Abstract
Muscle tissue is the major site for insulin-stimulated glucose uptake in vivo, due primarily to the recruitment of the insulin-sensitive glucose transporter (GLUT4) to the plasma membrane. Surprisingly, virtually all cultured muscle cells express little or no GLUT4. We show here that adenovirus-mediated expression of the transcriptional coactivator PGC-1, which is expressed in muscle in vivo but is also deficient in cultured muscle cells, causes the total restoration of GLUT4 mRNA levels to those observed in vivo. This increased GLUT4 expression correlates with a 3-fold increase in glucose transport, although much of this protein is transported to the plasma membrane even in the absence of insulin. PGC-1 mediates this increased GLUT4 expression, in large part, by binding to and coactivating the muscle-selective transcription factor MEF2C. These data indicate that PGC-1 is a coactivator of MEF2C and can control the level of endogenous GLUT4 gene expression in muscle.
Keywords: MEF2, diabetes
Type 2 diabetes mellitus, the most common endocrine disorder, potentially affects up to 5% of the western population (1). Patients with type 2 diabetes generally suffer both from reduced insulin secretion and from resistance to the actions of insulin. Hyperglycemic-hyperinsulinemic clamp analyses of human type 2 diabetic patients show that insulin resistance in muscle is caused by a defect in glucose transport (2). The principal insulin-sensitive glucose transporter in muscle is the insulin-sensitive glucose transporter (GLUT4), which is recruited to the sarcolemma following insulin stimulation. Definitive evidence that GLUT4 is the primary mediator both of basal and insulin-stimulated glucose transport in muscle comes from muscle-specific GLUT4 knockout mice (3). Ex vivo glucose transport assays of isolated muscle tissue from these mice demonstrate an 80% reduction in basal glucose transport and a complete loss of insulin-stimulated glucose uptake. Overexpression of GLUT4 in muscle of genetically diabetic mice (db/db) alleviates insulin resistance and improves glycemic control by elevating both basal and insulin-stimulated glucose transport (4). Together, these data point to skeletal muscle glucose transport as the rate-limiting step for whole body glucose disposal and suggest that the regulation of GLUT4 expression is a potential target for treatment of diabetes mellitus.
We have previously described a coactivator of PPARγ and other nuclear receptors termed PGC-1 that plays a key role in several aspects of thermogenesis and oxidative metabolism. PGC-1 also stimulates mitochondrial biogenesis per se through coactivation of nuclear respiratory factor-1 (NRF-1) (5, 6). Indeed, PGC-1 expression in both muscle and fat cells activates the expression of several genes of the oxidative phosphorylation pathway, including cytochrome c oxidase (COX) subunits II and IV, and ATP synthetase. This coactivator also stimulates the induction of a mitochondrial uncoupling protein (UCP), UCP-1 in fat cells and UCP-2 in muscle cells (6).
The chronically increased respiration and thermogenesis induced by PGC-1 must be ultimately balanced by increased fuel uptake. However, there have been no studies to date investigating the role of PGC-1 in the regulation of fuel uptake. This question is of particular interest for the diabetes problem because there have been studies suggesting that rates of fuel oxidation and mitochondrial function can affect glucose uptake (7–9). Also of interest is the fact that virtually all cultured skeletal muscle cell lines have been found to be deficient in GLUT4 gene expression, and we have recently found that cultured muscle cells also express far less PGC-1 than is expressed in vivo. This has led us to investigate the hypothesis that PGC-1 might be an important regulator of GLUT4 gene expression. In this report we show that PGC-1 powerfully induces the expression of the endogenous GLUT4 gene in cultured myotubes, resulting in expression comparable to that seen in muscle in vivo. Both basal glucose transport and insulin-stimulated glucose transport are increased by ectopic expression of PGC-1.
Materials and Methods
Materials.
2-Deoxy-d-[1,2-3H]glucose was purchased from New England Nuclear. Nonradioactive 2-deoxy-d-glucose, cytochalasin B, and insulin were purchased from Sigma.
Cell Culture.
C2C12 and L6 myoblast cell lines were maintained in DMEM supplemented with 10% FBS in an atmosphere of 5% CO2 in subconfluent culture. For experiments using differentiated myotube cultures, the cells were grown to confluence, at which time the medium was changed to DMEM supplemented with 2% equine serum (ES). The myoblasts were permitted to fuse into multinucleate myotubes for 24 h, and the cells were then infected with adenovirus preparations at the indicated multiplicity of infection (moi) for 12 h. Following infection, the DMEM, 2% ES medium was changed daily for the subsequent 4 days.
Glucose Transport.
Assessment of 2-[3H]deoxyglucose uptake was performed as described (30). Briefly, differentiated L6 myotube cultures were grown in 6-well plates, and were starved in DMEM containing 25 mM glucose for 5 h before the assay. After rinsing the cultures twice in HBS (140 mM NaCl/20 mM Hepes/2.5 mM MgSO4/1 mM CaCl2/5 mM KCl), glucose uptake was measured by incubating the cells in 10 μM 2-[3H]deoxyglucose for 3 min. Nonspecific cell-associated radioactivity was measured by the addition of 10 μM cytochalasin B. Cells were washed three times with ice-cold saline and lysed in 0.05 N NaOH. Radioactivity was measured by liquid scintillation counting. Each condition was assayed in triplicate, and data are representative of more than three independent experiments.
