Abstract
Single-stranded antisense oligonucleotides (SSOs) are used to modulate the expression of genes in animal models and are being investigated as potential therapeutics. To better understand why synthetic SSOs accumulate in the same intracellular location as the target RNA, we have isolated a novel mouse hepatocellular SV40 large T-antigen carcinoma cell line, MHT that maintains the ability to efficiently take up SSOs over several years in culture. Sequence-specific antisense effects are demonstrated at low nanomolar concentrations. SSO accumulation into cells is both time and concentration dependent. At least two distinct cellular pathways are responsible for SSO accumulation in cells: a non-productive pathway resulting in accumulation in lysosomes, and a functional uptake pathway in which the SSO gains access to the targeted RNA. We demonstrate that functional uptake, as defined by a sequence-specific reduction in target mRNA, is inhibited by brefeldin A and chloroquine. Functional uptake is blocked by siRNA inhibitors of the adaptor protein AP2M1, but not by clathrin or caveolin. Furthermore, we document that treatment of mice with an AP2M1 siRNA blocks functional uptake into liver tissue. Functional uptake of SSO appears to be mediated by a novel clathrin- and caveolin-independent endocytotic process.
INTRODUCTION
Antisense oligonucleotides (ASOs), designed to selectively bind to a targeted RNA through Watson–Crick base pairing, are widely used to modulate gene expression. ASOs can be designed to modulate RNA function by multiple mechanisms, with the siRNA and RNase H mechanisms most commonly exploited (1). In both cases, the ASO, once bound to the target RNA, promotes degradation of the RNA through endogenous nucleases, Ago 2 and RNase H, respectively. ASOs designed to work through the RNase H pathway are single-stranded oligonucleotides (SSO) that bind to the target RNA, recruiting the cellular enzyme RNase H1, which then catalyzes the cleavage of the targeted RNA (2,3). Chemically modified SSOs, delivered systemically, rapidly distribute out of plasma and are taken up by cells in tissues, demonstrating robust antisense effects without the need for formulations (4–9). Though the mechanisms for SSO distribution from the central compartment into tissues is poorly understood, recent studies demonstrate at least two distinct pathways are operant by which SSO accumulate in liver cells. We refer to these as a non-productive uptake pathway and productive uptake pathway. The productive uptake pathway, which delivers SSO to the target RNA cellular compartment, accounts for <20% of the total SSO delivered to liver tissue (10). The non-productive uptake pathway, accounting for the bulk SSO accumulating in cells is saturable, and the SSO does not appear to have access to the target RNA.
Most mammalian cells will readily accumulate SSOs, so the lack of antisense effects cannot be attributed to the lack of transport into cells (11–14). However, in most cases, this accumulation in cells does not appear to be functional without transfection agents (15–17). Primary keratinocytes and rodent hepatocytes functionally accumulate SSOs when added to the growth media (18,19). However, functional uptake is lost in primary hepatocytes after 24–36 h in culture, making it difficult to study the process. It has been reported that incubation of some cells with high concentrations of SSOs for several days results in functional uptake (17), but the timing of maximal antisense effects does not correlate with the results observed following systemic administration (20), bringing into question whether SSO uptake into these cultured cells is much less efficient than in liver hepatocytes, or conversely alternate pathway(s) for SSO uptake are being exploited in these cultured cells compared to pathways being utilized in liver hepatocytes.
We now describe a mouse hepatocellular carcinoma cell line that efficiently accumulates SSOs in a functional compartment and maintains this ability to efficiently accumulate SSOs for several years. Maximal antisense effects are achieved within 24 h of adding nanomolar concentrations of SSOs to the cell culture medium, consistent with maximal antisense effects observed in mouse liver (20). We have used this cell culture model to investigate the mechanisms by which SSOs accumulate in liver cells, demonstrating that both a functional and non-functional pathway exist. Both SSO uptake pathways appear to be endosomal with the functional uptake pathway being clathrin and caveolin independent, but dependent on the adaptor protein AP2M1. These effects were confirmed in mouse liver following systemic delivery. Thus, this cell culture model recapitulates many features of SSO uptake found in normal mouse liver and allows further characterization of the mechanisms of functional SSO accumulation in hepatocytes.
MATERIALS AND METHODS
Cells and reagents
MHT cells were cultured in DMEM supplemented with 10% fetal calf serum, streptomycin (0.1 µg/ml), and penicillin (100 U/ml). siRNA treatment was performed using Opti-MEM containing 5 µg/ml Lipofectamine 2000 at the indicated amount of siRNA for 4 h at 37°C, as described previously (21,22). Heparan sulfate, 10K dextran sulfate, chloroquine, methyl-b-cyclodextrin, Brefeldin A were purchased from Sigma (St. Louis, MI, USA).
Isolation of MHT cells
Cells were isolated from the liver of a transgenic mouse (∼3-month old) bearing SV40 large T-antigen gene under the CRP promoter (23). The liver was perfused with 0.5 mg/ml collagenase solution (24), and cells were placed in collagen coated 12-well plates (±collagen) with either DMEM containing 10% FBS or with William’s Medium E containing 10% FBS, 10 mM HEPES and 2 mM glutamine. Medium was changed every 3 days. Wells containing growing cells were expanded into 6-well plates. Distinct population of cells began rapidly growing after ∼1 month of culture, and SV40 mRNA expression was monitored in those cells. Two groups of cells, one from cells grown in Dulbeccos medium (DM), and one from cells grown in Williams Medium E (WE) were serially diluted and plated in 96-well collagen coated plates to obtain <1 cell per well. Cells were identified that grew under both growth conditions. Four clones from each growth condition (DM and WE) were selected, further expanded and frozen down. For the studies described in this article, we used clone 4 W.
Preparation of synthetic siRNA and transfection
Synthetic unmodified siRNAs were purchased from Dharmacon Research Inc. (Boulder, CO, USA). siRNA duplexes were formed according to the manufacturer’s instructions. The sequences are listed in Supplementary Table S1. Cells were transfected with siRNAs in Opti-MEM and 5 µg/ml Lipofectamine 2000 (Invitrogen) for 4 h. Medium was then changed to complete growth medium. The siRNA transfection was repeated the following day.
Oligonucleotide synthesis
Oligonucleotides were phosphorothioate-modified chimeric oligonucleotides composed of five 2′-O-(2-methoxy) ethyl modifications on both the 5′- and 3′-ends and 10 oligodeoxynucleotides in the center to support RNase H activity (Supplementary Table S1) (25). Oligonucleotides were prepared similar to that described previously (26) on either an Amersham AKTA 10 or AKTA 100 oligonucleotide synthesizer. Modifications from the reported procedure include: a decrease in the detritylation time to 1 min, as this step was closely monitored by UV analysis for complete release of the trityl group; phosphoramidite concentration was 0.1 M; 4,5-dicyanoimidazole catalyst was used at 0.7 M in the coupling step; 3-picoline was used instead of pyridine for the sulfurization step, and the time decreased from 3 to 2 min. The oligonucleotides were purified by ion-exchange chromatography on an AKTA Explorer and desalted by reverse phase high performance liquid chromatography (HPLC). Oligonucleotides were characterized by ion-pair-HPLC-MS analysis (IP-HPLC-MS) with an Agilent 1100 MSD system. The purities of the oligonucleotides were >90%. Fluorescent dyes were attached to oligonucleotides at the 5′-ends using either fluorescein or Cy3 phosphoramides (Glen Research).
