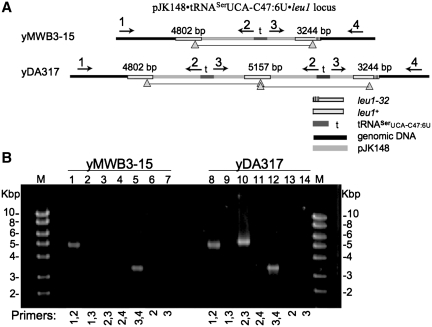
Partial duplicated state of the pJK148•tRNASerUCA-C47:6U•leu1 locus in the yDA317 mutant. (A) Overview of the pJK148•tRNASerUCA-C47:6U•leu1 locus in yMWB3-15 (upper) and yDA317 (lower), illustrating the relative positions of PCR primers (horizontal arrows labeled 1–4) used in (B). DNA lengths between the primer pairs are indicated in bp. Lower case t indicates position of the tRNASerUCA-C47:6U gene. The horizontal line connecting the two gray-filled triangles reflects the region duplicated in yDA317. A key depicting different components of the loci is shown in lower right. (B) EtBr stained gel of PCR products obtained with the different primers indicated under the lanes, using genomic DNA from yMWB3-15 (lanes 1–7) and yDA317 (lanes 8–14) as template. Primers 2 and 3 would produce a product only from the partially duplicated locus. Size markers are indicated on the left and right in kb.
RESULTS
Isolation of spontaneous phenotypic mutants
Our laboratory strains yAS95 and yAS99 containing ade6-704, were derived in 1996 after crossing sp1190 (62) (a.k.a., yAS50) with spDJV1 (63) derived from spB67, a diploid with the ade6-M210/ade6-216 alleles (64). The strain generated for the present study, yMWB3-15 (parent) harbors ade6-704, which contains a UGA stop codon (65) that leads to accumulation of red pigment when grown in media with limiting adenine (66). Suppressor tRNASerUCA can suppress this nonsense stop codon and lead to production of functional Ade6p enzyme, which suppresses red pigment. yMWB3-15 contains the suppressor tRNASerUCA-C47:6U allele which carries a nucleotide substitution in the variable arm, and the sla1-rrm mutant allele sla1-Y157A,F201A (Figure 1B). The tRNASer substitution changes a G:C basepair to a G:U basepair in the tRNASerUCA variable arm. This substitution prevents tRNASerUCA-C47:6U-mediated suppression in sla1-Δ as well as sla1-Y157,F201A cells (32). yMWB3-15 harbors ade6-704, tRNASerUCA-C47:6U and chromosomal sla1-Y157A,F201A, and expresses Sla1p-Y157A,F201A at wild-type levels (data not shown), but accumulates red pigment due to lack of tRNA-mediated suppression (TMS).
yMWB3-15 was plated on limiting adenine media in order to isolate spontaneously arising white colonies. Multiple isolated colonies were purified and subjected to conventional sequencing of the tRNASer-C47:6U, sla1-Y157A,F201A and ade6-704 loci to screen for mutants that had not reverted these alleles to their functional counterparts (no reversions were found, data not shown). The mutant designated yDA317 was chosen for Solexa sequencing because it exhibited higher levels of mature suppressor tRNASerUCA-C47:6U, than the parent strain (data not shown, see below).
Whole genome Solexa sequencing reveals known SNP mutations
yMWB2-4 is a strain generated during the development of yMWB3-15 that was known to harbor mutations in and around the sla1+ locus that differ from the parent strain. yMWB2-4 was subjected to whole genome sequencing in parallel with parent (yMWB3-15) and mutant (yDA317) to serve as a test and to help train our analytical methods. SNPs relative to the parent were determined blindly by MAQ alignment software (56) utilizing S. pombe genome 972h- (GenBank) with modifications (1,51) as the alignment reference (Table 1). This led to identification of all of the yMWB2-4 mutations at the nhp6-sla1-tim40 locus (Table 1, known), as well as three additional differences (unknown), all of which were verified by conventional sequencing.
We then compared the parent (yMWB3-15) and spontaneous mutant yDA317 and identified only one SNP, which represents a synonymous mutation to valine 74 of the erg3+ gene open reading frame (Table 1). Analysis of this SNP in erg3+ predicted no significant change in valine codon usage or local RNA structure. Furthermore, no reverse strand RNA or open reading frame (ORF) was annotated nor predicted. Given the number of adventitious synonymous SNPs discovered in our test strain (and in yFS101, below), it seemed unlikely that the erg3+ synonymous SNP caused the TMS phenotype in yDA317. We therefore sought to examine the whole genome sequences of our parent and mutant strains for mutations other than SNPs by further analysis of the Solexa data.
Identification of a tRNASerUCA-C47:6U gene duplication in yDA317
Insertions and deletions were sought by identifying sequence reads that flanked regions of zero mapped coverage in the ref. genome, followed by verification by PCR sequencing. This approach identified a single base deletion in an annotated miscRNA, at chromosome 2, Sanger position T603280 (Table 1). However, this deletion relative to the reference was shared by the parent and mutant strains (Table 1) and is not responsible for the mutant phenotype.
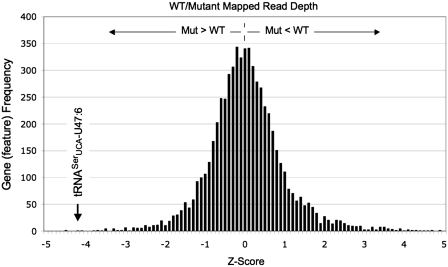
The next approach was to determine if a change in copy number of a gene or region had occurred, and, for this, two strategies were employed. At the positions of known and predicted genes, the mapped read depth was averaged across the entire length of the annotated feature, as an annotation context-dependent approach. These averages led to ratios of mapped read depth for the parent and mutant at each known transcription/annotation unit. These ratios were corrected slightly to reflect the difference in average read depth for the entire genomes of the strains, 40 for the parent and 45 for the mutant. When the ratios were plotted in a simple statistical distribution, the copy number varied along a standard bell curve at a mean of a 1-to-1 ratio (Figure 2). One significant outlier, however, was in the number of reads mapping to the suppressor tRNASerUCA-C47:6U sequence, suggesting gene duplication. The position on the bell curve of this copy number variation was more than 4 SD away from the median, toward increased copy number in the mutant strain (Figure 2).
