Abstract
Inversion polymorphisms have been linked to a variety of fundamental biological and evolutionary processes. Yet few studies have used large-scale genomic sequencing to directly compare the haplotypes associated with the standard and inverted chromosome arrangements. Here we describe the targeted genomic sequencing and comparison of haplotypes representing alternative arrangements of a common inversion polymorphism linked to a suite of phenotypes in the white-throated sparrow (Zonotrichia albicollis). More than 7.4 Mb of genomic sequence was generated and assembled from both the standard (ZAL2) and inverted (ZAL2m) arrangements. Sequencing of a pair of inversion breakpoints led to the identification of a ZAL2-specific segmental duplication, as well as evidence of breakpoint reusage. Comparison of the haplotype-based sequence assemblies revealed low genetic differentiation outside versus inside the inversion indicative of historical patterns of gene flow and suppressed recombination between ZAL2 and ZAL2m. Finally, despite ZAL2m being maintained in a near constant state of heterozygosity, no signatures of genetic degeneration were detected on this chromosome. Overall, these results provide important insights into the genomic attributes of an inversion polymorphism linked to mate choice and variation in social behavior.
Keywords: chromosomal polymorphism, evolutionary genetics, haplotype-based sequencing, inversion
Chromosomal inversions have been identified and studied in a variety of species (White, 1973). A key feature of inversions is their ability to suppress recombination within the inverted region between the standard and inverted chromosomes. The suppression of recombination between the alternative arrangements will thus maintain linkage between alleles captured by the inversion and can eventually lead to the genetic differentiation of the inverted and standard chromosomes. Consequently, some older and complex inversions that are strong suppressors of recombination can be associated with unusually large and distinct haplotypes that represent the inverted and standard arrangements (e.g., Fox et al., 1985; Dyer et al., 2007). Inversions can therefore foster selection for co-adapted allele complexes, also referred to as supergenes (Dobzhansky, 1970) or confer a selective advantage by capturing a set of locally adapted alleles (Kirkpatrick and Barton, 2006). Inversions can also have a functional impact on the genome by the physical disruption of a gene at an inversion breakpoint (e.g., Lakich et al., 1993) or through a position effect in which the regulation of a gene near an inversion breakpoint is altered due to a loss and/or gain of cis-regulatory elements (reviewed in Kleinjan and van Heyningen, 1998).
Clone-based sequencing has proven to be a useful method for comparing the sequence and long-range structure of haplotypes. For example, clone-based haplotype assemblies of the major histocompatibility complex have proven valuable for investigating this genetically diverse and complex segment of the genome (e.g., Raymond et al., 2005; Horton et al., 2008). However, despite the long-standing biological and theoretical interest in chromosomal inversions, besides the sequencing of alternative haplotypes associated with a human inversion polymorphism on chromosome 17 (Zody et al., 2008), we are unaware of clone-based haplotype sequencing having been applied toward characterizing the molecular evolution and genomic properties of other inversion polymorphisms.
The first avian inversion polymorphism was discovered in a North American migratory songbird, the white-throated sparrow (Zonotrichia albicollis) (Thorneycroft, 1966), and is linked to a suite of remarkable phenotypic traits (e.g., Lowther, 1961; Thorneycroft, 1975; Knapton and Falls, 1983; Kopachena and Falls, 1993a,1993b; Maney et al., 2008). Specifically, the second chromosome in this species has 2 arrangements, termed ZAL2 and ZAL2m, which differ at a minimum by a pair of pericentric inversions that together span >100 Mb (Thorneycroft, 1966; Thomas et al., 2008). Approximately half of the population is homozygous for the ZAL2 arrangement, whereas the other half of the population is heterozygous for the ZAL2 and ZAL2m chromosomes (reviewed in Falls and Kopachena, 1994). The polymorphism is maintained in the population by balancing selection through a strong pattern of negative assortative mating in which 96% of the breeding pairs consist of ZAL2/ZAL2 × ZAL2/ZAL2m individuals (reviewed in Falls and Kopachena, 1994). As a result, ZAL2m homozygotes comprise <0.25% of the population (Thorneycroft, 1975; Falls and Kopachena, 1994; Romanov et al., 2009). Although the specific biological cues that drive the negative assortative mating pattern have yet to be established, the choice of mates is likely to be influenced, at least in part, by the dominant and sex-independent plumage and behavioral differences that are known to be linked to ZAL2m (e.g., Lowther, 1961; Thorneycroft, 1975; Knapton and Falls, 1983; Kopachena and Falls, 1993a,1993b; Maney et al., 2008).
Recent cytogenetic mapping and sequencing of a limited number of small loci linked this chromosomal polymorphism have established that the ZAL2/2m inversions capture ∼10% of the white-throated sparrow genome and ∼1000 genes (Thomas et al., 2008). Gene flow between the 2 chromosome types appears to be restricted to a small region outside the inversions and is strongly suppressed inside the inversions. As a consequence, ZAL2 and ZAL2m have become genetically differentiated inside but not outside the inversions (Thomas et al., 2008; Huynh et al., 2010a). However, these studies were based on sample sequencing of ∼60 small, ∼500 bp, loci. Thus, a number of open questions remain with respect to the global genomic features and molecular evolution of this chromosomal polymorphism. Here, we describe our mapping and haplotype-based sequencing efforts designed to address those questions as well as characterize the structural differences between the ZAL2 and ZAL2m chromosomes.
Materials and Methods
Cytogenetic Mapping
Primary cell cultures, metaphase spreads, and fluorescence in situ hybridization (FISH) were carried out as described in (Thomas et al., 2008). All hybridizations were done on metaphase chromosomes from white-stripe individuals (ZAL2/ZAL2m) and included a control probe, TG_Ba-55A1, which localizes to the long arm of both ZAL2 and ZAL2m (Thomas et al., 2008). Zebra finch and white-throated sparrow BAC clones used in the FISH studies were selected based on their orthologous position in the zebra finch genome assembly (taeGut1, Warren et al., 2010). A summary of the FISH probes and mapping results are provided in Supplementary Table 1.
