Abstract
Little is known about whether components of the RNA-induced silencing complex (RISC) mediate the biogenesis of RNAs other than miRNA. Here, we show that depletion of key proteins of the RISC pathway by antisense oligonucleotides significantly impairs pre-rRNA processing in human cells. In cells depleted of Drosha or Dicer, different precursors to 5.8S rRNA strongly accumulated, without affecting normal endonucleolytic cleavages. Moderate yet distinct processing defects were also observed in Ago2-depleted cells. Physical links between pre-rRNA and these proteins were identified by co-immunoprecipitation analyses. Interestingly, simultaneous depletion of Dicer and Drosha led to a different processing defect, causing slower production of 28S rRNA and its precursor. Both Dicer and Ago2 were detected in the nuclear fraction, and reduction of Dicer altered the structure of the nucleolus, where pre-rRNA processing occurs. Together, these results suggest that Drosha and Dicer are implicated in rRNA biogenesis.
INTRODUCTION
Drosha and Dicer are RNase III family endonucleases required for miRNA maturation. In mammalian cells, miRNA genes are initially transcribed as mono- or polycistronic precursors (pri-miRNA). The pri-miRNAs are processed in the nucleus by microprocessor, a protein complex containing Drosha, to create 60–70 nt pre-miRNAs. Pre-miRNAs are then exported to the cytoplasm, where they are processed by the cytoplasmic protein, Dicer, into 21–24 nt miRNAs (1–4). Finally, miRNAs are incorporated into RNA-induced silencing complex (RISC) that contains Ago2, another endonuclease. The RISC complex mediates gene expression by either down-regulating mRNA levels or modulating mRNA translation (3,5).
The roles of Drosha and Dicer in miRNA biogenesis have been well studied; however, little is known about whether these RNase III enzymes participate in the biogenesis of other types of RNAs, in addition to miRNAs. Our group has previously shown that in human cells Drosha is required for processing of pre-ribosomal RNA (pre-rRNA), especially for maturation of 5.8S rRNA (6). This finding was further confirmed by a later study performed in mouse cells demonstrating that down-regulation of Drosha or Drosha-associated RNA helicases (P68 and P72) by siRNA significantly reduced the level of 5.8S rRNA (7). These observations prompted us to explore in more detail the potential roles of protein components in the RISC pathway in pre-rRNA processing.
In eukaryotes, 18S, 5.8S and 28S rRNAs are transcribed by RNA polymerase I into a polycistronic molecule. This precursor is sequentially processed in the nucleolus (and nucleus) by multiple steps of endonucleolytic cleavage and exonucleolytic trimming reactions to produce mature rRNAs (8–10). In vertebrates, the longest detectable transcript, a 47S pre-rRNA containing the three rRNAs, 5′ and 3′ external transcribed spacers (ETS), and two internal transcribed spacers (ITS1 and ITS2), is processed by two alternative pathways to separate small and large subunit rRNAs (Figure 1A).
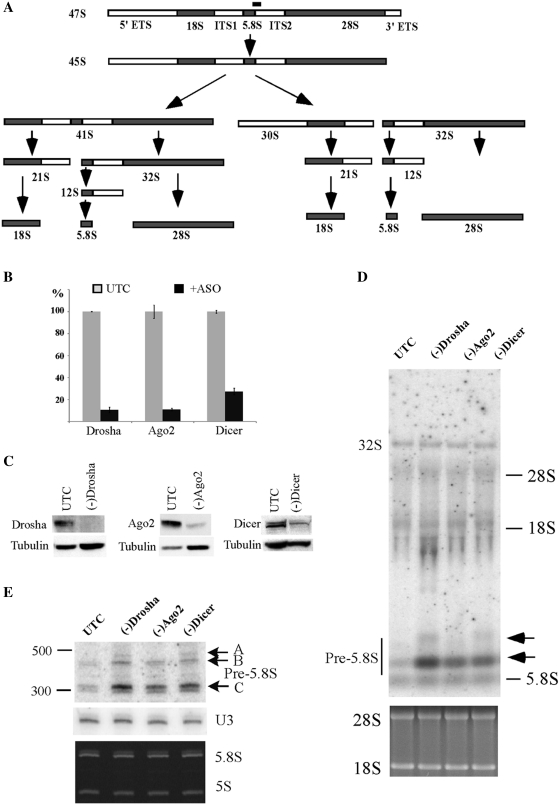
Figure 1.
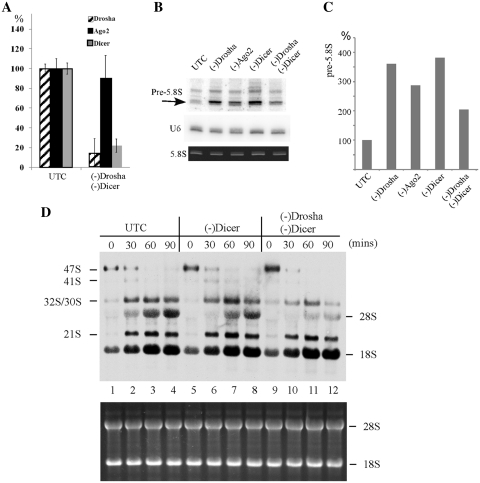
Pre-rRNA accumulation in cells depleted of RISC pathway proteins. (A) Pre-rRNA processing pathway in mammals. ETS and ITS are external and internal transcribed spacers, respectively. The position of the hybridization probe used in (D and E) is shown as a solid bar above 47S pre-rRNA. (B) mRNA levels were dramatically reduced 48 h after treatment with 50 nM ASOs targeting Drosha (ISIS25690), Ago2 (ISIS136764) or Dicer (ISIS138648), as determined by qRT–PCR. The error bars indicate standard deviation from two independent experiments with three replicates. UTC, untreated cells; +ASO, cells treated with ASOs. (C) The levels of targeted proteins were significantly reduced by ASO treatment, as determined by western analysis. Alpha-tubulin was used as a control for loading. (D) Northern hybridization for pre-rRNA species using a probe specific to the boundary of 5.8S rRNA/ITS2. Total RNA prepared from test cells 48 h after ASO treatment was separated on a 1.2% agarose gel, and the blot was subjected to hybridization. The arrows indicate precursors to 5.8S rRNA. The positions of mature rRNAs are indicated. Lower panel shows ethidium bromide staining of rRNAs in the same gel. (E) Pre-5.8S rRNA accumulated in cells depleted of Drosha, Ago2 or Dicer. Total RNA as used in (C) was separated in an 8% polyacrylamide, 7M urea gel and the blot was hybridized using the same probe as in (D). The arrows indicate different pre-5.8S rRNA species (marked as A, B and C). U3 snoRNA was probed to serve as a loading control. Lower panel shows ethidium bromide staining of rRNAs in the same gel.
Maturation of 5.8S rRNA is one of the most complicated pre-rRNA processing events. In yeast, the 5′-end of 5.8S rRNA is formed by two pathways. The major pathway involves endonucleolytic cleavage within ITS1, followed by 5′→3′ trimming (by Rat1 and Xrn1) to generate a shorter form of 5.8S rRNA (5.8S-S) (8,11). In a minor pathway, endonucleolytic cleavage occurs adjacent to the ‘normal’ 5′-end of 5.8S rRNA, to produce a longer form of rRNA containing additional 7 nt at its 5′-end (5.8S-L) (12). In vertebrates, two forms of 5.8S rRNA (5.8S-S and 5.8S-L) also co-exist (10), suggesting that the pathway(s) for 5′-end formation is consistent with the yeast model. In rat, cleavage was mapped to ∼160 nt upstream to the 5′-end of 5.8S rRNA. However, no cleavage sites within ITS1 corresponding to the yeast A2 and A3 sites have been mapped in human cells, and it was proposed that cleavage occurs at the 5′-end of 5.8S rRNA (10).