Taqman RT–PCR
Total mRNA was isolated using a QIAGEN RNAeasy kit (QIAGEN, Valencia, CA, USA). Reduction of target mRNA expression was determined by real time RT–PCR (27) using StepOne RT–PCR machines (Applied Biosystems). The sequences used in the RT–PCR reaction are listed in Supplementary Table S1. The expression data was normalized to ribogreen (Invitrogen). Data are mean values ±SDs of three replicates.
Quantitation of SSOs from cell lysates
Following treatment with SR-B1 SSO, cells were tryspinized, counted, and lysed in 300 μl RIPA buffer (150 mM NaCl, 1.0% T X-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris, pH 8.0). A calibration curve was prepared in control RIPA cell lysate ranging from 0.01 to 1 μg SR-B1 SSO. Half microgram of a 27-mer control SSO was added as an internal standard (IS) to cell lysates and the calibration standards. SSO was extracted from cell lysates and calibration standards using a liquid–liquid extraction with phenol /chloroform (25,24) followed by solid-phase extraction of the aqueous phase using phenyl-functionalized silica sorbent (Biotage, Uppsala, Sweden). Extracts were dried down using a warm-air evaporator and reconstituted in 100 μl 4 M urea, 25 mM EDTA. Samples were analyzed by LC/MS using a modification of a previously described method (28). Briefly, separation was accomplished using an 1100 HPLCMS system (Agilent Technologies, Wilmington, DE, USA) consisting of a quaternary pump, UV detector, a column oven, an autosampler and a single quadrupole mass spectrometer. Samples were injected on an Xbridge OST C18 column (2.1 mm × 50 mm, 2.5 μm particles; Waters, Milford, MA, USA) equipped with a SecurityGuard C18 guard column (Phenomenex, Torrance, CA, USA). The columns were maintained at 55°C. Tributylammonium acetate buffer (5 mM) and acetonitrile were used as the mobile phase at a flow rate of 0.3 ml/min. Acetonitrile was increased (gradient) from 20 to 70% over 11 min. Mass measurements were made online using a single quadrupole mass spectrometer and selected ion monitoring (SIM) of masses corresponding to the −4 charge state of full-length SR-B1 SSO and −5 charge state of the IS. Peak areas from extracted ion chromatograms were determined for SR-B1 SSO and IS and a trendline established (R2 = 0.99) using the calibration standards, plotting microgram SR-B1 SSO against the ratio of the peak areas SRB-1 SSO: IS. Total microgram SSO in cell lysates was determined using the established trendline and reported as molecules/cell, where molecules/cell = Avogadro constant * (total microgram SR-B1 SSO/cell number)/(molecular mass SR-B1 SSO).
Western blotting
Cells were lysed in RIPA lysis buffer. Equal amounts of protein were resolved on a SDS–PAGE gel and transferred to membranes. Proteins were detected using AP2M1 antibodies from Abcam (Cambridge, MA, USA) and SR-B1 antibodies from Novus (Littleton, CO, USA). Secondary antibodies (Lincoln, NE, USA) were conjugated to IR800. Blots were scanned using Odyssey from LI-COR.
Flow Cytometry
A fluorescein-labeled SSO was added to MHT cells with or without competitors for 24 h. Cells were trypsinized, resuspended in PBS with 5 mM EDTA and analyzed on FACS Calibur. Transferrin-AF488 (50 µg/ml) was added to cells for 7 h before trypsinization and analysis by flow cytometry.
Immunofluorescence
Cells were grown in glass-bottom dishes (MatTek). Cells were washed three times with 1× PBS, fixed at room temperature for 15 min with 4% formaldehyde and permeabilized for 5 min with 0.05% Saponin in 1× PBS. Cells were then incubated with 1× PBS with 0.05% Saponin containing a rabbit polyclonal antibody (6653) that recognizes phosphorothioate backbone containing oligonucleotides (1:200) or a rat anti-mouse LAMP1 antibody (1:1000, clone 1D4B, BD, Bioscience). After three washes (5 min each) with 1× PBS, cells were incubated for 1 h with PBS containing secondary antibodies against rabbit and mouse. After three washes, slides were mounted with Dapi Fluoromount G (Southern Biotech). Cells were imaged with a confocal microscope (Olympus, Fluoview 1000) and images were processed using software FV10-ASW 2.1. In some experiments the localization of the fluorescent labeled SSO was determined in unfixed cells using the confocal Olympus Fluoview 1000 or the Leica DM4000 B upright microscope.
siRNA formulation into LNP
Stock solutions of lipidoid 98N12-5 (1)·4HCl, cholesterol and mPEG2000-DMG MW 2660 (synthesized by Alnylam) were prepared in ethanol and mixed to yield a molar ratio of 42:48:10 (29). siRNA was incorporated in the nanoparticles at 1:7.5 (wt:wt) siRNA:total lipids. Resulting particles had a mean particle diameter of ∼50 nm and siRNA entrapment efficiency >95%.
In vivo mouse dosing
Balb/C male mice were purchased from Charles River Laboratories (Wilmington, MA, USA). siRNA were formulated with LNP01 and housed five per cage in box cages. Mice were maintained on feed and water ad libitum. All animal studies were conducted utilizing protocols and methods approved by the Institutional Animal Care and Use Committee (IACUC) and carried out in accordance with the Guide for the Care and Use of Laboratory Animals adopted and promulgated by the US National Institutes of Health. siRNA were formulated with LNP01. On day 1 and 7, 10 mg/kg AP2M1 and luciferase siRNA or PBS were administered with intravenous (i.v.) injection (tail vein). On day 14, 20 mg/kg SR-B1 SSO (ISIS 353382) was administered as a subcutaneous bolus injection. On day 17, mice were sacrificed. Liver were collected at sacrifice, 72 h after dosing was completed. A portion of each liver was homogenized in guanidine isothiocyanate buffer for mRNA extraction and RT–PCR analysis. The remaining liver was flash frozen and stored at −70°C until assayed for protein reduction.
