Abstract
Different populations suffer from different rates of obesity and type-2 diabetes (T2D). Little is known about the genetic or adaptive component, if any, that underlies these differences. Given the cultural, geographic, and dietary variation that accumulated among humans over the last 60,000 years, we examined whether loci identified by genome-wide association studies for these traits have been subject to recent selection pressures. Using genome-wide SNP data on 938 individuals in 53 populations from the Human Genome Diversity Panel, we compare population differentiation and haplotype patterns at these loci to the rest of the genome. Using an “expanding window” approach (100 to 1,600 kb) for the individual loci as well as the loci as ensembles, we find a high degree of differentiation for the ensemble of T2D loci. This differentiation is most pronounced for East Asians and sub-Saharan Africans, suggesting that these groups experienced natural selection at loci associated with T2D. Haplotype analysis suggests an excess of obesity loci with evidence of recent positive selection among South Asians and Europeans, compared to sub-Saharan Africans and Native Americans. We also identify individual loci that may have been subjected to natural selection, such as the T2D locus, HHEX, which displays both elevated differentiation and extended haplotype homozygosity in comparisons of East Asians with other groups. Our findings suggest that there is an evolutionary genetic basis for population differences in these traits, and we have identified potential group-specific genetic risk factors.
Keywords: obesity, type-2 diabetes, genetics, natural selection, population differentiation
Introduction
Obesity and type-2 diabetes (T2D) are major health concerns, and are of growing concern across most human populations (Deitel, 2003; Seidell, 2000). The rates, types and consequences of T2D and obesity have been found to differ between populations (Diamond, 2003; Goran, 2008; Haslam et al., 2005; Seidell, 2000; Wang et al., 2007), possibly reflecting genetic differences between these populations (Cheng et al., 2009; Fernandez et al., 2003; Tang et al., 2006; Williams et al., 2000). It is still unclear whether these putative genetic differences arose due to natural selection, and what specific genetic factors, if any, are responsible for these population differences.
Several evolutionary hypotheses have been proposed to explain the disproportionate burden of obesity and T2D among some populations (Benyshek et al., 2001; Benyshek et al., 2006; Diamond, 2003; Hancock et al., 2008; Neel, 1962; Wells, 2007). These range from environmentally induced epigenetic modifications, to longer term adaptation to different climates and/or subsistence modes. For instance, current-day genetic susceptibility to obesity and T2D is often attributed to specific selective pressures in our evolutionary past, favoring behavioral or physiological traits that buffered our ancestors from starvation in times of food shortage (Neel, 1962). Since the spread of modern humans to other parts of the globe, some groups have adopted sedentary agricultural lifestyles, whereas others have maintained a hunter-gatherer lifestyle. This along with other ecological factors (eg. climate, geography, diet) may have resulted in selection pressures in some populations for or against behavioral or physiological parameters associated with greater body weight and glucose regulation. Although it is often hypothesized that these population differences arose due to different selective pressures, there is very little genetic evidence to support this argument (Hindorff et al., 2009; Myles et al., 2008; Pickrell et al., 2009; Southam et al., 2009).
With the convergence of recent increases in genotyping capacity, the ability to conduct large-scale genome-wide association studies (GWAS), worldwide panels of DNA samples, and methodological advances enabling the detection of natural selection from genetic data, these hypotheses are becoming more easily testable. We therefore examined the worldwide pattern of differentiation and recent natural selection in the regions surrounding a total of 32 GWAS-identified risk alleles for obesity and T2D (16 each), using genome-wide SNP data on the individuals in the HGDP-CEPH panel of world-wide populations (Li et al., 2008). We hypothesize that humans have undergone selection pressures within the last 60,000 years, related to body weight regulation and blood glucose regulation, resulting in population differentiation and unusually extended haplotypes at these loci. We also hypothesize that these signatures of selection will be restricted to specific groups. By using several overlapping window sizes for the FST analysis (100 to 1,600 kb), we also hypothesize that if differentiation occurred in any given case, that the extent of differentiation will decay as the window size gets larger. This pattern can be expected because if selection is acting on or near the locus of interest, but not in nearby regions, then the pattern of between-population differentiation will be broken down by recombination at greater distances from the region under selection.
Methods
HGDP-CEPH populations and genotypes
We used the publicly available data from the HGDP/CEPH collection of 938 individuals from 53 different populations, genotyped at over 650,000 SNPs on the Illumina 650Y platform, and phased using fastPHASE (Li et al., 2008). Following recent publications on this dataset (Myles et al., 2008; Pickrell et al., 2009) we decided to group individual populations into seven broad geographical regions: Sub-Saharan Africa (n=102), Middle East (n=160), South Asia (n=200), Europe (n=156), East Asia (n=229), Oceania (n= 28), and America (n= 63). A listing of the populations in each group can be found in the Supplementary Material.
Obesity- and T2D-associated loci from GWAS studies
The set of loci utilized in this study was determined from published reviews on obesity and T2D GWASs (Florez, 2008; Hofker et al., 2009; O'Rahilly, 2009; Walley et al., 2009), and risk alleles were obtained from the online Catalog of Published Genome-Wide Association Studies (http://www.genome.gov/gwastudies/; accessed 10/25/2009). We examined a total of 28 variants in 16 obesity-associated loci and a total of 30 variants in 16 T2D-associated loci. Chimpanzee and Macaque reference alleles were obtained from the UCSC Genome browser (Kuhn et al., 2009). We examined the frequency of each obesity and T2D risk allele in each of the 7 population groups. A listing of the SNPs examined can be found in Supplementary Tables 1 and 2.
