Abstract
The p53 protein is the cell’s principal bastion of defense against tumor-associated DNA damage. Commonly referred as a “guardian of the genome”, p53 is responsible for determining the fate of the cell when the integrity of its genome is damaged. The development of tumors requires breaching this defense line. All known tumor cells either mutate the p53 gene, or in a similar number of cases, use internal cell p53 modulators, Mdm2 and Mdmx proteins, to disable its function. The release of functional p53 from the inhibition by Mdm2 and Mdmx should in principle provide an efficient, nongenotoxic means of cancer therapy. In recent years substantial progress has been made in developing novel p53-activating molecules thanks to several reported crystal structures of Mdm2/x in complex with p53-mimicking peptides and nonpeptidic drug candidates. Understanding the structural attributes of ligand binding holds the key to developing novel, highly effective, and selective drug candidates. Two low-molecular-weight compounds have just recently progressed into early clinical studies.
Keywords: anticancer agents, drug discovery, inhibitors, proteins, structure–activity relationships
1. Introduction
Discovered over 30 years ago, the p53 protein remains at the center of scientific interest: in 2009 roughly 4200 publications referred to p53.[1–5] The significance of p53 stems not only from its pivotal role in DNA-repair and cell-cycle control but also from its overarching regulating role in carcinogenesis.[6,7] Therefore p53 has become one of the most important therapeutic targets in cancer treatment in recent years.[6–9] It is now believed that all cancers contain either a mutated form of p53 or are subject to inactivation of the components of the p53 pathway.[1,8,9] In tumors in which p53 is not mutated, its function is usually inhibited by overexpression of its two negative protein regulators Mdm2 and Mdmx.[6,10] Mdm2 and Mdmx bind the N-terminal trans-activation domain of p53,[9] disabling its transcriptional function. Additionally, Mdm2 displays E3 ubiquitin ligase activity and is able to target p53 for proteasomal degradation.[11,12] The Mdm2/x–p53 system creates a negative feedback loop, which precisely controls the level and activity of p53.[6,13,14]
The restoration of the impaired function of the p53 protein by disrupting the Mdm2–p53 or Mdmx–p53 interaction offers a fundamentally new avenue for the treatment of a broad spectrum of cancers.[1–6,15] Cancer cells have been shown to be extremely sensitive to the restoration of p53 function, verifying the expectation of highly effective therapies from this approach.[15] Many currently used genotoxic chemotherapeutics rely on DNA-damage-dependent activation of p53 to mount an apoptotic response. However, high doses of genotoxic drugs can also induce p53-independent pathways and thus may cause severe toxicities in normal tissues eventually leading to secondary malignancies. Therefore, selective, nongenotoxic activation of p53, targeting the p53–Mdm2/x interaction, should be an important alternative to current cytotoxic chemotherapy. In addition, the combination of various drugs that target multiple p53 pathways may be a useful strategy to achieve synergistic drug efficacy.
Mdm2/x antagonists may have also important utility in protecting normal proliferating tissues during antimitotic chemotherapy of tumors expressing mutant p53. Normal cells possess wild-type functional p53, and pretreatment with Mdm2/x antagonists will arrest their proliferation and may protect them from the toxicity of chemotherapy. They may resume proliferation after drug removal. Cancer cells with defective mutant p53 will be insensitive to Mdm2/x antagonists and thus be selectively vulnerable to the mitotic poison.[1,16]
The search for efficient p53–Mdm2 inhibitors has led to several small-molecule ligands. The most thoroughly studied among them is Nutlin-3,[16,17] which has approximately 30 nM affinity towards the p53 binding site of Mdm2. Experiments with Nutlin-3 and other inhibitors performed in vivo confirmed the “proof of concept” that small-molecule inhibitors of the p53–Mdm2 interaction are able to induce either the cell-cycle arrest or apoptosis in tumor cells, while not affecting healthy cells.[18–22]
The Mdmx protein has emerged only recently as a critical p53 regulator and antitumor target.[23,24] Overexpression of Mdmx prevents the Nutlin-3 antitumor activity.[25] While no high-affinity p53–Mdmx binding inhibitors currently exist, peptide and knockout studies confirm that such an inhibitor should have high therapeutic value.[10,24] As mentioned above, efficient scaffolds have been developed for the Mdm2–p53 interaction.[21,26–28] For Mdmx, only few peptides and pepti-domimetics have been published that have affinities towards the p53 binding site of Mdmx in the nanomolar range.[29–32] Recently, the first effective inhibitor of the Mdmx–p53 interaction, called WW298, was described.[33]
In this Minireview we summarize all small-molecule Mdm2- and Mdmx-binding inhibitors for which crystallographic structures are available. Knowledge of precise ligand-binding interactions delivered by crystallographic analysis is an ultimate source of information on the structure–activity relationship (SAR). Therefore such structures should be of great help in developing novel ligand scaffolds that are expected to have optimized binding and pharmacological properties. We additionally review the recently published structures of both Mdm2/x proteins in complex with p53-derived peptides, which have 10–100 times stronger affinity to Mdm2/x than the native p53.