Figure 2.
A simple statistical distribution of annotated gene copy number as estimated from mapped read depth. A Z-score of zero represents a 1:1 ratio of wild type, (WT, yMWB3-15) and mutant (Mut, yDA317) read depth at an annotated genome feature. A negative Z-score reflects greater read depth or copy number in the mutant strain, and vice versa. The downward arrow indicates the suppressor tRNASerUCA-C47:6U gene, representing a 2-fold greater copy number in the mutant strain.
The potential for gene duplication was then assessed by a second strategy that also yielded size information as part of a context-independent analysis of mapped read depth. For each base of all three chromosomes, the average read depth of that base and 200 bases on either side was compared between parent and mutant strains using a cutoff of ≥1.8 fold difference in read depth between strains. This approach identified four regions of ∼2-fold increase in mapped reads in the mutant relative to the parent, two intergenic, one corresponding to a 43-bp region of the USP coding sequence and a large section (5.8 kb) corresponding to the suppressor tRNASerUCA-C47:6U allele plus associated flanking DNA used for its prior integration in ySH18, a precursor of yMWB3-15 (Table 2). While both strategies identified few differences in copy number at loci other than the suppressor tRNASerUCA-C47:6U gene, the others were not as well supported statistically as the tRNASerUCA-C47:6U gene duplication (see ‘Discussion’ section), nor could they as readily explain the suppression phenotype. Thus, the tRNASerUCA-C47:6U gene duplication was an excellent candidate as causative of the suppression phenotype.
Table 2.
Annotation context-insensitive computational screen for change in copy number
| Chr | Position | Length | MUT/WT | WT Cover | Mut Cover | Locus |
|---|---|---|---|---|---|---|
| 1 | 1552075 | 43 | 1.9 | 40 | 74 | USP (Universal Stress) family |
| 2 | 1974364 | 5809 | 2.1 | 54 | 110 | Suppressor-tRNA + flanking DNA |
| 3 | 565558 | 108 | 1.9 | 30 | 56 | Intergenic miscRNA/SPCC1672.03c |
| 3 | 997392 | 23 | 1.9 | 43 | 83 | Intergenic (vcx1/mad3) |
An averaging window of 400 nt was scanned across the three chromosomes (Chr) comparing average mapped read depth/coverage between parent (WT) and mutant (MUT) strains.
Base positions exceeding a 1.8-fold change in coverage were flagged and combined into continuous units.
The ratio of mutant/wild-type coverage is indicated (MUT/WT); the average coverage is indicated for WT and Mut.
Deliberate introduction of an additional tRNASerUCA-C47:6U gene into yMWB3-15 produces suppression phenotype
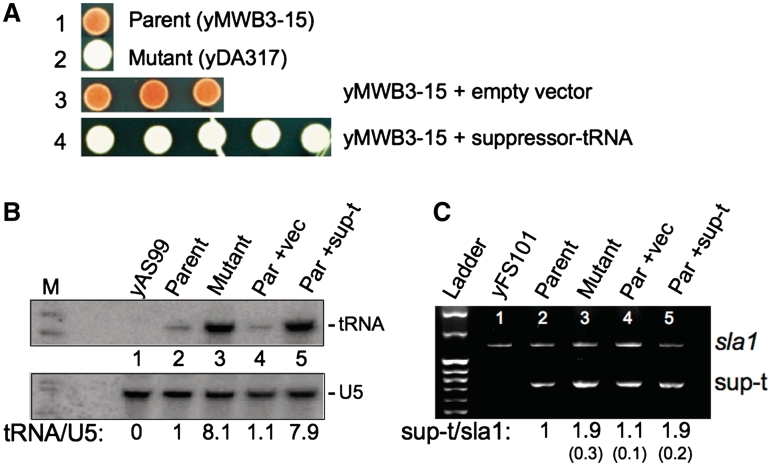
Since two analytical approaches suggested duplication of the suppressor tRNASerUCA-C47:6U gene, we tested if introduction of an additional copy of this gene would result in the suppression phenotype. The parent strain was transformed by chromosomal integration of linearized kanR-containing pFA6A vector harboring either a suppressor tRNASerUCA-C47:6U insert of ∼300 bp, or no insert. Transformed cells were plated on YES, then replica plated onto YES + G418. G418-resistant colonies were randomly picked from both the no-insert and the suppressor tRNASerUCA-C47:6U-insert transformants, streak purified and transferred to limiting adenine for comparison to each other, parent and mutant, for TMS phenotype. None of the three analyzed transformants derived from the empty vector showed suppression (Figure 3A, row 3). By contrast, all five analyzed transformants derived from the suppressor tRNASerUCA-C47:6U gene clearly demonstrated suppression greater than in the parent strain yMWB3-15 (Figure 3A, compare rows 1 and 4) and to a similar degree as in the spontaneous mutant yDA317 (Figure 3A, row 2). This demonstrated that introduction of a suppressor tRNASerUCA-C47:6U gene into our parent strain results in a phenocopy of the mutant phenotype. Moreover, although yDA317 contained an extra tRNASerUCA-C47:6U gene as well as associated pJK148 vector DNA, the present result isolated the phenotypic segment to the extra tRNASerUCA-C47:6U gene. It also showed that the synonymous erg3-SNP in yDA317 was not required for the suppression phenotype. The data argue that the suppressor tRNASerUCA-C47:6U gene duplication detected by whole genome sequencing is responsible for the suppression phenotype in the yDA317 mutant.