FST
FST partitions the total genetic variance into within- and between- population components, thereby quantifying the extent of population differentiation. An elevated FST at a given locus suggests that selection has driven differentiation between populations. Previous research has shown that single allele estimates of FST are highly variable and may therefore be unreliable indicators of differentiation at a genomic locus (Gardner et al., 2007; Weir et al., 2005). Therefore, we considered that a more conservative approach would be to calculate an average of FST values for all SNPs contained in varying window sizes, from 100 kb to 1.6 Mb, each centered on the obesity and T2D-associated SNP. In order to account for differences in recombination patterns across the genome, we also performed these analyses with cM (centimorgan) distance (0.1 to 1.6 cM) instead of kb distance. Although we are examining overlapping windows, we expect that by “zooming out” by a factor of two, we would see a slow decay in FST much as haplotype-based tests of selection test for a slow decay of haplotypes. In some cases, several risk SNPs have been identified in the same region, usually concentrated in a relatively small region (<50 kb), so we defined the window of interest based on the center-most SNP. For the 100 kb windows, the number of SNPs contained in a window ranges from 7 to 42 SNPs. To calculate FST, we used the method of Weir and Cockerham (Weir B.S. et al., 1984; Weir et al., 2002). We calculated a single global estimate of FST (based on all 7 population groups), as well as all 21 pair-wise estimates of FST. FST was not calculated for a SNP if it is monomorphic in the groups being compared. Negative values of FST were given a value of 0 since negative values are biologically meaningless.
To control for population-specific demographic effects on the genome, we compared the FST in the risk window to a null distribution of random windows. Random windows were chosen in the following way. For each risk locus, we randomly chose 1000 equally sized (in bp) windows along the same chromosome, with similar genic/non-genic content (± 10%), since FST tends to be slightly higher for genic SNPs (Coop et al., 2009). The genic/non-genic classification was performed according to the annotation provided by Sullivan et al. (https://slep.unc.edu/evidence/?tab=Downloads) which classifies a SNP based on whether it is in the transcribed region of a gene. SNP annotations were created using the TAMAL database (Hemminger et al., 2006) based chiefly on UCSC genome browser files (Hinrichs et al., 2006), HapMap (Altshuler et al., 2005), and dbSNP (Wheeler et al., 2006). For the 7-way FST as well as all 21 possible pair-wise FST values, we obtained percentile ranks of the obesity and T2D SNP-centered window, compared to the 1000 randomly centered windows. For testing each of the risk loci separately, we used a Bonferroni correction for multiple testing. A p-value cutoff of 0.0031 (0.9969th percentile) keeps the nominal type I error rate at 0.05. Since we are interested in examining the 16 obesity and 16 T2D regions as ensembles, as well as each risk region separately, we first averaged the 16 percentiles of the 7-way FST. In order to obtain a group-specific FST percentile for a given group, which we will refer to as GSFST, we averaged the FST percentiles of the six pair-wise comparisons that contain the group in question, and averaged the GSFST percentiles over all 16 loci to examine the loci as ensembles. In order to determine whether this average is an outlier, we simulated a null distribution by generating a random number between 0 and 1 representing the FST percentile rank for each locus and window size, and averaged these over 16 simulated loci. The percentile ranks of FST are distributed uniformally, hence the uniform distribution of random numbers between 0 and 1 is a suitable representation of the distribution of the FST percentile ranks. We repeated this averaging 10,000 times and determined the 95th, 97.5th, and 99th percentile cut-off values.
In order to estimate variances of the percentile ranks, we used bootstrapping to generate confidence intervals on the FST estimates and subsequent percentiles. We generated 1000 bootstrap samples, calculated FST for each, and examined the 95% confidence intervals for the GSFST percentiles. Due to computational limitations we restricted this analysis to a test case (HHEX) in order to get a general idea of the variance in our estimate.
REHH
Extended haplotype homozygosity (EHH) is defined as the probability that two randomly chosen chromosomes carrying the core haplotype of interest are identical by descent, and the relative EHH (REHH) is the factor by which EHH decays on the tested core haplotype compared to that of other core haplotypes combined (Sabeti et al., 2002). The REHH thus corrects for local variation in recombination rates. We obtained REHH values using Sweep software v1.1 (Sabeti et al., 2002), (downloaded from http://www.broadinstitute.org/mpg/sweep). Using the same phased haplotype data as above, we examined REHH at haplotypes containing the obesity risk SNP, and all haplotypes contained in the surrounding 400 kb region (200 kb in either direction) of each risk SNP. Core haplotypes were defined according to the definition of a haplotype block in Gabriel et al. (Gabriel et al., 2002), and REHH was measured 300 kb in either direction of each core. For each region and each population group, we compared the REHH in the risk SNP region to the entire chromosome on which the risk SNP resides, to determine if the candidate region contains haplotypes with exceptionally high REHH, binning by haplotype frequency. Empirical significance was therefore determined separately in each of 20 bins of core haplotype frequency. We only considered core haplotypes with frequency greater than 5%. We counted instances of extreme REHH (p<0.01, and p<0.001) values for each gene region and for each population group. We used a generalized version of Fisher's exact test to determine if there are differences among groups in the number of loci with at least one extreme REHH value (p<0.01). We then tested specific pair-wise comparisons, using a two-sided Fisher's exact test, and corrected for multiple testing using a False Discovery Rate method as implemented in SAS (Cary, NC). We also noted instances when a risk SNP was in the core, has an REHH above the 95th percentile, and whether it contained the risk or the non-risk allele.