2. Structures of Mdm2 and Mdmx with p53 Peptide Fragments
The very first structure of the p53 binding domain of Xenopus and human Mdm2 in complex with a 15-residue peptide of human p53 was published 14 years ago by Kussie et al[34] (Figures 1a, 2a, and 3a).[34] Mdmx was not recognized as important for p53 regulation and cancer control at that time. Structures of Mdmx with p53 were published only recently (Figure 2g and Figure 3g).[35,36] Both Mdm2 and Mdmx bind p53 through interactions that are almost entirely hydrophobic, with p53 forming a short helix inside the Mdm2/x binding clefts. The three p53 residues that principally contribute to the binding are Phe19, Trp23, and Leu26. These residues are located on the same side of the amphipathic p53 helix, with their side chains positioned deeply in the binding cavity of Mdm2/x. The Trp23 ε nitrogen atom forms a solvent-protected hydrogen bond with Leu54 in Mdm2 (Met53 of Mdmx). The p53–Mdm2 and p53–Mdmx complexes display nearly identical binding features (Figures 2a,g and 3a,g). The major difference is the shape of the Leu26 pocket. Firstly, it is smaller in Mdmx because of the Met53 side chain located there; this residue corresponds to and is larger than Leu54 of Mdm2. Secondly, the Pro95–Tyr99 regions in Mdm2 and Mdmx have different shapes.[36,37] Another important difference between the binding of p53 to Mdm2 and to Mdmx is the presence of a secondary hydrophobic area next to the Leu26 binding site in the latter. It is formed by Leu33, Val52, and Leu106 and separated from the Leu26 binding site by Met53 and Leu102 side chains. The p53 protein does not bind here.[36] This additional binding site is approximately 10 Å long but rather flat and could play an essential role in the discovery of high-affinity Mdmx ligands in the future.
Figure 1.
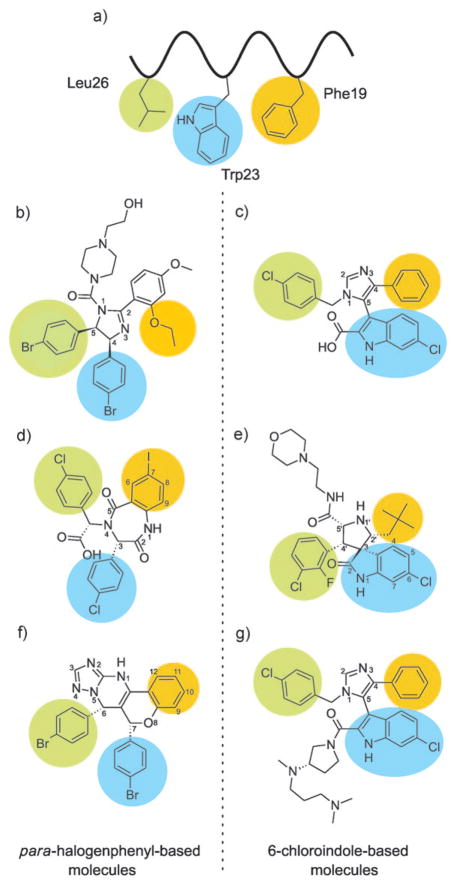
Low-molecular-weight inhibitors of p53–Mdm2/x binding. a) The p53 protein binds to Mdm2/x using a short helix with three hydrophobic residues (Phe19 (orange), Trp23 (blue), and Leu26 (green)) which fills the binding cleft. b) Nutlin-2 is a close analogue of the most-studied Mdm2 inhibitor Nutlin-3. c) Imidazole-indole compound WK23 in complex with Mdm2. WK23 possesses a 6-chloroin-dole group which is bound to Mdm2 in the same way as the Trp23 side chain of p53. d) Benzodiazepinedione inhibitors utilize para-halogenated phenyl rings similar to those of the Nutlins. The Phe19 pocket is filled by the 7-iodobenzene ring. e) A diastereomer of MI-63 positions the 6-chlorooxindole group in the Trp23 pocket. The Phe19 pocket interacts with the neopentyl group of the inhibitor and the 2-fluoro-3-chlorophenyl is situated in the Leu26 pocket. f) Chromenotria-zolopyrimidines are also equipped with two halogenated phenyl rings that fill Trp23 and Leu26 pockets in a “Nutlin-like” fashion. g) The imidazole-indole compound WW298 in complex with Mdmx. The 6-chloroindole group binds to Mdmx in the same way as the Trp23 side chain of p53 does. Note that in (c), (e), and (g) the 6-chloroindole group is used to bind in the Trp pocket, and that in (b), (d), and (f) a 4-halogenphenyl serves the same purpose.
Figure 2.
