Abstract
BACKGROUND
Carbamazepine causes various forms of hypersensitivity reactions, ranging from maculopapular exanthema to severe blistering reactions. The HLA-B★1502 allele has been shown to be strongly correlated with carbamazepine-induced Stevens–Johnson syndrome and toxic epidermal necrolysis (SJS–TEN) in the Han Chinese and other Asian populations but not in European populations.
METHODS
We performed a genomewide association study of samples obtained from 22 subjects with carbamazepine-induced hypersensitivity syndrome, 43 subjects with carbamazepine-induced maculopapular exanthema, and 3987 control subjects, all of European descent. We tested for an association between disease and HLA alleles through proxy single-nucleotide polymorphisms and imputation, confirming associations by high-resolution sequence-based HLA typing. We replicated the associations in samples from 145 subjects with carbamazepine-induced hypersensitivity reactions.
RESULTS
The HLA-A★3101 allele, which has a prevalence of 2 to 5% in Northern European populations, was significantly associated with the hypersensitivity syndrome (P = 3.5×10−8). An independent genomewide association study of samples from subjects with maculopapular exanthema also showed an association with the HLA-A★3101 allele (P = 1.1×10−6). Follow-up genotyping confirmed the variant as a risk factor for the hypersensitivity syndrome (odds ratio, 12.41; 95% confidence interval [CI], 1.27 to 121.03), maculopapular exanthema (odds ratio, 8.33; 95% CI, 3.59 to 19.36), and SJS–TEN (odds ratio, 25.93; 95% CI, 4.93 to 116.18).
CONCLUSIONS
The presence of the HLA-A★3101 allele was associated with carbamazepine-induced hypersensitivity reactions among subjects of Northern European ancestry. The presence of the allele increased the risk from 5.0% to 26.0%, whereas its absence reduced the risk from 5.0% to 3.8%. (Funded by the U.K. Department of Health and others.)
Carbamazepine is one of the most commonly prescribed drugs for the treatment of epilepsy, as well as trigeminal neuralgia and bipolar disorder. A minority of treated persons have hypersensitivity reactions that vary in prevalence and severity,1 with some forms associated with substantial morbidity and mortality. The mildest form, maculopapular exanthema, occurs in 5 to 10% of treated persons of European ancestry and resolves spontaneously after drug discontinuation. More severe reactions, such as the hypersensitivity syndrome, are associated with mortality of up to 10%2 and include symptoms such as rash, fever, eosinophilia, hepatitis, and nephritis. The most severe reactions, such as the Stevens–Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN), are characterized by a blistering rash affecting a variable percentage of the body-surface area. The rate of death increases with the degree of epidermal detachment. TEN is the rarest of these phenotypes and is associated with mortality of up to 30%. According to the labeling of carbamazepine, as mandated by the Food and Drug Administration (FDA), the estimated incidence of SJS–TEN is 1 to 6 cases in 10,000 persons of European ancestry who are exposed to the drug.
Genomewide approaches are increasingly used to identify genetic predisposing factors for drug-induced hypersensitivity reactions and drug-induced liver injury.3 For example, the HLA-B★1502 allele has been identified as a clinically important predictor of SJS–TEN in Asians of Han Chinese descent who are candidates for treatment with carbamazepine.4 This finding led the FDA to require a warning label for carbamazepine to indicate the need for genotyping for HLA-B★1502 before the drug is prescribed. This change seems warranted, given the prospective study described in this issue of the Journal,5 which shows that withholding carbamazepine from subjects of Han Chinese ancestry who carry the HLA-B★1502 allele substantially decreased the rate of the development of SJS–TEN. The value of HLA-B★1502 in predicting the risk of SJS–TEN has been shown in other Asian populations, including those of Thailand, Malaysia, and India.4,6–8 This effect is specific for SJS–TEN, as compared with the full spectrum of carbamazepine-related hypersensitivity reactions, which led to the suggestion that different phenotypes of carbamazepine-induced hypersensitivity may have distinct genetic predictors. Furthermore, HLA-B★1502 is rare (with a prevalence of less than 2%) in populations of European descent, in which carbamazepine-induced SJS–TEN occurs at a lower incidence than in Asian populations.9–12 Using a candidate-gene approach, previous studies have shown weak or moderate signals of association between maculopapular exanthema or the DRESS (drug reaction with eosinophilia and systemic symptoms) syndrome and single-nucleotide polymorphisms (SNPs) in the major histocompatibility complex (MHC) region,13–15 and these associations have not been consistently replicated.13,14
We present data from two independent studies that suggest the relevance of an HLA variant to the clinical spectrum of carbamazepine-related hypersensitivity reactions, including maculopapular exanthema, the hypersensitivity syndrome, and SJS–TEN.