Figure 3.
Deliberate introduction of an additional chromosomal copy of the suppressor tRNASerUCA-C47:6U gene into yMWB3-15 is sufficient to produce the suppressed phenotype with increased tRNASerUCA-C47:6U levels. (A) Suppression phenotypes of parent and mutant strains as compared to parent transformed with either the integrated vector without a suppressor tRNASerUCA-C47:6U gene (Parent + empty vector, 3 out of 3 isolates shown), or the integrated vector with the suppressor tRNASerUCA-C47:6U gene insert (Parent + suppressor-tRNA, 5 out of 5 isolates shown) (see text). (B) Northern blot of RNA purified from various strains and probed for mature suppressor tRNASerUCA-C47:6U (upper panel). Parent, mutant and one each of the ‘parent + empty vector' and ‘parent + suppressor-tRNA’ strains from (A) are shown; yAS99 is a negative control strain that does not carry a suppressor tRNASerUCA gene. Lower panel shows the same blot probed for U5 snRNA used as a loading control for normalizing quantitation; the ratio of tRNA/U5 snRNA is indicated below the lanes. (C) Semiquantitative PCR of the suppressor tRNASerUCA-C47:6U gene and the sla1+ locus used here as a single copy gene control. After ensuring that amplification was in the quantitative linear range of the assay (data not shown), duplex PCR was performed under linear quantitative conditions. Band densities were quantitated and normalized to parent, and indicated under the lanes; numbers in parentheses reflect the range obtained from quadruplicate experiments.
Elevated tRNASerUCA-C47:6U levels in TMS mutant yDA317
As noted in the ‘Introduction’ section, preliminary characterization indicated that yDA317 exhibited higher levels of tRNASerUCA-C47:6U than the parent, yMWB3-15. To evaluate if introduction of an extra tRNASerUCA-C47:6U gene also led to elevated tRNASerUCA-C47:6U levels, we performed northern blots. Figure 3B demonstrates increased suppressor tRNASerUCA-C47:6U levels in both yDA317 mutant and yMWB3-15 transformed with the suppressor tRNASerUCA-C47:6U gene, relative to the nontransformed parent strain. Using U5 snRNA for calibration, suppressor tRNASerUCA-C47:6U levels were reproducibly elevated roughly 8-fold in the yDA317 mutant relative to the yMWB3-15 parent (Figure 3B, compare lanes 2 and 3). Similar increase occurred in yMWB3-15 transformed with the suppressor tRNASerUCA-C47:6U gene (lane 5) but not with the vector only (lane 4).
Verification of the additional copy of the suppressor tRNASerUCA-C47:6 gene
This verification was achieved by semi-quantitative duplex PCR amplification of genomic DNA at the suppressor tRNASerUCA-C47:6U and the sla1 loci, the latter as a single copy gene control in the same reaction (Figure 3C). PCR products from four independent reactions were quantitated for the tRNASerUCA-C47:6U bands and normalized to the sla1 PCR bands in the same lanes with the results shown below the lanes of Figure 3C. The tRNASerUCA-C47:6U fragment was 2-fold increased in yDA317 relative to the yMWB3-15 parent strain (lanes 2 and 3), consistent with estimation derived from the Solexa sequence read coverage. A similar increase was observed for the tRNA-transformed parent (Figure 3C, lane 5) relative to the empty vector-transformed parent strain (lane 4). The cumulative data establish that a suppressor tRNASerUCA-C47:6U gene duplication causes elevated tRNASerUCA-C47:6U levels and increased tRNASerUCA-C47:6U-mediated suppression.
Tandem duplication of the tRNASer-C47:6U gene and surrounding DNA
The Solexa data indicated duplication of the suppressor tRNA gene and flanking DNA. We suspected a tandem duplication and tested this using a four-primer PCR approach (61). Genomic DNA was amplified using different combinations of four primers as diagramed in Figure 4A. The expected PCR fragments from primer pairs 1, 2 and 3, 4 should occur regardless of copy number and can be considered as positive controls for the primers (Figure 4B, lanes 1, 5, 8 and 12). Primer 2 alone or primer 3 alone would produce products only if tandem duplication occurred in opposite orientation, which was not observed (Figure 4B, lanes 6 and 7, and 13 and 14). However, combining primers 2 and 3 produced a ∼5.2-kb fragment from yDA317 but not yMWB3-15 (Figure 4B, compare lanes 3 and 10). This fragment would arise only if tandem duplication occurred in the same orientation. As confirmation, this 5157-bp fragment was subjected to standard sequencing and found to have the anticipated sequence including an intact leu1+ (data not shown). (Although yDA317 contains two leu1+ genes these cells do not exhibit growth advantage over yMWB3-15, which contains a single leu1+ gene, in media lacking leucine, data not shown.) From these data, it appeared that the yMWB3-15 region demarcated by the gray filled triangles (Figure 4A) containing the suppressor tRNA gene was duplicated in yDA317.
tRNA gene duplication as a common source of our spontaneous TMS mutants
Using the PCR tRNA gene duplication assay we examined genomic DNA from 30 additional mutants isolated at the same time as yDA317 and stored at −80°C. Upon reevaluation, three of the mutants had weak or no suppression phenotype and exhibited a single copy tRNASerUCA-C47:6U gene. Of the remaining 27 suppressed mutants, 21 had two copies of the tRNASerUCA-C47:6U gene and four had three or more copies. Thus, 25 of 28 spontaneous mutants (90%) with increased TMS had undergone tRNASerUCA-C47:6U gene duplication or additional amplification. The three suppressed mutants with a single tRNASerUCA-C47:6U gene did not express elevated levels of tRNASerUCA-C47:6U (data not shown) and are thus of questionable interest.