RESULTS
A mouse hepatocellular carcinoma cell line demonstrates specific antisense effects by free uptake
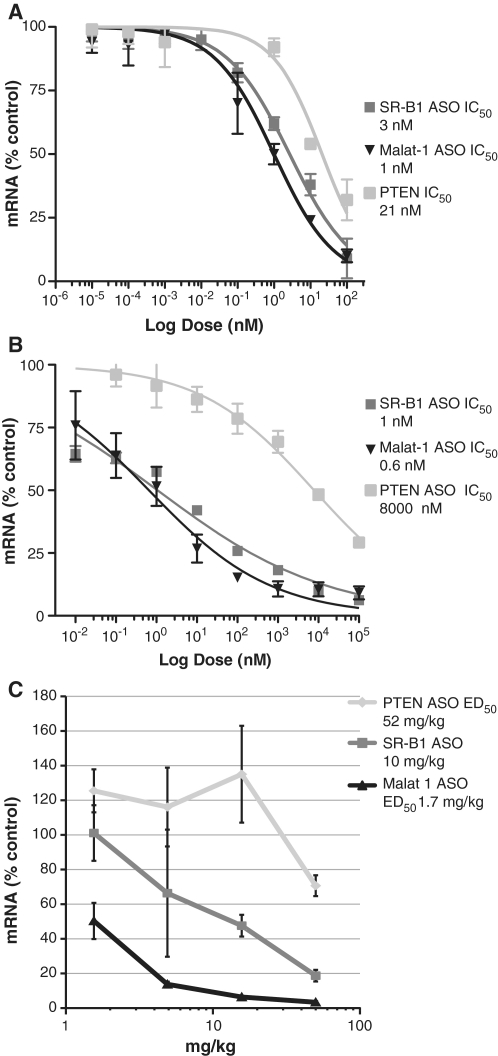
A stable mouse cell line, called MHT, was isolated from a hepatocellular carcinoma tumor which developed in a transgenic mouse expressing SV40 large T-antigen under the CRP promoter (23). To characterize the uptake of SSO, MHT cells were treated with 2′-MOE modified SSOs that were designed to work through RNase H1 (Supplementary Table S1) targeting the nuclear retained non-coding RNA, Malat1 (30), scavenger receptor B1 (SR-B1) and phosphatase and tensin homolog (PTEN) (5) in the presence or absence of a cationic lipid. The IC50 values for the Malat1, SR-B1 and PTEN SSO were 1, 3 and 21 nM, respectively, using Lipofectin reagent (Figure 1A). In the absence of cationic lipid, each of the SSOs produced a dose-dependent reduction of their respective targets with IC50 values for the Malat1, SR-B1 and PTEN SSO of 0.6, 1 and 8000 nM, respectively, when incubated for 48 h (Figure 1B). The cells have maintained their ability to efficiently accumulate SSO in a functional compartment for prolonged periods of time in culture, although we do find that over time in culture there is some loss in sensitivity. Therefore, we thaw an early passage of the cells after 20–25 passages. Mice were treated with a single injection of all three SSOs and effects on targeted gene expression in liver were determined (Figure 1C). The Malat1 and the SR-B1 SSOs exhibited an ED50 of 1.7 and 10 mg/kg, respectively, while the ED50 for the PTEN SSO was ∼52 mg/kg. The rank-order potency for the three different SSOs was the same by cationic lipid-mediated transfection, free uptake, or systemic administration to mice. The target reduction was sequence-specific and had no effect on the transcripts of the other two genes (Supplementary Figure S1).
Figure 1.
SSOs decrease SR-B1, PTEN and Malat1 expression in MHT cells and mouse liver. (A) MHT cells were transfected with SSOs using Lipofectin reagent for 4 h. Approximately 20 h after transfection, cells were harvested and the respective mRNA measured by real time RT–PCR. (B) SSOs were added to the growth medium, in the absence of cationic lipid for 48 h for free uptake. (C) Mice were given a single dose of SSOs (50, 16, 5 and 1.6 mg/kg). Seventy-two hours after dosing, animals were sacrificed and livers removed. Total RNA was extracted from the liver. mRNA levels for the respective genes was quantified using real-time RT–PCR. Mean values ± SDs (n = 5).
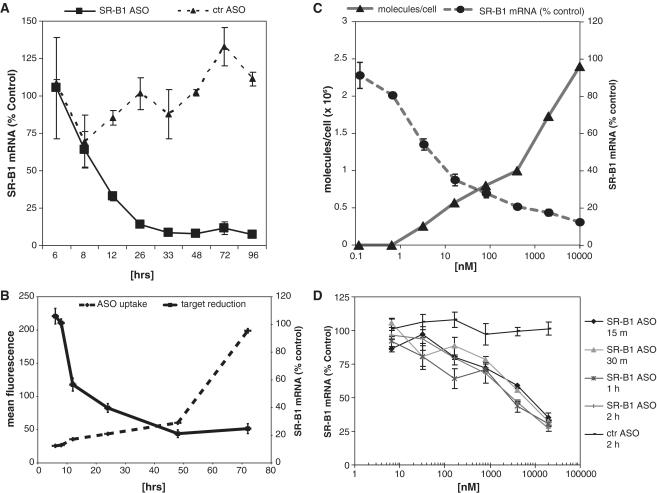
The maximal effects of 1 µM SR-B1 SSO on SR-B1 expression occurred ∼30 h after adding the SSO to the media (Figure 2A). Maximal effects of lower concentrations of SSO occurred at 48–72 h (data not shown). A control SSO failed to modulate SR-B1 expression, further demonstrating that the reduction in SR-B1 expression was sequence specific (Figure 2A). Attachment of the fluorescein dye to the 5′- or 3′-end of a SSO did not affect potency for reduction of the target RNA (data not shown), suggesting that the fluorescent dye did not interfere with functional uptake, hybridization to the target mRNA or RNase H cleavage. Using a fluorescein labeled SSO to quantitate cellular uptake, we demonstrate that cellular uptake was time-dependent and did not saturate over 70 h (Figure 2B). Similar to unlabeled SSO, the fluorescein-labeled SR-B1 SSO produced maximal effects on SR-B1 expression between 24 and 48 h after addition to the cells. We used mass spectrometry to more precisely quantitate the amount of unlabeled SSO in cells to correlate the number of SSO molecules in the cell with target mRNA reduction. Approximately 0.3 × 106 SSO molecules/cell were required to reduce SR-B1 mRNA by 50%, which was achieved by exposing the cells to 7 nM SR-B1 SSO (Figure 2C).
Figure 2.
Characterization of free uptake in MHT Cells. (A) The kinetics of SR-B1 SSO effects were determined by incubating cells in the presence of 1 µM SR-B1 SSO for the indicated times. (B) Target reduction and SSO uptake were compared using 10 µM fluorescein-conjugated SSO for the indicated times. Cell accumulation of SSO was determined by flow cytometry using the fluorescein labeled SSO. (C) Dose-dependent effects of SSO. The effects of the SR-B1 SSO on SR-B1 expression was determined by incubating MHT cells with increasing concentration of SSO for 24 h. The number of SSO molecules per cell was determined by mass spectrometry. (D) Kinetics of SSO association with MHT cells. MHT cells were incubated with increasing concentrations of SSOs for 15 min to 2 h, after which the cells were washed and incubated an additional 24 h before measuring target reduction. Mean values and SD were measured from triplicate samples.