The 3′-end formation of yeast 5.8S rRNA is initiated by an endonuclease cleavage within ITS2, followed by 3′→5′ trimming performed by exosome (8,9). In mammals, the 3′-end maturation probably involves at least two endonucleolytic cleavages, as two 5.8S rRNA precursors containing ∼156 or ∼250 nt of ITS2 sequence have been detected in human cells (13). As in yeast, 3′-end formation of the vertebrate 5.8S rRNA also requires the exosome (14).
Although pre-rRNA undergoes multiple endonucleolytic cleavages, the endonucleases responsible for each cleavage reaction are still largely unknown. In yeast, it has been shown that the endonuclease MRP complex is required for cleavage at site A3 within ITS1, whereas Rnt1, an RNase III protein, is required for the cleavage at 3′ ETS (8,9,15). The observation that the RNase III protein, Drosha, is required for 5.8S rRNA processing in mammals (6) raises an interesting possibility that another RNase III protein in the RISC pathway, Dicer, or even the RISC pathway itself, may also be involved in rRNA maturation. Indeed, in this study, we show that reduction of Dicer or Drosha or both in HeLa cells causes strong accumulation of aberrant precursors for 5.8S rRNA and significantly reduces the processing rate of 28S rRNA. Depletion of Ago2, the core protein in the RISC complex, also leads to moderate pre-rRNA processing defects that are distinct from those caused by loss of Dicer or Drosha. Our data suggest that Drosha and Dicer are implicated in pre-rRNA processing, in a manner most likely unrelated to the RISC pathway. However, the RISC pathway may also be involved in the maturation of pre-rRNA. Consistent with the observations that these proteins affect pre-rRNA processing, nuclear localization of Dicer and Ago2 is shown.
MATERIALS AND METHODS
Antibodies
Anti-Dicer (ab14601), anti-Ago2 (ab57113), anti-hnRNP A2 (ab6102), anti-GAPDH (ab8245) and anti-nucleolin (ab22758) antibodies were purchased from Abcam. Anti-SF3B3 antibody (SC-21324) was from Santa Cruz Biotechnology. Anti-α tubulin antibody (T-5168) was from Sigma. Anti-Drosha antibody was raised in rabbit using synthetic peptide (amino acids 266–284), as described previously (6). Anti-rabbit secondary antibody conjugated to Texas Red (ab6719) and anti-mouse antibody conjugated to FITC (ab6785) were purchased from Abcam.
Antisense oligonucleotides
Synthesis and purification of phosphorothioate/2′-methoxylethyl (2′MOE) oligonucleotides were carried out as described previously (16). The sequences of 2′-MOE ASOs, or oligonucleotides used for northern hybridization, primer extension, and RT–PCR are listed in Supplementary Data.
Cell culture and transfection
HeLa cells were grown on plates in DMEM medium supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin at 37°C in 5% CO2 incubator. For knock-down of targeted genes, sub-confluent cells were treated with ASOs at 50 nM final concentration in Opti-MEM medium containing 4 µg/ml Lipofectamine 2000 (Invitrogen) for 4 h. Cells were harvested 48 h after transfection. For double knock-down, the second ASO was transfected 24 h after first transfection, and cells were harvested 30 h later.
RNA preparation and manipulation
Forty-eight hours after ASO transfection, cells were washed once with PBS, and RNA prepared directly from culture dishes using Tri-Reagent (Sigma), based on manufacturer’s instruction. Northern analysis was performed as described previously (17). Primer extension was carried out essentially as in Ref. (18), except that extension reactions were performed at 45°C for 1 h. Reverse transcription–PCR and quantitative real-time PCR (qRT–PCR) were performed as described in Supplementary Data.
Sub-cellular fractionation
Cytoplasmic and nuclear fractions were separated from ∼5 × 107 cells using Nuclear Extract Kit (Qiagen), following the manufacturer’s instruction. Five percent of cytoplasmic and 20% of nuclear fractions were separated by SDS–PAGE for western analysis.
Immunoprecipitation
Whole cell extracts prepared in Buffer A [25mM Tris–Cl pH 8.0; 5 mM MgCl2; 150 mM KCl; 10% glycerol; 0.5 mM PMSF; 5 mM β-mercaptoethanol; and one tablet of Protease Inhibitor Cocktail/50ml (Roche)] were incubated at 4°C for 4 h with Protein A beads (Roche) pre-coated with antibodies against Drosha, Dicer or Ago2. After seven washes with wash buffer (50 mM Tris–Cl, pH 7.5; 150 mM NaCl; 5 mM EDTA; 0.1% NP-40; 0.05% SDS), the co-selected RNAs were prepared directly from the beads using Tri-Reagent.
In vivo pulse-chase labeling
Cells grown in 6-well dishes were pre-incubated with serum-free, methionine-free medium at 37°C for 30 min, followed by incubation for 30 min in 1.5 ml of the same medium containing 75 µCi L-[methyl-3H] methionine (Perkin-Elmer). The cells were then chased in the same medium containing 30 µg/ml unlabeled methionine for 0, 30, 60 or 90 min. Next, total RNA was isolated using Tri-Reagent, separated on a 1.2% agarose gel, and transferred to nylon membrane. The membrane was sprayed with Enhance Spray® (Perkin-Elmer), and exposed to X-ray film.
Western analysis and immunofluorescence
Western analysis was performed as described in Supplementary Data. For immunofluorescence, cells grown in Glass Bottom Culture Dishes (MatTek) were washed twice with 1× PBS, fixed with 4% formaldehyde in PBS for 30 min at room temperature, and permeabilized for 5 min using 0.15% Triton X-100 in PBS. Following three washes with 1× PBS, cells were treated with blocking buffer (1 mg/ml BSA in 1× PBS) at RT for 30 min, and incubated with first antibodies for Nucleolin (ab22758, 1:150, Abcam), Dicer (ab14601, 1:200, Abcam) or Ago2 (ab57113, 1:200, Abcam) at 4°C for ON, or at room temperature for 3 h. After three washes with wash buffer (0.1% NP-40 in 1× PBS), cells were incubated with secondary antibodies (1:200) in blocking buffer at RT for 2 h, and washed three times with wash buffer. For double staining (Figure 7), two antibodies were used together, as indicated in figure legends. Finally, cells were mounted using Prolong Gold anti-fade reagent with DAPI (Invitrogen) and covered. Image was taken using a confocal microscopy (Olympus) and analyzed with Fluoview Ver. 2.0b Viewer (Olympus).