The sla1-rrm allele is required for TMS in the tRNA gene duplication mutant
The northern blot data were somewhat surprising because prior experience with TMS suggested that an increase in suppressor tRNASerUCA levels necessary to cause a TMS phenotype would be greater than a 2-fold increase expected from a gene duplication.
We considered two models by which a tRNASerUCA-C47:6U gene duplication can lead to a disproportionately large increase in tRNASer-C47:6U levels, which could potentially be distinguished experimentally. It was assumed for both models that the increase in pre-tRNASerUCA-C47:6U transcription that results from gene duplication would overcome a concentration-dependent process that would allow some of the pre-tRNASerUCA-C47:6U to evade decay for at least long enough to be engaged by a more productive maturation process that promotes accumulation. In the first model, a 2-fold increase in the concentration of nascent pre-tRNA substrate might enhance the rate of association with the Sla1-Y157A,F201A protein, whereas in the second model, the increase in pre-tRNA might exceed the capacity of a degradative enzyme. The former was more appealing for multiple reasons including because as noted in the ‘Introduction’ section, Sla1p-rrm is expected to exhibit only a subtle deficiency in RNA binding relative to wild type Sla1p (32), which might be over ridden by a 2-fold increase in the concentration of nascent pre-tRNASerUCA-C47:6U ligand. In the second model, the increased pre-tRNASerUCA-C47:6U substrate might exceed the capacity of a degradative nuclease in the absence of Sla1p-rrm. It might be expected that in the absence of Sla1p more pre-tRNASerUCA-C47:6U would be targeted for decay than in its presence, and the increased pre-tRNA produced from a second copy of the tRNASerUCA-C47:6U gene would further exceed the capacity of the decay pathway and lead to an increased TMS phenotype.
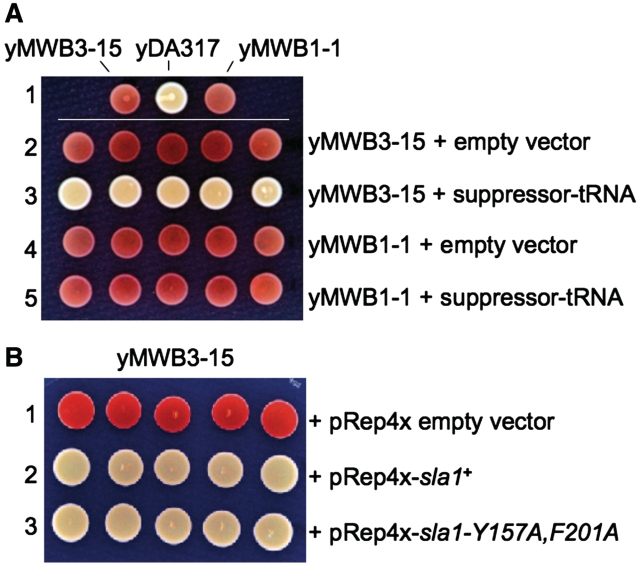
We did an experiment to ask if sla1-rrm was required for the TMS phenotype upon increase in tRNASerUCA-C47:6U gene copy number. We examined yMWB1-1 (sla1-Δ) which is isogenic with yMWB3-15 except that it is devoid of Sla1-rrm protein (Table 3 and data not shown). Neither yMWB1-1 nor yMWB3-15 exhibits TMS (Figure 5A, row 1). yMWB1-1 was transformed with empty vector or vector containing the tRNASerUCA-C47:6U gene, and then examined for TMS, with parallel transformation of yMWB3-15 to serve as control. Individual transformants were picked randomly and then examined for TMS (Figure 5A, rows 2–5). Introduction of vector alone or vector plus additional copy of suppressor tRNA gene into yMWB1-1 did not produce suppression (Figure 5A rows 4 and 5). This was in contrast to yMWB3-15 (sla1-Y157A,F201A) into which an extra copy of the suppressor tRNA gene produced increased TMS in all five transformants, whereas empty vector did not (Figure 5A, rows 2 and 3, respectively). These data would appear to distinguish between the two models considered above and suggest that the mechanism by which tRNASerUCA-C47:6U gene duplication leads to the observed TMS phenotype requires Sla1-rrm protein (see ‘Discussion’ section).
Figure 5.
The sla1-Y157A,F201A allele is required for TMS due to tRNA gene duplication. (A) TMS phenotypes of parent (yMWB3-15), mutant (yDA317) and isogenic test (yMWB1-1) strains (row 1) as compared to yMWB3-15 transformed with empty vector (row 2), or with the suppressor tRNASerUCA-C47:6U gene insert (row 3) and yMWB1-1 transformed with empty vector (row 4), or with suppressor tRNASerUCA-C47:6U gene insert (row 5). (B) yMWB3-15 transformed with empty expression vector pRep4x (row 1), pRep4x-sla1+ (row 3) and pRep4x-sla1-Y157A,F201A (row 3). Five transformants of each are shown.
The data are consistent with the expectation that an increase in nascent pre-tRNASerUCA-C47:6U produced from the second copy gene would be enough to increase its rate of association with the Sla1p-rrm, with resulting positive effects on TMS. If so, we might expect that overexpression of ectopic Sla1p-rrm in yMWB3-15 would lead to TMS. Indeed, ectopic expression of either wild-type sla1+ or sla1-Y157A,F201A, led to TMS (Figure 5B). The data in Figure 5 support the contention that increasing the association of pre-tRNASerUCA-C47:6U with Sla1p-Y157A,F201A, is sufficient to lead to TMS.