To characterize the kinetics of cell association, we incubated the MHT cells for 15 min to 2 h in the presence of 6 nM to 20 µM SR-B1 SSO to allow the SSO to bind to the cell surface and then washed the cells to remove free SSO and incubated for an additional 24 h. The potency was decreased compared to cells that were continually exposed to SSO for 24 h (Figure 2D). However, incubation of cells with SSO for as short as 15 min resulted in a concentration dependent reduction in SR-B1 expression, suggesting that SSOs quickly associated with cells. Continued exposure of SSOs to cells up to 2 h resulted in only a modest increase in SR-B1 mRNA reduction compared to 15 min incubation (Figure 2D). These results suggest that the SSOs initially bind to the cell surface and then are internalized into the functional pool. Pretreatment of the cells with trypsin dramatically reduced the specific antisense effects, suggesting that the SSOs interact with proteins expressed on the surface of MHT cells (Supplementary Figure S2).
Functional uptake of SSO in MHT cells is chemistry dependent
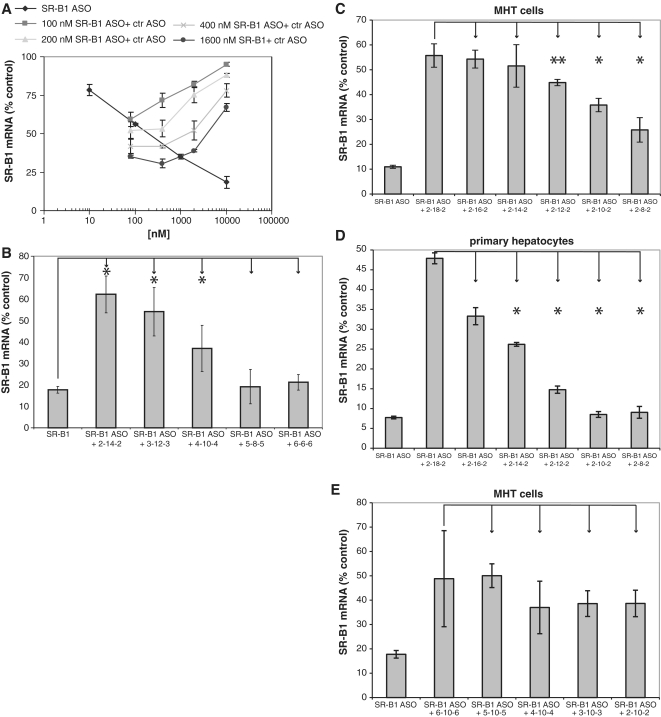
Although we were unable to demonstrate saturation of cellular uptake of oligonucleotide at concentrations up to 10 µM, we reasoned that binding to the cell surface should be a saturable event, therefore we used a control SSO to compete for cell uptake into the functional pool (Figure 3A). The antisense effects of the SR-B1 SSO were attenuated in the presence of increasing concentrations of a control SSO (Figure 3A). The control SSO also decreased the amount of fluorescently labeled SSO associated with cells consistent with the loss in pharmacological activity (Supplementary Figure S3). SSOs have three major chemical pharmacophores: the heterocycle base, the pentose sugar and the phosphate internucleosidic linkage. To determine the chemical structures recognized by this functional uptake pathway, we used different chemically modified oligonucleotides as competitors for functional uptake. The data suggest that SSOs containing more DNA were better competitors for functional uptake. As an example, maintaining the same oligonucleotide length and varying the amount of 2′-MOE and DNA modification demonstrates that increasing the 2′-MOE content from 4 to 10 nt and conversely decreasing the amount of DNA in the SSO resulted in decreased competition for functional uptake (Figure 3B). Similarly, if the length of the SSO was varied by increasing the DNA gap size and maintaining two 2′-MOEs at each end, there was a good correlation between DNA content and ability to compete (Figure 3C and D). This was true for MHT cells and primary hepatocytes. Keeping the DNA gap constant, but increasing the length of the SSO by increasing the 2′-MOE wings, resulted in a similar degree of competition for the different length SSOs (Figure 3E), further demonstrating that DNA content is more important than length.
Figure 3.
Functional uptake in MHT Cells is saturable. (A) MHT cells were incubated with increasing concentrations of SR-B1 SSO alone or the indicated concentration of SR-B1 SSO and increasing concentration of control (competitor) SSO. Twenty-four hours after adding the SSO to the cells, reduction in SR-B1 mRNA was measured by qRT–PCR. (B) DNA containing competitor SSOs are effecting functional uptake of SR-B1 SSO. MHT cells were incubated with 1 µM SR-B1 SSO in the presence of 10 µM competitor SSOs with increasing gap length. The more DNA present in the gap, the better the competition. (C) MHT cells and primary hepatocytes (D) were incubated with 100 nM SR-B1 SSO alone or in the presence of 5 µM competitor SSO of different lengths. The competitor SSOs had 2 MOE nucleotides on the 5′- and 3′-ends to help protect against nuclease degradation. The length of the DNA in the gap was varied as indicated. Twenty-four hours after adding the SSOs to the cells, SR-B1 mRNA was measured with qRT–PCR. SSOs with more DNA content were found to be better competitors. (E) Effect of different length competitors on functional uptake of SR-B1 SSO. MHT cells were incubated with 1 µM of SR-B1 SSO in the presence of 1 µM competitor SSOs. The length of the SSOs was varied by increasing the MOE nucleotides on the 5′- and 3′ wings with the number of DNA nucleotides in the center gap held constant at 10 nt. Mean values and SD were measured from triplicate samples, *P < 0.01, **P < 0.05, unpaired Student’s t-test.
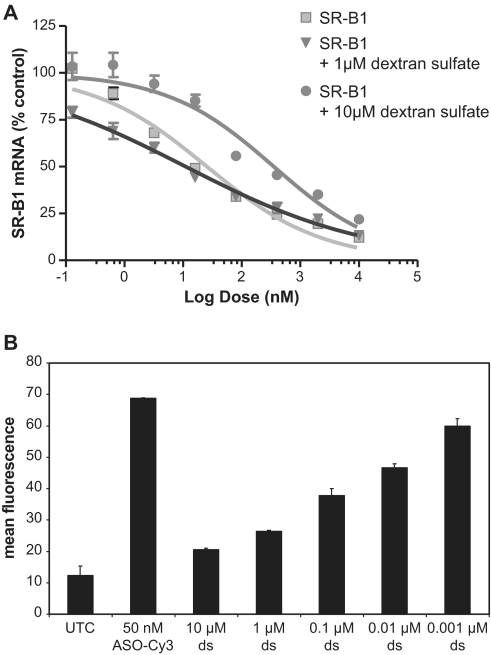
An abasic phosphorothioate deoxyribose oligomer (N = 20) failed to compete for antisense activity suggesting that the hydrophobic bases may contribute to cell uptake into the functional pool (data not shown). Also, unmodified and phosphorothioate modified duplexed RNAs in which the bases are base paired do not compete for uptake (data not shown). We investigated the ability of other polyanions to compete for functional cell uptake. Dextran sulfate at high concentrations attenuates the effect of the SSO, while low concentrations of dextran sulfate either had no effect or slightly enhanced the effect of the SSO (Figure 4A). Dextran sulfate decreased the amount of SSO associated with cells in a concentration dependent manner (Figure 4B). Suramin also weakly inhibited functional cell uptake (Supplementary Figure S4A), and heparan sulfate inhibited functional cell uptake only at high concentrations (Supplementary Figure S4B). These results demonstrate that functional uptake into MHT cells is not just due to polyanions interacting with membrane proteins, but appears to be also due to recognition of both the base and sugar.