Immunoblotting.
Whole cell extracts were prepared by lysis in an SDS-urea lysis buffer (50 mM Tris, pH 7.8/2% SDS/10% glycerol/10 mM Na4P2O7/100 mM NaF/6 M urea/10 mM EDTA). Proteins were separated by SDS/PAGE and transferred to Immobilin P membrane (Millipore). The membranes were probed with antibodies recognizing PGC-1 or cytochrome c (PharMingen). Immune complexes were detected either by enhanced chemiluminescence or by [125I]-Protein A.
Subcellular membrane fractions were prepared as described (31). Briefly, L6 myotubes were harvested in HES buffer (20 mM Hepes, pH 7.4/250 mM sucrose/1 mM EDTA/10 μg/ml aprotinin/1 μg/ml leupeptin/2 mM PMSF) and homogenized using a Teflon pestle in a glass homogenizer. The supernatant from a 19,000 × g centrifugation was centrifuged at 48,000 × g to obtain a high-density membrane (HDM) pellet. The 48,000-g supernatant was centrifuged at 210,000 × g to obtain the low-density microsomal (LDM) pellet. Equal amounts of protein from LDM fractions were solubilized, separated by SDS/PAGE, transferred to nitrocellulose, and immunoblotted with antisera against GLUT4. Immunoreactive material was detected by incubation with [125I]-labeled protein A.
Transient Transfection and Adenovirus Infection.
C2C12 myoblasts were seeded at 1.5 × 106 cells per 24-well plate and were grown for 12 h in DMEM supplemented with 10% FBS. The cells were transfected by using Superfect reagent (Qiagen, Chatsworth, CA) for 3 h followed by a 12-h recovery period in DMEM supplemented with 10% FBS. Cells were permitted to differentiate an additional 24 h in DMEM supplemented with 2% ES. Luciferase reporter activity was measured, and data are representative of at least three experiments performed in triplicate.
The GLUT4 2.4-kb promoter-luciferase reporter plasmid was generated by subcloning the SacI/XbaI fragment from the GLUT4 2.4-kb promoter–chloramphenicol acetyltransferase (CAT) plasmid into SacI/NheI digested pGL3 Basic. The MEF2 recognition sequence (-473 to -464) of the GLUT4 promoter was mutated from CTAAAAATAG to CTAAGGCTAG by site-directed mutagenesis (Stratagene).
Adenoviruses were constructed to express either green fluorescent protein (GFP) or both GFP and PGC-1 (29). L6 myoblasts were grown to confluence, induced to differentiate for 48 h, and then infected with adenovirus at varying mois. Cells were allowed to express the recombinant proteins for 96 h before assay.
Affinity Chromatography and Coimmunoprecipitation.
GST fusion proteins were expressed in Escherichia coli and purified on glutathione Sepharose beads. Protein binding assays were performed as described (5).
For each 10-cm dish of 293T cells, 8 μg Flag-PGC-1 and/or 8 μg pcDNA3.1 MEF2C was transfected by Fugene 6 (Boehringer Mannheim). Cells were allowed to express the proteins for 48 h posttransfection and were lysed (20 mM Hepes, pH 7.9/125 mM NaCl/0.1% Nonidet P-40/1 mM EDTA/1 mM PMSF) by freeze-thaw method. Cellular debris was pelletted for 10 min at 14,000 × g. Protein was quantitated by Bradford assay, and 500 μg was subjected to immunoprecipitation with an M2 agarose anti-FLAG affinity resin (Sigma) for 2 h at 4°C. The resin was washed extensively with lysis buffer, and proteins were separated by SDS/PAGE. Coimmunoprecipitated PGC-1 and MEF2C were detected by immunoblot analysis.
Results
Ectopic Expression of PGC-1 in Myotubes Increases GLUT4 Gene Expression.
The rat L6 muscle cell line expresses very little, if any, PGC-1 and far less GLUT4 than is present in striated muscle in vivo. We used an adenovirus vector to express PGC-1 protein in the L6 myotube cultures. PGC-1 protein was detectable by Western blot analysis in myotube cultures that were infected with an adenovirus expressing PGC-1 at a moi greater than or equal to 100, but was undetectable in myotubes infected with lower concentrations of this virus or any concentration of the control adenovirus expressing GFP (Fig. 1A). As expected and as shown earlier, PGC-1 induced the expression of mRNA of key components of the respiratory chain, such as cytochrome c and cytochrome c oxidase subunit IV (5, 6). Both of these inductions were readily detectable at a moi of 100. Strikingly, ectopic expression of PGC-1 activated expression of the GLUT4 mRNA from undetectable levels up to levels that are equivalent to those observed from adult muscle tissue (Fig. 1A). The elevation of GLUT4 mRNA in myotubes was observed in cultures that were infected with PGC-1 adenovirus at an moi of 30, where PGC-1 protein is still below detection limits on these Western blots. No induction of GLUT4 mRNA was observed with the GFP-expressing virus at any moi used.
Figure 1.