Accumulation of 24 nuclear DNA mutations in separated S. pombe strains
The original Sanger Institute sequence assembly of 972h- (1) was recently compared to that obtained using Solexa technology for a single 972h- isolate designated yFS101 (51). We compared our sequence to the yFS101 sequence obtained by Solexa technology and identified 24 differences (Table 4). At each of these positions, yDA317 and yMWB3-15 were identical. We prepared DNA from yFS101 (kindly provided by N. Rhind) and verified these differences in yMWB3-15 and yFS101 in parallel by conventional sequencing. The 24 SNPs consisted of four coding mutations that changed identities of single amino acids in each of four protein sequences, one miscRNA of unknown function, four synonymous coding and 14 intergenic or intronic mutations (Table 4). In addition to these, the Solexa data confirmed mutation in ade6-704 allele in our strain, a T→A that changes a cysteine codon at position 215 of Ade6p to a stop codon (Table 4) as reported (65), as well as mutations in the ura4+ locus (data not shown).
Mitochondrial mutations
After establishing our modified reference genome for yMWB3-15 and obtaining and verifying the nuclear DNA SNPs relative to yFS101, we further modified it by incorporating the available S. pombe mt DNA reference sequence (54,55). This had two remarkable effects on the analysis. Overall mapped read coverage increased from 52% to 96%, with 44% of the reads mapping to the mtDNA. Accordingly, read depth for the mt DNA was uniformly very high, with a range of 5000–10 000 coverage (e.g. see Table 4). Assuming ∼100 copies of mtDNA per S. pombe cell (67), this degree of observed coverage is congruent with our nuclear DNA coverage.
With a cutoff of 85% identity, we observed 13 SNPs and two single-nucleotide insertion/deletions in the mtDNAs of the reference mtDNA sequence (54,55) as compared to our parent and mutant strains (Table 4, Mt). In contrast to the observed SNPs in the nuclear chromosomes, the majority of these mt mutations result in coding mutations. The source(s) of these mtDNA differences is unknown (see ‘Discussion’ section).
We then examined the available yFS101 data set which had not previously been analyzed for mtDNA (51), comparing it to the reference mtDNA (54,55). The same set of 13 SNPs and two insertion/deletions was identified in yFS101 as in our strain, revealing 100% identity of yMWB3-15 and yFS101 throughout their mtDNA (Table 4, Mt). In conclusion, our strain and yFS101 differ in 24 SNPs in their nuclear DNA but are identical in their mtDNA.
DISCUSSION
One conclusion that can be drawn from this work is that comparative whole genome sequencing of S. pombe can be used to detect and map mutations in spontaneous mutants selected for a phenotype, in this case related to tRNA expression, after one plating. The analytical tools described here can detect insertions, deletions, differences in gene copy number and SNPs. This led to the discovery that duplication of a suppressor tRNASerUCA-C47:6U gene is responsible for the selected phenotype.
To compare our strains with each other we first generated an electronic version of our parent strain genome using the original S. pombe reference as a platform (1). Having this in place, we could then compare our strain with the reference 972h- yFS101 genome that had been recently updated using Illumina (Solexa) technology (1,51). This revealed that our strain differs from yFS101 by only 24 SNPs (in addition to expected difference in marker alleles such as ade6-704) (Table 4) (65).
A second conclusion is that the RNA chaperone-like activity of La protein appears to be able to activate its target pre-tRNA ligands for subsequent accumulation of the mature functional tRNA. As will be discussed in more detail below, consistent with other RNA chaperones that function via low-affinity and nonspecific interactions with their RNA ligands, our data suggest that the deficiency in nonspecific RNA binding activity of the mutated Sla1-rrm protein can apparently be overcome by the increase in nascent pre-tRNA that would result from a 2-fold increase in gene dosage. Although a 2-fold increase in a nascent pre-tRNA may appear modest, the effects should be considered in the context of the increased concentration of a single pre-tRNA in a diverse mixture of other pre-tRNAs that compete with variable affinity for La. Accordingly, as a result of a 2-fold increase in suppressor pre-tRNA it should more competitively associate with the Sla1-rrm protein and thereby benefit from its chaperone-like activity.
tRNA gene duplication
Our genetic screen was designed to select for the appearance of mutants that exhibited a gain-of-function in codon-specific TMS of ade6-704. Indeed the output of the screen reported here was found to be an increase in the codon-specific TMS which occurred in association with increased suppressor tRNASerUCA-C47:6U levels. Analysis led to the discovery that duplication of the suppressor tRNASerUCA-C47:6U gene is responsible for the selected phenotype. Although at first glance this might appear to have been likely, it was unexpected for the following reasons. First, on the basis of other genetic screens of yeast, SNPs appear to be more often uncovered than gene duplication. Second, prior and current evidence indicate that the deficiency in tRNASerUCA-C47:6U expression in our parent strain is at the posttranscriptional level, due to degradation of the precursor-tRNA in the absence of protection by La activity-2 (32). Thus, our expectation was that we might find a debilitating mutation in a gene that contributes to a surveillance-related degradation of the pre-tRNASerUCA-C47:6U in the absence of La activity-2. Finally, prior experience suggested that the increase of tRNASerUCA levels required to cause increased TMS phenotype was anticipated to be greater than the 2-fold increase expected from a gene duplication. On this issue, we were surprised by the disproportionately high (8-fold) level of increase in mature suppressor tRNASerUCA-C47:6U in yDA317 and upon intentional introduction of a second copy of the tRNASerUCA-C47:6U gene.