XP-EHH
XP-EHH refers to a cross population comparison of EHH and is generally more powerful for detecting selection events that have gone to fixation (Sabeti et al., 2007). This method may therefore complement EHH and/or FST by detecting genomic regions that have experienced older selection events than those detectable by EHH. To determine XP-EHH for each obesity risk region, we used the HGDP Selection Browser (http://hgdp.uchicago.edu/cgi-bin/gbrowse/HGDP/). We entered the risk SNP, viewed the surrounding 500 kb window, and noted which population groups had XP-EHH that exceeded 2.5 (the −log10 of the p-value for a window centered at the SNP) at least once in that 500 kb region. This value reflects the degree to which a SNP is an outlier compared to the rest of the genome. As described by Pickrell et al. (Pickrell et al., 2009) since XP-EHH is the comparison of EHH between populations, the comparisons used were between Bantu (sub-Saharan Africa) and each of the non-African groups, whereas Europe was used as the reference for the Bantu group.
Results
Risk alleles
Out of 28 obesity risk alleles, 14 are derived since they differ from the reference allele in Chimpanzee (and Macaque, in most cases) (see Supplementary Table 1). Out of the 30 T2D risk alleles, 14 are derived (see Supplementary Table 2). We also find that the frequencies for some of the risk alleles differ substantially between populations, especially in FTO, SH2B1, NEGR1, and KCTD15 among the obesity SNPs, and HHEX, THADA, and KCNQ1 among the T2D SNPs (see Supplementary Table 1).
Global and pair-wise FST
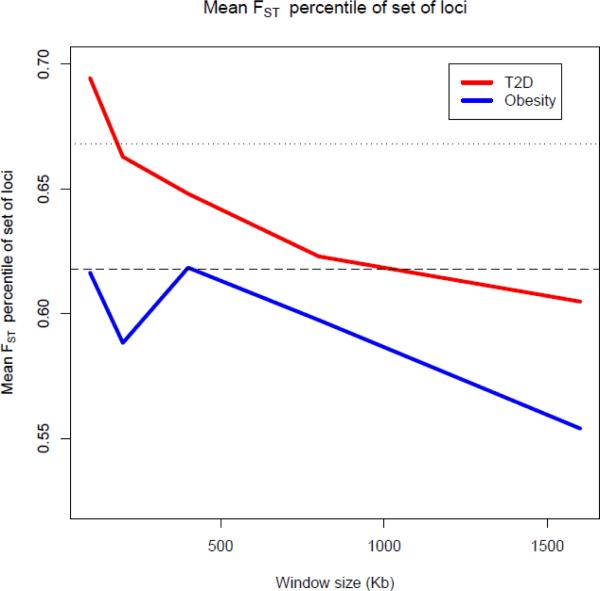
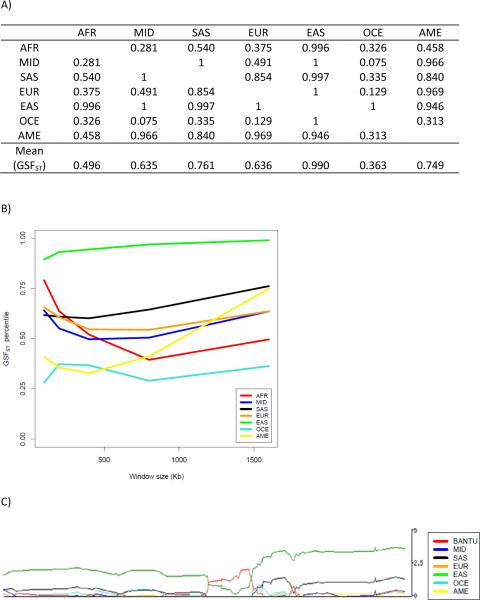
The percentile for the 7-way FST, averaged separately over the 16 obesity and 16 T2D loci, is shown in Figure 1 for varying window sizes. We find that, as an ensemble, the degree of differentiation at the 16 obesity loci slightly exceeds the 95th percentile (see Figure 1). The ensemble of T2D loci exceeds the 99th percentile for 100 kb windows, and the 95th percentile for 200–800 kb windows (see Figure 1). For the 100 kb window of T2D loci, the FST percentiles for 4 out of 16 loci are between the 90th and 100th percentile (TSPAN8, TCF7L2, HHEX, KCNQ1), and 4 are between 80th and 90th percentile. Therefore half of the 16 loci are above the 80th percentile. The rest of the loci lie above the 31st percentile, resulting in an average FST percentile of 69.4, which is higher than the 66.8 threshold representing the 99th percentile of the null distribution. Notably, we find that the degree of differentiation generally decays as the window size gets larger, further suggesting the localized action of natural selection. Upon examining the mean GSFST percentiles across each set of loci, we find evidence of elevated levels of differentiation (i.e. mean of 16 loci is above 50th percentile) at the ensemble of obesity loci (see Figure 2A) for most of groups, although none reach the 95th percentile. Among the T2D loci, we find that sub-Saharan Africans are highly differentiated (exceeding the 95th percentile) from other groups for the 100 kb window size. East Asians are highly differentiated from other groups for all window sizes and their mean GSFST percentile exceeds the 95th percentile for all window sizes greater than 100 kb (see Figure 2B). The high level of differentiation between East Asians and other groups, and between sub-Saharan Africans and other groups, as shown in Figure 2B, is responsible for the pattern we observe for the 7-way FST for T2D loci, as shown in Figure 1. For all above analyses, we find similar results using cM distance as opposed to kb distance, with the main exception being that the GSFST for East Asians at the set of T2D loci exceeds the 95th percentile only for the 0.1 and 0.8 cM windows.
Figure 1.
Mean percentile of global (7-way) FST across all 16 loci by window size. Horizontal dashed line indicates 95th percentile cut-off (0.614), and horizontal dotted line indicates 99th percentile cutoff (0.668).
Figure 2.