Figure 4.
Dextran sulfate competes for bulk uptake and functional SSO uptake. Increasing doses of SR-B1 SSO was added to complete medium with or without the indicated concentrations of dextran sulfate for 24 h. (A) High concentrations (10 µM) dextran sulfate decrease the pharmacological effects of the SR-B1 SSO while 1 µM dextran sulfate does not inhibit the pharmacological effects of the SR-B1 SSO. (B) Dextran sulfate inhibits SSO accumulation in MHT cells. MHT cells were incubated with 50 nM of fluorescein labeled SR-B1 SSO in the presence of increasing concentrations of dextrans sulfate for 24 h. At the end of the incubation period cells were collected and the amount of fluorescein labeled SSO determined by flow cytometry. Mean values and SD were determined from triplicate samples.
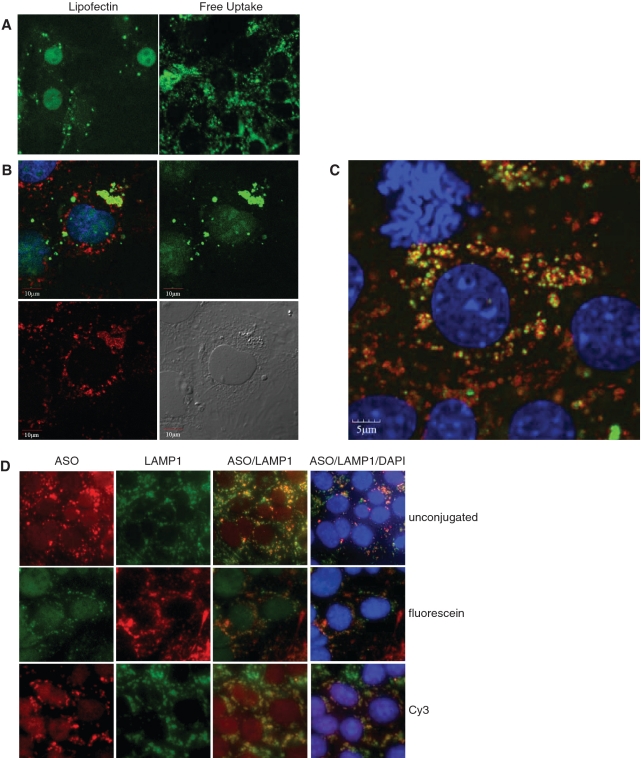
The bulk of SSOs accumulating in MHT cells localizes in vesicular structures
The localization of a fluorescein-labeled SSO was determined in the presence or absence of cationic lipid. Similar to previous results (15), we found that the SSO primarly localized in the nucleus when delivered into the cells with cationic lipids (Figure 5A). In the absence of cationic lipid, the highest concentration of SSO was found in punctuate peri-nuclear structures (Figure 5A–D). The same SSO distribution was seen in fixed and live cells (data not shown). We used antibodies to the lysosomal membrane protein 1 (LAMP1) to determine if the SSO co-localized in lysosomal structures. Two hours after adding the fluorescein-conjugated SSO to cells, SSOs localized to punctuate structures in the cytoplasm but did not co-localize with lysosomes (Figure 5B). However, after 24 h most of the fluorescent SSO co-localized with lysosomes (Figure 5C), suggesting that SSO accumulation in lysosomes is a slow process in these cells. Unconjugated SSO also localized in punctuate peri-nuclear structures as demonstrated using a polyclonal antibody that recognizes phosphorothioate modified SSO. The localization of the unlabeled SSO was similar to SSO labeled with fluorescein or Cy3 (Figure 5D).
Figure 5.
Localization of fluorescently labeled SSO in MHT cells. (A) Fluorescein labeled SSO was transfected into MHT cells using lipofectin reagent for 4 h followed by 20 h of incubation in the absence of the cationic lipid in complete medium (left panel) or delivered to cells by adding to complete medium for 24 h (right panel). SSO localization is shown in formaldehyde fixed cells. (B) MHT cells were incubated with 100 nM fluorescein labeled SR-B1 SSO (green) for 2 h and cells were fixed and permeabilized. Cells were stained with a LAMP1 antibody (red) to label lysosomal structures and visualized using confocal microscopy. Nuclei of the cells were counterstained with DAPI. (C) MHT cells were incubated with 100 nM fluorescein labeled SR-B1 SSO (green) for 24 h and cells were fixed and permeabilized. Cells were stained with a LAMP1 antibody (red) to label lysosomal structures and visualized using confocal microscopy. Nuclei of the cells were counterstained with DAPI (blue). (D) MHT cells were incubated with 100 nM unlabeled SR-B1 SSO (top row), fluorescein conjugated SR-B1 SSO or Cy3 labeled SR-B1 SSO for 24 h. At the end of the incubation period, cells were fixed and stained for LAMP1 using an unlabeled LAMP1 monoclonal antibody, followed by either a fluorescein (top and bottom rows) or Texas red (middle row) labeled goat anti-mouse antibody. Cells were counterstained with DAPI (blue dye). The unlabeled antibody, was detected using a rabbit polyclonal antibody that recognizes the phosphorothioate backbone present in the SSO followed by a Texas red goat anti-rabbit antibody.
Functional SSO uptake in MHT cells is an endocytotic process
We used pharmacological inhibitors of various endocytotic processes to further define the pathways which SSO accumulate in the functional compartment of cells. Chloroquine, an inhibitor of lysosome acidification, and Brefeldin A (BFA), a lactone antibiotic that blocks retrograde protein transport from the Golgi apparatus to the endoplasmic reticulum (31,32), both antagonized the antisense effects of the SR-B1 SSO (Supplementary Figure S5A and B). Although choroquine was very effective in blocking the antisense effects of the SSO, it had no effect on cellular uptake and, in fact, increased the total SSO in cells (data not shown), suggesting that choroquine did not block cell uptake but affected intracellular dynamics of the SSO. BFA slightly inhibited SSO uptake (data not shown). Incubation of MHT cells with methyl-β-cyclodextrin (MβCD), which depletes cells of cholesterol and 1 µg/ml lovostatin (33,34), modestly inhibited the antisense activity of the SR-B1 SSO (Supplementary Figure S5C). Finally, dynasore, a small molecule dynamin inhibitor, and a dynamin siRNA both failed to inhibit functional uptake (Supplementary Figure S6). In these experiments, dynamin mRNA levels were reduced by 80% compared to control siRNA-treated cells (data not shown).