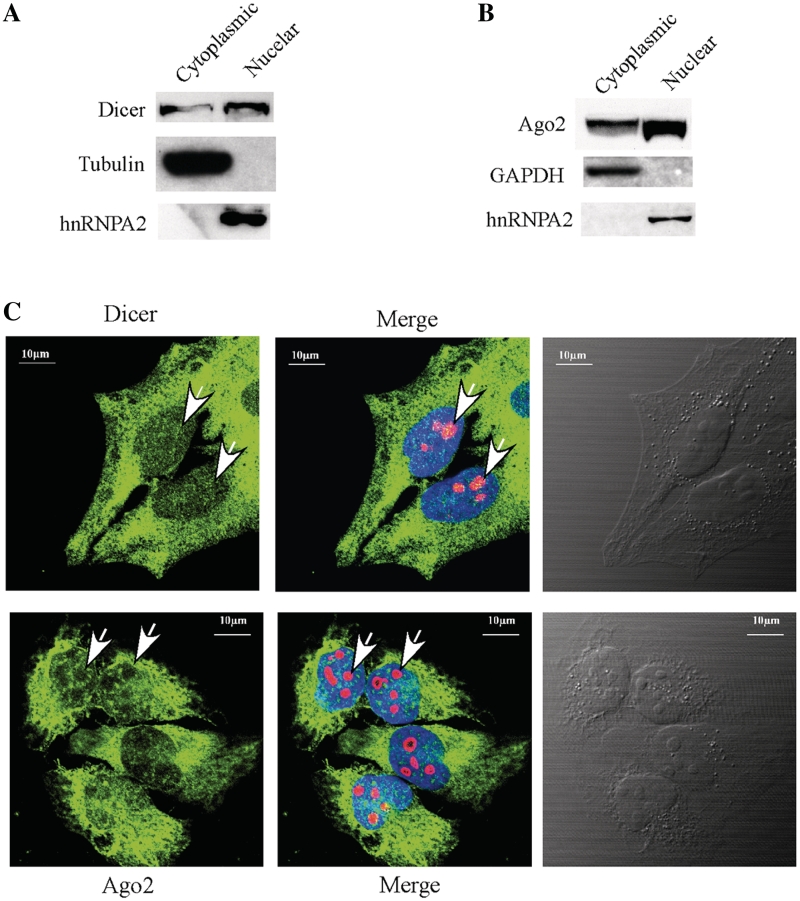
Figure 7.
Nuclear localization of Dicer and Ago2. (A) Western analysis of Dicer in cytoplasmic and nuclear fractions. The 5% cytoplasmic and 20% nuclear fractions prepared from HeLa cells were loaded in a 4–12% SDS–PAGE gel, transferred to a membrane and proteins were detected using antibodies. Alpha-tubulin and hnRNP A2 were used as controls for cytoplasmic and nuclear proteins, respectively. (B) Ago2 can be found in both cytoplasmic and nuclear fractions. Western analysis was performed using the same samples as in (C), and the membrane was probed using different antibodies. GAPDH and hnRNPA2 were detected and used as controls for cytoplasmic and nuclear proteins, respectively. (C) Localization of Dicer (upper panel) and Ago2 (lower panel) in Hela cells. Cells grown on glass-bottom dishes were fixed, stained with first antibodies against Dicer (1:200, ab14601, from mouse, Abcam) and Nucleolin (1:200, ab22758, from rabbit, Abcam), or against Ago2 (1:150, ab57113, from mouse, Abcam) and Nucleolin, as described in ‘Materials and Methods’ section. Secondary antibodies were anti-mouse antibody (1:200, ab6785, Abcam, conjugated with FITC (green) and anti-rabbit antibody (1:200, Ab6719, conjugated with Texas Red). Nucleolin (red) was detected and served as a nucleolar marker. The nucleus was stained with DAPI (blue). The arrow indicates the positions of nucleoli. The scale bars: 10 µm.
RESULTS
Depletion of Dicer or Drosha leads to strong accumulation of 5.8S rRNA precursors
To determine the potential involvement of Dicer and Drosha in pre-rRNA processing, HeLa cells were treated for 48 h with RNase H-dependent, chimeric phosphorothioate/2′-O-methyloxylethyl modified antisense oligonucleotides (ASOs) targeting the mRNAs of these two proteins, as previously described (16,19). Ago2 mRNA was also targeted to examine if the RISC pathway is involved. As expected, ASO treatment resulted in ∼90% reduction in the levels of Drosha and Ago2 mRNAs, and ∼75% reduction for the Dicer mRNA, as determined using qRT–PCR (Figure 1B). The levels of corresponding proteins were also dramatically reduced, as detected by western analysis (Figure 1C).
The effect of depleting the targeted proteins on pre-rRNA processing was analyzed by northern hybridization, using oligonucleotide probes complementary to different regions of the pre-rRNA. Significant accumulation of low molecular weight pre-rRNAs was found in cells depleted of Drosha, Dicer or Ago2 (Figure 1D), as determined using a probe complementary to the boundary of 5.8S rRNA and ITS2. This probe recognizes pre-rRNA species containing both 5.8S rRNA and the ITS2 sequence, including the known 47S, 45S, 32S and 12S pre-rRNAs. 47S and 45S pre-rRNA were not detected in this experiment, due to their low abundance, but 47S pre-rRNA was detected in other experiments with no significant accumulation in the treated cells, similar to 32S pre-rRNA (data not shown). No significant changes in the levels of mature rRNAs and other pre-rRNAs were observed (Figure 1D and E; and data not shown). These results suggest that depletion of Drosha, Dicer or Ago2 impairs pre-rRNA processing at a late stage, most likely at the stage of 5.8S rRNA maturation (Figure 1A).
To confirm the defects on 5.8S rRNA maturation with higher resolution, total RNA was separated by polyacrylamide gel electrophoresis, and pre-5.8S rRNA species were detected using the same probe as in Figure 1D. Indeed, a precursor (band C) in the size of ∼300 nt was found to be strongly accumulated, with ∼350%, ∼110% and ∼230% increase in cells lacking Drosha, Ago2 or Dicer, respectively, as compared with control cells. Another larger precursor (band B region, ∼400 nt) was detected in all cells, but appeared to accumulate moderately in Drosha or Dicer-depleted cells. In addition, at least one aberrant precursor (band A, ∼500 nt) was observed in cells depleted of Drosha or Dicer, but not detected in control or Ago2-depleted cells. These results suggest that reduction of Ago2 caused either a different phenotype or less severe defects on pre-rRNA processing than loss of Drosha or Dicer.
Although ribosome production is well regulated in response to physiological conditions (20), the observed defects on pre-rRNA processing are not due to a non-specific cellular response to transfection of ASOs, nor due to off-target activities of the ASOs that unexpectedly down-regulate the expression of other genes. Evidence includes: (i) transfection of several non-relevant control ASOs had no effect on the level of pre-5.8S rRNA (Supplementary Figure S1 and dada not shown); (ii), transfection of other ASOs targeting different regions of Drosha, Dicer or Ago2 mRNA caused similar defects on 5.8S rRNA processing (Supplementary Figure S2); and (iii) accumulation of pre-5.8S rRNA strongly correlates with the extent and the kinetics of depletion of the targeted genes (Supplementary Figures S3–S6). For example, pre-5.8S rRNA had already significantly accumulated 24 h after ASO treatment, correlating with the time when the target proteins were dramatically reduced (Supplementary Figures S3–S5). In addition, the weaker accumulation of pre-5.8S rRNA in Ago2-depleted cells is not due to potential compensatory effects of other Ago proteins, since depletion of Ago1, Ago3 or Ago4 alone did not result in accumulation of pre-5.8S rRNA, and co-depletion of Ago2 and either one of the other three Ago proteins did not increase the level of pre-5.8S rRNA beyond that caused by depletion of Ago2 alone (Supplementary Figure S7). Finally, the processing defect is not due to impaired biogenesis of processing small nucleolar RNAs (snoRNAs) caused by depletion of these endonucleases, since the levels of the snoRNAs known to be involved in pre-rRNA processing, namely U3, U14, U17, U8, U22 and MRP RNA (21), were not affected (data not shown). Taken together, these data suggest that depletion of Dicer, Drosha or Ago2 impairs pre-rRNA processing, albeit to different extents.
Different pre-5.8S rRNA species accumulate in cells depleted of the RISC pathway proteins
All the pre-5.8S rRNA species detected in Figure 1 contain 3′ extensions, since the hybridization probe recognizes only this subset of pre-rRNAs. The two major bands (∼300 and ∼400 nt) are identical in size to the two previously reported precursors, which contain mature 5′-end of 5.8S rRNA and 3′ extensions with ∼156 or ∼250 nt ITS2 sequence, respectively (13,22). These data suggest that the two precursors we detected here also contain only the 3′ extensions. To confirm this, and to determine if loss of these proteins leads to accumulation of precursors containing 5′ extension, high resolution northern hybridization was performed using probes specific to 5.8S rRNA (5.8S), or the 5′, or the 3′ boundaries of the 5.8S rRNA (Figure 2A). The 5′ or 3′ probes recognize precursors containing ITS1 sequence at 5′-end of 5.8S rRNA, or ITS2 sequence at 3′-end of 5.8S rRNA, respectively. However, the 5.8S probe can detect precursors containing either 5′ or 3′ extension, or both.