Physical Mapping of ZAL2/ZAL2m
The white-throated sparrow BAC library, constructed from a ZAL2/ZAL2m female (http://bacpac.chori.org/library.php?id=469), was screened with overgo hybridization probes designed from sequences conserved between zebra finch and chicken or from previously published white-throated sparrow sequence (Thomas et al., 2008). Specifically, segments of the chicken and zebra finch genomes orthologous to the regions likely to contain the ZAL2m inversion breakpoints or genes of interest linked to ZAL2/ZAL2m were identified and aligned. The chicken–zebra finch genomic sequence alignments were then used as the template to design “universal” overgo probes for screening the white-throated sparrow library using the Custom option on Uprobe Web site (Sullivan et al., 2008). Batches of 16–33 overgo probes were then hybridized to the white-throated sparrow library and probe-content and restriction-enzyme fingerprint maps of each targeted region were constructed using previously described methods (Thomas et al., 2002). BAC-end sequences (BESs) were generated from the isolated clones by the BC Cancer Agency Genome Sciences Center, Vancouver, Canada, using Sanger sequencing (ABI 3730xl). The BESs were then mapped to the zebra finch genome as described in (Davis et al., 2010) to anchor the clones to the zebra finch genome assembly and identify discordant paired BESs. A summary of the comparative mapping of the BESs orthologous to zebra finch chromosome 2 (TGU2, designated as chromosome 3 in the taeGut1 assembly) is provided in Supplementary Tables 2 and 3, and all the BESs sequences have been deposited in GenBank (Ac. FI592759–FI596471).
Sequence Tiling Path Selection and BAC Sequencing
The probe-content and restriction-enzyme fingerprint maps combined with the comparative mapping data for the BESs were used to guide the selection of sequence tiling paths with maximal representation of both haplotypes. In particular, whenever possible polyPhred (Nickerson et al., 1997) was used to identify SNPs in overlapping BES, and pairs of BACs representing the alternative alleles were then selected for sequencing. In addition, we used previously described and novel PCR-based and PCR/sequencing assays that could distinguish between the ZAL2 and ZAL2m alleles based on fixed differences between the chromosomes to phase and select clones representing both the chromosomes. The selected BAC clones were then Sanger shotgun sequenced, assembled, ordered, and oriented as described in Blakesley et al. (2004). Note, the sequence of all the individual BAC clones were deposited in the HTG division of GenBank and can be retrieved using the query: “Zonotrichia albicollis[Organism] AND HTG[Keyword] AND NISC.”
BAC Clone Phasing and ZAL2 and ZAL2m Specific Assemblies
The following evidence-based strategy was used to assign the sequenced BACs to either the ZAL2 or ZAL2m chromosomes. Previously published PCR-based genotyping/sequencing assays that detected fixed genetic differences between the chromosome types inside the inversion linked to VIP, DSE1, FAM83B, ESR1, and CGA were used to phase clones spanning those loci (Michopoulos et al., 2007; Thomas et al., 2008), and analogous PCR/sequencing assays were developed as needed to phase other unlinked clones (Supplementary Table 4). The phase of the neighboring clones was then inferred by sequence overlaps (i.e., an identical match was indicative of the same haplotype whereas mismatches indicated an alternative haplotype) and extended on a clone-by-clone basis. In instances where neighboring clones did not overlap, sequence alignments of paired BESs that spanned the gap were used to extend the haplotype phasing to the nonoverlapping clones. Alternatively, in cases where a clone gap in one haplotype was spanned in the alternative haplotype, we were able to infer the phase of the neighboring but nonoverlapping clones because both represented the opposite haplotype compared with the nongapped sequence assembly. The arrangement of the clones spanning the ZAL2 and ZAL2m breakpoints was also used as a means to directly phase the clones. Outside the inversion, where there is no genetic differentiation between ZAL2 and ZAL2m, we assigned the clones to the alternative or same haplotypes based on sequence identity or differences (clone overlaps, paired BESs that spanned gaps, etc.). Potential phase switches outside the inversion are indicated in the GenBank records. The phased clones were then merged into a single assembly representing each chromosome based on local physical mapping information and the chromosome-wide cytogenetic mapping data (GenBank Ac. DP001173.2 and DP001174.2, also see Supplementary Data File 1 for the orthologous positions in TGU2 spanned by the assemblies). Note these assemblies are updates of much smaller, <1 Mb, previously published assemblies of regions associated with a pair of evolutionary breakpoints (Davis et al., 2010).
Sequence Annotation and Analyses
Gene annotation was done by orthologous sequence alignments of zebra finch and other species complementary DNA sequences to the white-throated sparrow assemblies using SPIDEY (Wheelan et al., 2001). Repetitive elements were characterized using REPEATMASKER (http://repeatmasker.org) with the repeat library used to annotate the zebra finch genome assembly (Warren et al., 2010), as well as with a custom library of white-throated sparrow repetitive elements generated by RETROTECTOR (Sperber et al., 2009) and PILER (Edgar and Myers, 2005). In addition, WINDOWMASKER (Morgulis et al., 2006) was used as an independent method to estimate repetitive DNA content. DNA sequence alignments were generated with BLASTZ (Schwartz et al., 2003) or CLUSTALW (Thompson et al., 1997) and DNA divergence calculations done with BASEML (Yang, 1997) and MEGA (Kumar et al., 2004) using the Kimura 2-parameter model of nucleotide substitution (Kimura, 1980). Estimates of Ka/Ks were calculated with codeml (Yang, 1997) based on alignments between the ZAL2 and ZAL2m alleles and the zebra finch ortholog on a gene-by-gene basis and as a concatenated alignment of 62 genes that included 27 969 synonymous and 72 906 nonsynonymous sites. Note that all alignments were quality masked to exclude sites with a phred score <50.