Induction of GLUT4 expression by PGC-1. (A) Ectopic expression of PGC-1 potentiates GLUT4 gene expression. L6 myoblasts were grown to confluence and were induced to differentiate. Myotube cultures were infected with varying moi, as indicated, of adenovirus expressing either GFP (as control) or PGC-1. The myotubes were permitted to express the proteins for 96 h, at which time total RNA or total cellular proteins were isolated. Thirty micrograms of protein were subjected to immunoblot analysis using either anti-PGC-1 or anti-cytochrome c antisera. Twenty micrograms of total RNA was subjected to Northern blot analysis using either GLUT4 or COX IV cDNA as probe. (B) PGC-1 induction of GLUT4 gene expression is cell-type dependent. Cos 1 cells were infected with varying moi, as indicated, of adenovirus expressing either GFP (as control) or PGC-1. Total RNA was subjected to Northern blot analysis using either PGC-1 or GLUT4 cDNA as probe, and anti-cytochrome c antiserum was used as described above.
The expression of GLUT4 is very tissue-selective in vivo; we performed a similar series of experiments in the COS-1 kidney epithelial cell line to assess whether the effect of PGC-1 on GLUT4 gene expression was cell-type-dependent. PGC-1 mRNA was detectable in cells infected at a moi of 10 with PGC-1 adenovirus (Fig. 1B). Expression of cytochrome c was elevated in cells infected with PGC-1 adenovirus at moi greater than or equal to 30, but the GLUT4 gene was unresponsive to ectopic expression of PGC-1 at all concentrations tested. These results demonstrate that the GLUT4 gene is powerfully regulated by PGC-1 with some apparent specificity for muscle cells.
PGC-1 Stimulates Both Basal and Insulin-Stimulated Glucose Transport.
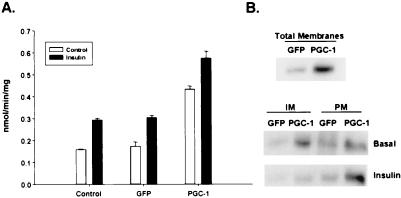
The effects of PGC-1 on GLUT4 gene expression suggested a possible effect on glucose transport. Uninfected cultures of L6 rat myotubes have a basal glucose uptake of ≈1.5 nmol/min/mg of protein, and insulin stimulated a 2-fold increase in glucose transport activity (Fig. 2A). Infection of myotube cultures with the control GFP adenovirus had no effect either on basal or insulin-stimulated glucose. In contrast, adenovirus expression of PGC-1 induced a 3-fold increase in basal rates of glucose uptake in myotubes. Insulin-stimulated glucose uptake in PGC-1 infected myotubes was increased by an additional 50% to 6 nmol/min/mg of protein (Fig. 2A). The finding that increases in GLUT4 expression results in higher rates of basal glucose transport are not entirely surprising because muscle-specific overexpression of GLUT4 in transgenic mice also raises basal glucose transport by 50% (10, 11).
Figure 2.
PGC-1 induces glucose transport in muscle cells. (A) PGC-1 induces both basal and insulin-stimulated glucose transport. Differentiated L6 myotubes were infected either with GFP or PGC-1 expressing adenovirus at moi of 100 or they were grown in the absence of adenovirus (control) for 96 h. The cultures were serum-deprived for 6 h before the addition of 100 nM insulin (black bars) and were incubated an additional 20 min. Cells were rinsed, and glucose uptake was measured as described in Materials and Methods. Results shown are representative of more than three independent experiments that were performed in triplicate. (B) Ectopic expression of PGC-1 in myotubes increases total and plasma membrane-associated GLUT4 protein. Differentiated L6 myotubes were infected either with GFP or PGC-1 expressing adenovirus at moi of 100 or they were grown in the absence of adenovirus (control) for 96 h. The cells were stimulated with 100 nM insulin, as indicated, and cellular homogenates were fractionated either into total membranes, plasma membrane (PM), or intracellular membranes (IM). Equivalent amounts of protein were fractionated by SDS/PAGE and assayed for GLUT4 protein by immunoblotting.
One possible explanation why the robust effects of PGC-1 increase glucose uptake would be increased numbers of GLUT4 transporters localized to the plasma membrane. To investigate the effects of PGC-1 on GLUT4 protein levels and GLUT4 trafficking, we isolated membrane fractions from infected myotube cultures. Western blot analysis of proteins from the total membrane fraction revealed a 3.7-fold increase in GLUT4 protein from cells expressing PGC-1 (Fig. 2B). This corresponds well to the 3-fold increase in basal glucose transport seen in Fig. 2A. Under basal conditions, PGC-1 expressing myotubes contain a 4.4-fold elevated GLUT4 protein in the intracellular membrane compartments and a 2.7-fold elevation on the plasma membrane, compared with the GFP control myotubes. Insulin stimulation of both cultures promotes the translocation of the GLUT4 protein from the intracellular membrane compartments to the plasma membrane (Fig. 2B). This results in a 2.9-fold increase in GLUT4 protein on the plasma membrane in the PGC-1-infected cells compared with the controls. Thus, PGC-1-induced expression of the GLUT4 gene results in elevated functional plasma membrane-associated GLUT4 protein in the basal state, which is further increased after maximal stimulation with insulin. However, expression of PGC-1 does not increase the fold stimulation of glucose transport by insulin.