Mean percentile of GSFST across all 16 loci by window size for obesity (A) and T2D (B). Dashed horizontal line indicates 95% percentile cutoff (0.614), and horizontal solid line indicates 99th percentile cutoff (0.668). (AFR: Sub-Saharan Africa, MID: Middle East, SAS: South Asia, EUR: Europe, EAS: East Asia, OCE:Oceania, AME:America)
We next examined patterns of differentiation among the specific loci. None of the loci exhibit empirical p-values that reach the statistical significance threshold after Bonferroni correction. The regions that are above the 95th percentile for the overall 7-way FST for at least one window size are NEGR1 (100 kb window), FTO (200–400 kb window), FAIM2 (800–1600 kb window), SH2B1 (200–400 kb window), and for T2D: TSPAN8 (100–400 kb), PPARG (800 kb), HHEX (1600 kb), and WFS1 (1600 kb) (data not shown). Using cM instead of kb distance, we observe the same pattern for FAIM2 in which the global FST reaches statistical significance (100th percentile) for the 0.8 cM window. For the other obesity loci mentioned above, we observe similar patterns, although not reaching the 95th percentile. In addition, we observe that the global FST at the 1.6 cM window surrounding the PTER locus nearly reaches statistical significance (99.6th percentile) according to the Bonferroni correction, and that the 0.4 cM window of KCTD15, and the 1.6 cM window surrounding NPC1 exceed the 95th percentile. Among the T2D loci, we find similar results using cM-based distance with the exception that PPARG and TSPAN8 do not quite exceed the 95th percentile, JAZF1 exceeds the 95th percentile for the 0.4 and 0.8 cM windows, and TCF7L2 exceeds the 95th percentile for the 0.1 and 0.2 cM windows.
Finally, we examined GSFST (group-specific FST) for specific loci. The region surrounding the NEGR1 and PTER loci are highly differentiated (above 95th percentile in some windows) in sub-Saharan Africans (see Figure 3 and Supplementary Figure 1). The region surrounding SEC16B appears to have undergone more differentiation among Oceanians than among other groups (see Supplementary Figure 2). Also of note among obesity loci is that the region surrounding NCR3 has universally experienced little differentiation, especially for sub-Saharan Africans, Oceanians, and East Asians (see Supplementary Figure 2). Among T2D loci, we find that HHEX and THADA are highly differentiated between East Asians and other groups (see Figure 4). Using bootstrap re-sampling, we find no overlap between the East Asia and any of the other groups' 95% confidence intervals for the 200 kb window size surrounding HHEX, suggesting that this magnitude of difference is statistically significant. We also observe that the region surrounding CDC123 appears to have undergone more differentiation among Oceanians than other groups (see Supplementary Material 3). For the above analyses, we observe similar results using cM-based distance.
Figure 3.
A) Pair-wise FST percentiles for the 100 kb window surrounding NEGR1, B) GSFST across different window sizes for NEGR1. (AFR: Sub-Saharan Africa, MID: Middle East, SAS: South Asia, EUR: Europe, EAS: East Asia, OCE:Oceania, AME:America)
Figure 4.
A) Pair-wise FST percentiles for the 1600 kb window surrounding HHEX, B) GSFST across different window sizes for HHEX. C) XP-EHH in 500 kb window surrounding HHEX risk SNP, from the HGDP Genome Browser, where the y-axis represents the −log10 of the empirical p-value for a window centered at the SNP. (AFR: Sub-Saharan Africa, MID: Middle East, SAS: South Asia, EUR: Europe, EAS: East Asia, OCE:Oceania, AME:America)
REHH
First, we counted all instances in which the SWEEP-identified haplotypes in the 400 kb surrounding each risk SNP had REHH values that were in the top 1% for each population group and for the respective chromosome (see Tables 1 and 2). We also counted all instances where a risk allele was contained in a haplotype that was in the top 0.1% of the respective chromosome (see Supplementary Table 3). Among the obesity loci, we find that there is a significant difference among groups in the number of loci with at least one REHH value in the top 99th percentile (p=0.0066; see Table 1). Pair-wise comparisons show that South Asians (9) and Europeans (7) have more loci having at least one 99th percentile REHH value (with haplotype frequency >0.05) than Sub Saharan Africans (1) and Native Americans (1), although this difference is not statistically significant (p=0.059 for South Asians; p=0.019 for Europeans) after correction for 21 possible two-sided pair-wise comparisons. However, we consider this statistical test to be conservative because we do not take into account the fact that at several loci, South Asians and Europeans exhibit single loci with anywhere from 2 to 9 extreme REHH values. Among the T2D loci, we find no significant differences among groups in the number of loci having at least one 99th percentile REHH value (p=0.15) (see Table 2). We also examined instances in which the risk SNP is in the core of a haplotype that has an extreme REHH value (top 5%), and noted whether the risk or non-risk allele is contained in that particular core. For the obesity loci, among the 21 cases in which we observe such a haplotype in a given group and for a given locus, 15 of the cores of these haplotypes contain the risk allele, while the rest contain the non-risk allele. For the T2D loci, among the 7 cases, 5 haplotype cores contain the risk allele. (see Supplementary Tables 4 and 5). This result suggests that there has been recent positive selection for variants that are associated with higher body weight and insulin resistance.
Table 1.