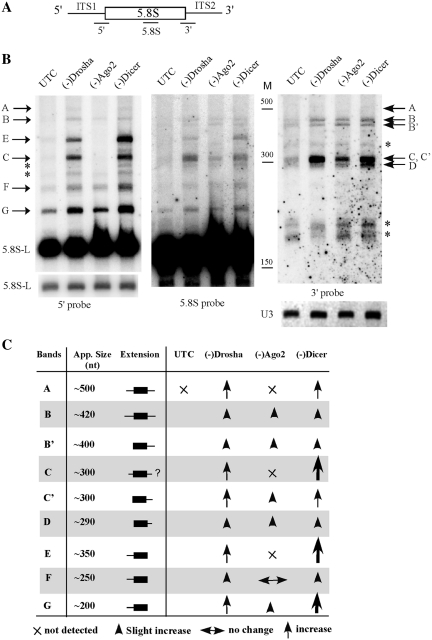
Figure 2.
Loss of RISC proteins leads to accumulation of different pre-5.8S rRNA species. (A) Depiction of probe positions around the 5.8S rRNA region. The probe names are shown under the pre-rRNA. (B) Total RNA was prepared from test cells 48 h after ASO treatment, and subjected to northern hybridization. The same membrane was hybridized sequentially using probes specific to 5′ (left panel, 5′ probe), 5.8S (middle panel, 5.8S probe) and 3′ regions (right panel, 3′ probe), respectively. U3 snoRNA was used as a loading control. Different pre-5.8S rRNA species are indicated. The asterisks indicate RNA species that were only detected with one probe. M, size maker in nucleotides (low range ssRNA ladder, Biolabs); 5.8S-L, the longer version of mature 5.8S rRNA; shorter exposure for 5.8S-L rRNA is shown in left panel, lower part. (C) Summary of changes in the levels of different pre-5.8S rRNA species in cells depleted of the RISC pathway proteins. App. size, estimated nucleotide length of the RNA. Extension, types of 5′ or 3′ flanking sequences in pre-5.8S rRNAs. The larger arrows indicate stronger accumulation.
For convenience, the detected RNA species were named and labeled. Again, when the 3′ probe was used (Figure 2B, right panel), strong accumulation of the 300 nt precursor was detected in Drosha- or Dicer-depleted cells, and moderate accumulation in Ago2-depleted cells. At least eight bands were detected with the 3′ probe, namely band A (∼500 nt), B (∼420 nt), B′ (∼400 nt), C (∼300 nt), D (∼290 nt), one larger (∼340 nt) and two shorter bands (∼170 nt). The latter three (indicated by asterisks) were only detected with the 3′ probe but not with 5.8S probe, suggesting that these bands are the result of non-specific hybridization or represent pre-rRNA species lacking 5.8S rRNA sequence complementary to the 5.8S probe, thus we eliminate them from further discussion.
When the 5.8S probe was used (Figure 2B, middle panel), the bands (B–D) were again detected. Band A was not visible here but was detected in other experiments (data not shown). In addition to 5.8S rRNA, three new products (bands E, F and G) were detected to be strongly increased in Drosha- or Dicer-depleted cells that were not identified with the 3′ probe. This observation indicates that these three precursors do not contain 3′ extensions, but have 5′ extensions. Indeed, when the 5′ probe was used (Figure 2B, left panel), the three bands were clearly detected and were enriched in Drosha- or Dicer-depleted cells, but band E and F were not enriched in Ago2-depleted cells. These data indicate that depletion of these proteins causes accumulation of different 5.8S rRNA precursors with either 5′ or 3′ extensions, or both. However, the level of mature 5.8S or 5.8S-L rRNA was not significantly affected (Figures 1E and 2B, left panel), suggesting that processing was not fully blocked.
For comparison, the hybridization results are summarized in Figure 2C. Band A contains both 5′ and 3′ extensions and was accumulated only in Drosha- or Dicer-depleted cells. Band B contains both 5′ and 3′ extensions, as it was detected by both 5′ and 3′ probes. Band B′ contains only a 3′ extension, since it was detected by either the 3′ or the 5.8S probe in all cells, but not by the 5′ probe. The most accumulated product, band C, appears to contain two different RNA species. One (C′) contains only a 3′ extension but not a 5′ extension, since no RNA was detected in Ago2-depleted cells using the 5′ probe at this position (Figure 2B, middle and right panels), but a product was readily found using either the 3′ probe or the 5.8S probe. However, the 5′ probe detected a product at this position (band C) in Drosha- or Dicer-depleted cells, indicating that band C has a 5′ extension, with or without a 3′ extension. Product D, which slightly increased in cells depleted of these proteins, contains only a 3′ extension. Finally, two pre-rRNAs, bands E and F, contain 5′ but not 3′ extensions and accumulated only in cells lacking Dicer or Drosha. Band G, which has only a 5′ extension, was strongly increased in Drosha- or Dicer-depleted cells and moderately increased in Ago 2-depleted cells. The detection of products containing 5′ extension (bands E, F and G) in control cells indicates that in normal processing pathway, endonucleolytic cleavage occurs upstream to the 5′-end of 5.8S rRNA, as is the case in yeast (10). Together, these results indicate that depletion of these proteins impairs processing of pre-5.8S rRNA, causing accumulation of abnormal intermediates. Ago2 reduction causes moderate accumulation of precursors mainly with a 3′ extension, whereas depletion of Drosha or Dicer leads to accumulation of precursors with extensions at either 5′- or 3′-end, or both ends, but preferentially with 5′ extensions. Although reducing either Drosha or Dicer results in similar defects, certain difference does exist. For example, band E is much more abundant in Dicer-reduced cells than in Drosha-reduced cells.
Depletion of Drosha or Dicer does not block normal endonucleolytic cleavages for production of 5.8S rRNA
The accumulation of abnormal pre-5.8S rRNA species in Dicer- or Drosha-depleted cells may stem from impaired exonucleolytic trimming, or from impaired endonucleolytic cleavage events. Two endonucleolytic cleavage sites within ITS1 (A2 and A3) region have been identified in yeast, however, no such sites have been mapped in human cells. In addition, two processing intermediates have been detected in human cells that contain the mature 5′-end of 5.8S rRNA and ∼156 and ∼250 nt of ITS2 sequence, respectively (13), yet it is not known if these products are generated from endonucleolytic cleavage or exonucleolytic trimming. To identify potential cleavage events in ITS regions and to determine if depletion of Dicer or Drosha impairs endonucleolytic cleavages, primer extension was performed using six different primers specific to ITS1 or ITS2 regions (Figure 3A). Two major extension stops were detected in control cells using probe 2 located in the junction of ITS1/5.8S rRNA, indicating the existence of two precursors containing 5.8S rRNA coding region with 109–110 nt or 152–153 nt of ITS1 sequence, respectively (Figure 3B). No strong extension stop was detected further upstream using probe 1 (data not shown). The two extension stops may represent structural stops in the primer extension assay. However, this seems unlikely, since most other weaker extension stops, upstream or downstream, exhibited comparable signal strength in control and Dicer-depleted cells, whereas these two stops appear stronger in Dicer-depleted cells. Thus, these two products may stem from direct endonucleolytic cleavage at these positions, or from 5′→3′ trimming of precursors cleaved somewhere upstream. Despite this uncertainty, it is clear that endonucleolytic cleavage occurs within ITS1 sequence either at or upstream to these positions. Significantly stronger signals were found at these two positions in Dicer-depleted cells, but not in Drosha-depleted cells, suggesting that loss of Dicer or Drosha did not affect the normal endonucleolytic cleavages within ITS1 regions, yet downstream processing events are slower in Dicer-depleted cells, leading to accumulation of precursors containing these 5′-ends. Interestingly, two additional stops (∼120–123 or ∼138–143 nt upstream to the 5′-end of 5.8S RNA) in between the two normal extension stops appeared in Dicer-depleted cells, and to a lesser extent also in Drosha-depleted cells. This observation indicates that depletion of Dicer or Drosha causes accumulation of pre-rRNAs containing an ITS1 sequence with abnormal 5′-ends. The accumulation of these precursors is consistent with the northern hybridization results (Figure 2), which showed stronger accumulation of pre-5.8S rRNA containing 5′-extensions in Dicer-depleted cells than in Drosha-depleted cells.