Insulin stimulation of cells initiates a protein phosphorylation cascade that ultimately leads to the translocation of GLUT4-containing vesicles from intracellular compartments to the plasma membrane (12, 13). To evaluate whether PGC-1 affects insulin signaling per se, we performed phosphorylation-specific Western blot analysis of key target proteins, such as the insulin receptor (IR), insulin-receptor substrate-1 (IRS-1), and Akt (PKB) as a function of insulin concentration. Stimulation of either uninfected myotubes or myotubes infected with GFP or PGC-1 adenoviruses with a concentration range of insulin from 1 to 100 nM resulted in nearly identical patterns of phosphorylation (data not shown). These results demonstrate that PGC-1 increases glucose transport in muscle by inducing GLUT4 expression, but not by sensitizing or substantially altering the proximal steps of the insulin signaling pathway.
PGC-1 Induces GLUT4 Gene Expression via Coactivation of MEF2C.
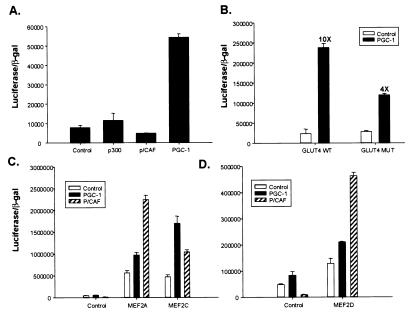
The effects of PGC-1 on GLUT4 gene expression may be direct or indirect. To study this, we analyzed the effects of PGC-1 on wild type and mutant GLUT4 promoters. Extensive promoter analysis of the GLUT4 gene by using transgenic mice has shown that 2.4 kb of 5′-flanking DNA is sufficient to confer tissue-specific expression of the gene in muscle, adipose, and heart (14). By using a reporter construct consisting of 2.4 kb of 5′-flanking DNA of the GLUT4 gene fused to luciferase, we analyzed the specificity of the GLUT4 promoter region to coactivation by PGC-1 and by other well known transcriptional coactivators, such as p300 and p/CAF. Transfected C2C12 myoblasts were induced to differentiate for at least 24 h before reporter assays. As shown in Fig. 3A, the GLUT4-luciferase reporter gene was unresponsive to ectopic expression of either p300 or p/CAF; in contrast, expression of PGC-1 induced a 6- to 10-fold increase in transcriptional response. Hence, PGC-1 activates the GLUT4 promoter with some degree of specificity relative to some other well known transcriptional coactivators.
Figure 3.
PGC-1 specifically transactivates the GLUT4 promoter via a MEF2 element. (A) PGC-1 increases transcriptional activity of the GLUT4 promoter. C2C12 myoblast cells were transfected in a 24-well format with 150 ng GLUT4 2.4-kb promoter-luciferase plasmid, 50 ng CMV βgal plasmid, and 700 ng of CMV-driven expression vectors either for p300, p/CAF, or full-length PGC-1. The cells were transfected for 3 h and 10% FBS/DMEM was replaced overnight. The transfected cells were induced to differentiate for at least 24 h before lysis. Luciferase activity was normalized by β-galactosidase activity. (B) PGC-1 coactivates GLUT4 promoter activity via the MEF2 enhancer element. The MEF2 recognition sequence (-473 to -464) of the 2.4-kb GLUT4 promoter was mutated from CTAAAAATAG to CTAAGGCTAG by site-directed mutagenesis. C2C12 myoblast cells were transfected as described above. (C and D) PGC-1 preferentially coactivates MEF2C. 293 cells were transfected in a 24-well format with 150 ng 3× MEF2 recognition sequence-luciferase plasmid, 50 ng CMV β-gal plasmid, 700 ng of CMV-driven expression vectors either for p/CAF or PGC-1, and either pRC-CMV MEF2A, MEF2C, or MEF2D. Cells were grown for 36 h posttransfection, before lysis.
The 2.4-kb promoter region of the GLUT4 gene contains numerous transcription factor binding sites, such as thyroid hormone response elements and E-box target sequences, but special attention has been focused on a conserved binding site for the MEF2 family of transcription factors that is necessary for full promoter activity (15–17). This is of particular interest here because MEF2 factors are regulated during muscle differentiation and play important roles in muscle-selective gene expression, and because, as was shown in Fig. 1, the effects of PGC-1 on this promoter have some apparent selectivity for muscle cells. Fig. 3B illustrates that mutation of the MEF2 element located at -473 to -464 of the GLUT4 promoter causes a 60% decrease in PGC-1-mediated activation of the GLUT4 reporter gene. Thus, regulation of MEF2 amount or activity by PGC-1 may be a significant mechanism of interaction with this promoter, although other productive interactions with transcription factors also appear likely.
Several isoforms of MEF2 exist. To elucidate whether PGC-1 can coactivate MEF2 factors, we transiently transfected the heterologous 293 cell line with a 3× MEF binding site–luciferase reporter gene, along with expression vectors encoding the three individual MEF2 proteins. We compared coactivation between PGC-1 and p/CAF because PGC-1 activates the full GLUT4 promoter and p/CAF does not (Fig. 3A). PGC-1 was a poor coactivator of MEF2A and MEF2D, but stimulated a 4-fold increase in transcription via MEF2C (Fig. 3C). In contrast, p/CAF was a powerful coactivator of MEF2A and MEF2D, but had a much smaller effect on MEF2C. Taken together, these data indicate that much of PGC-1's effect on the GLUT4 promoter occur via the MEF2 site and that PGC-1 preferentially coactivates the MEF2C isoform.