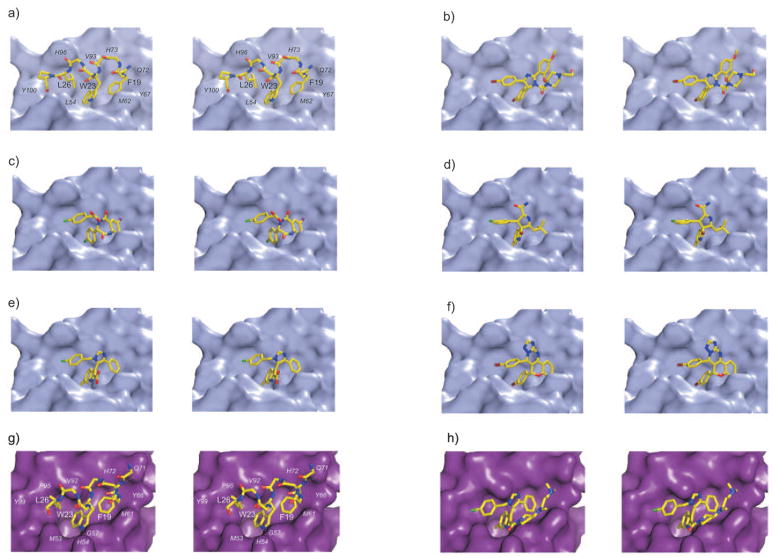
Structural details of the Mdm2/x–p53 binding inhibitors shown as stereograms. p53 residues are labeled in bold and Mdm2/x in italics. Hydrogen bonds are indicated as yellow dashed lines. Color scheme: yellow C, dark red Br, green C, light blue F, purple I, blue N, red O. a) p53 forms a short helix that positions Phe19, Trp23, and Leu26 in the binding cleft of Mdm2. b) Nutlin-2. c) Benzodiazepinedione. d) MI-63 diastereomer (no electron density is observed for the morpholinoethyl part). e) WK23. f) Chromenotriazolopyrimidine. g) The structure of human Mdmx in complex with the p53 peptide. The overall interaction mechanism of p53 is nearly identical to that seen in Mdm2 (a). h) The WW298 inhibitor binds to Mdmx in a similar way as WK23; additionally, the N,N-dimethylminopropylfragment folds over the Phe19 pocket extending hydrophobic interactions.
Figure 3.
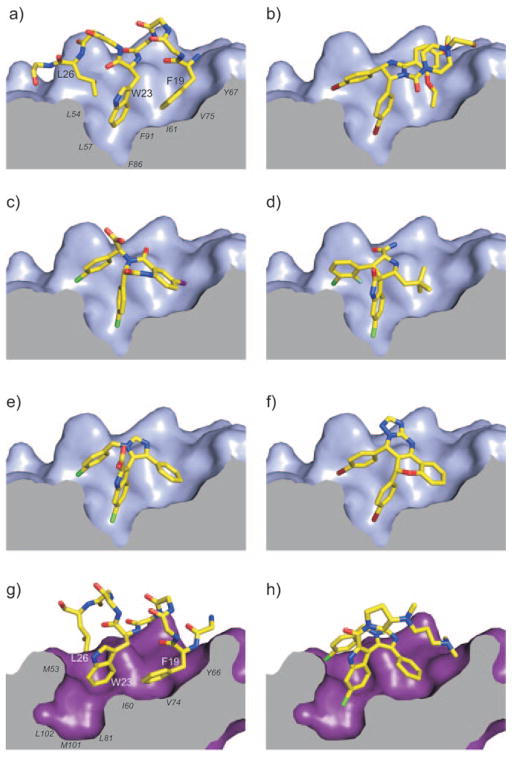
Enlarged views of the complexes (color scheme given in Figure 2). a) Mdm2 in complex with p53 (for clarity, side chains other than Phe19, Trp23, and Leu26 are omitted). p53 residues are labeled in bold and Mdm2/x in italics. b) Nutlin-2, c) benzodiazepineione, d) the MI-63 diastereomer, e) WK23, f) chromenotriazolopyrimidine, g) Mdmx in complex with p53 (the shape of Leu26 pocket differs from that in Mdm2), h) WW298.
3. Low-Molecular-Weight Inhibitors of Mdm2–p53 and Mdmx–p53 Interactions
Small-molecule inhibitors of enzymes, receptors, or protein–protein complexes are highly favored as drug candidates by the pharmaceutical industry. It is therefore not surprising that all major pharmaceutical companies as well as many small biotech companies have projects on p53–Mdm2/x inhibitors. The most progress has been made with inhibitors in the Nutlin and MI series, which inhibit Mdm2–p53 binding. In the following we describe all the compounds with high-resolution X-ray crystal structures currently available, starting with Roche’s Nutlins. Essentially, in all published, potent inhibitors either a set of p-halogenated phenyl groups or a combination of 6-chloro(ox)indole are used to fill the Trp23 pocket, and other with diverse bulky hydrophobic moieties are located in the Phe19 and Leu26 pockets (Figure 1).
3.1. Nutlins
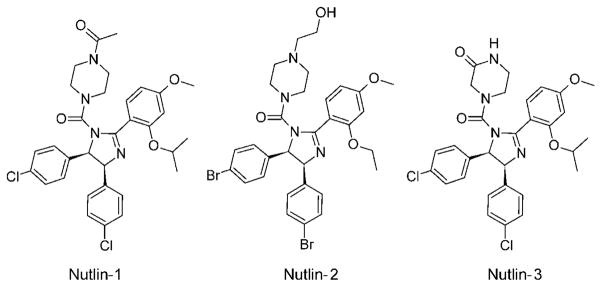
The Nutlin scaffold based on a tetrasubstituted imidazoline unit was discovered by high-throughput screening (HTS), and this family of compounds is the best characterized class of compounds so far, with more than 900 literature references to date.[16,21,38] Nutlin-2 was also the first published inhibitor with a crystallographic structure in complex with Mdm2 (PDB ID: 1V1).[38] Out of the three Nutlins published, the most potent and most studied is Nutlin-3, with a Ki value of 36 nM.[39] Nutlin-1 and Nutlin-2 have approximately three- and twofold lower affinity towards Mdm2, respectively. Nutlin-3 binds to Mdmx too but with a roughly 1000-fold lower affinity of about 25 μM.[35] Nutlin-3 is based on a cis-4,5-dihydroimidazole scaffold (Figure 1b), with four attached substituents. The details of the binding configuration of Nutlin-2 are shown in Figures 2b and 3b. The two para-bromophenyl substitutents directly insert into two pockets of the Mdm2-binding site (Trp23 and Leu26), whereas a third phenyl substitutent reaches the third pocket (Phe19) only indirectly by means of an ortho-isopropoxy group. The para-bromophenyl ring at position 5 submerges deeply into the Trp23 pocket, while the second para-bromophenyl substituent, attached to position 4 of its scaffold ring, fills the Leu26 pocket. The chlorine atom, positioned at the very bottom of the Trp23 pocket, fills a small cavity visible on the molecular surface of Mdm2, which is not occupied by the indole ring of the p53 Trp23. Filling this space with a hydrophobic atom, usually a halogen, seems to be a critical feature of an efficient Mdm2 inhibitor and was recognized already in early peptide studies.[40] The Phe19 pocket is filled with the ortho-isopropoxy entity. Interestingly, the Phe19 pocket could be expected to require an aromatic pharmacophore, but this is not the case with the Nutlin and MI molecules (see Section 3.3). The fourth imidazoline substitutent, the N-2-hydroxyethylpiperazine ring, does not penetrate the p53 cleft directly, but instead covers the Phe19 pocket near the Met62 side chain of Mdm2. This heterocyclic motif, with two nitrogen atoms and one OH group, likely increases water solubility considerably; in addition, its three hydrophobic ethylene fragments enhance hydrophobic contacts, whereas the hydroxy group forms polar contacts with the side chain of Gln72. Altogether those features provide the complex with a continuous hydrophilic surface.