Evidence that nonspecific, low-affinity interactions contribute to the RNA chaperone-like activity of La protein
We hoped to identify a trans-acting factor whose mutation would allow the structurally defective pre-tRNASer-C47:6U to overcome decay as in wild type sla1+ cells. Surprisingly, tRNASerUCA-C47:6U gene duplication led to disproportionately high levels of tRNASerUCA-C47:6U accumulation. An initial idea that the tandem arrangement of tRNASerUCA-C47:6U genes in yDA317 contributes to activated transcription due to increased local concentration of pol III was dismissed after finding that non-targeted introduction of a second tRNASerUCA-C47:6U gene also led to increased suppression in all of the multiple transformants examined, with a 5–8-fold increase in tRNASerUCA-C47:6U levels (Figure 3, data not shown). Toward gaining insight into the mechanism by which the gene duplication led to unexpectedly high levels of tRNASerUCA-C47:6U we determined that this was dependent on the presence of sla1-rrm allele which carries the Y157A,F201A substitutions (or wild type sla1+) since introduction of an extra copy tRNASer-C47:6U gene into the sla1-null strain yMWB1-1 did not cause suppression (Figure 5A). Likewise overexpression of sla1-Y157A,F201A (or wild type sla1+) also led to increased TMS in yMWB3-15 (and yMWB1-1, not shown) (Figure 5B).
The question is how a 2-fold increase in synthesis of a structurally deficient pre-tRNA, as would be expected to occur upon gene duplication, can lead to a greater-fold increase in tRNA accumulation in a La-dependent manner. We believe that a 2-fold increase in nascent pre-tRNA is sufficient to overcome the slight decrease in the RNA-binding affinity of Sla1p caused by the Y157A,F201A mutations to the β-sheet surface of RRM1. By overcoming this binding deficiency, the pre-tRNASerUCA-C47:6U would benefit from the RNA chaperone-like activity of Sla1p and evade nuclear decay during a critically vulnerable period in its maturation. Avoiding decay during a vulnerable period, even transiently, could allow a fraction of the tRNASerUCA-C47:6U to accumulate, e.g. by export to the cytoplasm or other locale away from the nuclear decay system.
To better understand the Sla1p-rrm related aspects of this model, some of the RNA-binding characteristics of La protein and other RRM proteins as well as RNA chaperone activities should be considered. Different RRM-containing proteins use their β-sheets and connecting loops in a variety of ways to engage their target RNAs (68). In some RRM proteins, the aromatic residues equivalent to Sla1p Y157 and F201 interact with RNA via stacking in a sequence-nonspecific manner and contribute significantly to overall affinity, whereas in others their contribution is much less (68). For La protein, the LaM confers high-affinity UUU-3′OH recognition, while the RRM1 β-sheet surface appears to confer nonspecific binding to other regions of the pre-tRNA as might be expected for interaction with a variety of RNAs that differ in sequence (47). Whereas mutation of the conserved aromatic residues on the RRM1 β-sheet of some other RRM proteins decrease RNA binding, for La RRM1 no significant reduction in pre-RNA binding was observed in vitro, although the negative effects on TMS were apparent in vivo (32). Thus, mutation of the RRM1 β-sheet aromatic residues appears to minimally decrease the nonspecific RNA-binding activity of La protein (32,47). Accordingly, a 2-fold increase in the concentration of pre-tRNA ligand may be sufficient to increase the rate of association with Sla1p-Y157,F201 and overcome this minimal deficiency. Consistent with this model is that nonspecific, low-affinity interactions indeed characterize other RNA chaperones that assist RNAs during folding (69). That sla1p-rrm confers this activity advances our understanding of the RNA chaperone-like activity of La protein.
Finally, as noted in the third paragraph of the ‘Discussion’ above, we should also consider the effect of a 2-fold increase in the concentration of a single pre-tRNA on the potential competitive strength for La binding in the presence of all of the other nascent pre-tRNAs. Accordingly, a 2-fold increase in nascent suppressor pre-tRNA should confer better competition for the Sla1-rrm protein and thereby afford it the benefit of its chaperone-like activity.
Genetic ‘mutation’ in the S. pombe strains examined here is relatively low
The number of discrepancies detected in our parent and mutant strain was low, consisting of a synonymous SNP and a duplication of >500 bp, the latter of which was causative of the selective phenotype. We note that in addition to the suppressor tRNA gene, three other duplications were detected (Table 2), although the relatively small size of these regions at 23, 43 and 108 bp, and the intergenic locale of two of them, made them unlikely candidates as causative of the phenotype. Moreover, for these loci the ratio of mutant-to-parent reads was <2.0, whereas for the suppressor tRNA gene the ratio was >2.0. Perhaps, more significant, the read depth or coverage at each of these three loci was lower than for the suppressor tRNA gene (Table 2), and likely result from relative mapped read depth noise exceeding the averaging window. On the basis of the last two points, we suspect that these may not reflect true duplications in the yDA317 strain, although we have not attempted to verify this.
The original sequence assembly of 972h- (1) was recently compared to that obtained using Solexa technology for a single 972 isolate designated yFS101 (51). In that case, 200-fold coverage provided a very low false positive rate. Significantly, 190 discrepancies were observed and noted to be possibly due to errors in the reference 972 assembly, sequence polymorphisms in isolates of 972 or systematic errors (51). Our sequence matched the yFS101 sequence in all of these previously noted discrepancies, but did not match either the original reference nor the yFS101 at 24 other positions (Table 2). At each of these 24 positions, yDA317 and yMWB3-15 were identical. These differences in yMWB3-15 and yFS101 were verified by PCR of the loci from both strains (yFS101 provided by N. Rhind) and conventional sequencing (Table 4, last two columns).
The 24 SNPs observed in our strain and yFS101 consisted of five coding mutations that changed the identity of an amino acid in a protein sequence, one miscRNA of unknown function, four synonymous coding and 14 intergenic or intronic SNPs (Table 4). Functional consequences of these SNPs are unknown. These could, in some cases, reflect mutations that confer a phenotype or be completely neutral, we do not know.