METHODS
CASE SUBJECTS
Recruitment
We recruited case subjects at centers collaborating with the University of Liverpool and Walton Centre for Neurology (both hereinafter referred to as “the Liverpool collaborators”) or at centers affiliated with the EPIGEN consortium (for details, see the Supplementary Appendix, available with the full text of this article at NEJM.org). All case subjects and control subjects were of European ancestry as determined by either self-report or genetic-marker analysis.13 Many of these subjects have been described previously in the context of hypersensitivity reactions.9,13,16
Hypersensitivity Syndrome
The group with the hypersensitivity syndrome consisted of 26 subjects: 23 who were recruited through the Liverpool collaborators and 3 who were recruited through the EPIGEN consortium. The hypersensitivity syndrome was defined as the presence of rash or liver involvement within 3 months after the initiation of carbamazepine treatment, accompanied by a minimum of two of the following manifestations: a prolonged recovery phase, despite withdrawal of the drug; fever; involvement of other internal organs (liver, kidney, lung, central nervous system, heart, muscle, thyroid, or lymphoid system); or the presence of hematologic abnormalities, such as eosinophilia and atypical lymphocytosis.17 We considered the DRESS syndrome to be the same as the hypersensitivity syndrome.
Acute Generalized Exanthematous Pustulosis
We recruited one subject with acute generalized exanthematous pustulosis (AGEP), who had a typical pustular reaction with high fever,18 from a collaborating clinic in Switzerland. We then analyzed the data from this subject together with data from the subjects with the hypersensitivity syndrome.
Maculopapular Exanthema
The group with maculopapular exanthema consisted of 106 subjects: 57 who were recruited through the Liverpool collaborators and 49 who were recruited through the EPIGEN consortium. Maculopapular exanthema was defined as rash without systemic symptoms that required the discontinuation of carbamazepine within 3 months after the initiation of drug therapy.
SJS–TEN
The SJS–TEN group consisted of 12 subjects: 8 who were recruited through the Liverpool collaborators and 4 who were recruited through the EPIGEN consortium. The diagnostic criteria suggested by the RegiSCAR Project (a research network for severe cutaneous adverse drug reactions)11,19 were used for this group (see the Supplementary Appendix).
CONTROL SUBJECTS
Population Controls
Genotype data from the Wellcome Trust Case Control Consortium (WTCCC) (www.wtccc.org.uk) were used as a population control group. The Liverpool collaborators used data from a genome-wide association study of samples from 2691 subjects in the U.K. National Blood Services Collection. EPIGEN used samples from a homogeneous subgroup of 1296 subjects in the 1958 British Birth Cohort that were selected by principal component analysis. We did not screen either set of population controls for carbamazepine-related adverse drug reactions. (For details about our access to these data, see the Supplementary Appendix.)
Clinical Controls
The clinical control group consisted of 257 subjects with epilepsy who had been taking carbamazepine for at least 3 months with no clinical or biochemical signs of hypersensitivity. Of these subjects, 44 were recruited by the Liverpool collaborators and 213 were recruited from clinics affiliated with EPIGEN.
GENOMEWIDE GENOTYPING
Genomewide analysis of samples that were initially available from the 22 Liverpool subjects with carbamazepine-induced hypersensitivity syndrome was performed with the use of the Illumina Infinium 1.2M chip at the Wellcome Trust Sanger Institute. Samples from 43 case subjects and 200 control subjects from the EPIGEN consortium were genotyped with the use of the Illumina 610K Quad platform at the Duke University Center for Human Genome Variation. (Details regarding quality controls and principal-components analysis are provided in the Supplementary Appendix.)
IMPUTATION OF HLA ALLELES
We based the imputation of classic HLA alleles in samples from the 22 Liverpool subjects with the hypersensitivity syndrome and control subjects from the U.K. National Blood Service on a dense reference panel of SNP data and four-digit HLA types in 2767 unrelated subjects of European descent, according to a method described recently.20 We imputed classic HLA alleles in the samples from the EPIGEN consortium, using MACH 1.0 on the Web site of the Center for Statistical Genetics (www.sph.umich.edu/csg/abecasis/MACH).21 The 1458 subjects in the HLA-typed 1958 British Birth Cohort constituted the reference population. (See the Supplementary Appendix for further details.) We used different approaches to impute the HLA alleles in the Liverpool and EPIGEN subjects, because the two studies were originally independent.
HLA-A HIGH-RESOLUTION GENOTYPING
To confirm the accuracy of imputation, high-resolution, sequence-based HLA-A typing of samples from 22 subjects with the hypersensitivity syndrome and from a subgroup of 44 clinical control subjects (i.e., subjects who did not have adverse reactions to carbamazepine) was performed by Histogenetics. We determined the HLA-A alleles in samples from 49 subjects with maculopapular exanthema and a subgroup of 213 clinical control subjects from the EPIGEN consortium, using sequence-specific polymerase-chain-reaction assay primers (details available on request). Direct genotyping showed 100% concordance with the imputed alleles.