Identification of Domains Involved in the PGC-1/MEF2C Interaction.
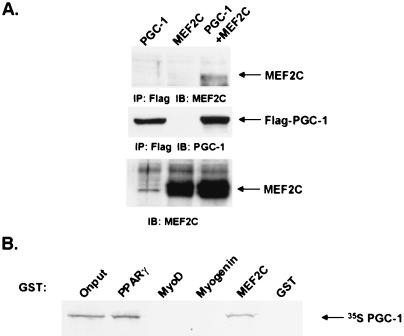
To investigate the biochemical characteristics of the interaction between PGC-1 and MEF2C, we first coexpressed these proteins in cells. The 293 cell line was transiently transfected with a Flag-tagged PGC-1 expression vector in the absence or presence of a MEF2C expression vector. Following anti-Flag immunoprecipitation, the interaction between Flag-PGC-1 and MEF2C was assessed by immunoblotting the precipitates for the presence of MEF2. MEF2C protein was detected in the coimmunoprecipitation reaction from cell lysates that expressed both Flag-PGC-1 and MEF2C (Fig. 4A), but not from those expressing either protein alone.
Figure 4.
PGC-1 interacts specifically with MEF2C in vivo and in vitro. (A) Interaction between PGC-1 and MEF2C in cells. Expression vectors for Flag-PGC-1 and MEF2C were transfected into 293T cells, as indicated. Total cellular lysate was collected 48 h posttransfection. Coimmunoprecipitation using a Flag affinity resin and subsequent Western blot analysis was performed as described in Materials and Methods. (B) Specific interaction between PGC-1 and MEF2C in vitro. GST fusion proteins for PPARγ, MyoD, Myogenin, and MEF2C were expressed in E. coli. Purified GST fusion proteins were bound to beads and incubated with [35S]-labeled PGC-1. After extensive washing, [35S]-labeled PGC-1/activator interaction was detected by autoradiography following SDS/PAGE separation.
To examine the interaction of PGC-1 interaction with MEF2C in vitro, we generated PGC-1 as an [35S]-radiolabeled reticulocyte translation product and assayed their interaction in binding reactions with immobilized GST fusion protein for MEF2C and for similar fusion proteins for PPARγ, MyoD, and myogenin. As described (5), we found that ≈20% of the radiolabeled PGC-1 interacted with the nuclear receptor, PPARγ. Furthermore, ≈20% of the radiolabeled PGC-1 interacted with MEF2C, but not with two other transcription factors important in muscle-selective gene expression, MyoD and myogenin (Fig. 4B). These data together suggest that the effects of PGC-1 on the MEF2 promoter site occur via a physical interaction with MEF2C.
PGC-1 contains several distinct domains: an N-terminal transcriptional activation region within amino acids (aa) 1–200, a domain known to interact with several transcription factors at aa 200–400, and a C-terminal domain within aa 550–797 involved in processing newly transcribed RNA. The major domains within PGC-1 that interact with PPARγ and NRF proteins have been mapped to PGC-1 aa 338–403 and 180–403, respectively (5, 6). To determine how MEF2C interacts with PGC-1, we generated various carboxy-terminal deletions of PGC-1 as [35S]-radiolabeled reticulocyte translation products and established binding reactions with immobilized GST-MEF2C fusion protein. We detected weak interactions between MEF2C and PGC-1 aa 1–187, 1–292, and 1–403. However, PGC-1 1–570, as well as full-length PGC-1, interacted strongly with MEF2C as 23% and 20% of onput was recovered from the affinity chromatography, respectively (Fig. 5A). Functional interaction between MEF2C and PGC-1 deletion constructs was determined by transient transfection assays using GLUT4 luciferase as reporter. We found that PGC-1 aa 1–400, which contain transcriptional activation function and interact with transcription factors PPARγ and nuclear respiratory factor-1 (NRF-1), were ineffective in eliciting a transcriptional response from the GLUT4 promoter (Fig. 5B). PGC-1 aa 1–550 promoted a 2.5-fold increase in GLUT4 transcription, which is in agreement with the physical interaction that was detected between MEF2C and PGC-1 1–570 deletion protein. The carboxy-terminal amino acids that encode the putative RNA processing domain of PGC-1 (aa 550–797) are necessary for a full transcriptional response because full-length PGC-1 elicited a 6-fold increase in GLUT4 promoter activity. These observations allow us to assign aa 403–570 of PGC-1 as a domain that is necessary to mediate interactions with MEF2C. But these experiments do not rule out the possibility that amino acids within other domains of PGC-1 may also contribute to the interactions with MEF2C. Interestingly, this is the first identified function for this region.
Figure 5.