Results
Refined Mapping of the ZAL2/2m Inversions
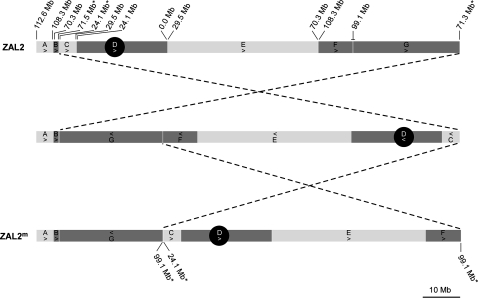
Previously we reported that ZAL2 and ZAL2m differed by a pair of inversions (Thomas et al., 2008). To further map the inversion breakpoints and other regions of interest linked to the chromosomal polymorphism, we cytogenetically mapped an additional 39 BAC clones to ZAL2 and ZAL2m using the assembly of the orthologous chromosome in the zebra finch (TGU2, which is called chromosome 3 in the published genome assembly, taeGut1, of the zebra finch) (Warren et al., 2010) as a comparative point of reference. A summary of the mapping results, which were consistent with the previous 2 inversion model, is illustrated in Figure 1, and the mapping results of the individual BAC clones are provided in Supplementary Table 1.
Figure 1.
Comparative cytogenetic maps of ZAL2 and ZAL2m. The cytogenetic location of a total of 58 BAC clones anchored to the zebra finch genome assembly of TGU2 (designated chromosome 3 in taeGut1) on the ZAL2 and ZAL2m chromosomes was determined by FISH and used to develop chromosome-wide comparative maps of both chromosome types. Maps of ZAL2 (top) and ZAL2m (bottom) are shown along with an inferred intermediate chromosome arrangement (middle). Dashed lines between the chromosomes indicate the edges of the 2 predicted inversions. In order to highlight the differences in arrangement between ZAL2 and ZAL2m, the chromosomes were split into blocks according to their orthologous positions on TGU2: block A (112.6–108.3 Mb), B (70.3–71.5 Mb), C (24.1–29.5 Mb), D (24.1–0.0 Mb), E (29.5–70.3 Mb), F (108.3–99.1 Mb), G (99.1–71.3 Mb). Note one boundary of block G differs between the ZAL2, 71.3 Mb, and ZAL2m, 71.5 Mb, due to a ZAL2-specific duplication (see Results and Figure 2). The relative orientation of the blocks is indicated by an arrow. The orthologous TGU2 positions of block boundaries are shown above ZAL2 (asterisk marks the inversion breakpoint boundaries). Only the orthologous positions on TGU2 of the inversion breakpoints are labeled on the ZAL2m.
Localization of ZAL2/2m Inversion Breakpoints
In order to localize and sequence inversion breakpoints associated with this chromosomal polymorphism, we aligned n = 1231 pairs of BESs from white-throated sparrow BAC clones isolated as part of our physical mapping efforts targeting the inversion breakpoints and other regions of interest along the chromosome. (Supplementary Figure S1 and Supplementary Tables 2 and 3). Note that the comparative mapping of n = 480 of these BESs were previously reported in Davis et al. (2010). Because the BAC library we screened was generated from a single ZAL2/ZAL2m bird (Davis et al., 2010), we expected that the detection of discordant pairs of BESs could be an indication of a structural rearrangement between TGU2 and both white-throated sparrow arrangements or a structural difference between TGU2 and either ZAL2 or ZAL2m. In the first scenario, we expected that all the white-throated sparrow paired BESs spanning a discrete orthologous position on TGU2 would be discordant. In contrast, a difference in arrangement between ZAL2 and ZAL2m would result in a mixed pattern of concordant and discordant paired BESs spanning a common position.
Two clusters of discordant clones mapped to predicted inversion breakpoint intervals orthologous to ∼71.5 and 99.1 Mb on TGU2 displayed the interlaced pattern of concordant and discordant clones expected at the site of a structural difference between ZAL2 and ZAL2m (Supplementary Figure S1 and Supplementary Tables 2 and 3). Interestingly, the mate pair of the discordant BESs from both of the above regions mapped to a common ∼130-kb interval near position 24.1 Mb on TGU2, suggesting one side of both inversion breakpoints mapped to a similar region (Supplementary Figure S1 and Supplementary Table 3). The other clusters of discordant clones visible in Supplementary Figure S1 represented an interspecies indels and the positions of evolutionary breakpoints between the white-throated sparrow and zebra finch reported in Davis et al. (2010) and are not described further. Integrating the cytogenetic and comparative BESs mapping data indicated that the pair of ZAL2/ZAL2m inversion breakpoints we had cloned corresponded to those mapping to the p-arm of both chromosome arrangements (Figure 1 and Supplementary Tables 1–3), but not the 2 q-arm breakpoints. The relative cytogenetic positions of BAC clones mapped by FISH to ZAL2 and ZAL2m that flanked the inversion breakpoints indicated that arrangement in which the regions orthologous to ∼71.5 and 24.1 Mb on TGU2 were adjacent in the white-throated sparrow corresponded to ZAL2, whereas the regions orthologous to 99.1 and 24.1 Mb on TGU2 were adjacent on ZAL2m (Figure 1). Thus, assuming the zebra finch arrangement represents the ancestral “noninverted” state, both ZAL2 and ZAL2m represent derived and distinct inverted arrangements of the ancestral chromosome.
Genomic Properties of ZAL2 and ZAL2m
In order to characterize the inversion breakpoints and compare the genomic properties associated with ZAL2 and ZAL2m inside and outside the inversion, the BAC-based physical maps were used to select 110 BACs from 13 discrete regions distributed along the chromosome for sequencing. After the sequenced BAC clones were assigned to either the ZAL2 or ZAL2m chromosomes (see Materials and Methods), we generated separate multi-BAC assemblies representing both chromosome arrangements that reflected the order and orientation of the clones predicted by our combined cytogenetic and physical maps. The resulting assemblies included 7.47 and 7.44 Mb of ZAL2 and ZAL2m sequence, respectively (Table 1). Four additional clones from the ZAL2 or ZAL2m chromosomes were also sequenced but could not be readily assigned to either haplotype and thus were not included in subsequent analyses.