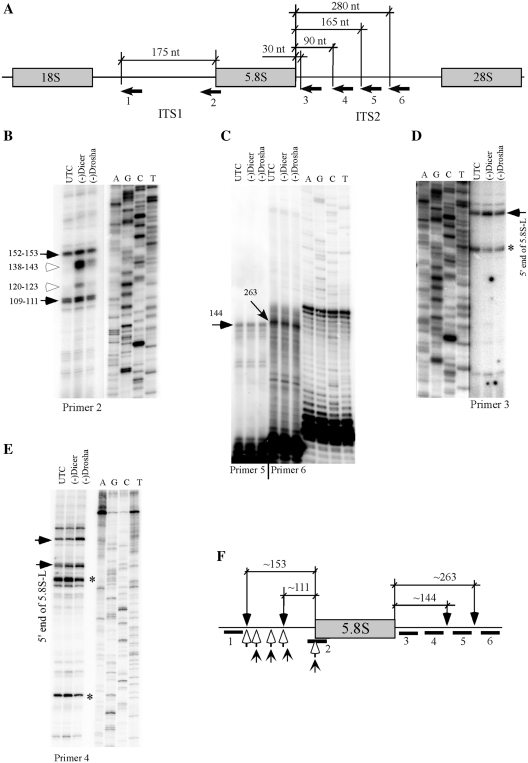
Figure 3.
Depletion of Dicer or Drosha does not block normal endonucleolytic cleavages in pre-rRNA. (A) Relative positions of primers used in primer extension assays. The numbered arrows below the pre-rRNA represent the primers. Positions relative to 5.8S rRNA are shown. (B) Primer extension analysis for ITS1 region using primer 2. The extension products were separated in an 8% sequence gel, next to a primer extension sequencing ladder performed with a primer specific to 18S rRNA (XL066). The same sequencing ladder was used for all gels in this figure. The calculated distances relative to the 5′-end of 5.8S rRNA are given in nucleotides. The solid arrows indicate normal processing products, whereas open arrows indicate products accumulated only in Dicer- or Drosha-depleted cells. (C) Primer extension for ITS2 regions using primers 5 and 6, as in (B). The two potential cleavage products are marked by arrows and the distances relative to the 3′-end of 5.8S rRNA are shown in nucleotides. (D and E) Primer extension using primers 3 and 4, respectively. The arrows indicate pre-rRNA species with 5′-ends that exist in normal cells, but accumulated in Dicer- or Drosha-depleted cells. The asterisks indicate extension stops that occurred within the 5.8S rRNA coding regions. (F) Summary of the mapping data. The solid bars below the pre-rRNA represent positions of primers, as indicated in (A). The potential cleavage positions in normal cells are marked with downward arrows and the relative distances to 5.8S rRNA are shown. The open arrows indicate 5′ positions of pre-rRNAs that accumulated in Dicer-depleted cells, whereas the upward sharp arrows indicate 5′ positions of pre-rRNAs that accumulated in Drosha-depleted cells.
Two significant stops were also detected within ITS2 regions, using primers 5 and 6, respectively (Figure 3C). These two stops were mapped to ∼144 nt and ∼263 nt downstream from the 3′-end of 5.8S rRNA, indicating that endonucleolytic cleavages occur around these two regions, since precursors containing 5.8S rRNA and ∼156 and ∼250 nt ITS2 sequences have been identified previously using a RNase protection assay (13). However, depletion of Dicer or Drosha did not cause significant changes in the signal strength or cleavage positions, indicating that endonucleolytic cleavages at these two regions were not affected. Accumulation of precursor(s) containing a mature 5′-end of 5.8S rRNA and an ITS2 sequence was found in Dicer or Drosha-depleted cells, as determined using primer 3 located 31 nt downstream to the 3′-end of 5.8S rRNA (Figure 3D). Reduced signal strength was observed in Dicer- or Drosha-depleted cells for a primer extension stop located within 5.8S rRNA coding region (indicated by an asterisk), which may represent either a structural or a modification stop. Accumulation of similar precursor(s) containing mature 5′-end of 5.8S rRNA and ITS2 was also detected using primer 4 (Figure 3E), consistent with the northern hybridization results showing accumulation of pre-5.8S rRNA with 3′ extension (Figure 2). Interestingly, a product containing the ITS1 sequence was accumulated in Drosha-depleted cells, but not in Dicer-depleted cells (Figure 3E), indicating that depletion of Drosha or Dicer caused different processing defects. However, no new extension stop was detected using primer 4 in cells depleted of Drosha or Dicer. In addition, the extension stops detected in Figure 3B were not observed using primers 3 and 4 (Figure D and E), suggesting that the products may not contain a 3′ extension.
The primer extension results were summarized in Figure 3F. The data suggest that in normal cells, endonucleolytic cleavage occurs within ITS1 and ITS2 regions in human cells. Depletion of Dicer or Drosha did not affect endonucleolytic cleavages at normal sites; rather, downstream processing events of pre-5.8S rRNA appeared to be impaired, as indicated by the accumulation of abnormal pre-rRNAs containing various 5′-ends.
Simultaneous reduction of Dicer and Drosha leads to different pre-rRNA processing defects
Reduction of Dicer or Drosha causes similar defects of pre-5.8S rRNA maturation, without fully blocking pre-rRNA processing. This observation raises an interesting question: do the two proteins play redundant roles in pre-rRNA processing? To evaluate this possibility, the two proteins were depleted simultaneously by transfection of ASOs targeting the two mRNAs. We reasoned that if the two proteins play redundant roles, simultaneous loss of the two proteins should cause a greater defect than what was observed for a single depletion.
Double depletion caused reduction in the levels of Drosha and Dicer mRNAs by ∼85% and 80%, respectively (Figure 4A). However, simultaneous depletion of Drosha and Dicer significantly decreased the elevated level of pre-5.8S rRNA in response to reduction of a single protein (Figure 4B). In cells depleted of Drosha or Dicer alone, pre-5.8S rRNA level increased by ∼2.5- to 3-fold, whereas only 1-fold increase was found in cells depleted of the two proteins (Figure 4C). This observation suggests that depletion of the two proteins either leads to a different defect, or suppresses the processing defects caused by single depletion.
Figure 4.
Simultaneous depletion of Drosha and Dicer leads to processing defects different from loss of a single protein. (A) mRNA levels in control cells or in cells depleted of Drosha and Dicer were determined using qRT–PCR, as described in Figure 1. The error bars indicate standard deviation from three independent experiments. (B) Northern hybridization for pre-5.8S rRNA, as in Figure 1. The arrow indicates the pre-5.8S rRNA product used for quantification in panel C. 5.8S rRNA was detected by ethidium bromide staining. U6 snRNA was used as a loading control. (C) The levels of pre-5.8S rRNA detected in (B) was normalized to U6 snRNA and plotted. (D) Double depletion of Dicer and Drosha reduced the processing rates for 32S pre-rRNA and 28S rRNA. In vivo pulse-chase labeling was performed as described in ‘Materials and Methods’ section. Various rRNA species are indicated. The chase times (in minutes) are given above each lane. The lower panel shows ethidium bromide staining of rRNAs in the same gel.