Interaction between PGC-1 and MEF2C maps to discrete domains. (A) The interaction domain of PGC-1 for MEF2C binding is located between aa 403 and 570. GST-MEF2C fusion protein was immobilized on glutathione beads and incubated with [35S]-labeled PGC-1 deletion fragments. After extensive washing, [35S]-labeled PGC-1 protein interaction with GST-MEF2C was detected by autoradiography following SDS/PAGE separation. (B) Functional interaction between MEF2C and PGC-1 deletion constructs. C2C12 myoblast cells were transfected in a 24-well format with 150 ng GLUT4 2.4-kb promoter-luciferase plasmid, 50 ng CMV βgal plasmid, and 700 ng of CMV-driven expression vectors either for PGC-1 aa 1–400, 1–550 or full-length PGC-1. The cells were transfected for 3 h and 10% FBS/DMEM was replaced overnight. The transfected cells were induced to differentiate for at least 24 h before lysis. Luciferase activity was normalized by β-galactosidase activity. (C) Diagram summarizing the protein fragments of PGC-1 that were examined.
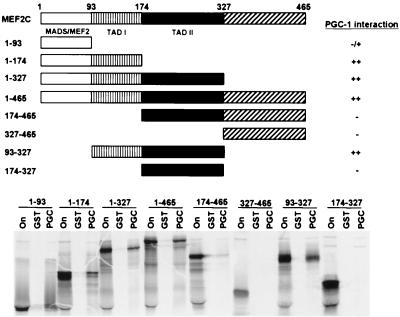
Recently, p300/CBP and GRIP-1 have been shown to function as coactivators for MEF2C by direct physical interaction with the MADS domain of MEF2C at aa 1–100 (18, 19). To determine the domain of MEF2C that directly contacts PGC-1, we constructed various carboxy-terminal and amino-terminal deletions of MEF2C as [35S]-labeled proteins and evaluated their ability to bind an immobilized GST-PGC-1 fusion protein. The MEF2C protein truncations that interacted strongly with PGC-1 contained aa 1–174, 1–327, 1–465, and 93–327 (Fig. 6). MEF2C truncations encoding aa 174–465, 327–465, and 174–327 lacked all ability to bind PGC-1. Only a weak interaction with PGC-1 was detected by using [35S]-labeled MADS box (aa 1–93). These observations point to aa 93–174 of MEF2C as the primary PGC-1 interaction domain. Of interest, this domain has been noted for sustaining 76% of MEF2C's transcriptional activation function in 10T1/2 cells when fused to the GAL4 DNA binding domain, and has been termed transactivation domain I (TADI).
Figure 6.
MEF2C interaction domain for PGC-1. The interaction domain of MEF2C for PGC-1 binding is located between aa 93 and 174. The top panel summarizes the protein fragments of MEF2C that were examined and the results of the GST affinity chromatography experiment that is shown below. GST-PGC-1 fusion protein was immobilized on glutathione beads and incubated with [35S]-labeled MEF2C deletion fragments. After extensive washing, [35S]-labeled MEF2C protein interaction with GST-PGC-1 was detected by autoradiography following SDS/PAGE separation.
Discussion
The transcriptional coactivator PGC-1 has been shown to be a potentially important molecule in the process of adaptive thermogenesis in that it is induced by exposure of animals to cold (5) or refeeding after a fast (unpublished data). In addition, expression of PGC-1 powerfully induces multiple components of a thermogenic response: increased mitochondrial biogenesis, elevated expression of enzymes of the electron transport system and uncoupling proteins, and an overall increase in rates of respiration. Unless cells expressing PGC-1 rapidly catabolize themselves, this increased energy expenditure must be balanced by an increase in fuel uptake. The fact that obese individuals often have excessive fuel levels (glucose and lipids) in their blood, and may have relatively reduced rates of resting energy expenditure (20), suggests that an increased understanding of the connection between these two parameters may ultimately provide benefit in obesity and type 2 diabetes.
Considerable research has been devoted to finding ways to increase glucose uptake and GLUT4 transporter expression in muscle. However, there has been no data to date illustrating means of robustly activating the endogenous GLUT4 gene in muscle cells. In this study, we show that repletion of the coactivator PGC-1 restores the expression of the GLUT4 gene in cultured myotubes from undetectable levels to the levels found in muscle tissue.
The effects of PGC-1 in activating GLUT4 gene expression are reflected in an increased ability of the muscle cells to transport glucose. In addition to the data shown here for L6 cells, we have found that expression of PGC-1 also greatly increased expression of GLUT4 and glucose transport rates in murine C2C12 myotubes. However, in both cell types, though absolute glucose transport rates are greatly increased, the proportion of this transport that is insulin stimulated does not increase. This is almost certainly due to the fact that this increased expression of GLUT4 protein becomes localized to the plasma membrane even in the absence of insulin treatment. This is similar to what has been seen previously when GLUT4 itself is ectopically expressed in these cells or in transgenic mice. Upon analysis of insulin signaling pathways, particularly the tyrosine phosphorylation of the insulin-receptor substrate-1 (IRS-1), or the active phosphorylation of Akt, we have observed no increase in cellular sensitivity to insulin in PGC-1 overexpressing cells. Hence, although PGC-1 is an apparently powerful activator of GLUT4 gene expression, there is no evidence that it sensitizes these cells to insulin.