Table 1.
Summary of ZAL2 and ZAL2m sequence assemblies
| Outside inversiona | Inside inversion |
|||||||
| Mb | # Genes | % Repetitiveb | % GC | Mb | # Genes | % Repetitiveb | % GC | |
| ZAL2 | 2.12 | 40 | 12.1 (10.7) | 41.2 | 5.36 | 79 | 6.7 (9.8) | 41.6 |
| ZAL2m | 1.89 | 37 | 13.1 (10.3) | 41.4 | 5.55 | 86 | 6.9 (9.2) | 41.9 |
The alternative sequence assemblies outside the inversion represent the 2 haplotypes present in the individual from which the library was made. However, because gene flow between the chromosome arrangements occurs outside the inversion, there are no fixed differences between ZAL2 or ZAL2m in this region of the chromosome. Thus, assignment as ZAL2 or ZAL2m outside the inversion is arbitrary.
Repetitive DNA content was estimated using REPEATMASKER and WINDOWMASKER.
The ZAL2 and ZAL2m sequence assemblies each included ∼2 Mb and ∼40 genes that mapped outside the inversion, and ∼5.4 Mb and ∼80 genes that mapped within the inversion (Table 1). Most of the sequence outside (∼1.8 Mb) and inside (∼4.5 Mb) the inversion were represented in both assemblies. The repetitive element content estimated using an augmented REPEATMASKER library (see Materials and Methods) was similar between ZAL2 and ZAL2m outside and inside the inversion, although it was nonuniform across the chromosome (i.e., ∼12–13% repetitive element content outside vs. ∼7% inside the inversion, see Table 1). Those general trends were also observed when using an alternative method for identifying repetitive sequence (Morgulis et al., 2006), though the difference in repetitive sequence content between outside and inside the inversion was more subtle (Table 1). GC content was relatively uniform along the chromosome and between the chromosomal arrangements (41.2–41.9%, Table 1). Thus, we observed no prominent differences in sequence composition between the ZAL2 and ZAL2m chromosomes.
Genomic Features of the Sequenced Inversion Breakpoints
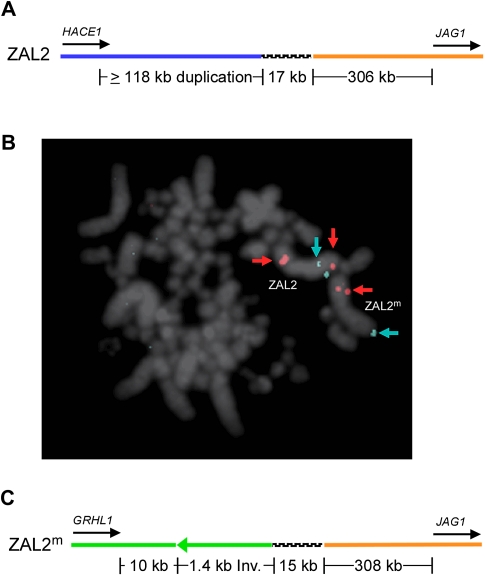
Our combined cytogenetic and BESs mapping indicated we had identified clones containing the breakpoints of the inversions that distinguish the ZAL2 and ZAL2m chromosomes. We therefore examined alignments between the alternative assemblies and zebra finch to further localize and characterize the pair of sequenced inversion breakpoints that mapped to the p-arm of ZAL2 and ZAL2m. In the case of the ZAL2 inversion breakpoint, we were able to determine that it maps ∼112 kb downstream of HACE1 and ∼323 kb upstream of JAG1 (Figure 2A). The most notable feature of the sequence flanking this inversion breakpoint is a large (≥118 kb) ZAL2-specific segmental duplication that includes at least part of the HACE1 locus (Figure 2A and B). The other copy of this duplication maps near the telomere of the q-arm, which is in close proximity to the predicted location of the opposite end of the ZAL2 inversion (Figures 1 and 2B). This pair of duplicons are ∼99% identical (0.012 ± 0.0003 substitutions/site, sample size of 115 805 sites), suggesting they are the product of a relatively recent duplication. With respect to the ZAL2m inversion breakpoint, it maps ∼12-kb downstream of GRHL1 and ∼323-kb upstream of JAG1 (Figure 2C). The most notable feature of the sequence flanking the ZAL2m inversion breakpoint was ∼1.4-kb microinversion (Figure 2C).
Figure 2.
Location and genomic features of the ZAL2 and ZAL2m p-arm inversion breakpoints. (A) The ZAL2 p-arm inversion breakpoint maps between HACE1 and JAG1 and corresponds to the juxtaposition of blocks B and C illustrated in Figure 1. (B) Cytogenetic mapping of the segmental duplication that is specific to the ZAL2 chromosome and maps near the predicted location of the other edge of the ZAL2 inversion. The red signal is BAC CH264-321F19, and the blue signal is BAC TG_Ba-55A01, which maps to the q-arm of both ZAL2 and ZAL2m (see Supplementary Table 1 for the precise orthologous positions in zebra finch for these clones). (C) The ZAL2m p-arm inversion breakpoint maps between GRHL1 and JAG1, corresponds to the juxtaposition of blocks G and C illustrated in Figure 1, and includes a microinversion directly adjacent to the breakpoint (green arrow). Colored lines designate the regions orthologous to ∼71 Mb (blue), ∼24 Mb (orange), and ∼99 Mb (green) with respect to chromosome 3 in the zebra finch genome assembly (taeGut1). Black hashed lines indicate the location of sequence that does not align to either adjacent orthologous regions in zebra finch. The inversion breakpoints most likely map to the positions corresponding to where the orthology with zebra finch ends or within the intervening sequence.