To distinguish these possibilities, pre-rRNA processing was analyzed using in vivo pulse-chase labeling with [methyl-3H]methionine, which labels pre-rRNA through nucleotide methylation during rRNA maturation. Different precursors (47S, 41S, 32S and 21S) as well as the newly produced 28S and 18S rRNAs were clearly detected (Figure 4D). However, the signal for 5.8S rRNA was extremely low (data not shown), most likely due to the fact that this RNA contains only a few modified nucleotides. In Dicer-depleted cells, maturation of 28S rRNA was moderately slower than control cells, as evidenced by lower level of 28S rRNA at time points 30, 60 and 90 min after the onset of chase, as compared with control cells. In cells depleted of both Dicer and Drosha, maturation of 28S rRNA was much slower, even than Dicer-depleted cells. However, the level of 32S pre-rRNA, the common precursor of large subunit rRNAs (5.8S and 28S), was also lower than control and Dicer-depleted cells, indicating that accumulation of 32S pre-rRNA is impaired by depletion of the two proteins. The difference in signal strength of 32S pre-rRNA is not due to impaired methylation (labeling) of rRNA, since the early precursor (47S pre-rRNA) and mature rRNA (18S rRNA) were normally labeled. No significant defect was found for processing of 28S and 18S rRNA in cells depleted of Drosha with 50 nM ASO as used in this study (pulse-chase labeling, data not shown), consistent with our previous finding that depletion of Drosha using low ASO concentration (75 nM) did not cause accumulation of 32S and 12S pre-rRNA, which occurred with high concentration (150 or 300 nM), as observed in the previous work (6) and current study (data not shown). These results indicate that the under-accumulation of 32S rRNA in double-depleted cells is due to combined loss of both proteins. As an internal control, the processing rate for 18S rRNA was not impaired in cells depleted of Dicer or Dicer and Drosha together. These data suggest that the decrease in the level of pre-5.8S rRNA in cells depleted of both Dicer and Drosha stems from reduced production of the upstream precursor (32S), and that combined depletion of both proteins caused somewhat different processing defects, with slower production of 32S pre-rRNA and its downstream products.
Nucleolar structure is altered in cells lacking Dicer
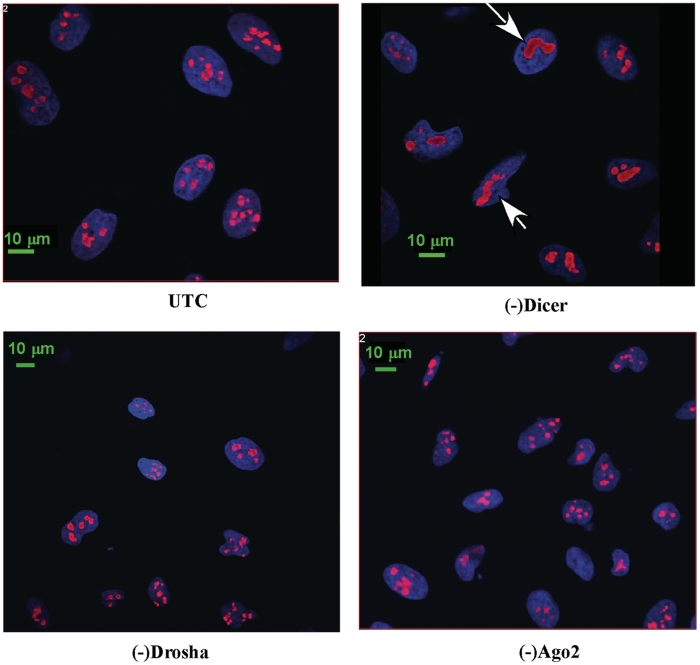
In eukaryotes, pre-rRNA is transcribed and processed in the nucleolus, which is a tripartite structure in terms of known functions. Pre-rRNA is transcribed in the fibrillar center (FC), and initially processed in a surrounding domain known as dense fibrillar center (DFC). Late processing events occur in a third structure, the granular component (GC) (23). Certain step(s) of pre-rRNA processing also occurs in the nucleoplasm (24). Since loss of Drosha or Dicer affects rRNA processing, we reasoned that nucleolar structure might be affected as well. Thus, indirect immunofluorescence was performed to stain a nucleolar marker protein, Nucleolin, which localizes in DFC and GC. This protein is also known to be required for pre-rRNA processing (25). Significant difference in the nucleolar structure was found in cells depleted of Dicer, which exhibit fewer, but larger, aggregated nucleoli than control cells (Figure 5). This difference is significant, since ∼65% of cells treated with the Dicer ASO contained aggregated nucleoli, whereas <10% of control cells showed similar nucleolar structures (data not shown). No significant difference in nucleolar structure was observed in cells depleted of Ago2 or Drosha, although reduction of Drosha caused processing defects similar to depletion of Dicer. Together, these results indicate that reduction of Dicer alters nucleolar structure, directly or indirectly, and that Dicer and Drosha may affect pre-rRNA processing in different ways.
Figure 5.
Cells lacking Dicer exhibit altered nucleolar structure. Indirect immunofluorescence was performed for cells depleted of Dicer, Drosha or Ago2 using first antibody against nucleolin and secondary anti-rabbit antibody conjugated to Texas Red (red). DNA was stained with DAPI (blue). Arrows indicate aggregated nucleoli in cells depleted of Dicer.
Co-immunoprecipitation of pre-rRNA with RISC pathway proteins
Dicer, Drosha and Ago2 proteins are all required for different steps of the biogenesis or function of the RISC pathway, which modulates expression of numerous genes. Thus, the defects on pre-rRNA processing caused by depletion of these proteins may be a secondary effect. To address this possibility, we examined if pre-rRNA could be co-immunoprecipitated with these proteins.
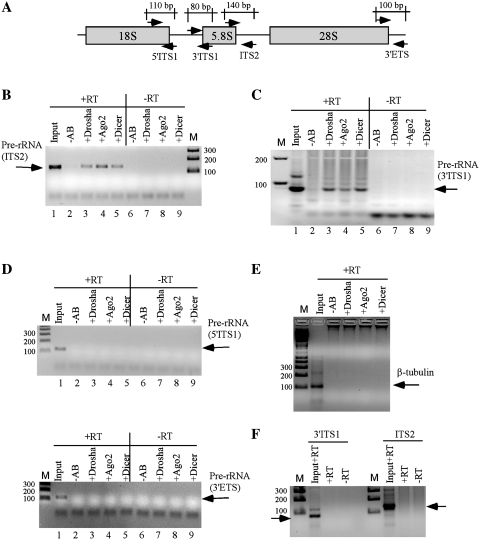
Immunoprecipitation was carried out at 150 mM salt concentration using antibodies against Dicer, Drosha or Ago2. The co-selection of pre-rRNAs was examined by RT–PCR. Four sets of primers were used that are specific to different regions of pre-rRNA: the boundaries of 18S rRNA/ITS1 (5′ITS1), ITS1/5.8S rRNA (3′ITS1), 5.8S rRNA/ITS2 (ITS2) and 28S rRNA/3′ ETS (3′ETS) (Figure 6A). A single RT–PCR product specific to 5.8S/ITS2 primers was detected at the expected size with the precipitated RNAs using Drosha, Ago2 or Dicer antibodies (Figure 6B, upper panel, lanes 3–5), but no signal was detected in control experiments without antibody. Similarly, an RT–PCR product specific to the junction of ITS1/5.8S rRNA was also detected using 3′ITS1 primers (Figure 6C). The relative recovery of the co-precipitated pre-rRNA was ∼1–3% of total, as estimated based on the PCR signal detected with the RNA sample prepared from 10% of whole-cell extract used in immunoprecipitation experiment, suggesting that only a fraction of pre-rRNA is linked with these proteins. However, neither pre-rRNA(s) containing 18S/ITS1 or 3′ ETS sequence nor β-tubulin mRNA was detected, as determined using primers specific to 5′ITS1, 3′ETS or tubulin mRNA, respectively (Figure 6D and E). Importantly, the pre-rRNA(s) co-selected with the RISC proteins were not co-precipitated with an antibody against splicing factor SF3B3 (Figure 6F), suggesting that pre-rRNA could be specifically precipitated with the RISC pathway proteins. Together, the results suggest that these proteins are physically linked to pre-rRNA species containing the 5.8S rRNA and flanking sequences, either directly or mediated by other components. In addition, these data also strongly argue that the defect on pre-rRNA processing is a direct effect caused by loss of the RISC pathway proteins.