MEF2C is a member of the MADS family of transcription factors that has been implicated in the regulation of muscle-selective gene expression, including the GLUT4 gene. Mutation of the consensus MEF2 site located at -437/-428 bp (or -473/ -464 in human) resulted in a virtual loss of reporter gene expression in myotubes and in all tissues of transgenic mice (16, 21). However, the independent MEF2 binding sequence alone is insufficient to activate GLUT4 gene expression in myotubes because a 75-bp promoter fragment that encompasses the MEF2 site but omits two downstream thyroid hormone responsive elements (TRE) caused a 60% loss of myotube-specific reporter gene expression. Therefore, the MEF2-binding site of the GLUT4 promoter is necessary but not sufficient for the normal tissue-specific expression of GLUT4. As shown here, ectopic expression of PGC-1 in myotubes activates a 2.4-kb GLUT4 promoter reporter gene by 6- to 10-fold over basal levels of expression, and we found that PGC-1 requires an intact MEF2 site of the GLUT4 promoter for full activation of the GLUT4 promoter. It is likely that the striking ability of PGC-1 to powerfully activate GLUT4 gene transcription relies on combinatorial activities of multiple transcriptional activators, such as thyroid hormone receptor (TR) and E-box binding proteins, in addition to MEF2.
Although the identity of the protein(s) that bind to the MEF2 sequence of the GLUT4 gene in muscle cannot be determined with absolute certainty, our data suggest PGC-1 is likely working by coactivation of MEF2C. To date, four vertebrate genes have been identified to give rise to distinct MEF2 gene products: MEF2-A, -B, -C, and -D (reviewed in ref. 22). In skeletal muscle in culture, MEF2D is expressed in proliferating myoblasts, before the onset of differentiation. MEF2A is expressed at the onset of differentiation, whereas MEF2C is expressed late in the course of differentiation. We found that both the endogenous GLUT4 gene and the GLUT4 reporter construct are induced maximally by PGC-1 in differentiated myotube cultures. In agreement with the expression patterns of MEF2 proteins in myotube culture and with the observation made by Thai et al. (17) that MEF2C binds specifically to the GLUT4 MEF2-binding sequence, we believe that MEF2C is responsible, at least in part, for the activation of the GLUT4 gene by PGC-1. At least a portion of the coactivation of MEF2C by PGC-1 is through the transactivation domain I (TADI) domain of MEF2C (18, 19).
MEF2C has been shown to be involved in several important biological functions, particularly skeletal muscle differentiation and gene expression, cardiac-selective gene expression, and vascular development. In addition to the effects shown here, it is highly likely that the MEF2C-PGC-1 interaction is involved in other MEF2C-regulated processes, particularly where PGC-1 is highly expressed—e.g., skeletal muscle and heart. However, it is important to note that PGC-1 has been found to have promoter-selective effects; for example, some but not all PPARγ-regulated genes are increased by ectopic expression of PGC-1 in the adipose lineages (5). Hence, it will be important to examine the specific effects of PGC-1 on a variety of MEF2C target genes.
GLUT4 gene expression is regulated by a variety of stimuli, both physiological and pharmacological. For example, streptozotocin-induced diabetes and denervation result in decreased GLUT4 mRNA in skeletal muscle; whereas cold exposure, exercise training, and triiodo-l-thyronine treatment increase GLUT4 mRNA in skeletal muscle (11, 23–28). The ability of PGC-1 to regulate GLUT4 gene expression in muscle suggests that PGC-1 potentially could be a therapeutic target in various diabetic states.
Acknowledgments
We thank Drs. Kohjiro Ueki, Peter Bailey, Pasha Sarraf, and Elisabetta Mueller for helpful discussions. We gratefully acknowledge the gift of L6 myocyte cultures and advice from Drs. Phil Bilan and Amira Klip. We thank Dr. Eric Olson for MEF2 expression plasmids and reporter constructs, Dr. Jeffrey Pessin for the GLUT4 promoter construct, and Dr. Andrew Lassar for GST-MyoD, -Myogenin, and -MEF2C expression plasmids. This work was supported by a grant from the National Institutes of Health (DK54477 to B.M.S.). L.F.M. was supported by a postdoctoral fellowship from the National Institutes of Health (GM19680).