400 kb windows surrounding obesity risk SNP with at least one REHH p-value below 0.01, and haplotype frequency above 5%. Listed is the number of qualifying haplotypes and their frequencies or range of frequencies, indicated in parentheses.(AFR: Sub-Saharan Africa, MID: Middle East, SAS: South Asia, EUR: Europe, EAS: East Asia, OCE:Oceania, AME:America)
| AFR | MID | SAS | EUR | EAS | OCE | AME | Total groups | Total haplotypes | |
|---|---|---|---|---|---|---|---|---|---|
| NEGR1 | 1 (0.08) | 1 (0.06) | 2 | 2 | |||||
| SEC16B | 1 (0.24) | 1 (0.08) | 2 (0.34, 0.36) | 3 (0.10 – 0.32) | 4 | 7 | |||
| TMEM18 | 2 (0.12, 0.15) | 4 (0.22 – 0.31) | 4 (0.054 – 0.18) | 5 (0.07 – 0.28) | 4 | 15 | |||
| ETV5 | 2 (0.20, 0.25) | 1 | 2 | ||||||
| GNPDA2 | 0 | 0 | |||||||
| NCR3 | 5 (0.11 – 0.15) | 9 (0.11–0.37) | 3 (0.054 – 0.21) | 7 (0.07–0.18) | 2 (0.27, 0.27) | 5 | 26 | ||
| PTER | 2 (0.085, 0.07) | 1 (0.57) | 2 | 3 | |||||
| BDNF | 1 (0.21) | 1 | 1 | ||||||
| MTCH2 | 0 | 0 | |||||||
| FAIM2 | 0 | 0 | |||||||
| SH2B1 | 0 | 0 | |||||||
| MAF | 2 (0.23, 0.21) | 1 (0.63) | 1 (0.79) | 2 (0.071, 0.054) | 4 | 6 | |||
| FTO | 2 (0.15, 0.10) | 1 (0.38) | 2 | 3 | |||||
| NPC1 | 1 (0.59) | 1 (0.09) | 2 | 2 | |||||
| MC4R | 2 (0.14, 0.17) | 1 (0.71) | 1 (0.054) | 4 (0.13 – 0.92) | 4 | 8 | |||
| KCTD15 | 0 | 0 | |||||||
|
| |||||||||
| Total genes | 1 | 3 | 9 | 7 | 6 | 4 | 1 | ||
| Total haplotypes | 1 | 9 | 24 | 13 | 18 | 6 | 4 | ||
Table 2.
400 kb windows surrounding T2D risk SNP with at least one REHH p-value below 0.01, and haplotype frequency above 5%. Listed is the number of qualifying haplotypes and their frequencies or range of frequencies, indicated in parentheses. (AFR: Sub-Saharan Africa, MID: Middle East, SAS: South Asia, EUR: Europe, EAS: East Asia, OCE:Oceania, AME:America)
| AFR | MID | SAS | EUR | EAS | OCE | AME | Total groups | Total haplotypes | |
|---|---|---|---|---|---|---|---|---|---|
| NOTCH2 | 1 (0.16) | 1 | 1 | ||||||
| THADA | 1 (0.22) | 1 | 1 | ||||||
| ADAMTS9 | 2 (0.15, 0.14) | 1 (0.47) | 3 (0.058 – 0.13) | 8 (0.055 – 0.36) | 2 (0.24, 0.24) | 5 | 16 | ||
| IGF2BP2 | 1 (0.14) | 1 | 1 | ||||||
| PPARG | 1 (0.11) | 1 | 1 | ||||||
| WFS1 | 2 (0.09, 0.12) | 8 (0.06 – 0.48) | 2 | 10 | |||||
| CDKAL1 | 1 (0.23) | 1 | 1 | ||||||
| JAZF1 | 4 (0.12 – 0.15) | 1 (0.20) | 1 (0.10) | 5 (0.055 – 0.08) | 4 | 11 | |||
| SLC30A8 | 4 (0.23 – 0.62) | 1 | 4 | ||||||
| CDKN2A | 1 (0.1) | 1 (0.36) | 2 | 2 | |||||
| CDC123 | 1 (0.19) | 1 (0.14) | 3 (0.06 – 0.20) | 3 | 5 | ||||
| HHEX | 1 (0.23) | 1 | 1 | ||||||
| TCF7L2 | 1 (0.33) | 2 (0.06, 0.24) | 1 (0.07) | 2 (0.56, 0.56) | 4 | 6 | |||
| KCNJ11 | 1 (0.54) | 3 (0.22–0.33) | 1 (0.07) | 3 | 5 | ||||
| KCNQ1 | 1 (0.06) | 1 (0.053) | 2 (0.15, 0.26) | 3 | 4 | ||||
| TSPAN8/LGR5 | 4 (0.25 – 0.37) | 1 (0.30) | 2 | 5 | |||||
|
| |||||||||
| Total genes | 4 | 9 | 3 | 7 | 2 | 5 | 5 | ||
| Total Haplotypes | 8 | 16 | 5 | 10 | 13 | 5 | 17 | ||
XP-EHH
Using the HGDP genome browser, we find very few instances of outlier XP-EHH in the regions surrounding the obesity risk alleles. There does not appear to be an excess of instances of elevated XP-EHH in any particular population group (see Supplementary Table 6A). On a gene by gene basis, there does appear to be an excess of elevated XP-EHH in the MAF, PTER and MC4R regions (see Supplementary Table 6B).
For the T2D-associated regions, we find a slight excess of elevated XP-EHH among East Asians, specifically at KCNJ11, HHEX, and THADA, although the difference between groups is not statistically significant (Supplementary Table 6B). The region surrounding KCNJ11 appears to have extended haplotypes in all groups compared to sub-Saharan Africans (Bantu). THADA also shows evidence of extended haplotypes among Native Americans.
Discussion
We have tested the hypothesis that some groups of humans have recently experienced more evolutionary change at loci found to be associated with obesity and T2D compared to the rest of the genome. We have examined FST, a measure of population differentiation, and measures of shared extended haplotypes indicative of recent positive selection on new variation. Although our findings are not entirely consistent across tests, they have uncovered general as well as population- and gene-specific patterns.