HLA-A*3101 PROXY SNP GENOTYPING
Genotyping of the rs1061235 SNP in samples from all Liverpool subjects (including case subjects with maculopapular exanthema, the hypersensitivity syndrome, or SJS–TEN and clinical control subjects) was performed by KBioscience. This allele has previously been shown to be in complete linkage disequilibrium (r2 = 1) with the HLA-A★3101 allele in populations of European descent.22
STATISTICAL ANALYSIS
Statistical analyses were performed with the use of PLINK (version 1.05),23 RevMan (version 5) (http://ims.cochrane.org/revman), and Haploview24 software packages. For the genomewide association studies, both the Liverpool collaborators and the EPIGEN consortium used logistic regression with an additive model of inheritance. They adjusted for population stratification by including significant principal components as covariates in the logistic-regression model and used a P value of less than 5.0×10−8 to indicate genomewide significance. Pooled analysis of the different groups of subjects was performed to estimate odds ratios and confidence intervals from summary data with the use of a random-effects model. Between-study heterogeneity was tested by calculating I2 and Tau.2 The z-score directionality or estimated risk was checked for consistency with observed data.
RESULTS
ASSOCIATION WITH THE HYPERSENSITIVITY SYNDROME
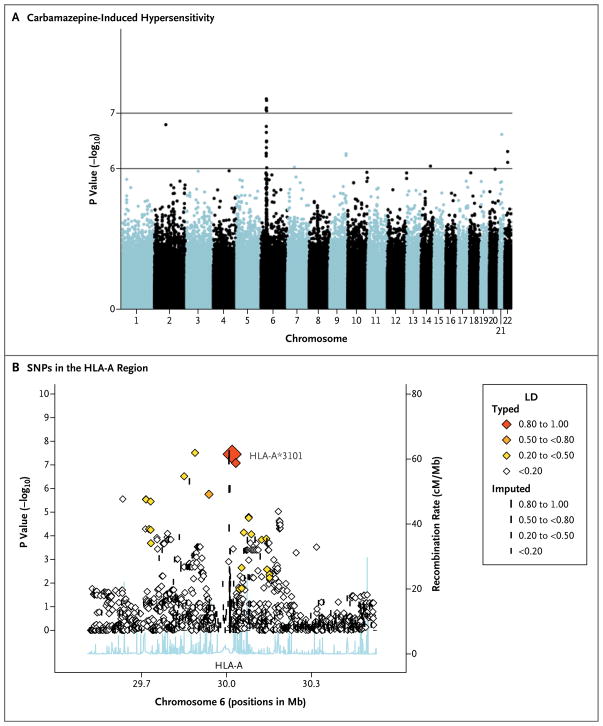
We first conducted an association analysis involving 1.2M SNPs in samples from the 22 Liver-pool subjects with carbamazepine-induced hypersensitivity syndrome and from 2691 healthy controls from the U.K. National Blood Services Collection. (The full set of results can be obtained from the European Genotype Archive, access number EGAS00000000037.) We identified a strong signal in the MHC region on chromosome 6, with several SNPs around HLA-A reaching genome-wide significance (P = 3.5×10−8) (Fig. 1A and 1B). This variant was seen in 40.0% of case subjects but in only 4.9% of control subjects. The top hit (rs1061235) had previously been shown to be a proxy for the HLA-A★3101 allele in persons of European descent.22 Therefore, we focused on this locus as a predictor of response through high-resolution, sequence-based HLA-A typing, which confirmed the absolute correlation (r2 = 1) between rs1061235 and HLA-A★3101 that had been observed previously.22 No other classic HLA-A alleles reached nominal significance (Table 1 in the Supplementary Appendix).
Figure 1. Results of a Genomewide Association Study of Samples from Case Subjects with Carbamazepine-Induced Hypersensitivity Syndrome and Control Subjects.
Panel A shows a Manhattan plot for logistic regression with a strong signal in the HLA-A region on chromosome 6. Each dot represents a P value for the comparison of 22 case subjects with the hypersensitivity syndrome and 2691 healthy control subjects from the Wellcome Trust Case Control Consortium. Panel B shows P values for single-nucleotide polymorphisms (SNPs) in the HLA-A region. There is a strong association between carbamazepine-induced hypersensitivity syndrome and the HLA-A*3101 allele, with identification of the allele in 40.0% of case subjects and only 4.9% of control subjects. The x axis shows the SNP position on chromosome 6 (National Center for Biotechnology Information build 36). The left y axis shows the negative log10 of P values for the comparison between case subjects and control subjects, as calculated with logistic regression. The right y axis shows the recombination rate on chromosome 6 between 29 and 31 Mb. The diamonds show the degree of linkage disequilibrium (LD) in the samples.