Small structural differences between Nutlin-1 and Nutlin-3, basically finetuning of the hydrophilic side of the molecule, are responsible for almost three times better binding of Nutlin-3 and can be used to improve affinity and other target-independent properties (Table 1, Scheme 1).
Table 1.
Binding constants of Mdm2/x–p53 inhibitors with known complex structures.
| Inhibitor | Mdm2 Ki [μm] |
Mdmx Ki [μm] |
Reference |
|---|---|---|---|
| p53 | 0.89 | 0.21 | [35] |
| Nutlin-1 | 0.26 (IC50) | – | [38] |
| Nutlin-2 | 0.14 (IC50) | – | [38] |
| Nutlin-3[a] | 0.036, 0.09 (IC50) | 9.38 | [38,39] |
| WK23 | 0.916 | 36 | [33] |
| benzodiazepinedione | 0.080 | – | [41] |
| MI-63 (analogue) | 0.005 (0.036) | 55 | [33,39] |
| chromenotriazolopyrimidine | 1.23 (IC50) | – | [51] |
| WW298 | 0.109 | 11 | [33] |
No crystal structure has been published, but it is expected to be virtually identical to the Nutlin-2 complex.
Scheme 1.
Comparison of Nutlin-1, Nutlin-2, and Nutlin-3.
It is evident that while the hydrophilic fragment of the Mdm2-binding molecule does not interact directly with Mdm2, it provides the solvent-exposed “cover” of the binding interface. Preventing water accession to the hydrophobic interface has an evidently beneficial influence on binding energy. Thus, the final optimization of the ligand has to take into account the hydrophilic properties of the molecule too, as this part is more than a “solubility-tag” and has an important influence on the whole complex.
The structure of Mdm2 in complex with Nutlin-2 does not show significant induced-fit changes relative to the p53–Mdm2 or apoprotein structures (the all-atom root-mean-square deviation between the Mdm2–Nutlin and Mdm2–p53 complexes is 0.85 Å). The only noteworthy difference occurs at the side chain of Tyr100, which points outside the Leu26 pocket in the structure with the wt-p53 peptide bound (the Tyr100 “open” conformation),[33,34] while it is directed towards the inside of the pocket (the “closed” conformation)[33,38] in the structure of the Nutlin–Mdm2 complex.
3.2. Benzodiazepinediones
Shortly after the Nutlin inhibitors had been described, Grasberger et al. reported a complex containing a benzodiazepinedione inhibitor (PDB ID: 1T4E; Figure 1d).[41] The scaffold was found by HTS with the temperature-dependent protein-unfolding assay ThermoFluor.[42] Over 300000 compounds were tested. The strongest Mdm2-binding compound from this family was optimized and later named TDP222669.[43,44] TDP222669 has a Ki value of 80 nM and was confirmed to be active in vitro (Table 1). The compound suffered, however, from low bioavailability and rapid clearance and was therefore subsequently optimized at the cost of a slightly lower binding constant.[42,43] The structure of TDP222669 bound to Mdm2 reveals that, in a similar way to the Nutlins, the para-chlorophenyl group is attached to position 3 of a saturated 1,4-diazepinedione ring, the main scaffold. The para-chlorophenylglycine group, attached to the nitrogen atom at position 4 of the benzodiazepinedione ring, penetrates the Leu26 cavity, again with the chlorine atom positioned in the deepest part of the Leu26 pocket. It should be noted that the positions of these two para-chlorophenyl rings are nearly identical to those seen in the Nutlin complex (Figures 2c and 3c). When the two complexes are superimposed, the chlorine atoms differ only by 0.32 Å in the Trp23 pocket and 0.44 Å in the Leu26 pocket. The 7-iodophenyl group fused to the diazepinedione ring is located in the Phe19 pocket. Noticeably, this element does not insert itself into this subpocket as deeply as the Phe19 phenyl ring does, but apparently forms a high-affinity interaction. As recently noted the iodine atom in position 7 makes contacts to the backbone carbonyl group of Gln72 that are shorter than van der Waals contacts.[45] The binding data for the isomorphic replacement of this iodo group by other substitutents (H, halogens, acetylene, Me, CN etc) suggest that the C–I···H bond is comparable in strength to a weak hydrogen bond and contributes considerably to the overall affinity.