Clustering of Breakpoints Associated with the ZAL2 and ZAL2m Inversions
The localization of one side of the ZAL2 and ZAL2m inversion breakpoints to a common interval orthologous to 24.1 Mb on TGU2 suggested these inversions might represent an example of breakpoint reusage, that is where the same sequence is present at the breakpoint of 2 or more independent chromosomal rearrangements (e.g., Pevzner and Tesler, 2003; Murphy et al., 2005). The alignments of the common side of the ZAL2 and ZAL2m inversion breakpoint to zebra finch terminate at precisely the same orthologous position (Figure 2A and C). Strikingly, by comparing the arrangement of this chromosomal segment in the chicken, zebra finch, and other sequenced genomes, we were able to infer that the breakpoint of an evolutionary rearrangement that occurred in the zebra finch lineage mapped within ∼1 kb of the inferred location of the common side of the ZAL2 and ZAL2m inversions (24 117 463 vs. 24 116 641 bp on taeGut1 chromosome 3). Note that the derived arrangement in the zebra finch compared with chicken and other species was supported by the presence of multiple concordant pairs of paired-end sequences from large-insert clones (data not shown, Warren et al., 2010).
At both the ZAL2 and ZAL2m inversion breakpoints, there was ∼15- to 17-kb block of sequence that did not align with the predicted orthologous zebra finch intervals (i.e., either near 24.1, 71.5, or 99.1 Mb on TGA2, Figure 2A and C), and a BLAST search (Altschul et al., 1997) of this white-throated sparrow sequence also failed to identify a clear orthologous region anywhere else in the zebra finch genome. Similarly, ∼19 kb of sequence at the breakpoint in zebra finch was neither clearly orthologous to any sequence in the chicken genome nor similar to the sequence at the breakpoints in the white-throated sparrow. In addition, although ZAL2 and ZAL2m white-throated sparrow-specific sequences did partially align to one another and were somewhat similar (0.06 ± 0.003 substitutions/site), both sequences included tandem repeats, the structure of which clearly differed between the chromosome types (Supplementary Figure S2). Thus, one side of the ZAL2 and ZAL2m inversion breakpoints mapped in close proximity to one another, as well as an evolutionary breakpoint, and also colocalized with sequences of unknown origin.
Genetic Differentiation between ZAL2 and ZAL2m
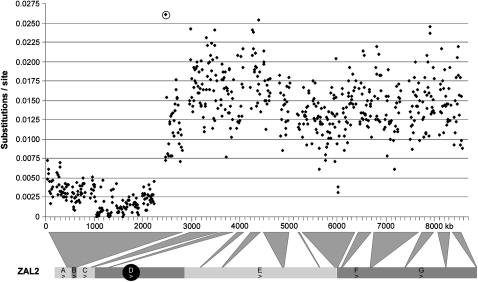
Previous small-scale sequencing efforts indicated that the genetic differentiation between ZAL2 and ZAL2m is elevated inside versus outside the inversions (Thomas et al., 2008; Huynh et al., 2010a). To visualize the pattern of genetic differentiation between ZAL2 and ZAL2m on a more global scale, we aligned the assemblies representing the 2 chromosome types and plotted the single-nucleotide divergence between them using a sliding window approach (Figure 3 and Supplementary Table 5). In line with previous studies, the divergence (calculated after excluding annotated protein-coding exons and UTRs, as well as low-quality sites) outside and inside the inversions were 0.00256 ± 0.00004 substitutions/site (sample size of 1 729 417 bp) and 0.01443 ± 0.00006 substitutions/site (sample size of 4 253 200 bp), respectively. Likewise, synonymous differences between alternative alleles of the genes located outside or inside the inversions showed a similar pattern: Ks = 0.00172 ± 0.00035 (sample size of 13 634 sites) outside and Ks = 0.01416 ± 0.00056 (sample size of 31 758 sites) inside the inversions. The transition from the low to high divergence between ZAL2 and ZAL2m mapped distal of the ZAL2 p-arm inversion breakpoint (Figure 3, circled data point marks the position of the inversion breakpoint). More precisely, taking into account gaps in our assemblies, the shift from low to high divergence between the chromosomes occurred within a 160-kb region just upstream of the ZAL2 p-arm inversion breakpoint. Note that this stratification of genetic differentiation was not reflected in the interspecies divergence between the white-throated sparrow and zebra finch outside and inside the inversion, which were 0.0886 ± 0.0003 and 0.0929 ± 0.0002 substitutions/site, respectively (Supplementary Figure S3). Thus, an intrinsic difference in mutation rate between the regions inside versus outside the inversion is unlikely to have contributed to the pattern of genetic differentiation observed across the chromosome.
Figure 3.
Divergence between the ZAL2 and ZAL2m. The ZAL2 and ZAL2m assemblies were aligned, and after excluding low-quality sites (phred <50) and annotated protein-coding exons and UTRs, divergence (Kimura 2-parameter) between the chromosomes was calculated in nonoverlapping 10-kb windows. Only windows that contained at least 5000 aligned sites are shown in the graph. To generate the most reliable alignment between ZAL2 and ZAL2m, the ZAL2m assembly was reordered to reflect the arrangement of ZAL2 and then aligned using the single-strand and chaining options of BLASTZ. The divergence values are plotted with respect to their position in the ZAL2 assembly (that including gaps is ∼8.7 Mb). The chromosomal locations of the regions included in the assemblies are indicated below the graph. The divergence value for the window that spans the ZAL2 inversion breakpoint is circled. Data points to the left of the circled value are outside the inversion and those to the right are inside the inversion.
The relative frequency of indels between ZAL2 and ZAL2m paralleled that seen for the single nucleotide substitutions (0.0005 and 0.0017 indels/all aligned sites, outside and inside the inversions, respectively). As has been observed in mammals and other birds (e.g., Brandstrom and Ellegren, 2007; Levy et al., 2007), the indel size spectrum was heavily skewed toward small indels, with ∼40–50% of all observed indels being a single base (Supplementary Figure S4). Notably, we did detect 136 large indels (i.e., ≥100–8143 bp) and found that ∼65% of the bases within the large indels corresponded to white-throated sparrow lineage-specific repetitive elements, suggesting that active transposable elements are contributing to the genetic variation in this bird.