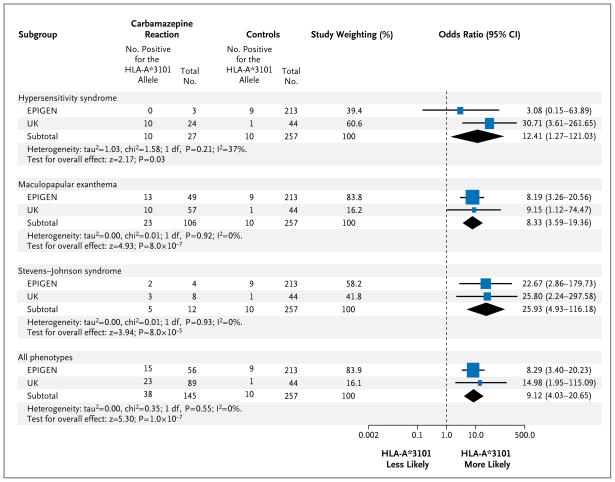
During the course of this study, samples from 4 additional subjects with the hypersensitivity syndrome and from 1 additional subject with AGEP became available from the Liverpool and EPIGEN centers. Of these 5 subjects, 2 (including the single subject with AGEP) tested positive for HLA-A★3101. A comparison of all 27 subjects with the 257 control subjects without adverse drug reactions produced an odds ratio of 12.41 (95% confidence interval [CI], 1.27 to 121.03; P = 0.03) (Fig. 2).
Figure 2. Distribution of the HLA-A*3101 Allele across the Spectrum of Clinical Phenotypes of Case Subjects with Carbamazepine- Induced Hypersensitivity and Control Subjects without Adverse Reactions to Carbamazepine.
Subjects were recruited at centers collaborating with the University of Liverpool and Walton Centre for Neurology (UK) or centers affiliated with the EPIGEN consortium. The sample obtained from one patient with acute generalized exanthematous pustulosis (AGEP) was analyzed with the samples for the group of case subjects with the hypersensitivity syndrome. Since there were no observations of the HLA-A*3101 allele in the three case subjects with the hypersensitivity syndrome in the EPIGEN cohort, 0.5 was added to each value in a two-by-two contingency table to estimate an odds ratio. One patient with the Stevens–Johnson syndrome and toxic epidermal necrolysis (SJS–TEN) was of mixed European and Thai ancestry. Study weighting (indicated by different sizes of squares) refers to the proportion of subjects who were recruited from each study cohort. Diamonds indicate pooled odds ratios. The horizontal lines indicate 95% confidence intervals. The abbreviation df denotes degrees of freedom, and I2 is the percentage of total variation that is due to heterogeneity rather than chance.
ASSOCIATION WITH MACULOPAPULAR EXANTHEMA
EPIGEN investigators performed a genomewide association study of samples from 43 subjects with maculopapular exanthema and 1296 healthy control subjects from the 1958 British Birth Cohort. (The full set of results can be obtained from dbGAP, access number EGAS00000000036.) HLA-A★3101 was the most strongly associated allele (P = 1.11×10−6) and was observed in 27% of case subjects and 4% of control subjects, although the effect was not significant genomewide (Fig. 3 in the Supplementary Appendix). However, we had access to another 63 case subjects with maculopapular exanthema and 257 control subjects without adverse drug reactions. Our combined analysis of all 106 subjects with maculopapular exanthema and the 257 clinical control subjects showed a significant allelic association with HLA-A★3101 (P=8.0×10−7), generating an odds ratio of 8.33 (95% CI, 3.59 to 19.36) (Fig. 2). Although the analyses of samples from subjects with maculopapular exanthema and those with the hypersensitivity syndrome were carried out in the Netherlands and Liverpool, respectively, the effect size of HLA-A★3101 on risk was consistent across the EPIGEN and U.K. groups.
ASSOCIATION WITH SJS–TEN
On the basis of our hypothesis that the HLA-A★3101 allele is associated with carbamazepine-induced SJS–TEN, we genotyped HLA-A★3101 in a group of 12 subjects with SJS–TEN who were recruited from both the Liverpool and EPIGEN-affiliated centers. Of these 12 subjects, 5 (42%) carried the allele, as compared with 10 (4%) of the 257 clinical control subjects (odds ratio, 25.93; 95% CI, 4.93 to 116.18; P = 8.0×10−5) (Fig. 2).
A pooled analysis of all 145 subjects with carbamazepine-induced hypersensitivity and 257 control subjects without adverse drug reactions showed a strong association between hypersensitivity and the HLA-A★3101 allele (odds ratio, 9.12; 95% CI, 4.03 to 20.65; P = 1.0×10−7). On the basis of this pooled collection, we calculated that the presence of HLA-A★3101 had a sensitivity of 26% and a specificity of 96% as a predictor of carbamazepine-associated hypersensitivity. Given that carbamazepine hypersensitivity has a prevalence of 5% (1:20), application of the test criteria from our pooled analysis would increase the post-test probability to 26% (approximately 1:4) on the basis of the positive likelihood ratio of 6.74 (95% CI, 3.51 to 13.00). Since the negative likelihood ratio was 0.77 (95% CI, 0.67 to 0.84), a negative HLA-A★3101 test would reduce the probability of hypersensitivity from 5.0% to 3.8% (1:26 ratio).