Of the four coding changes, we note the following. For Rpb2p, L872R changes a conserved Leu in the several yeast sequences examined and therefore appears to be a mutation in our strain or its predecessor. For Puf3p, I709M occurs close to the end of the protein; the surrounding sequence is highly conserved among Schizosaccharomyces species (although not S. cerevisiae or human) and, therefore, appears to be a mutation in our strain or its predecessor.
Twenty-four SNPs observed in two strains that were separated about 30 years ago suggest genetic drift or possibly inadvertent selection if a SNP conferred a phenotype. In any case, we would caution against the use of this data for the purpose of calculating a ‘mutation rate’ because the length of time these strains have been continuously growing (versus frozen storage) is unknown as are other aspects of their propagation. Nonetheless, the perspective that arises is that the overall mutation burden in laboratory strains of S. pombe appears not to be very high. Indeed, it is low enough to make whole genome sequencing of spontaneous mutants a plausible endeavor.
We note that this study is to be distinguished from a previous report of whole genome sequencing for mutant mapping in S. pombe (70). In that case, sequencing was done after cells were mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine (MNG) and the mutant was isolated. Comparative analyses led to a relatively large number of SNPs, 73 of which were present in the selected mutant that had been mutagenized (70). The presence of so many associated mutations required a significant amount of genetic techniques before the selected phenotype could be attributed to the causative mutation (70). By contrast, our analyses of whole genome sequencing after spontaneous mutation led to one synonymous SNP plus a gene duplication that was shown to be causative of the selected phenotype, with relatively far less requirement for genetic techniques.
That we isolated a tRNA gene duplication rather than SNPs as a common mutation (90%) in our TMS mutants may reflect an idiosyncrasy of our genetic screen, or perhaps a characteristic of tRNA genes. As noted above, SNPs would appear to be more often uncovered from genetic screens of yeast than gene duplication. However, since mutants obtained from genetic screens involve characterization prior to detailed analysis, it seems possible that typical prerequisite examination might disfavor characterization of gene duplication mutants. The ability to map phenotype causative mutations by whole genome sequencing without the need for extensive prerequisite characterization may allow a wider variety of mutations to be isolated.
Software availability and accessibility
The toolset of programs compiled for this purpose is designated MATCH-G (Mutational Analysis Toolset Comparing wHole Genomes) and is available with description and instructions for free download at http://science.nichd.nih.gov/confluence/display/smcb/MATCH-G+Program. Some of the programs preexisted and some were developed specifically for this purpose. Use requires minimal basic computer knowledge (folder structure, file formats) and is written to be able to scale to any size genome for the purpose of finding differences between two sequence data sets where a pre-existing related reference genome exists. It is run either from the command prompt or with a simple graphic-user interface. No coding or knowledge of programming languages is required for use. Use in MacOSX or UNIX-like environments is most straightforward where terminal windows and Perl are present by default. Use in windows operating systems may be more complicated.
CONCLUDING REMARKS
Our work demonstrates that whole-genome sequencing of S. pombe has the ability to detect single-nucleotide mutations, base insertions and deletions, and changes in gene copy number. The tools developed here should be applicable toward identifying factors and pathway components in high-throughput screens that produce spontaneously arising mutants. This work expands the tools available for the S. pombe community. The tools developed here for comparative whole genome sequencing could readily be applied to S. cerevisiae and other model systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Intramural Research Program of the NICHD; National Institutes of Health. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Sergei Gaidamakov and Steven Salzberg for contributions to this work, and Jim Kennison for the tandem duplication PCR strategy. We also thank Valerie Wood and Nick Rhind for information and materials related to the reference strain.
REFERENCES
- 1.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 2.Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgs PG, Ran W. Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage. Mol. Biol. Evol. 2008;25:2279–2291. doi: 10.1093/molbev/msn173. [DOI] [PubMed] [Google Scholar]
- 4.Lowe TM, Eddy SR. A Genomic tRNA Database. 2011 http://rna.wustl.edu/GtRDB. [Google Scholar]
- 5.Rogers HH, Bergman CM, Griffiths-Jones S. The evolution of tRNA genes in Drosophila. Genome Biol. Evol. 2010;2:467–477. doi: 10.1093/gbe/evq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 7.Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H. The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 2006;34:1816–1835. doi: 10.1093/nar/gkl085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. Silencing near tRNA genes requires nucleolar localization. J. Biol. Chem. 2005;280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen VC, Clelland BW, Hockman DJ, Kujat-Choy SL, Mewhort HE, Schultz MC. Replication stress checkpoint signaling controls tRNA gene transcription. Nat. Struct. Mol. Biol. 17:976–981. doi: 10.1038/nsmb.1857. [DOI] [PubMed] [Google Scholar]
- 10.McFarlane RJ, Whitehall SK. tRNA genes in eukaryotic genome organization and reorganization. Cell Cycle. 2009;8:3102–3106. doi: 10.4161/cc.8.19.9625. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 14.Begley U, Dyavaiah M, Patil A, Rooney JP, Direnzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maraia RJ, Blewett NH, Bayfield MA. It's a mod mod tRNA world. Nat. Chem. Biol. 2008;4:162–164. doi: 10.1038/nchembio0308-162. [DOI] [PubMed] [Google Scholar]