TDP222669 also exhibits a carboxylic acid group—a polar side chain—pointing into water. The role of this group is again dual: enduing the compound with some water solubility and also increasing its affinity to the binding pocket. Extensive SAR studies have clearly defined the role of this additional surface-exposed group. Additionally, it was found that the nature of the “solubilizing” side chains has a delicate influence on the cellular activities of the compounds, likely effecting the permeation of different cell-membranes.[42,43]
The binding of the benzodiazepinedione inhibitor to Mdm2 again does not cause pronounced induced-fit changes relative to the structure of the p53–Mdm2 complex. The position of Tyr100 is similar to that in the Nutlin–Mdm2 complex, although the influence of the extra Gln16–Gln24 sequence present in the Mdm2 construct crystallized by Grasberger et al.[41] cannot be excluded. This sequence is not present in the Mdm2 used for the crystallization of the Nutlin-bound complex.
3.3. The MI-219 Family
MI-219 and its homologue MI-63 were the first inhibitors of the Mdm2–p53 interaction that utilize a 6-chlorooxindole unit to mimic the native Trp23 configuration (Figure 1e).[39,46] They are the second most studied Mdm2–p53 inhibitors after the Nutlins.[21] The scaffold was designed de novo by expanding the oxindole group. The design was based on known natural products that contain this moiety.[46] The affinities of MI-219 and MI-63 towards Mdm2 are 5 and 3 nM, respectively. The affinities towards Mdmx are >10000-fold lower, with Ki values of over 50 μM (Table 1). The core of this class of molecules is built upon spirooxindole-3,3′-pyrrolidine, which 1) fills the Trp23 pocket, 2) serves as a scaffold for positioning further substitutents to fill the Leu26 and Phe19 pockets, and 3) ensures sufficient water solubility. According to modeling studies by Ding et al.[46] the neopentyl substituent attached to position 2′ of the pyrrolidine ring fills the Leu26 pocket, and the halogen-substituted phenyl ring attached to position 4′ is located in the Phe19 pocket. This configuration is realized by 2′R,3S,4′R,5′R diastereomer. Interestingly, in the recently published structure, the 2′R,3R,4′S,5′R diastereomer was crystallized (PDB ID: 3LBL), which has a measured affinity similar to that of the former diastereomer.[33] The details of the binding are shown on Figures 2d and 3d. The structure of the former diastereomer was recently mentioned by Jacoby et al.[47] It is not possible to analyze this structure as the coordinates are not available. Since the p53-binding pocket of Mdm2 is almost symmetrical along the Trp23 indole plane, it is probable that both diastereomers bind to Mdm2 with similar, high affinities. In the published crystal structure, the 6-chlorooxindole group is located in the Trp23 pocket and forms a hydrogen bond with the Mdm2 Leu54 carbonyl oxygen atom. This interaction is identical to that predicted by Ding et al.[46] In the crystal structure, however, the 2-fluoro-3-chlorophenyl ring is located in the Leu26 pocket, in a similar mode to the para-chlorophenyl ring of Nutlin. The configurations of both the 2-fluoro-3-chlorophenyl group and the neopentyl group in this structure are an exact mirror image of the binding model presented by Ding et al.[46]
Owing to the high symmetry of the p53-binding pocket along the indole plane of Mdm2, the molecule can bind in two different modes. Each mode can be realized by a different enantiomer or diastereoisomer. So far there has been no systematic study of the binding properties of different isomers of the same molecule to Mdm2. Certainly this unusual aspect must still be explored. Most experiments are usually performed on racemic mixtures of the p53–Mdm2 binding inhibitors. It is therefore important to treat the binding data cautiously as the possibility exists that more than one diastereomer interacts.
The Tyr100 residue remains in an “open” conformation, thus allowing enough space for a halogen atom.[33] The Phe19 pocket is filled by the neopentyl group. Here a significant induced-fit change can be observed: the Tyr67 side chain of Mdm2 bends towards the inside of the pocket, displacing the His73 residue. Additionally, the pyrrolidine ring of the inhibitor extends over Val93. The amide group at position 5 forms a hydrogen bond between its carbonyl oxygen and the His96 side chain.
The MI-63 derivative features a morpholinoethylamide side chain which is not visible in the crystal structure, probably because of its flexibility. However, based on the pyrrolidine amide position this side chain must extend along the Phe19 binding pocket and the morpholino ring is found in the region between Gln72 and Lys94. Again, this side chain provides water solubility and additional binding interactions.
The X-ray structure explains well why MI molecules are poor Mdmx inhibitors: Mdmx does not undergo induced-fit changes in the Phe19 pocket, Tyr99 is fixed in the “closed” conformation, and the spirooxindole backbone used in the MI series is stiff. Based on these findings a new set of pharmacophores can be proposed for further ligand optimization.