DISCUSSION
We found that a variation in the presence of the HLA-A★3101 allele is an important predictor of the full spectrum of carbamazepine-induced hypersensitivity reactions in persons of European descent. Such reactions range from the relatively mild but nevertheless troublesome maculopapular exanthema to more severe reactions, such as the hypersensitivity syndrome and SJS–TEN. Although the presence of the HLA-A★3101 allele is neither necessary nor sufficient for the development of hypersensitivity to carbamazepine, it is associated with a significantly increased risk.
Our data add to growing evidence of the role of different HLA alleles in predisposing patients to immune-mediated adverse reactions, such as drug-induced hypersensitivity4,25–27 and liver injury.3,28 The most prominent example thus far is the strong predictive value of the HLA-B★5701 allele for hypersensitivity to the drug abacavir, used for the treatment of human immunodeficiency virus infection.25,26,29 FDA guidelines now recommend HLA-B★5701 testing in advance of abacavir prescription. Testing results in a reduction in the incidence of abacavir hypersensitivity27,30 and is cost-effective.29,31
Our study now provides strong evidence suggesting that HLA-A★3101 is a predictor of carbamazepine hypersensitivity in Europeans. First, despite the small number of subjects who were included in the analysis, this signal had genome-wide significance. Second, the signal was observed across independent groups of case subjects with different phenotypes of carbamazepine hypersensitivity, with the use of independent sets of control subjects. Third, HLA-A★3101 has previously been shown to be associated with multiple carbamazepine-hypersensitivity phenotypes, including SJS–TEN in Japanese subjects15,32 and with carbamazepine-induced maculopapular exanthema in subjects of Han Chinese ancestry.14 The one subject in our study who had AGEP, a reaction characterized by sterile pustules, neutrophilic inflammation, and high levels of interleukin-8 in skin,33 also was found to carry the HLA-A★3101 allele. The prevalence of this allele is 2 to 5% in Northern European populations, 2% in Han Chinese populations, and 9% in Japanese populations.32,34,35 The finding that HLA-A★3101 is associated with carbamazepine-induced hypersensitivity in populations of different ancestries parallels the finding of HLA-B★5701-associated abacavir hypersensitivity.30,36
HLA-B★1502, a predictor of carbamazepine hypersensitivity reactions in Asian populations, seems to be phenotype-specific in that it predicts the development of SJS–TEN but not the development of the hypersensitivity syndrome or maculopapular exanthema.14 This finding has led to the suggestion that the pathogenesis of carbamazepine-induced SJS–TEN is distinct from that of the hypersensitivity syndrome. This hypothesis is based on the fact that class I alleles are usually associated with specific CD8+ cytotoxic T-cell reactions, which is consistent with the known pathologic features of SJS–TEN.37 However, our data suggest that the class I allele HLA-A★3101 may be important for an increased range of phenotypes, including maculopapular exanthema and the hypersensitivity syndrome, at least in European populations. Persons with carbamazepine-induced hypersensitivity syndrome have been shown to have several types of drug-specific T cells in peripheral blood, including CD4+, CD8+, and CD4−CD8+ cells.38 Although there is no obvious relationship between HLA-A★3101 and HLA-B★1502, reports have indicated a possible overlap in peptide-binding specificity.39 Given the degree of linkage disequilibrium across the extended MHC region, we cannot be certain that either HLA-B★1502 or HLA-A★3101 is causal.
We used samples from healthy volunteers in the WTCCC as control subjects for both genomewide association studies. Given that the hypersensitivity syndrome occurs in 1 in 5000 to 1 in 10,000 persons, it is unlikely that more than one or two population control subjects would be at risk for hypersensitivity with exposure to carbamazepine. The larger WTCCC collections therefore afforded increased power over that provided by comparison of case subjects with the clinical control subjects. Nevertheless, in analyses involving the 257 clinical control subjects, the association with HLA-A★3101 remained strong for both the hypersensitivity syndrome (odds ratio, 12.41; 95% CI, 1.27 to 121.03) and SJS (odds ratio, 25.93; 95% CI, 4.93 to 116.18), although the effect size was smaller with maculopapular exanthema (odds ratio, 8.33; 95% CI, 3.59 to 19.36).