Patterns of Molecular Evolution Associated with ZAL2 and ZAL2m
Based on the strong suppression of recombination between ZAL2 and ZAL2m inside the inversions, and the low frequency of ZAL2m homozygotes in the population (<0.25%) (Thorneycroft, 1975; Falls and Kopachena, 1994; Romanov et al., 2009), the ZAL2m inverted segment is predicted to have very limited opportunities to recombine. As such, it might be prone to the accumulation of deleterious mutations or genetic degeneration (Charlesworth B and Charlesworth D, 2000; Betancourt et al., 2009). However, previous analyses of a limited number of informative sites suggested that recombination between ZAL2m chromosomes has occurred inside the inversion and that this chromosome is not associated with signatures of genetic degeneration (Huynh et al., 2010a). Given this much larger data set, we next asked whether ZAL2m had accumulated potentially deleterious mutations at a higher rate inside the inversion than ZAL2.
As a first pass to detect a signature of genetic degeneration, we scanned the predicted protein coding sequences within the inversion for protein truncating mutations. No such mutations or gross disruptions of any genes that mapped within the inversion on either ZAL2 or ZAL2m were detected. In addition, a likelihood ratio test (Yang, 1998) based on a concatenated alignment of 62 genes did not reveal a significant difference in the overall Ka/Ks ratio between the ZAL2 and ZAL2m lineages (M1 vs. M2; 2Δl = 0.496, degree of freedom [df] = 1, P value = 0.48, for details, see Materials and Methods). Moreover, only a single gene, CCDC88A2, was found to have a significant difference in Ka/Ks between the ZAL2 and ZAL2m lineages (M1 vs. M2; 2Δl = 6.958, df = 1, uncorrected P value = 0.008), with an estimated 5 nonsynonymous substitutions having accumulated in the ZAL2m lineage versus none in ZAL2.
An excess accumulation of repetitive elements is another expected feature of nonrecombining portions of the genome (reviewed in Charlesworth et al., 2005). We therefore compared the repetitive element content between ZAL2 and ZAL2m inside the inversion from the subset of the regions that were sampled in both assemblies. Whereas the repetitive element content estimated by REPEATMASKER was slightly higher inside the inversions on ZAL2m versus the ZAL2 (7.0% vs. 6.0%), the opposite was true for repetitive content estimated by WINDOWMASKER, that is, 9.3% on ZAL2m versus 9.9% on ZAL2. Thus, we found no obvious signatures for a reduction in the efficacy of selection on ZAL2m versus the ZAL2.
Discussion
The ZAL2/2m Inversion Breakpoints
The ZAL2 and ZAL2m chromosomes have been hypothesized to differ by a minimum of 2 pericentric inversions (Thomas et al., 2008 and see Figure 1). The cloning and sequencing of the ZAL2 and ZAL2m inversion p-arm inversion breakpoints and the corresponding regions on the alternative arrangements allowed us to precisely define 2 of the 4 edges of the proposed inversions. The exact location of the q-arm breakpoints was not determined, but the cytogenetic mapping data were consistent with them mapping near the telomeres. Regions adjacent to telomeres in other species have been associated with higher than average frequency of chromosomal breakpoints (e.g., Rudd, 2007); however, because we were unable to clone and sequence the q-arm breakpoints, it is unclear if the fragile nature of subtelomeric regions contributed to this polymorphism.
The sequence of the p-arm inversion breakpoints revealed that neither directly disrupted any genes. Note that since we have not ruled out a position effect on the neighboring genes or functional consequences at the unsequenced inversion breakpoints, we cannot exclude the possibility of functionally distinct allelic forms of genes in close proximity to the breakpoints. The sequenced inversion breakpoints were associated with 2 notable features that have been observed at the breakpoints of chromosomal rearrangements in other species: segmental duplications and breakpoint clustering. Segmental duplications tend to be enriched at the breakpoints of chromosomal rearrangements (e.g., Murphy et al., 2005) and can either mediate the rearrangements via nonallelic homologous recombination (reviewed in Stankiewicz and Lupski, 2002) or can be the direct result of the DNA repair process at the breakpoints (e.g., Kehrer-Sawatzki et al., 2005; Ranz et al., 2007). Our finding of a large segmental duplication at the ZAL2 p-arm inversion breakpoint is reminiscent of what has been observed in other species and adds another example to the growing list of segmental duplications detected in avian genomes (Hillier et al., 2004; Warren et al., 2010). In addition, the absence of this duplication on ZAL2m combined with the predicted localization of the second copy of this sequence near the ZAL2 q-arm inversion breakpoint suggest this duplication is likely to represent a common copy-number variant in white-throated sparrows and may have originated as a consequence of the inversion.
Secondly, previous studies have shown that a subfraction of mammalian genomes are enriched for breakpoints of chromosomal rearrangements, suggesting that there are evolutionarily conserved fragile genomic regions (e.g., Pevzner and Tesler, 2003; Murphy et al., 2005). The clustering of the ZAL2 and ZAL2m inversion breakpoints and their colocalization with an evolutionary breakpoint in the zebra finch suggest that chromosomal breakage at a genomic fragile site conserved in songbirds may in part be responsible for the origin of this chromosomal polymorphism. The potentially rapidly evolving sequence associated with the breakpoints is unclear but we speculate it may have been generated as part of the DNA breakage and repair process or may contribute to the apparent fragility of this region. However, it should be pointed out that a definitive interpretation of the sequence composition at this breakpoint is confounded by the presence of gaps in the assemblies.