To further define the pathways involved we chose to inhibit the expression of specific genes using siRNA. We chose to use siRNA for these experiments as they exploit a different molecular mechanism than SSOs and therefore should not result in competition for RNase H or other components required for the SSOs used in these studies, potentially complicating interpretation of the data. We screened a mouse siRNA library targeting membrane trafficking proteins for effects on antisense activity of the SR-B1 SSO (35). As a positive control we demonstrated that RNAse H1 siRNAs (Figure 6A) decreased the activity of the SSOs, indicating that they work through an RNAse H1 dependent mechanism. In this experiment the RNase H siRNAs reduce the amount of RNase H mRNA by 75%. The strongest and most reproducible hit for inhibiting the effects of the SR-B1 SSO was the adaptor protein AP2M1. Inhibition of AP2M1 with either a pool of four siRNAs or individual siRNAs in the pool attenuated the antisense effects of SR-B1 SSO (Figure 6B). Treatment with the AP2M1 siRNA pool resulted in an 80% inhibition of AP2M1 mRNA levels (Figure 6C). Inhibition of AP2M1 expression decreased fluorescein labeled SSO uptake by 50% (Supplementary Figure S7A). AP2M1 is an adaptor protein that recognizes a peptide sequence on the cytoplasmic face of cell membrane proteins linking the cell membrane cargo to clathrin (36,37). Surprisingly, inhibition of clathrin expression using siRNAs (80% inhibition, data not shown) had no effect on the SSO activity (Figure 6A and Supplementary Figure 6B), but did decrease the uptake of fluorescently labeled transferrin uptake in MHT cells, as did the AP2M1 siRNA and chlorpromazine (Figure 6D). These results demonstrate that functional uptake of SSO into MHT cells is AP2M1-dependent, but clathrin independent. Inhibition of caveolin expression also had no effect on functional SSO uptake (data not shown).
Figure 6.
SSO mediated functional uptake in MHT cells is AP2M1 dependent and clathrin independent. (A) MHT cells were treated with 25 nM control, AP2M1, clathrin or RNAse H1 siRNAs 2 days prior to adding 80 nM SR-B1 SSO. Cells were incubated an additional 24 h with the SR-B1 SSO and SR-B1 mRNA levels determined by qRT–PCR. SR-B1 mRNA reduction in AP2M1 siRNA-treated cells are inhibited compared to control siRNA-treated cells. (B and C) MHT cells were treated with 25 nM control siRNA, AP2M1 siRNA or no siRNA. Forty-eight hours later, increasing concentrations of the SR-B1 SSO were added to the cells in complete medium for an additional 24 h. Total cell RNA was isolated and SR-B1 (B) and AP2M1 (C) mRNAs were measured using qRT-PCR. (D) MHT cells were treated with 25 nM clathrin, AP2M1 or control siRNAs for 48 h, after which the cells were incubated with 100 nM fluorescently labeled transferrin-AF488 for 24 h. Uptake of Transferrin-AF488 was measured using flow cytometry. Cells were incubated with 10 µM chlorpromazine for 2 h prior to adding Transferrin-AF488 for 7 h. Mean values and SD were measured from triplicate samples, *P < 0.001, unpaired Student’s t-test.
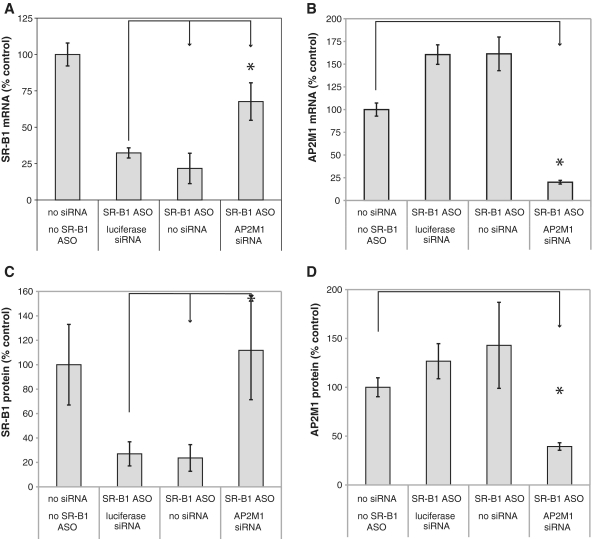
We extended these observations to mouse liver. AP2M1 or luciferase siRNAs were administered to mice on day 1 and 7 using a lipidoid nanoparticle (LNP) formulation (38). An unformulated SSO targeting SR-B1 was injected subcutaneously on day 14 and mice were sacrificed on day 17. As observed in MHT cells, inhibition of AP2M1 in mouse liver blunted the antisense effects of the SR-BI SSO while the luciferase siRNA had no effect on SR-B1 reduction (Figure 7A). The AP2M1 siRNA resulted in 80% reduction of mouse AP2M1 mRNA (Figure 7B). Inhibition of AP2M1 also inhibited SR-B1 protein reduction by the SR-B1 SSO (Figure 7C). The siRNA reduced AP2M1 protein by 60% (Figure 7D).
Figure 7.
Inhibition of AP2M1 expression in mouse liver attenuates SR-B1 target reduction. (A–D) Balb/C mice were treated with 10 mg/kg nanoparticle formulated siRNAs on days 1 and 7 before dosing 20 mg/kg of the unformulated SR-B1 SSO on day 14. Mice were euthanized on day 17 (3 days after treating with SSO) and livers removed. (A) Total RNA was isolated and SR-B1 mRNA quantitated using qRT-PCR. Mean values ± SDs (n = 5). Reduction of SR-B1 levels in AP2M1 siRNA-treated mice are inhibited compared to luciferase siRNA-treated mice. (B) AP2M1 mRNA was determined in liver samples treated as indicated using qRT-PCR. (C) SR-B1 and (D) AP2M1 protein levels were determined using western blotting. Mean values ± SDs (n = 5), *P < 0.05, unpaired Student’s t-test.
DISCUSSION
ASOs show promise as therapeutic agents, while also being widely used to functionalize gene targets (27). The chemistry of SSO drugs has evolved to improve potency, pharmacokinetic properties and safety. Although many cell types will accumulate SSOs in a functional pool, it is not well understood how polyanionic SSOs gain access to the cytoplasmic and/or nuclear compartments of the cell where they can bind to the target RNA. Understanding how SSOs traffic in cells can lead to more efficient design of SSOs and better dosing paradigms.
Unfortunately, there has been a paradox in how SSOs behave in cell culture and how they behave in the intact organism. For most cultured cells, transfection agents are required to produce robust antisense effects. In contrast, systemic dosing of SSOs results in antisense effects in a broad range of peripheral tissues and administration into the CSF provides access to a variety of neuronal cell types (21,39–43). We report the isolation of a cell line, MHT, from livers of a transgenic mouse expressing SV40 large T-antigen that maintain functional SSO uptake. Cellular uptake pathways appear to be similar in this cell line compared to the mechanisms of uptake in mice, supporting their use to gain a better understanding of SSO trafficking in cells. SSOs reduce their target RNAs at nanomolar concentrations, effects are observed within 24 h of administration and are similar to in vivo observations (10). The bulk SSO taken up by the cells is eventually found in lysosomes.