Additional studies in populations of European descent and other ancestry are required to further characterize this association. The International Serious Adverse Event Consortium is coordinating a global effort to recruit subjects with drug-induced severe adverse events (www.saeconsortium.org). Efforts such as this will facilitate the analysis of larger groups of subjects, which may well uncover other genetic variants associated with a smaller effect size or population-specific variants. Additional work is also required to determine whether the effect of the HLA-A★3101 allele is specific to carbamazepine or whether it also applies to other drugs. A prospective study that is based on the results we present here would facilitate the routine clinical use of HLA-A★3101 testing in carbamazepine treatment. In this issue of the Journal, a report describes how prospective HLA-B★1502 typing can prevent carbamazepine-induced SJS–TEN in Han Chinese populations.5
We propose that HLA-A★3101 is clinically relevant as a marker to predict hypersensitivity reactions. On the assumption that the prevalence of carbamazepine-induced hypersensitivity is 5.0%, the presence of the HLA-A★3101 allele increases the risk of hypersensitivity to 26.0%, whereas its absence reduces the risk to 3.8%. Among Japanese patients, the estimated reduction is from 2.9% to 1.1%. The sensitivity and specificity values estimated here for Europeans (26% and 96%) and by others for Japanese populations (61% and 88%)32 vary. Among a number of possible reasons for these differences are differences in study size, in the frequency of HLA-A★3101 (and potential interacting variation), and in the effect size of the allele across the two populations. Nonetheless, these numbers are similar to those underlying the test criteria calculated for the presence of the HLA-B★5701 allele and abacavir hypersensitivity (sensitivity, 45.5%; specificity, 97.6%; and a risk reduction from 7.8% to 2.7%).
On the basis of our data, we have calculated that 83 patients would need to be screened to prevent one case of carbamazepine hypersensitivity; the number in the Japanese population is 56. However, it is important to note that this calculation is based on a conservative estimate of the prevalence of carbamazepine hypersensitivity (5%). The prevalence of carbamazepine-induced hypersensitivity that was determined in the context of the Standard and New Antiepileptic Drugs (SANAD) trial (Current Controlled Trials number, ISRCTN38354748) (also involving subjects of European descent) was 10%.40 On the basis of this prevalence, the number of persons who would need to be screened to prevent one instance of carbamazepine-induced hypersensitivity is 39. We therefore suggest that consideration be given to adding the association with HLA-A★3101 to the drug label for carbamazepine.
Supplementary Material
Acknowledgments
Supported by grants to the Liverpool collaborators from the Department of Health, the National Health Service Chair of Pharmacogenetics, the Medical Research Council Centre for Drug Safety Science, the Wolfson Foundation, the Wellcome Trust Sanger Institute, and the National Institute for Health Research (to Dr. Pirmohamed); by grants to the EPIGEN consortium from the Medical Research Council (G0400126), the Well-come Trust (084730), University College London Hospitals Charity, Clinical Research and Development Committee (F136), and the National Institute for Health Research (08–08-SCC); by an award (2009/001) from Brainwave–the Irish Epilepsy Association; by the Medical Research Charities Group of Ireland, the Health Research Board, the National Society for Epilepsy, Fonds National de la Recherche Scientifique, and Fonds Erasme pour la Recherche Médicale, Université Libre de Bruxelles (Belgium); by a Translational Research Scholars award from the Health Research Board of Ireland (to Mr. McCormack); by the Department of Health National Institute for Health Research Biomedical Research Centres; and by a contract from the National Cancer Institute (HHS-N261200800001E) and the Intramural Research Program at the National Cancer Institute, National Institutes of Health.
We thank the subjects and the physicians who recruited them; U.K. Eudragene co-coordinator Paul McKeigue; Drs. Ines Salado, Alfonso Carvajal, Luisa Ibanez, Jean-Louis Montastruc, Maryse Lapeyre-Mestre, Emmanuelle Bondon-Guitton, Aatif M. Husain, William B. Gallantine, and Mohamad Mikati for their help in recruiting subjects; the pharmacovigilance centers involved in case ascertainment in France and Spain, the Association Française des Centres Régionaux de Pharmacovigilance and Sistema Español de Farmacovigilancia; and Agencia Española de Medica-mentos y Productos Sanitarios.