3.4. Imidazole-Indoles
The imidazole-indole family is the latest development in the field of inhibitors of the p53-Mdm2/x interaction. These compounds were independently and simultaneously disclosed by several research groups (Figure 1c,g).[48–50] In this family only one compound, called WW298 or Novartis-101, has been characterized crystallographically in complex with Mdmx protein (PDB ID: 3LBJ).[33] The same publication also presents the structure of another member of this family, called WK23, bound to the Mdm2 protein (PDB ID: 3LBK; Figures 2e and 3e). While the affinity of WW298 towards Mdmx is in the low micromolar range (109 nM for Mdm2; Table 1), the inhibitor–Mdmx structure provides first insight into the details of Mdmx–inhibitor binding (Figures 2h and 3h).[33] The basic scaffold comprises a planar aromatic imidazole ring with an attached phenyl substitutent at position 4, a 2-carboxy-6-chloroindole substitutent at position 5, and a 4-chlorobenzyl substitutent at position 1. Compounds in this family differ mostly in the amide substituents of the carboxylic group of the 2-carboxy-6-chloroindole unit. The overall mechanism of binding of these compounds is similar for both Mdm2 and Mdmx. The indole group is anchored at the bottom of the Trp23 pocket in a “tryptophan-mimicking” fashion. The chlorine atom in this group induces significant alterations in the bottom of the Mdmx pocket to accommodate its larger volume. There is no need for such a rearrangement in Mdm2, as sufficient space is already provided. In both structures, the indole nitrogen atoms form hydrogen bonds to Met53 (Mdmx) or Leu54 (Mdm2), mimicking the p53 peptide interactions. The Phe19 pocket is filled in a similar fashion in both proteins by the phenyl ring, which is, however, oriented perpendicular to the Phe19 ring. It is noteworthy that the Phe phenyl ring does not submerge itself as deeply into the pocket as Phe19 of p53. This seems to be a general feature of most Mdm2 inhibitors which, however, does not affect the high affinity. The Leu26 pocket is filled by the para-chlorobenzyl group in a “Nutlin-like” fashion. These interactions are seen for both Mdm2 and Mdmx. It is clear, however, that the greater flexibility of the imidazo-indole family (relative to the spirooxindole compounds) allows small adjustments of the ligand to different features of the binding pockets.
Additionally, in the Mdmx structure the N,N-dimethyl-aminopropyl part of WW298 is seen folded over the Phe19 pocket similar to the piperazine positioning seen in the Nutlin structure. The N,N-dimethylaminopropyl pyrrolidine fragment is responsible for the strong water solubility of WW298. At the same time it protects the Phe19 binding site from solvent, shielding the hydrophobic region formed by Met61 and Tyr66. It also provides additional hydrophobic and electrostatic interactions and makes the surface of the complex continuously hydrophilic. The amide oxygen atom at the 2-position of the indole moiety forms a hydrogen bond to the His54 side chain of Mdmx. Despite the structural similarity of the Mdm2 and Mdmx binding pockets and these additional interactions, WW298 achieves only moderate affinity towards Mdmx (11 μM),[33] while WK23, being only the essential “core” of WW298, binds Mdmx with an affinity of 36 μM and still has nanomolar binding affinity to Mdm2. Taking into account that a 6-chloroindole-based peptide can achieve low nanomolar affinity for both Mdm2 and Mdmx, the recued Mdmx affinity of the imidazole-indole family can be explained by the suboptimal substituent-filling of the Leu26 or Phe19 pockets, or the lack of conformational flexibility in the inhibitor to position these elements in energetically optimal positions. The Mdmx–WW298 structure enables, however, detailed analysis and suggests optimization strategies for achieving a ligand with high affinity.[33]
3.5. Chromenotriazolopyrimidines
Another recently published class of compounds with structural data is based on the chromenotriazolopyrimidine scaffold (PDB ID: 3JZK).[51] These compounds were found by HTS and were subsequently optimized. They were found to be active on various cell assays, causing elevated levels of p53 and proapoptotic effects. Their binding to Mdm2 is in submicromolar range (Table 1). The compounds are based on a flat heteroaromatic chromenotriazolopyrimidine scaffold with two para-brominated phenyl rings attached to positions 6 and 7 (Figure 1f). Typical for most known of the inhibitors of the p53–Mdm2 interaction, only specific stereoisomers show good affinity. The two phenyl rings of the inhibitor occupy the Trp23 and Leu26 pockets in a Nutlin-like fashion, although they are located approximately 0.5 Å deeper in the pockets than in the Nutlin structure. The benzene ring of the chromene group is located in the Phe19 pocket, in a similar way to the benzene fragment in the benzodiazepinedione family. The details of the binding are shown in Figures 2f and 3 f. The triazolopyrimidine fragment folds over Val93 and His96 to provide additional hydrophobic and π–π interactions.
4. Optimized Peptides and Peptide-Based Miniproteins
The phage display and structure-based optimization have led to the development of a large number of p53-like peptides[29–32,41,52–55] and miniproteins[54,56] that have enhanced binding to Mdm2/x compared to the wild-type p53 analogue. All of these peptides contain the identical hot-spot triad of Phe19, Trp23, and Leu26. The binding mechanism of these peptides is also very similar to the binding of the native p53 described by Kussie et al.[34] (Figures 1a 3). The peptides form a short helix, orienting the Phe-Trp-Leu triad towards the binding cleft of Mdm2/x. The principal feature of these peptides is the substitution of Pro27 with other amino acids. This single change is sufficient to improve the binding of a peptide to Mdm2 from 0.7 μM for the wild-type p53 peptide to approximately 4.7 nM.[31,53] Interestingly, we found that Pro27 seems to be highly conserved among all UV-exposed vertebrates and may be essential in maintaining the physiological level of p53. The mutation of Pro27 allows extension of the p53 helix. This has been suggested to be required for high-affinity interactions.[57] The other important substitution is the replacement of Leu22 by Tyr,[31,41] extending hydrophobic interactions with Mdm2 and Mdmx. In contrast to small molecules that have a strong preference towards Mdm2 and only limited binding to Mdmx, the modified peptides achieve easily low nanomolar affinities towards both proteins.[31,52] It is conceivable thus that the Mdm2-optimized para-chloro-phenyl substituents, commonly used in these small-molecule inhibitors, are not optimal, especially in the Leu26 pocket, which shows most differences between Mdm2 and Mdmx.