The uptake of SSO into mouse liver is time and concentration dependent as was the finding in MHT cells (10,20,44). We demonstrate that the bulk of SSO which accumulates in MHT cells does not appear to be in the productive uptake pathway as evidenced by the competition studies with dextran sulfate in which we can reduce the total amount of SSO in cells by 60% and not antisense activity. Non-productive uptake pathways seem to exist in animal tissues as well. Delivery of SSOs with slow infusion led to increased SSO concentration in liver compared to i.v. bolus administration that was expected (10). Unexpectedly, however, an increase in liver drug concentration due to slower infusion did not confer improved pharmacodynamics, perhaps suggesting that the low plasma concentration favored an uptake pathway that is, in part, a non-productive pathway. This was further supported by studies using a control SSO as a competitor, in which the competitor decreased total SSO concentration in liver by 80%, yet paradoxically increased pharmacodynamic effects (10).
Our results suggest that functional uptake is a vesicular uptake process, since it is blocked by choroquine, brefeldin A and the inhibitor of adaptor protein subunit AP2M1. Interestingly, functional uptake does not appear to be mediated by clathrin or caveolin, but is dependent on the adaptor protein AP2M1. Most endocytotic pathways described in the literature that are AP2 dependent are also clathrin dependent. In general, clathrin-independent pathways are described but are poorly characterized. The minor group human rhinoviruses bind low-density lipoprotein receptors and are internalized by classical clathrin-dependent endocytosis (45,46). However, the virus can enter cells through a clathrin-, caveolin- and flotillin-independent pathway (46,47).
Recently, functional free uptake of a 16-mer LNA gapmer SSO targeting bcl-2 was reported in several cell lines (17). Cells were plated at low density and exposed to high concentrations of SSOs over days to produce the antisense effects. Contrary to our observations, the authors report that LNA SSO is taken up much slower and maximal target reduction does not occur until 5 days after SSO exposure. They localized the LNA SSOs in P-bodies, while we failed to observe MOE SSOs in P-bodies (Supplementary Figure S8). The LNA modified SSO used in the study were designed to work through an RNase H dependent mechanism. As the authors failed to show that the LNA modified SSOs used in their study worked through a RISC dependent or RNase H mechanism, it is not clear if the localization to P-bodies is functional. We looked at the effect of LNA SSOs in the competition assay and we found that LNA compete in a similar manner as MOE-gapmers. It is possible that the 16-mer LNA gapmers could have different uptake abilities than the 20-mer MOE-gapmer. However, based upon our preliminary results in MHT cells using LNA and related structures we do not believe that to be the case. More likely scenarios are that the human cell lines used to study uptake of ASOs by Stein et al. (17) retain a similar functional uptake pathway as the MHT cells but it is much less efficient. Alternatively, the MHT cells used in our study have a distinct SSO uptake pathway not found in the cells used by Stein et al. (17). Additional studies are needed to determine if these are distinct pathways for functional uptake of SSOs.
We have identified AP2M1 as an important adaptor protein for free uptake of SSOs and subsequent target reduction in MHT cells and in mouse liver. These results help validate MHT cells as a cell culture model to study SSO uptake and trafficking. We have tested other cells like mouse embryo fibroblasts and H2.35 liver cells where inhibition of AP2M1 inhibited SSO mediated target reduction. This shows that AP2M1 plays a similar role in cells other than MHT cells. Additional studies are needed to identify the cell surface proteins that the SSOs interact with, how the adaptor protein facilitates SSO internalization and how SSO escape endosomal vesicles. However, these studies are an important first step in characterizing how SSOs gain access to target RNAs in the nucleus and/or cytoplasm of cells. A better understanding of how SSOs enter cells and how they reach their target RNA will aid in the design of more potent SSO drugs and more specific targeting.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Isis Pharmaceuticals Inc.
Conflict of interest statement. E. Koller, T.M. Vincent, A. Chappell and C.F. Bennett are employees of Isis Pharmaceuticals. S. De and M. Manoharan are employees of Alnylam Pharmaceuticals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tracy Reigle for help preparing the figures and Jon K. Bodnar for additional manuscript edits.
REFERENCES
- 1.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Ann. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 2.Cerritelli SM, Crouch RJ. Ribonucelase H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 4.Dean NM, McKay RA. Inhibition of protein kinase C-alpha expression in mice after systemic administration of phosphorothioate antisense oligodeoxynucleotides. Proc. Natl Acad. Sci. USA. 1994;91:11762–11766. doi: 10.1073/pnas.91.24.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler M, McKay RA, Popoff I, Gaarde W, Witchell D, Murray S, Dean NM, Bhanot S, Monia BP. Specific inhibition of PTEN expression reverses hyperglycemia in diabetic mice. Diabetes. 2002:1028–1034. doi: 10.2337/diabetes.51.4.1028. [DOI] [PubMed] [Google Scholar]
- 6.Chi KM, Eisenhauer E, Fazli L, Jones EC, Goldenberg SL, Powers J, Gleave ME. A Phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2′-methoxyethyl antisense to clusterin, in patients with localized prostate cancer prior to radical prostatectomy. J. Natl Cancer Inst. 2005;97:1287–1296. doi: 10.1093/jnci/dji252. [DOI] [PubMed] [Google Scholar]
- 7.Kastelein JJP, Wedel MK, Baker BF, Su J, Bradley JA, Yu RZ, Chuang E, Graham MJ, Crooke RM. Potent reduction of Apolipoprotein B and LDL cholesterol by an antisense inhibitor of Apolipoprotein B. Circulation. 2006;114:1729–1735. doi: 10.1161/CIRCULATIONAHA.105.606442. [DOI] [PubMed] [Google Scholar]
- 8.Sazani P, Gemignani F, Kang SH, Maier MA, Manoharan M, Persmark M, Bortner D, Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 9.Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, Dean NM. Modification of MyD88 mRNA splicing and inhibition of IL-1beta signaling in cell culture and in mice with a 2′-O-methoxyethyl-modified oligonucleotide. J. Immunol. 2006;176:3652–3661. doi: 10.4049/jimmunol.176.6.3652. [DOI] [PubMed] [Google Scholar]