Abbreviations
- GLUT4
insulin-sensitive glucose transporter
- moi
multiplicity of infection
- GFP
green fluorescent protein
- MEF
myocyte enhancer factor
References
- 1.Warram J H, Rich S S, Krolewski A S. In: Joslin's Diabetes Mellitus. Kahn C R, Weir G C, editors. Philadelphia: Lea and Febiger; 1995. [Google Scholar]
- 2.Cline G W, Petersen K F, Krssak M, Shen J, Hundal R S, Trajanoski Z, Inzucchi S, Dresner A, Rothman D L, Shulman G I. N Engl J Med. 1999;341:240–246. doi: 10.1056/NEJM199907223410404. [DOI] [PubMed] [Google Scholar]
- 3.Zisman A, Peroni O D, Abel E D, Michael M D, Mauvais-Jarvis F, Lowell B B, Wojtaszewski J F P, Hirshman M F, Virkamaki A, Goodyear L J, et al. Nat Med. 2000;6:924–928. doi: 10.1038/78693. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs E M, Stock J L, McCoid S C, Stukenbrok H A, Pessin J E, Stevenson R W, Milici A J, McNeish J D. J Clin Invest. 1995;95:1512–1518. doi: 10.1172/JCI117823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 6.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, et al. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 7.Khayat Z A, Tsakiridis T, Ueyama A, Somwar R, Ebina Y, Klip A. Am J Physiol. 1998;275:C1487–C1497. doi: 10.1152/ajpcell.1998.275.6.C1487. [DOI] [PubMed] [Google Scholar]
- 8.Bashan N, Burdett E, Guma A, Sargeant R, Tumiati L, Liu Z, Klip A. Am J Physiol. 1993;264:C430–C440. doi: 10.1152/ajpcell.1993.264.2.C430. [DOI] [PubMed] [Google Scholar]
- 9.Lin B, Coughlin S, Pilch P F. Am J Physiol. 1998;275:E386–E391. doi: 10.1152/ajpendo.1998.275.3.E386. [DOI] [PubMed] [Google Scholar]
- 10.Hansen P A, Gulve E A, Marshall B A, Gao J, Pessin J E, Holloszy J O, Mueckler M. J Biol Chem. 1995;270:1679–1684. doi: 10.1074/jbc.270.5.1679. [DOI] [PubMed] [Google Scholar]
- 11.Ren J M, Marshall B A, Mueckler M M, McCaleb M, Amatruda J M, Shulman G I. J Clin Invest. 1995;95:429–432. doi: 10.1172/JCI117673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheatham B, Kahn C R. Endocr Rev. 1995;16:117–142. doi: 10.1210/edrv-16-2-117. [DOI] [PubMed] [Google Scholar]
- 13.Cushman S W, Goodyear L J, Pilch P F, Ralston E, Galbo H, Ploug T, Kristiansen S, Klip A. Adv Exp Med Biol. 1998;441:63–71. doi: 10.1007/978-1-4899-1928-1_6. [DOI] [PubMed] [Google Scholar]
- 14.Olson A L, Liu M L, Moye-Rowley W S, Buse J B, Bell G I, Pessin J E. J Biol Chem. 1993;268:9839–9846. [PubMed] [Google Scholar]
- 15.Ezaki O. Biochem Biophys Res Commun. 1997;241:1–6. doi: 10.1006/bbrc.1997.7587. [DOI] [PubMed] [Google Scholar]
- 16.Liu M L, Olson A L, Edgington N P, Moye-Rowley W S, Pessin J E. J Biol Chem. 1994;269:28514–28521. [PubMed] [Google Scholar]
- 17.Thai M V, Guruswamy S, Cao K T, Pessin J E, Olson A L. J Biol Chem. 1998;273:14285–14292. doi: 10.1074/jbc.273.23.14285. [DOI] [PubMed] [Google Scholar]
- 18.Sartorelli V, Huang J, Hamamori Y, Kedes L. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S L, Dowhan D H, Hosking B M, Muscat G E. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- 20.Ravussin E, Gautier J F. Int J Obes Relat Metab Disord. 1999;23, Suppl. 1:37–41. doi: 10.1038/sj.ijo.0800793. [DOI] [PubMed] [Google Scholar]
- 21.Richardson J M, Pessin J E. J Biol Chem. 1993;268:21021–21027. [PubMed] [Google Scholar]
- 22.Black B L, Olson E N. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 23.Napoli R, Hirshman M F, Horton E S. J Clin Invest. 1995;96:427–437. doi: 10.1172/JCI118053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garvey W T, Huecksteadt T P, Birnbaum M J. Science. 1989;245:60–63. doi: 10.1126/science.2662408. [DOI] [PubMed] [Google Scholar]
- 25.Block N E, Menick D R, Robinson K A, Buse M G. J Clin Invest. 1991;88:1546–1552. doi: 10.1172/JCI115465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wake S A, Sowden J A, Storlien L H, James D E, Clark P W, Shine J, Chisholm D J, Kraegen E W. Diabetes. 1991;40:275–279. doi: 10.2337/diab.40.2.275. [DOI] [PubMed] [Google Scholar]
- 27.Ploug T, Stallknecht B M, Pedersen O, Kahn B B, Ohkuwa T, Vinten J, Galbo H. Am J Physiol. 1990;259:E778–E786. doi: 10.1152/ajpendo.1990.259.6.E778. [DOI] [PubMed] [Google Scholar]
- 28.Weinstein S P, Watts J, Haber R S. Endocrinology. 1991;129:455–464. doi: 10.1210/endo-129-1-455. [DOI] [PubMed] [Google Scholar]
- 29.Lehman J J, Barger P M, Kovacs A, Saffitz J E, Medeiros D M, Kelly D P. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klip A, Guma A, Ramlal T, Bilan P J, Lam L, Leiter L A. Endocrinology. 1992;130:2535–2544. doi: 10.1210/endo.130.5.1572281. [DOI] [PubMed] [Google Scholar]
- 31.Volchuk A, Mitsumoto Y, He L, Liu Z, Habermann E, Trimble W, Klip A. Biochem J. 1994;304:139–145. doi: 10.1042/bj3040139. [DOI] [PMC free article] [PubMed] [Google Scholar]