Figure 6.
Pre-rRNA containing 5.8S rRNA and flanking ITS sequences can be co-immunoprecipitated with RISC proteins. Immunoprecipitation was carried out using antibodies against Drosha, Ago2 or Dicer. Co-selected RNAs were analyzed by RT–PCR. (A) The positions of probe sets in pre-rRNA used for RT–PCR reaction are indicated. The expected sizes of PCR product are given. Co-immunoprecipitated RNAs were subjected to reverse transcription with (+RT) or without (–RT) oligonucleotides complementary to different regions of pre-rRNA or tubulin mRNA. RT reactions were used as templates for PCR amplification. The PCR products were analyzed on 2% agarose gels. Input, RNA prepared from 10% of the material used for immunoprecipitation. –AB, control immunoprecipitation experiment without antibody; M, 1 kb plus DNA ladder. The sizes are shown in base pairs. (B and C) RT–PCR detection for pre-rRNAs containing 5.8S/ITS2 and ITS1/5.8S regions, respectively. (D) RT–PCR for 18S/ITS1 region (upper panel) or 28S/3′ETS region (lower panel). (E) RT–PCR for β-tubulin mRNA. (F) Immunoprecipitation was performed using an antibody against a splicing factor (SF3B3), and the precipitated RNA was subjected to RT–PCR for ITS1/5.8S (3′ITS1) or 5.8S/ITS2 (ITS2) regions.
Dicer and Ago2 localize in both the cytoplasm and the nucleus
It is known that pre-rRNA processing occurs mainly in the nucleolus, and that later stages of rRNA maturation, including processing of pre-5.8S rRNA into mature rRNA in yeast, may occur in the nucleoplasm or even in the cytoplasm (24,26,27). Drosha localizes in the nucleus and at S phase, it is translocated to the nucleolus (6). Thus, Drosha and pre-rRNA could co-localize in the nucleus. However, Dicer and Ago2 are known to be present predominantly in the cytoplasm (5). The observation that these two proteins associate with pre-rRNA and affect pre-rRNA processing raises a possibility that they may also localize in the nucleus. Indeed, recent studies showed that Ago2 and Dicer were also detected in the nucleus by immunofluorescence staining in mammalian cells (28).
To confirm the nuclear localization of Dicer and Ago2 biochemically, sub-cellular fractionation was performed to separate cytoplasmic and nuclear proteins, and the presence of Dicer and Ago2 in these fractions was determined by western analysis. Dicer was clearly detected in both cytoplasmic and nuclear fractions (Figure 7A). The nuclear signal of Dicer is not due to cross contamination of the fractionation experiment, since a cytoplasmic protein (alpha-tubulin) and a nuclear protein (hnRNP A2) were only detected in the corresponding fractions. Similarly, Ago2 was also clearly detected in both cytoplasmic and nuclear fractions, as compared with a cytoplasmic protein (GAPDH) and a nuclear protein (hnRNP A2), which were detected only in cytoplasmic and nuclear fractions, respectively (Figure 7B). Although the nuclear signals of Dicer and Ago2 appears to be higher than the cytoplasmic signals, it does not represent the quantitative levels in the same number of cells, since ∼20% of nuclear fraction and ∼5% of cytoplasmic fraction were loaded for western analysis.
To examine if Dicer and Ago2 are present in the nucleolus where major pre-rRNA processing events occur, immunofluorescence staining was performed using antibodies against Dicer (Figure 7C, upper panel, green) or Ago2 (Figure 7C, lower panel, green). Both proteins are enriched in the cytoplasm, however, weaker but detectable signal can also be found in the nucleus. No nucleolar enrichment was found for either protein; instead, it appears that Ago2 was excluded from the nucleolus, since the nucleolar signal is weaker than the nucleoplasmic signal. However, Dicer appears evenly distributed in the nucleus and nucleolus, suggesting that little amount of Dicer may be present in this sub-nuclear compartment. Despite that Dicer and Ago2 were not found to be enriched in the nucleolus, here we confirmed that both proteins are present in the nucleus. This is consistent with the possible involvement of these proteins in pre-rRNA processing, since maturation of 5.8S rRNA may occur in the nucleus.
DISCUSSION
In this study, we show that depletion of Drosha or Dicer impairs pre-rRNA processing. In addition to their roles in miRNA biogenesis, accumulating evidence suggests that these proteins may have multiple functions in other cellular processes, especially in RNA metabolism. The involvement of Drosha in pre-rRNA processing has been identified previously by our group (6). Recent studies showed that the microprocessor, Drosha/DGCR8 complex, can cleave the 5′ UTR of DGCR8 mRNA, thus negatively regulating DGCR8 expression (29,30). In addition, small RNAs similar in size to miRNAs that are derived from abundant non-coding RNAs, such as rRNA, small nuclear RNA, snoRNA, as well as tRNAs, are often found in cells (31,32). These small RNAs are generally considered to be degradation products; however, a recent study showed that processing of such a small RNA from a tRNA depends on Dicer (31). Other studies have also demonstrated that production of miRNAs derived from snoRNAs requires Dicer activity (33,34), indicating that Dicer may be involved in the metabolism of many types of cellular RNAs. Our finding that depletion of Drosha or Dicer impairs pre-rRNA processing suggests new functions for these versatile RISC pathway proteins. In addition, since these RISC pathway proteins are not found in budding yeast, the related pre-rRNA processing events may thus be different between human and yeast, although major processing steps are conserved.
The defects in pre-rRNA processing following treatment with Drosha or Dicer ASOs appear to be specific to loss of expression of the targeted genes. First, transfection of control ASOs had no effect. Second, different ASOs targeting the same mRNAs produce similar results, arguing against the possibility of off-target effects (35). Third, pre-rRNA can be co-selected with antibodies against these proteins, providing further evidence of a direct effect, although it is possible that the association of pre-rRNA with Ago2 and Dicer is actually mediated by each other, since these two proteins can be present in the same RNP complex (36). Finally, we have found that the levels of known processing snoRNAs were not changed by depletion of these proteins, arguing against the possibility that processing defects stem from impaired snoRNA production. It is unlikely that the processing defects caused by loss of Drosha or Dicer are solely secondary in response to impaired RISC function, since knockout of DGCR8, a Drosha partner protein required for miRNA biogenesis, has no effect on pre-rRNA processing (37). Importantly, we showed that depletion of Ago2, the core component of functional RISC complex, caused a different and much weaker processing defect than loss of Drosha or Dicer. Although it is possible that the presence of other Ago proteins (Ago1, Ago3 and Ago4) may partially compensate for loss of Ago2, this possibility seems unlikely, since co-depletion of Ago2 along with one of the other three Ago proteins did not increase the defects (Supplementary Figure S7). Together, these observations make it difficult to believe that the processing defects caused by depletion of Dicer or Drosha are merely due to impaired miRNA or snoRNA biogenesis, although we cannot completely rule out such a possibility. However, it is possible that Ago2 modulates pre-rRNA processing via the RISC pathway (see below).