- 10.Geary RS, Wancewicz E, Matson J, Pearce M, Siwkowski A, Swayze E, Bennett F. Effect of dose and plasma concentration on liver uptake and pharmacologic activity of a 2′-methoxyethyl modified chimeric antisense oligonucleotide targeting PTEN. Biochem. Pharmacol. 2009;78:284–291. doi: 10.1016/j.bcp.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Loke SL, Stein CA, Zhang XH, Mori K, Nakanishi M, Subasinghe C, Cohen JS, Neckers LM. Characterization of oligonucleotide transport into living cells. Proc. Natl Acad. Sci. USA. 1989;86:3474–3478. doi: 10.1073/pnas.86.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crooke RM, Graham MJ, Cooke ME, Crooke ST. In vitro pharmacokinetics of phosphorothioate antisense oligonucleotides. J. Pharmacol. Exp. Ther. 1995;275:462–473. [PubMed] [Google Scholar]
- 13.Krieg AM. Uptake and efficacy of phosphodiester and modified antisense oligonucleotides in primary cell cultures. Clin.Chem. 1993;39:710–712. [Google Scholar]
- 14.Iversen PL, Zhu S, Meyer A, Zon G. Cellular Uptake and Subcellular Distribution of Phosphorothioate Oligonucleotides into Cultured Cells. Antisense Res. Dev. 1992;2:211–222. doi: 10.1089/ard.1992.2.211. [DOI] [PubMed] [Google Scholar]
- 15.Bennett CF, Chiang MY, Chan H, Shoemaker JEE, Mirabelli CK. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol. Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- 16.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, Worm J, Hedtjarn M, Souleimanian N, Miller P, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nestle FO, Mitra RS, Bennett CF, Chan H, Nickoloff BJ. Cationic lipid is not required for uptake and selective inhibitory activity of ICAM-1 phosphorothioate antisense oligonucleotides in keratinocytes. J. Invest. Dermatol. 1994;103:569–575. doi: 10.1111/1523-1747.ep12396876. [DOI] [PubMed] [Google Scholar]
- 19.Giachetti C, Chin DJ. Increased oligonucleotide permeability in keratinocytes of artificial skin correlates with differentiation and altered membrane function. J. Invest. Dermatol. 1996;107:256–262. doi: 10.1111/1523-1747.ep12329755. [DOI] [PubMed] [Google Scholar]
- 20.Yu RZ, Zhang H, Geary RS, Graham M, Masarjian L, Lemonidis K, Crooke R, Dean NM, Levin AA. Pharmacokinetics and pharmacodynamics of an antisense phosphorothioate oligonucleotide targeting Fas mRNA in mice. J. Pharmacol. Exp. Ther. 2001;296:388–395. [PubMed] [Google Scholar]
- 21.Dean NM, McKay R, Condon TP, Bennett CF. Inhibition of protein kinase C-alpha expression in human A549 cells by antisense oligonucleotides inhibits induction of intercellular adhesion molecule 1 (ICAM-1) mRNA by phorbol esters. J. Biol. Chem. 1994;269:16416–16424. [PubMed] [Google Scholar]
- 22.Dean NM, Griffey RH. Identification and characterization of second-generation antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 1997;7:229–233. doi: 10.1089/oli.1.1997.7.229. [DOI] [PubMed] [Google Scholar]
- 23.Ruther U, Woodroofe C, Fattori E, Ciliberto G. Inducible formation of liver tumors in transgenic mice. Oncogene. 1993;8:87–93. [PubMed] [Google Scholar]
- 24.Neufeld DS. Isolation of rat liver hepatocytes. Methods Mol. Biol. 1997;75:145–151. doi: 10.1385/0-89603-441-0:145. [DOI] [PubMed] [Google Scholar]
- 25.McKay RA, Miraglia LJ, Cummins LL, Owens SR, Sasmor H, Dean NM. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 26.Ravikumar VT, Andrade M, Carty RL, Dan A, Barone S. Development of siRNA for therapeutics: efficient synthesis of phosphorothioate RNA utilizing phenylacetyl disulfide (PADS) Bioorg. Med. Chem. Lett. 2006;16:2513–2517. doi: 10.1016/j.bmcl.2006.01.081. [DOI] [PubMed] [Google Scholar]
- 27.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 28.Gaus HJ, Owens SR, Winniman M, Cooper S, Cummins LL. On-line HPLC electrospray mass spectrometry of phosphorothioate oligonucleotide metabolites. Anal. Chem. 1997;69:313–319. doi: 10.1021/ac960557q. [DOI] [PubMed] [Google Scholar]
- 29.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 32.Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 33.Neufeld EB, Cooney AM, Pitha J, Dawidowicz EA, Dwyer NK, Pentchev PG, Blanchette-Mackie EJ. Intracellular trafficking of cholesterol monitored with a cyclodextrin. J. Biol. Chem. 1996;271:21604–21613. doi: 10.1074/jbc.271.35.21604. [DOI] [PubMed] [Google Scholar]
- 34.Alberts AW, Chen J, Kuron G, Hunt V, Huff J, Hoffman C, Rothrock J, Lopez M, Joshua H, Harris E, et al. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl Acad. Sci. USA. 1980;77:3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolokoltsov AA, Deniger D, Fleming EH, Roberts NJ, Jr, Karpilow JM, Davey RA. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 2007;81:7786–7800. doi: 10.1128/JVI.02780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- 37.Wieffer M, Maritzen T, Haucke V. SnapShot: endocytic trafficking. Cell. 2009;137:382. doi: 10.1016/j.cell.2009.04.012. e381–e383. [DOI] [PubMed] [Google Scholar]
- 38.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl Acad. Sci. USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett CF, Chiang MY, Chan H, Shoemaker JE, Mirabelli CK. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol. Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- 40.Dean NM, McKay R. Inhibition of protein kinase C-alpha expression in mice after systemic administration of phosphorothioate antisense oligodeoxynucleotides. Proc. Natl Acad. Sci. USA. 1994;91:11762–11766. doi: 10.1073/pnas.91.24.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler M, McKay RA, Popoff IJ, Gaarde WA, Witchell D, Murray SF, Dean NM, Bhanot S, Monia BP. Specific inhibition of PTEN expression reverses hyperglycemia in diabetic mice. Diabetes. 2002;51:1028–1034. doi: 10.2337/diabetes.51.4.1028. [DOI] [PubMed] [Google Scholar]
- 42.Wahlestedt C. Antisense oligonucleotide strategies in neuropharmacology. Trends Pharmacol. Sci. 1994;15:42–46. doi: 10.1016/0165-6147(94)90107-4. [DOI] [PubMed] [Google Scholar]
- 43.Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsiger CS, Ward CM, McAlonis-Downes M, Wei H, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Cook J, Nickel J, Yu R, Stecker K, Myers K, Dean NM. Reduction of liver Fas expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nat. Biotechnol. 2000;18:862–867. doi: 10.1038/78475. [DOI] [PubMed] [Google Scholar]
- 45.Snyers L, Zwickl H, Blaas D. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J. Virol. 2003;77:5360–5369. doi: 10.1128/JVI.77.9.5360-5369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan AG, Pickl-Herk A, Gajdzik L, Marlovits TC, Fuchs R, Blaas D. Human rhinovirus 14 enters rhabdomyosarcoma cells expressing icam-1 by a clathrin-, caveolin-, and flotillin-independent pathway. J. Virol. 2010;84:3984–3992. doi: 10.1128/JVI.01693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayer N, Schober D, Huttinger M, Blaas D, Fuchs R. Inhibition of clathrin-dependent endocytosis has multiple effects on human rhinovirus serotype 2 cell entry. J. Biol. Chem. 2001;276:3952–3962. doi: 10.1074/jbc.M004722200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.