5. Optimization of Target-Unrelated Properties
A druglike compound used to evaluate a target pathway for drug development should address not only target-related features such as potency and selectivity but also target-unrelated properties such as water solubility, lipophilicity, and pKa.[58] Whereas the former properties can give rise to good pharmacological effects (e.g. efficacy, selectivity), the latter are as important to endow compounds with optimal ADME/Tox (e.g. penetration of the blood–brain barrier, metabolic stability, toxicology). A key property in this regard is water solubility. Sufficient water solubility ensures that the compound can be transported to the diseased tissue and that high enough concentration are available to exert biological activity. In contrast, poorly water-soluble compounds often display poor absorption and oral bioavailability, insufficient solubility for intravenous dosing; they are often difficult to develop and must be administered to patients frequently and in high doses. Additionally, low water solubility often leads to erratic assay results (false positives). Thus water solubility is a very important property in drug discovery and must be optimized by the medicinal chemist.
The interfaces of protein–protein interactions in general, and the p53/Mdm2/Mdmx interfaces in particular, are highly hydrophobic. This presents a challenge to the researchers who design inhibitors for these types of interactions. Therefore ligands addressing this binding site must have intrinsically high lipophilicity, which is unavoidably accompanied by low water solubility. Additionally, the ligands must possess a hydrophilic site to expose to the solvent upon binding to Mdm2/x so that the resulting complex has an uninterrupted hydrophilic surface. The differences in the binding constants of Nutlin-1 and -3 as well as of WK23 and WW298 confirm that the optimized, solvent-exposed part of the ligand is essential to achieving good binding. On the other hand, a certain lipophilicity is desired as it also helps compounds to enter cells through the membrane. This is crucial for the biological activity of p53 since the target is intracellular. The dilemma of target-required lipophilicity and the transportation-required hydrophilicity is well known to medicinal chemists and can be solved in the present case by studying the published co-crystal structures. Clearly, all Mdm2/x crystal structures indicate a strongly hydrophobic pocket which must be occupied by the ligand to obtain high affinity; however, it also shows a highly water-accessible convex binding surface (Figure 3). Approaches to specifically address the water solubility of Mdm2 antagonists have been published by Srivastava et al.[28]
6. Summary and Outlook
High-resolution X-ray structure data has been instrumental in optimizing compound classes to obtain clinical candidates with high affinity and selectivity and suitable physico-chemical properties for good pharmacokinetics and pharma-codynomics (PK/PD). Since the first structure of the p53–Mdm2 complex published in 1996, many structures with different peptides bound to Mdm2 or Mdmx have been described. Structural analysis of several classes of small molecules binding to Mdm2 has also led to a simple pharmacophore model known as “thumb–index–middle” finger that can explain the structural requirements of Mdm2 binders (Figures 1a, 2a, and 3a).[59]
Until very recently, the only characterized low-molecular-weight inhibitors of p53–Mdm2/x binding were Nutlin-2 and benzodiazepinedione inhibitors. This situation has, however, recently changed. With the structures of five different classes of inhibitors now published, it is possible to employ known, proven techniques—like scaffold variations and other rational structure-based approaches—to develop new, potent inhibitors of Mdm2 and Mdmx. Since most of the known inhibitors were deliberately “tuned” for Mdm2, their affinity towards Mdmx is limited. However, given the only subtle differences between both proteins, it should be now possible to systematically search the chemical combinatorial space for efficient Mdmx binders. It is even possible that a molecule rejected from lead-optimization process, because of its low Mdm2 affinity, might prove efficient towards Mdmx. It is therefore worthwhile to re-evaluate existing Mdm2-oriented libraries for Mdmx affinity. It has been postulated that efficient inhibitors of p53–Mdmx binding should also prove effective against Mdm2, but not vice versa.[35] Thus, it would be possible to efficiently free p53 from its Mdm2/x restrainers enabling its antitumor activity. The progress made in Mdm2 inhibitors suggests that the development of potent Mdmx-only or dual-action Mdm2/x binding inhibitors is possible. We thus hope that such compounds may become a base for the development of novel antitumor drugs.
Acknowledgments
This work was supported by the Deutsche Krebshilfe, (grant 108354 to T.A.H.) and by the NIH (grants 1R21M087617-01A and 1P41M094055-01), the NCI-RAND program, and the University of Pittsburgh (A.D.).
Biographies

Grzegorz Popowicz studied medical physics at the AGH University of Science and Technology, Krakow (Poland) and obtained his Master’s degree in 2002. In 2006 he completed PhD research at the Max Planck Institute for Biochemistry, Martinsried (Germany), where he is currently a research associate scientist. His research is focused on controlling protein–protein interactions for therapeutic purposes. Using NMR and X-ray crystallography techniques, he is developing small-molecule and peptidic inhibitors for proteins involved in tumorigenesis.