First, with respect to the derived vs. ancestral status of the risk alleles, we find no evidence that the risk alleles tend to be either ancestral or derived for either the obesity or the T2D loci. We expected that if the thrifty genotype hypothesis applied specifically to the entire human species as an outlier among other primates, a majority of risk alleles would be derived. However, it is difficult to make any firm conclusions on the basis of this finding, since we are only considering markers that are still polymorphic in humans, and since the risk alleles that are reported in GWASs are unlikely to be the causative alleles, and are instead likely to only be associated with the causative variants.
Given the above-mentioned limitation, and the fact that these specific variants have been found to explain a very small proportion of the expected genetic variance (Hofker et al., 2009; Willer et al., 2009), we chose to examine average FST and haplotype patterns in the surrounding regions of each reported risk SNP (up to 800 kb in either direction). This enabled us to take into account more of the variation that is associated with any particular SNP, and may give some indication as to the timing and strength of selection. For example, depending on the population, an elevated GSFST that stretches over a long stretch of DNA may indicate more recent positive selection. Also, averaging FST over many SNPs in a region may be more a more sensitive approach given the highly variable nature of FST across neighboring loci.
We have found that the regions harboring T2D loci, as an ensemble, have experienced unusually high levels of differentiation compared to random regions of the genome, as assessed by the genotyped SNPs. Differentiation decays with distance as expected. Obesity loci, as an ensemble, also show unusually high levels of differentiation, but to a lesser extent than T2D loci. Our results further suggest that East Asians and sub-Saharan Africans have experienced higher levels of group-specific differentiation than other groups at the ensemble of T2D loci. We also find, as expected, that the degree of differentiation quickly decays with larger window sizes for sub-Saharan Africans, given overall reduced LD in these groups. Pickrell et al. (2009) used the same dataset to examine the single SNP with the highest FST in each T2D-associated region (within a 100 kb window) and found that sub-Saharan Africans are significantly differentiated from East Asians and Europeans at these loci. Our results confirm this finding and also uncover a high degree of differentiation among East Asians at larger window sizes. Our results also confirm the results of Pickrell et al. and others (Helgason et al., 2007; Southam et al., 2009) that the loci TCF7L2, JAZF1, and TSPAN8 show signatures of natural selection
The reasons for a high degree of differentiation are usually interpreted as being due to natural selection. However, various types of natural selection could explain any given pattern of differentiation: purifying selection in one or several groups, or positive selection in one group but not others, or in all groups except for one. These could represent one of several evolutionary/historical scenarios. One is that there was selection either for or against insulin resistance among East Asians and sub-Saharan Africans. Another possibility is that East Asians and sub-Saharan Africans underwent a relaxation of selection pressures at these genes due to a diet that did not select for insulin resistance and gluconeogenisis.
Among the T2D-associated loci, we find that HHEX is the most strongly differentiated between groups, specifically between East Asians and other groups. HHEX (hematopoietically-expressed homeobox protein) encodes a transcriptional regulator involved in pancreatic development (Bort et al., 2004). The risk allele has been found to be associated with reduced pancreatic β-cell function (Pascoe et al., 2007), and there is evidence that HHEX belongs to a highly conserved “genomic regulatory block” (Ragvin et al., 2010). Among sub-Saharan Africans, no single locus explains the overall T2D trend, suggesting that it is the effect of many moderately differentiated loci that contributes to the overall pattern for the ensemble of T2D loci.
Among the obesity loci, NEGR1 (Neuronal growth regulator 1) was found to be highly differentiated among sub-Saharan Africans. This gene has a role in neuronal outgrowth (Schafer et al., 2005) and is highly expressed in the hypothalamus (Willer et al., 2009). The region surrounding this allele has also been found to contain a large copy number polymorphism that could be a causal variant (Willer et al., 2009).
Whereas FST is most powerful for detecting selection on already standing genetic variation, present on multiple haplotype backgrounds, becoming favoured in one geographic region, REHH is most powerful to detect recent strong positive selection on a novel mutation that has reached an intermediate haplotype frequency in the population. Our results show that there are differences among groups in the number of obesity loci that show evidence of recent positive selection according to the REHH test. It appears that South Asians and Europeans exhibit more such loci compared, most notably, to sub-Saharan Africans and Native Americans.
An interesting result from the FST and REHH tests is the case of the region near NCR3. The risk SNP is near both NCR3 (natural cytotoxicity triggering receptor 3 precursor) and AIF1 (allograft inflammatory factor 1). Being in the HLA region of chromosome 6, this region appears to be highly conserved among different human populations, since it shows very little differentiation compared to the rest of the genome (see Supplementary Figure 2). However, this same region has among the strongest evidence of extended haplotypes among South and East Asian populations. In instances of extended haplotypes containing the risk SNP, we have determined that it is the risk allele that is contained in these haplotypes, suggesting that selection has recently favored variation in this region that enables individuals to avoid an energy deficit.
Finally, for the XP-EHH test, we do not observe major differences in signals between groups for the obesity loci. However for the T2D loci, we find that East Asians exhibit more evidence of recent positive selection, most notably at HHEX and THADA, a result that converges with the extreme differentiation that East Asians exhibit at this locus.
Although we do observe some overlap of genetic and geographic regions identified by the three tests considered (FST. REHH, XP-EHH), the lack of overlap could be due to several factors. Haplotype-based measures, such as those based on EHH, test for very restricted and likely rare set of cases of positive selection acting on newly arisen variation, that is relatively recent, and in which the haplotype quickly rises to an intermediate frequency in a given population (best seen by EHH) or a high frequency in one population but not another (best seen by XP-EHH). Therefore, the congruence of these various tests will depend on the type, timing, and strength of selection for each particular genetic and geographical region.