Although strong accumulation of pre-5.8S rRNA was observed in Drosha- or Dicer-depleted cells, the processing defect does not appear to be severe, since the levels of mature rRNAs were not affected. This is consistent with the observation from a previous study, which showed that the level of mature rRNAs was not changed in Drosha deficient mouse T cells (38). Thus, the processing steps impaired by depletion of Drosha or Dicer may not be essential, and production of 5.8S rRNA can be achieved through unknown, alternative processing pathways, which often occur during pre-rRNA processing (8,39,40). Another possibility is that Drosha and Dicer may be required for degradation of excess amounts of pre-rRNA species. Depletion of these proteins then could lead to accumulation of different precursors that otherwise are normally degraded.
Despite the fact that multiple endonucleolytic cleavage sites in pre-rRNA have been identified, only the MRP complex and Rnt1 have been identified as endonucleases responsible for certain cleavage reactions (8,15). Here, we found that loss of Drosha and Dicer in human cells leads to accumulation of pre-5.8S rRNA species containing extensions at 5′, 3′ or both ends. In addition, Dicer is able to cleave pre-rRNA in vitro to produce distinct products, suggesting the existence of Dicer preferred cleavage sites/structures (Supplementary Figure S8). However, the two RNase III proteins appear not to be directly required for normal endonucleolytic cleavages of pre-rRNA, since no reduction of normal cleavage products was observed in Dicer- or Drosha-depleted cells (Figure 3), and pre-rRNA processing is not fully blocked by loss of these proteins. Thus, if Drosha and Dicer are involved in cleavage of pre-rRNA, upon depletion, other endonuclease(s) must compensate for the loss of these proteins, resulting in a sub-optimal kinetics with a slower processing rate. It is also possible that the presence of Drosha and Dicer alters the conformation of pre-rRNA, directly or indirectly, to facilitate the activities of enzymes absolutely required for pre-rRNA processing. Finally, we cannot completely rule out the possibility that the pre-rRNA processing detects are caused by impaired RISC pathway that misregulated other protein factor(s) required for pre-rRNA processing. Determining the underlying mechanisms remains a challenge for future studies.
The finding that loss of Ago2 moderately impairs pre-rRNA processing is interesting. We showed that the weaker defect in Ago2-depleted cells than in Dicer- or Drosha-depleted cells is not due to compensatory effects of other Ago proteins. The processing defects mimic those caused by loss of the exosome components, leading to accumulation of pre-5.8S rRNAs containing 3′ extensions only. Thus, it is possible that depletion of Ago2 may interfere with the function of the exosome, for example, by altering the structure of pre-ribosome thus changing the accessibility to the exosome. On the other hand, the involvement of Ago2 in pre-rRNA processing may be mediated by miRNAs (RISC pathway), which can either regulate gene(s) required for pre-rRNA processing, or directly regulate pre-rRNA processing. Recent studies have identified miRNAs in the nucleolus in rat cells with unknown function (34,41), and at least one miRNA (miRNA-206) was found to localize in the nucleolus, especially the granular center, where late stage of pre-rRNA processing occurs (42). Interestingly, we found that 62 potential miRNA interaction sites exist in the 5′ 400 nt sequence of human ITS2 (data not shown). Thus, it remains an interesting question as whether some of these miRNAs modulate pre-rRNA processing.
Although Dicer and Ago2 are generally believed to be cytoplasmic proteins, here we showed that these two proteins can be clearly detected in both cytoplasmic and nuclear fractions in HeLa cells. This nuclear localization is consistent with their potential involvements in pre-rRNA processing. Nuclear localization of Dicer has been proposed in a previous study based on immunofluorescence staining (43), and the nuclear functions of Ago proteins (RISC) have been demonstrated in various organisms, including fungi, plants, Drosophila and Caenorhabditis elegans (44). Recent studies also showed nuclear functions of the RISC pathway in mammals. For example, the nuclear RNAs, 7SK RNA and U6, can be down-regulated by siRNAs in human cells (45), suggesting that Ago2, the cleavage competent Ago protein in mammals, is present in the nucleus. Indeed, two recent studies have shown that Ago2 was evenly distributed between the cytoplasm and nucleus in human cells, as detected by immunofluorescence (28,46). Based on the fact that biogenesis of most RNAs takes place in the nucleus, the nuclear localization of these RISC pathway proteins may imply their additional functions. Determining the potential roles of RISC pathway in the metabolism of other types of RNAs remains intriguing questions for future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: ISIS Pharmaceuticals.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Timothy A. Vickers, Walt F. Lima, Hongjiang Wu, Frank Bennet, Andy Watt, Erich Koller, Frank Rigo and Sue Freier for stimulating discussions and help.
REFERENCES
- 1.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Bio.l. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Mallory AC, Bouche N. MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci. 2008;13:359–367. doi: 10.1016/j.tplants.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Seitz H, Zamore PD. Rethinking the microprocessor. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 8.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 9.Nazar RN. Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life. 2004;56:457–465. doi: 10.1080/15216540400010867. [DOI] [PubMed] [Google Scholar]
- 10.Gerbi SA, Borovjagin AV. In: The Nucleolus. Olson MOJ, editor. Texas: Landes Bioscience, Georgetown; 2004. pp. 170–198. [Google Scholar]
- 11.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faber AW, Vos HR, Vos JC, Raue HA. 5′-end formation of yeast 5.8SL rRNA is an endonucleolytic event. Biochem. Biophys. Res. Commun. 2006;345:796–802. doi: 10.1016/j.bbrc.2006.04.166. [DOI] [PubMed] [Google Scholar]
- 13.Schilders G, Raijmakers R, Raats JM, Pruijn GJ. MPP6 is an exosome–associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers TA, Lima WF, Wu H, Nichols JG, Linsley PS, Crooke ST. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 ‘Slicer’ independent and can be mediated by Ago1. Nucleic Acids Res. 2009;37:6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Liang XH, Liu L, Michaeli S. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 2001;276:40313–40318. doi: 10.1074/jbc.M104488200. [DOI] [PubMed] [Google Scholar]
- 19.Bennett CF. In: Antisense Drug Technology - Principles, Strategies, and Applicaitons. Crooke ST, editor. New York: CRC Press, Boca Raton, London; 2006. pp. 273–304. [Google Scholar]
- 20.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand E, Fournier MJ. In: The Nucleolus. Olson MOJ, editor. Georgetown, TX: Landes Bioscience Publishing; 2004. pp. 225–261. [Google Scholar]
- 22.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam YW, Trinkle-Mulcahy L, Lamond AI. The nucleolus. J. Cell Sci. 2005;118:1335–1337. doi: 10.1242/jcs.01736. [DOI] [PubMed] [Google Scholar]
- 24.Thomson E, Tollervey D. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010;30:976–984. doi: 10.1128/MCB.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 26.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 29.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triboulet R, Chang HM, Lapierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 33.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224, 1–10. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svoboda P. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr. Opin. Mol. Ther. 2007;9:248–257. [PubMed] [Google Scholar]
- 36.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 40.Torchet C, Hermann-Le Denmat S. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA. 2000;6:1498–1508. doi: 10.1017/s1355838200000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl Acad. Sci. USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels SM, Melendez-Pena CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Laine S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 45.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 46.Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.