- 16.Bermudez-Santana C, Attolini CS, Kirsten T, Engelhardt J, Prohaska SJ, Steigele S, Stadler PF. Genomic organization of eukaryotic tRNAs. BMC Genomics. 2010;11:270. doi: 10.1186/1471-2164-11-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parmley JL, Huynen MA. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genet. 2009;5:e1000548. doi: 10.1371/journal.pgen.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohli J, Munz P, Aebi R, Amstutz H, Gysler C, Heyer WD, Lehmann L, Schuchert P, Szankasi P, Thuriaux P, et al. Interallelic and intergenic conversion in three serine tRNA genes of Schizosaccharomyces pombe. Cold Spring Harb. Symp. Quant. Biol. 1984;49:31–40. doi: 10.1101/sqb.1984.049.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Munz P, Amstutz H, Kohli J, Leupold U. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature. 1982;300:225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- 20.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc. Nat Acad. Sci., USA. 2005;102:18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.French SL, Osheim YN, Schneider DA, Sikes ML, Fernandez CF, Copela LA, Misra VA, Nomura M, Wolin SL, Beyer AL. Visual analysis of the yeast 5S rRNA gene transcriptome: regulation and role of La protein. Mol. Cell. Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 23.Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 24.Pannone B, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maraia RJ. La protein and the trafficking of nascent RNA polymerase III transcripts [review] J. Cell. Biol. 2001;153:F13–F17. doi: 10.1083/jcb.153.4.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolin SL, Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 27.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. WIRES RNA. 2011 doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phizicky EM. Waste not, want not: a case for tRNA repair. Nat. Struct. Mol. Biol. 2008;15:783–784. doi: 10.1038/nsmb0808-783. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 33.Qiu H, Hu C, Anderson J, Björk G, Sarkar S, Hopper A, Hinnebusch AG. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eIF2. Mol. Cell. Biol. 2000;20:2505–2516. doi: 10.1128/mcb.20.7.2505-2516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinert T, Hopper AK. tRNA traffic meets a cell-cycle checkpoint. Cell. 2007;131:838–840. doi: 10.1016/j.cell.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 37.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator′s key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- 40.Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol. Cell. Biol. 2007;27:3303–3312. doi: 10.1128/MCB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell. 2002;9:1113–1123. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 44.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belisova A, Semrad K, Mayer O, Kocian G, Waigmann E, Schroeder R, Steiner G. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003;22:6562–6572. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayfield MA, Maraia RJ. Precursor-product discrimination by La protein during tRNA metabolism. Nat. Struct. Mol. Biol. 2009;16:430–437. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006 doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Intine RVA, Sakulich AL, Koduru SB, Huang Y, Pierstorrf E, Goodier JL, Phan L, Maraia RJ. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol. Cell. 2000;6:339–348. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 50.Wood VS. pombe gene database. 2011 http://old.genedb.org//genedb/pombe/ [Google Scholar]
- 51.Nusbaum C. Data generated at the Broad Institute. Chad Nusbaum personal communication. 2011 http://www.sanger.ac.uk/Projects/S_pombe/sequence_discrepancies.shtml. [Google Scholar]
- 52.Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki M, Idiris A, Tada A, Kumagai H, Giga-Hama Y, Tohda H. The gap-filling sequence on the left arm of chromosome 2 in fission yeast Schizosaccharomyces pombe. Yeast. 2008;25:673–679. doi: 10.1002/yea.1613. [DOI] [PubMed] [Google Scholar]
- 54.Lang BF. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe: highly homologous introns are inserted at the same position of the otherwise less conserved cox1 genes in Schizosaccharomyces pombe and Aspergillus nidulans. EMBO J. 1984;3:2129–2136. doi: 10.1002/j.1460-2075.1984.tb02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang BF. Complete S. pombe mitochondrial DNA sequence. 2011 http://www.ncbi.nlm.nih.gov/nucleotide/13639?report=genbank&log$=nucltop&blast_rank=1&RID=BYP3YFDS011. [Google Scholar]
- 56.Li K, Brownley A, Stockwell TB, Beeson K, McIntosh TC, Busam D, Ferriera S, Murphy S, Levy S. Novel computational methods for increasing PCR primer design effectiveness in directed sequencing. BMC Bioinform. 2008;9:191. doi: 10.1186/1471-2105-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Driscoll CT, Darlington GJ, Maraia RJ. The conserved 7SK snRNA gene localizes to human chromosome 6 by homolog exclusion probing of somatic cell hybrid RNA. Nucleic Acids Res. 1994;22:722–725. doi: 10.1093/nar/22.5.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooper MT, Conant AW, Kennison JA. Molecular genetic analysis of Chd3 and polytene chromosome region 76B-D in Drosophila melanogaster. Genetics. 2010;185:811–822. doi: 10.1534/genetics.110.115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Connolly T, Beach D. Interaction between the Cig1 and Cig2 B-type cyclins in the fission yeast cell cycle. Mol. Cell. Biol. 1994;14:768–776. doi: 10.1128/mcb.14.1.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 64.McLeod M, Shor B, Caporaso A, Wang W, Chen H, Hu L. Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase to regulate cell cycle progression and meiotic development. Mol. Cell. Biol. 2000;20:4016–4027. doi: 10.1128/mcb.20.11.4016-4027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park JM, Intine RV, Maraia RJ. Mouse and human La proteins differ in kinase substrate activity and activation mechanism for tRNA processing. Gene Expr. 2007;14:71–81. doi: 10.3727/105221607783417619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohli J, Kwong T, Altruda F, Soll D, Wahl G. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. J. Biol. Chem. 1979;254:1546–1551. [PubMed] [Google Scholar]
- 67.Chu Z, Li J, Eshaghi M, Karuturi RK, Lin K, Liu J. Adaptive expression responses in the Pol-gamma null strain of S. pombe depleted of mitochondrial genome. BMC genomics. 2007;8:323. doi: 10.1186/1471-2164-8-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 69.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 70.Irvine DV, Goto DB, Vaughn MW, Nakaseko Y, McCombie WR, Yanagida M, Martienssen R. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 2009;19:1077–1083. doi: 10.1101/gr.089318.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teplova M, Yuan Y-R, Ilin S, Malinina L, Phan AT, Teplov A, Patel DJ. Structural basis for recognition and sequestration of UUU-OH 3′-termini of nascent RNA pol III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.