Our findings along with other published evidence appear to be slightly more consistent with the hypothesis that cycles of feast and famine were as or more severe among agricultural populations (Benyshek et al., 2006; Cordain et al., 1999). The REHH results among South Asians suggesting recent natural selection favoring obesity risk alleles is consistent with evidence of major famines in South Asia (Wells, 2007). It may be that the adoption of agriculture, along with its associated features such as sedentary life-ways resulted in an inflexible over-reliance on a more highly variable food supply. We find that while Eurasian populations show REHH signatures of selection at several obesity loci, American and sub-Saharan African populations show signs of selection at only one locus each (Table 1). This is consistent with the fact that the relative isolated sub-Saharan and Native American populations adopted agriculture later than Eurasian populations. These findings should be interpreted with caution since the loci that we have examined have been associated with body weight only among European or European/derived populations. If agriculture did indeed select for thrifty genes, we are left with the puzzle of explaining why rates of obesity and T2D are relatively low among individuals of European ancestry, for example. It suggests that non-genetic factors could more readily explain population differences in obesity and T2D prevalence. These could be environmental factors that are not yet well understood (Gravlee et al., 2009; McAllister et al., 2009), including infectious agents (Ley et al., 2006; Vijay-Kumar et al., 2010; Wells, 2009; Whigham et al., 2006) that would perfectly track genetic admixture proportions.
A limitation of our findings is that we have tested whether these candidate loci are outliers compared to the rest of the genome with respect to population differentiation and extended haplotype homozygosity. It is presently difficult to determine with certainty whether such outlier loci are the result of natural selection, as opposed to other evolutionary forces such as genetic drift. Another limitation of our findings, as mentioned above, is that we have examined loci that have been found to be associated with these traits among Europeans. Although several GWASs have recently been conducted in other populations (Cho et al., 2009; Liu et al., 2010; Tsai et al., 2010), there is still some uncertainty as to whether the same loci explain variation in these traits in different populations. If, as we have shown, there has been differentiation and selection at these loci, it may be that the genetic architecture of these traits is different in different populations. There may be loci that affect these traits in non-Europeans that we have not considered in this analysis. Our findings of greater evidence of thrifty genes among Eurasians may therefore be biased by the possibility that these loci are found to be associated in GWASs in Europeans, precisely because they underwent recent selection in those groups. It should also be noted that our results could be influenced by a subset of the several populations within each broader group that we are using.
In conclusion, our results have shown that genetic regions surrounding loci associated with T2D, and to a lesser extent, obesity, have been subject to unusually high levels of change in the last 50,000 to 100,000 years. Most notably, sub-Saharan Africans and East Asians appear to have undergone selection at T2D loci. Identifying specific targets of recent selection in the human genome can aid in determining population-specific risk variants, especially insofar as prevalence differences differ between populations (Ayodo et al., 2007). We anticipate that future studies will be at a finer scale at both the population, genetic, and phenotypic level, potentially further elucidating the genetic basis of obesity and T2D, and the population-specific genetic or non-genetic mechanisms that lead to different rates, types, and consequences of obesity and T2D.
Supplementary Material
Acknowledgments
The authors thank the individuals in the HGDP sample, Vinodh Srinivasasainagendra for computational assistance, and the UAB High Performance Computing Center. The authors also thank Nick Pajewski, Guo-Bo Chen, Nathan Wineinger, Robert Makowsky, and Charity Morgan for help with analyses. This work was funded by NIH-T32HL007457.
Funded by: NIH T32HL007457 from the National Heart, Lung, and Blood Institute
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Reference List
- 1.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayodo G, Price AL, Keinan A, Ajwang A, Otieno MF, Orago AS, Patterson N, Reich D. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benyshek DC, Martin JF, Johnston CS. A reconsideration of the origins of the type 2 diabetes epidemic among Native Americans and the implications for intervention policy. Med Anthropol. 2001;20:25–64. doi: 10.1080/01459740.2001.9966186. [DOI] [PubMed] [Google Scholar]
- 4.Benyshek DC, Watson JT. Exploring the thrifty genotype's food-shortage assumptions: a cross-cultural comparison of ethnographic accounts of food security among foraging and agricultural societies. Am J Phys Anthropol. 2006;131:120–126. doi: 10.1002/ajpa.20334. [DOI] [PubMed] [Google Scholar]
- 5.Bort R, Martinez-Barbera JP, Beddington RS, Zaret KS. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development. 2004;131:797–806. doi: 10.1242/dev.00965. [DOI] [PubMed] [Google Scholar]
- 6.Cheng CY, Reich D, Coresh J, Boerwinkle E, Patterson N, Li M, North KE, Tandon A, Bailey-Wilson JE, Wilson JG, Kao WH. Admixture Mapping of Obesity-related Traits in African Americans: The Atherosclerosis Risk in Communities (ARIC) Study. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 8.Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordain L, Miller J, Mann N. Scant evidence of periodic starvation among hunter-gatherers. Diabetologia. 1999;42:383–384. doi: 10.1007/pl00002924. [DOI] [PubMed] [Google Scholar]
- 10.Deitel M. Overweight and obesity worldwide now estimated to involve 1.7 billion people. Obes Surg. 2003;13:329–330. doi: 10.1381/096089203765887598. [DOI] [PubMed] [Google Scholar]
- 11.Diamond J. The double puzzle of diabetes. Nature. 2003;423:599–602. doi: 10.1038/423599a. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, Nicklas B, Ryan AS, McKeigue PM, Hoggart CL, Weinsier RL, Allison DB. Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res. 2003;11:904–911. doi: 10.1038/oby.2003.124. [DOI] [PubMed] [Google Scholar]