APPENDIX
The authors’ affiliations are as follows: Molecular and Cellular Therapeutics, the Royal College of Surgeons in Ireland (M. McCormack, S.A., N.D., G.L.C.), and the Division of Neurology, Beaumont Hospital (G.D.O., E.C., N.D.) — both in Dublin; the Department of Molecular and Clinical Pharmacology, University of Liverpool (A.A., G.J.S., T.M., B.K.P., M. Pirmohamed), and Walton Centre for Neurology (T.M.) — both in Liverpool; Wellcome Trust Sanger Institute, Hinxton (S.B., P.D.); and the National Society for Epilepsy, Chalfont-St-Peter, Buckinghamshire (K.C., S.M.S.) — all in the United Kingdom; the Department of Medicine, Boston University (J.F.F.), Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University (M.C.), Harvard–MIT Division of Health Sciences and Technology (X.J.), and the Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School (P.I.W.B.) — all in Boston; the Department of Clinical and Experimental Epilepsy, University College London, Institute of Neurology, Queen Square (D.K., K.C., S.M.S.), the Department of Primary Care and Public Health Sciences, Division of Health and Social Care Research, King’s College (M. Molokhia), and the Centre for Neuroscience, Department of Medicine, Imperial College London (M.R.J.) — all in London; the Cancer and Inflammation Program, Laboratory of Experimental Immunology, SAIC–Frederick, National Cancer Institute at Frederick, Frederick, MD (M.C.); the Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA (P.I.W.B.); Julius Center for Health Sciences and Primary Care and the Department of Medical Genetics, Division of Biomedical Genetics, University Medical Center, Utrecht, the Netherlands (P.I.W.B.); the Center for Human Genome Variation (K.V.S., E.L.H., N.W., D.B.G.) and the Department of Medicine (Neurology) (R.A.R.), Duke University Medical School, Durham, NC; the Department of Neurology, Hôpital Erasme, Université Libre de Bruxelles, Brussels (M. Pandolfo, C.D.); and the Department of Rheumatology, Clinical Immunology, and Allergology, University of Bern, Bern, Switzerland (W.P.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Roujeau JC. Clinical heterogeneity of drug hypersensitivity. Toxicology. 2005;209:123–9. doi: 10.1016/j.tox.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Syn WK, Naisbitt DJ, Holt AP, Pirmohamed M, Mutimer DJ. Carbamazepine-induced acute liver failure as part of the DRESS syndrome. Int J Clin Pract. 2005;59:988–91. doi: 10.1111/j.1368-5031.2005.00550.x. [Erratum, Int J Clin Pract 2005;59: 1371.] [DOI] [PubMed] [Google Scholar]
- 3.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B★5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–9. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 4.Chung WH, Hung SI, Hong HS, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Lin J-J, Lu C-S, et al. Carbamazepine-induced toxic effects and HLA-B★1502 screening in Taiwan. N Engl J Med. 2011;364:1126–33. doi: 10.1056/NEJMoa1009717. [DOI] [PubMed] [Google Scholar]
- 6.Ding WY, Lee CK, Choon SE. Cutaneous adverse drug reactions seen in a tertiary hospital in Johor, Malaysia. Int J Dermatol. 2010;49:834–41. doi: 10.1111/j.1365-4632.2010.04481.x. [DOI] [PubMed] [Google Scholar]
- 7.Locharernkul C, Loplumlert J, Limotai C, et al. Carbamazepine and phenytoin induced Stevens-Johnson syndrome is associated with HLA-B★1502 allele in Thai population. Epilepsia. 2008;49:2087–91. doi: 10.1111/j.1528-1167.2008.01719.x. [Erratum, Epilepsia 2009;50:971.] [DOI] [PubMed] [Google Scholar]
- 8.Mehta TY, Prajapati LM, Mittal B, et al. Association of HLA-B★1502 allele and carbamazepine-induced Stevens-Johnson syndrome among Indians. Indian J Dermatol Venereol Leprol. 2009;75:579–82. doi: 10.4103/0378-6323.57718. [DOI] [PubMed] [Google Scholar]
- 9.Alfirevic A, Jorgensen AL, Williamson PR, Chadwick DW, Park BK, Pirmohamed M. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics. 2006;7:813–8. doi: 10.2217/14622416.7.6.813. [DOI] [PubMed] [Google Scholar]
- 10.Lonjou C, Borot N, Sekula P, et al. A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet Genomics. 2008;18:99–107. doi: 10.1097/FPC.0b013e3282f3ef9c. [DOI] [PubMed] [Google Scholar]
- 11.Lonjou C, Thomas L, Borot N, et al. A marker for Stevens-Johnson syndrome: ethnicity matters. Pharmacogenomics J. 2006;6:265–8. doi: 10.1038/sj.tpj.6500356. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell PB, Jr, McLeod HL. Carbamazepine, HLA-B★1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics. 2008;9:1543–6. doi: 10.2217/14622416.9.10.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfirevic A, Mills T, Harrington P, et al. Serious carbamazepine-induced hypersensitivity reactions associated with the HSP70 gene cluster. Pharmacogenet Genomics. 2006;16:287–96. doi: 10.1097/01.fpc.0000189800.88596.7a. [DOI] [PubMed] [Google Scholar]
- 14.Hung SI, Chung WH, Jee SH, et al. Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmacogenet Genomics. 2006;16:297–306. doi: 10.1097/01.fpc.0000199500.46842.4a. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwagi M, Aihara M, Takahashi Y, et al. Human leukocyte antigen genotypes in carbamazepine-induced severe cutaneous adverse drug response in Japanese patients. J Dermatol. 2008;35:683–5. doi: 10.1111/j.1346-8138.2008.00548.x. [DOI] [PubMed] [Google Scholar]