Implications of the Patterns of Molecular Evolution Associated with ZAL2/2m
The analyses and comparison of the BAC-based haplotype sequences were broadly consistent with previous smaller scale sequencing surveys of this chromosomal polymorphism (Thomas et al., 2008; Huynh et al., 2010a). For example, divergence was on average ∼5-fold higher inside versus outside the inversions and is consistent with a general lack of gene flow due to the suppression of recombination between the arrangements within the inversions. Despite the largely colinear but offset arrangements of the chromosome types (Figure 1), which one might expect could facilitate gene flow via gene conversion or double crossovers between the arrangements, the tendency for recombination to occur near the telomeres and not in the central segment of songbird macrochromosomes may be another important property that limits gene flow between ZAL2 and ZAL2m within the inversions (Backstrom et al., 2010). Nevertheless, some of the variation in the level of divergence between the chromosome types within the inversion could be an indication of historical gene flow. For example, ∼1.1-Mb segment centered around ∼6000 kb in the ZAL2 assembly exhibited a lower divergence (0.01272 ± 0.0001 substitutions/site) than the overall average divergence within the inversions and included a 19-kb region in which the genetic differentiation between ZAL2 and ZAL2m was similar to that observed outside the inversion (see pair of data points above 6000 kb in Figure 3 and Supplementary Table 6). In addition, unlike some other inversions where suppression of recombination extends up to 1 Mb into the flanking region immediately adjacent to the inversion breakpoints (Kulathinal et al., 2009), we estimate that gene flow between ZAL2 and ZAL2m has occurred in the flanking region as close as ∼160 kb from the breakpoint on p-arm.
Although the juxtaposition of segments of low and high genetic differentiation caused by the inversions is striking, the absolute levels of differentiation are within the range observed elsewhere in the genome. In particular, estimates of haplotype differentiation elsewhere in the genome using methods similar to those used here have been reported to be ∼0.001–0.016 substitutions/site (Huynh et al., 2010b). This does not mean the level of heterozygosity linked to the ZAL2/ZAL2m inversion is trivial. In fact, in the sample of genes we sequenced that map within the inversions, we observed ∼209 nonsynonymous substitutions between the ZAL2 and ZAL2m alleles. Furthermore, from our estimates of Ka for those genes, the number of genes that we infer map within the inversion based on annotation of the orthologous chromosome in the zebra finch (n = ∼1100), and assuming an average of 1000 nonsynonymous sites per gene, we estimate there are on the order of 1870 nonsynonymous differences between ZAL2 and ZAL2m alleles within the inversion. Thus, given that the majority of differences between ZAL2 and ZAL2m represent fixed genetic polymorphisms (Thomas et al., 2008; Huynh et al., 2010a), and considering the number of potential functional variants that could have accumulated in nonprotein coding regions, it is perhaps not surprising that a number of dominant traits have been linked to ZAL2m.
Finally, the frequency of recombination within the inversion is likely to be dramatically reduced on ZAL2m compared with ZAL2 due to the suppression of recombination within the inversions in ZAL2/ZAL2m heterozygotes and because of ∼200-fold lower frequency of ZAL2m homozygotes versus to ZAL2 homozygotes in the population (i.e., <0.25% vs. 50%) (Thorneycroft, 1975; Falls and Kopachena, 1994; Romanov et al., 2009). In a somewhat analogous genetic system, the well-studied t complex in mice, numerous recessive lethal mutations have been found to map within inversions that have limited opportunities to recombine (Klein et al., 1984). Within the inversions on ZAL2m, however, we observed no signatures of a decrease in efficacy of selection that would be expected for a region with greatly reduced recombination (Betancourt et al., 2009), or signatures of genetic degeneration expected to occur in the prolonged absence of recombination (Charlesworth B and Charlesworth D, 2000). Thus, we have found no evidence that wide-spread genetic degeneration of ZAL2m might contribute to the phenotypic variation linked to this chromosomal polymorphism.
Possible History of the ZAL2/ZAL2m Inversions
ZAL2m was previously presumed to be the derived arrangement and have originated since the divergence of the white-throated sparrow from other birds in the Zonotrichia genus (Thorneycroft, 1975). Our prior phylogenetic analyses and molecular dating were potentially at odds with that interpretation in that by those metrics the inversion appeared to predate the origin of the white-throated sparrow (Thomas et al., 2008). The model shown in Figure 1 (see also Thomas et al., 2008) is consistent with either independent inversions of an inferred ancestral chromosome giving rise to ZAL2 and ZAL2m or successive inversions of ZAL2 (or ZAL2m) giving rise to the alternative arrangements. Note that a comparison of the divergence estimates between chromosomal segments that were within just the large inversion versus the segment included in the small inversion did not reveal the presence of evolutionary strata (data not shown). The lack of detectable evolutionary strata is consistent with either the large inversion occurring first, the inversions occurring simultaneously, or that the small inversion occurred first but that the age difference between the inversions was too small to be detected by our divergence estimates. Future studies, including cytogenetic mapping and direct typing of the alternative chromosomal arrangements observed in the white-throated sparrow using PCR primers that flank the inversion breakpoints in additional species, will therefore be needed to better resolve the origins of this unique chromosomal polymorphism.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Institutes of Health (NIH) (1R21MH082046) and the Center for Behavioral Neuroscience to J.K.D., J.J.L., D.L.M, C.L.M., and J.W.T. Intramural Research Program of the National Human Genome Research Institute of the NIH to NIH Intramural Sequencing Center.