- 13.Florez JC. Clinical review: the genetics of type 2 diabetes: a realistic appraisal in 2008. J Clin Endocrinol Metab. 2008;93:4633–4642. doi: 10.1210/jc.2008-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, Defelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 15.Gardner M, Williamson S, Casals F, Bosch E, Navarro A, Calafell F, Bertranpetit J, Comas D. Extreme individual marker F(ST)values do not imply population-specific selection in humans: the NRG1 example. Hum Genet. 2007;121:759–762. doi: 10.1007/s00439-007-0364-9. [DOI] [PubMed] [Google Scholar]
- 16.Goran MI. Ethnic-specific pathways to obesity-related disease: the Hispanic vs. African-American paradox. Obesity (Silver Spring) 2008;16:2561–2565. doi: 10.1038/oby.2008.423. [DOI] [PubMed] [Google Scholar]
- 17.Gravlee CC, Non AL, Mulligan CJ. Genetic ancestry, social classification, and racial inequalities in blood pressure in Southeastern Puerto Rico. PLoS One. 2009;4:e6821. doi: 10.1371/journal.pone.0006821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G, Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 20.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, Benediktsson R, Hinney A, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Schafer H, Faruque M, Doumatey A, Zhou J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Sigurdsson G, Hebebrand J, Pedersen O, Thorsteinsdottir U, Gulcher JR, Kong A, Rotimi C, Stefansson K. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 21.Hemminger BM, Saelim B, Sullivan PF. TAMAL: an integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics. 2006;22:626–627. doi: 10.1093/bioinformatics/btk025. [DOI] [PubMed] [Google Scholar]
- 22.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A, Thomas DJ, Trumbower H, Weber RJ, Weirauch M, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofker M, Wijmenga C. A supersized list of obesity genes. Nat Genet. 2009;41:139–140. doi: 10.1038/ng0209-139. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 27.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, Chen Z, Yu L, Yang Y, Feng G, Li J, Zhang J, Liu S, Zhang Z, He L, Xu H. Meta-analysis Added Power to Identify Variants in FTO Associated With Type 2 Diabetes and Obesity in the Asian Population. Obesity (Silver Spring) 2010 doi: 10.1038/oby.2009.469. [DOI] [PubMed] [Google Scholar]
- 29.McAllister EJ, Dhurandhar NV, Keith SW, Aronne LJ, Barger J, Baskin M, Benca RM, Biggio J, Boggiano MM, Eisenmann JC, Elobeid M, Fontaine KR, Gluckman P, Hanlon EC, Katzmarzyk P, Pietrobelli A, Redden DT, Ruden DM, Wang C, Waterland RA, Wright SM, Allison DB. Ten putative contributors to the obesity epidemic. Crit Rev Food Sci Nutr. 2009;49:868–913. doi: 10.1080/10408390903372599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 32.O'Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 33.Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, Weedon MN, Mari A, Hattersley AT, McCarthy MI, Frayling TM, Walker M. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 34.Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, Pritchard JK. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes O, Engstrom PG, Alonso ME, Mustienes EL, Skarmeta JL, Tavares MJ, Casares F, Manzanares M, van H,V, Molven A, Njolstad PR, Argenton F, Lenhard B, Becker TS. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 2010;107:775–780. doi: 10.1073/pnas.0911591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 37.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, Schaffner SF, Lander ES, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Johnson TA, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer M, Brauer AU, Savaskan NE, Rathjen FG, Brummendorf T. Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol Cell Neurosci. 2005;29:580–590. doi: 10.1016/j.mcn.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Seidell JC. Obesity, insulin resistance and diabetes--a worldwide epidemic. Br J Nutr. 2000;83(Suppl 1):S5–S8. doi: 10.1017/s000711450000088x. [DOI] [PubMed] [Google Scholar]
- 40.Southam L, Soranzo N, Montgomery SB, Frayling TM, McCarthy MI, Barroso I, Zeggini E. Is the thrifty genotype hypothesis supported by evidence based on confirmed type 2 diabetes- and obesity-susceptibility variants? Diabetologia. 2009;52:1846–1851. doi: 10.1007/s00125-009-1419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang H, Jorgenson E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- 42.Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, Chang CC, Chen P, Chen CH, Fann CS, Chen YT, Wu JY. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science. 2010 doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Beydoun MA. The obesity epidemic in the United States--gender, age, socioeconomic, racial/ethnic, and geographic characteristics: a systematic review and meta-regression analysis. Epidemiol Rev. 2007;29:6–28. doi: 10.1093/epirev/mxm007. [DOI] [PubMed] [Google Scholar]
- 46.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 47.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 49.Wells JC. Commentary: Why are South Asians susceptible to central obesity?--the El Nino hypothesis. Int J Epidemiol. 2007;36:226–227. doi: 10.1093/ije/dyl287. [DOI] [PubMed] [Google Scholar]
- 50.Wells JC. Ethnic variability in adiposity and cardiovascular risk: the variable disease selection hypothesis. Int J Epidemiol. 2009;38:63–71. doi: 10.1093/ije/dyn183. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Geer LY, Helmberg W, Kapustin Y, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whigham LD, Israel BA, Atkinson RL. Adipogenic potential of multiple human adenoviruses in vivo and in vitro in animals. Am J Physiol Regul.Integr.Comp Physiol. 2006;290:R190–R194. doi: 10.1152/ajpregu.00479.2005. [DOI] [PubMed] [Google Scholar]
- 53.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O'Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams RC, Long JC, Hanson RL, Sievers ML, Knowler WC. Individual estimates of European genetic admixture associated with lower body-mass index, plasma glucose, and prevalence of type 2 diabetes in Pima Indians. Am J Hum Genet. 2000;66:527–538. doi: 10.1086/302773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.