- 16.Pirmohamed M, Lin K, Chadwick D, Park BK. TNFalpha promoter region gene polymorphisms in carbamazepine-hypersensitive patients. Neurology. 2001;56:890–6. doi: 10.1212/wnl.56.7.890. [DOI] [PubMed] [Google Scholar]
- 17.Kardaun SH, Sidoroff A, Valeyrie-Allanore L, et al. Variability in the clinical pattern of cutaneous side-effects of drugs with systemic symptoms: does a DRESS syndrome really exist? Br J Dermatol. 2007;156:609–11. doi: 10.1111/j.1365-2133.2006.07704.x. [DOI] [PubMed] [Google Scholar]
- 18.Speeckaert MM, Speeckaert R, Lambert J, Brochez L. Acute generalized exanthematous pustulosis: an overview of the clinical, immunological and diagnostic concepts. Eur J Dermatol. 2010;20:425–33. doi: 10.1684/ejd.2010.0932. [DOI] [PubMed] [Google Scholar]
- 19.Auquier-Dunant A, Mockenhaupt M, Naldi L, Correia O, Schröder W, Roujeau JC. Correlations between clinical patterns and causes of erythema multiforme majus, Stevens-Johnson syndrome, and toxic epidermal necrolysis: results of an international prospective study. Arch Dermatol. 2002;138:1019–24. doi: 10.1001/archderm.138.8.1019. [DOI] [PubMed] [Google Scholar]
- 20.The International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bakker PI, McVean G, Sabeti PC, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Hetherington S, Hughes AR, Mosteller M, et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359:1121–2. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]
- 26.Mallal S, Nolan D, Witt C, et al. Association between presence of HLA-B★5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–32. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 27.Mallal S, Phillips E, Carosi G, et al. HLA-B★5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–79. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 28.Donaldson PT, Daly AK, Henderson J, et al. Human leucocyte antigen class II genotype in susceptibility and resistance to co-amoxiclav-induced liver injury. J Hepatol. 2010;53:1049–53. doi: 10.1016/j.jhep.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 29.Hughes DA, Vilar FJ, Ward CC, Alfirevic A, Park BK, Pirmohamed M. Cost-effectiveness analysis of HLA B★5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics. 2004;14:335–42. doi: 10.1097/00008571-200406000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Saag M, Balu R, Phillips E, et al. High sensitivity of human leukocyte antigen-B★5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis. 2008;46:1111–8. doi: 10.1086/529382. [DOI] [PubMed] [Google Scholar]
- 31.Schackman BR, Scott CA, Walensky RP, Losina E, Freedberg KA, Sax PE. The cost-effectiveness of HLA-B★5701 genetic screening to guide initial antiretroviral therapy for HIV. AIDS. 2008;22:2025–33. doi: 10.1097/QAD.0b013e3283103ce6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozeki T, Mushiroda T, Yowang A, et al. Genome-wide association study identifies HLA-A★3101 allele as a genetic risk factor for carbamazepine-induced cutaneous adverse drug reactions in Japanese population. Hum Mol Genet. 2011;20:1034–41. doi: 10.1093/hmg/ddq537. [DOI] [PubMed] [Google Scholar]
- 33.Britschgi M, Steiner UC, Schmid S, et al. T-cell involvement in drug-induced acute generalized exanthematous pustulosis. J Clin Invest. 2001;107:1433–41. doi: 10.1172/JCI12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt AH, Baier D, Solloch UV, et al. Estimation of high-resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Hum Immunol. 2009;70:895–902. doi: 10.1016/j.humimm.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Wen SH, Lai MJ, Yang KL. Human leukocyte antigen-A, -B, and -DRB1 haplotypes of cord blood units in the Tzu Chi Taiwan Cord Blood Bank. Hum Immunol. 2008;69:430–6. doi: 10.1016/j.humimm.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Zucman D, Truchis P, Majerholc C, Stegman S, Caillat-Zucman S. Prospective screening for human leukocyte antigen-B★5701 avoids abacavir hypersensitivity reaction in the ethnically mixed French HIV population. J Acquir Immune Defic Syndr. 2007;45:1–3. doi: 10.1097/QAI.0b013e318046ea31. [DOI] [PubMed] [Google Scholar]
- 37.Lerch M, Pichler WJ. The immunological and clinical spectrum of delayed drug-induced exanthems. Curr Opin Allergy Clin Immunol. 2004;4:411–9. doi: 10.1097/00130832-200410000-00013. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Farrell J, Pirmohamed M, Park BK, Naisbitt DJ. Generation and characterization of antigen-specific CD4+, CD8+, and CD4+CD8+ T-cell clones from patients with carbamazepine hypersensitivity. J Allergy Clin Immunol. 2007;119:973–81. doi: 10.1016/j.jaci.2006.12.617. [DOI] [PubMed] [Google Scholar]
- 39.Chelvanayagam G. A roadmap for HLA-A, HLA-B, and HLA-C peptide binding specificities. Immunogenetics. 1996;45:15–26. doi: 10.1007/s002510050162. [DOI] [PubMed] [Google Scholar]
- 40.Marson AG, Al-Kharusi AM, Alwaidh M, et al. The SANAD study of effectiveness of carbamazepine, gabapentin, la-motrigine, oxcarbazepine, or topiramate for treatment of partial epilepsy: an un-blinded randomised controlled trial. Lancet. 2007;369:1000–15. doi: 10.1016/S0140-6736(07)60460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.