Supplementary Material
Acknowledgments
The authors thank Soojin Yi for her comments on the manuscript, Greg K. Tharp for computer support, the BC Cancer Agency Genome Sciences Centre, Vancouver, Canada, for generating the BESs reported here, and members of the National Institutes of Health Intramural Sequencing Center including E.D. Green, R. Blakesley, G. Bouffard, and J. McDowell.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Ost T, Schneider M, Kempenaers B. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010;20:485–495. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Welch JJ, Charlesworth B. Reduced effectiveness of selection caused by a lack of recombination. Curr Biol. 2009;19:655–660. doi: 10.1016/j.cub.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Blakesley RW, Hansen NF, Mullikin JC, Thomas PJ, McDowell JC, Maskeri B, Young AC, Benjamin B, Brooks SY, Coleman BI, et al. An intermediate grade of finished genomic sequence suitable for comparative analyses. Genome Res. 2004;14:2235–2244. doi: 10.1101/gr.2648404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstrom M, Ellegren H. The genomic landscape of short insertion and deletion polymorphisms in the chicken (Gallus gallus) genome: a high frequency of deletions in tandem duplicates. Genetics. 2007;176:1691–1701. doi: 10.1534/genetics.107.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Davis J, Lowman J, Thomas P, te Hallers B, Koriabine M, Huynh L, Maney D, de Jong P, Martin C Program NCS. Evolution of a bitter taste receptor gene cluster in a New World sparrow. Genome Biol Evol. 2010;2:358–370. doi: 10.1093/gbe/evq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of the evolutionary process. New York: Columbia University Press; 1970. [Google Scholar]
- Dyer KA, Charlesworth B, Jaenike J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc Natl Acad Sci U S A. 2007;104:1587–1592. doi: 10.1073/pnas.0605578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(1 Suppl):i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Falls JB, Kopachena JG. White-throated sparrow (Zonotrichia albicollis) In: Poole A, Gill F, editors. The birds of North America. Philadelphia (PA): The Academy of Natural Sciences.128; 1994. [Google Scholar]
- Fox HS, Martin GR, Lyon MF, Herrmann B, Frischauf AM, Lehrach H, Silver LM. Molecular probes define different regions of the mouse t complex. Cell. 1985;40:63–69. doi: 10.1016/0092-8674(85)90309-5. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh LY, Maney DL, Thomas JW. Chromosome-wide linkage disequilibrium caused by an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis) Heredity. 2010a doi: 10.1038/hdy.2010.85. doi: 10.1038/hdy.2010.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh LY, Maney DL, Thomas JW. Contrasting population genetic patterns within the white-throated sparrow genome (Zonotrichia albicollis) BMC Genetics. 2010b;11:96. doi: 10.1186/1471-2156-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Sandig CA, Goidts V, Hameister H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet Genome Res. 2005;108:91–97. doi: 10.1159/000080806. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Sipos P, Figueroa F. Polymorphism of t-complex genes in European wild mice. Genet Res. 1984;44:39–44. [Google Scholar]
- Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- Knapton RW, Falls JB. Differences in parental contribution among pair types in the polymorphic white-throated sparrow. Can J Zool. 1983;61:1288–1292. [Google Scholar]
- Kopachena JG, Falls JB. Aggressive performance as a behavioral correlate of plumage polymorphism in the white-throated sparrow. Behavior. 1993a;124:249–266. [Google Scholar]
- Kopachena JG, Falls JB. Re-evaluation of morph-specific variations in parental behavior of the white-throated sparrow. Wilson Bull. 1993b;105:48–59. [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MA. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH, Jr., Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5:236–241. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther JK. Polymorphism in the white-throated sparrow, Zonotrichia albicollis (Gmelin) Can J Zool. 1961;39:281–292. [Google Scholar]
- Maney DL, Lange HS, Raees MQ, Reid AE, Sanford SE. Behavioral phenotypes persist after gonadal steroid manipulation in white-throated sparrows. Horm Behav. 2008;55:113–120. doi: 10.1016/j.yhbeh.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Michopoulos V, Maney DL, Morehouse CB, Thomas JW. A genotyping assay to determine plumage morph in the white-throated sparrow, Zonotrichia albicollis. Auk. 2007;124:1330–1335. [Google Scholar]
- Morgulis A, Gertz EM, Schaffer AA, Agarwala R. WindowMasker: window-based masker for sequenced genomes. Bioinformatics. 2006;22:134–141. doi: 10.1093/bioinformatics/bti774. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, Ashburner M, Bergman CM. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Kas A, Paddock M, Qiu R, Zhou Y, Subramanian S, Chang J, Palmieri A, Haugen E, Kaul R, et al. Ancient haplotypes of the HLA Class II region. Genome Res. 2005;15:1250–1257. doi: 10.1101/gr.3554305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov MN, Tuttle EM, Houck ML, Modi WS, Chemnick LG, Korody ML, Mork EM, Otten CA, Renner T, Jones KC, et al. The value of avian genomics to the conservation of wildlife. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-S2-S10. (2 Suppl):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK. Subtelomeres; evolution in the human genome. In: Kehrer-Sawatzki H, editor. Encyclopedia of life sciences. Chichester. John Wiley & Sons, Ltd; 2007. pp. 1–9. [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber G, Lovgren A, Eriksson NE, Benachenhou F, Blomberg J. RetroTector online, a rational tool for analysis of retroviral elements in small and medium size vertebrate genomic sequences. BMC Bioinform. 2009;10 doi: 10.1186/1471-2105-10-S6-S4. (Suppl 6):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Sullivan RT, Morehouse CB, Thomas JW. Uprobe 2008: an online resource for universal overgo hybridization-based probe retrieval and design. Nucleic Acids Res. 2008;36:W149–W153. doi: 10.1093/nar/gkn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Caceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Prasad AB, Summers TJ, Lee-Lin SQ, Maduro VV, Idol JR, Ryan JF, Thomas PJ, McDowell JC, Green ED. Parallel construction of orthologous sequence-ready clone contig maps in multiple species. Genome Res. 2002;12:1277–1285. doi: 10.1101/gr.283202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneycroft HB. Chromosomal polymorphism in the white-throated sparrow, Zonotrichia albicollis (Gmelin) Science. 1966;154:1571–1572. doi: 10.1126/science.154.3756.1571. [DOI] [PubMed] [Google Scholar]
- Thorneycroft HB. A cytogenetic study of the white-throated sparrow, Zonotrichia albicollis. Evolution. 1975;29:611–621. doi: 10.1111/j.1558-5646.1975.tb00855.x. [DOI] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Church DM, Ostell JM. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. Animal cytology and evolution. London: Cambridge University Press; 1973. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.