Alexander Dömling studied chemistry and biology at the Technical University Munich. After completing his PhD under the supervision of Ivar Ugi, he spent a postdoctoral year at the Scripps Research Institute as a Feodor Lynen scholar in the group of Barry Sharpless. He is the founder of several biotech companies. In 2004 he completed his habilitation in chemistry at the Technical University Munich. Since 2006 he has been an associate professor at the University of Pittsburgh in the Department of Pharmacy with secondary appointments in the Departments of Chemistry and Computational Biology. His research interests are the design of multicomponent reactions and their application to the design of antagonists of protein–protein interactions.

Tad A. Holak is the head of the Biological NMR Structure Group at the Max Planck Institute for Biochemistry, Martinsried (Germany). His research focuses on discovering three-dimensional structures and structure–function properties of proteins using a combination of multinuclear NMR spectroscopy, biochemistry, and X-ray crystallography. From 1998 to 2009, he was a member of the Editorial Board of the European Journal of Biochemistry and the FEBS Journal.
Contributor Information
Dr. Grzegorz M. Popowicz, Max Planck Institute for Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany)
Prof. Alexander Dömling, Departments of Pharmaceutical Sciences and Chemistry, University of Pittsburgh, Pittsburgh, PA 15261 (USA)
Dr. Tad A. Holak, Max Planck Institute for Biochemistry, Am Klopferspitz 18, 82152 Martinsried (Germany).
References
- 1.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Nat Rev Cancer. 2009;9:862. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Lu H. Cancer Cell. 2009;16:369. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menendez D, Inga A, Resnick MA. Nat Rev Cancer. 2009;9:724. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Elf SE, Asai T, Miyata Y, Liu Y, Sashida G, Huang G, Giandomenico S, Koff A, Nimer SD. Cell Cycle. 2009;8:3120. doi: 10.4161/cc.8.19.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meek DW. Nat Rev Cancer. 2009;9:714. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 6.Wade M, Wang YV, Wahl GM. Trends Cell Biol. 2010;20:299. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dey A, Lane DP, Verma CS. Semin Cancer Biol. 2010;20:3. doi: 10.1016/j.semcancer.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Vousden KH, Lane DP. Nat Rev Mol Cell Biol. 2007;8:275. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 9.Joerger AC, Fersht AR. Annu Rev Biochem. 2008;77:557. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 10.Marine JC, Jochemsen AG. Cell Cycle. 2004;3:900. [PubMed] [Google Scholar]
- 11.Marine JC, Lozano G. Cell Death Differ. 2010;17:93. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 12.Lee JT, Gu W. Cell Death Differ. 2010;17:86. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo F, Wahl GM. Nat Rev Cancer. 2006;6:909. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 14.Toledo F, Wahl GM. Cell Biol. 2007;39:1476. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Marx J. Science. 2007;315:1211. doi: 10.1126/science.315.5816.1211. [DOI] [PubMed] [Google Scholar]; b) Martins CP, Brown-Swigart L, Evan GI. Cell. 2006;127:1323. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]; c) Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Nature. 2007;445:661. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]; d) Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Nature. 2007;445:656. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassilev LT. J Med Chem. 2005;48:4491. doi: 10.1021/jm058174k. [DOI] [PubMed] [Google Scholar]
- 17.Vassilev LT. Trends Mol Med. 2007;13:23. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa M, Aonuma M, Lee SH, Fukutake S, Mc-Cormick F. Oncogene. 2008;27:5303. doi: 10.1038/onc.2008.164. [DOI] [PubMed] [Google Scholar]
- 19.Tokalov SV, Abolmaali ND. BMC Cancer. 2010;23:57. doi: 10.1186/1471-2407-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng T, Wang J, Song X, Meng X, Pan S, Jiang H, Liu L. J Cancer Res Clin Oncol. 2010;136:1597. doi: 10.1007/s00432-010-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shangary S, Wang S. Annu Rev Pharmacol Toxicol. 2009;49:223. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurie NA, Shih CS, Dyer MA. Curr Cancer Drug Targets. 2007;7:689. doi: 10.2174/156800907782418266. [DOI] [PubMed] [Google Scholar]
- 23.Lam S, Lodder K, Teunisse AF, Rabelink MJ, Schutte M, Jochemsen AG. Oncogene. 2010;29:2415. doi: 10.1038/onc.2009.522. [DOI] [PubMed] [Google Scholar]
- 24.Marine JC, Dyer MA, Jochemsen AG. J Cell Sci. 2007;120:371. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 25.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. J Biol Chem. 2006;281:33030. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 26.Weber L. Expert Opin Ther Pat. 2010;20:179. doi: 10.1517/13543770903514129. [DOI] [PubMed] [Google Scholar]
- 27.Dudkina AS, Lindsley CW. Curr Top Med Chem. 2007;7:952. doi: 10.2174/156802607780906762. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava S, Beck B, Wang W, Czarna A, Holak TA, Dömling A. J Comb Chem. 2009;11:631. doi: 10.1021/cc9000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu B, Gilkes DM. J Chen Cancer Res. 2007;67:8810. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 30.Kallen J, Goepfert A, Blechschmidt A, Izaac A, Geiser M, Tavares G, Ramage P, Furet P, Masuya K, Lisztwan J. J Biol Chem. 2009;284:8812. doi: 10.1074/jbc.M809096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czarna A, Popowicz GM, Pecak A, Wolf S, Dubin G, Holak TA. Cell Cycle. 2009;8:1176. doi: 10.4161/cc.8.8.8185. [DOI] [PubMed] [Google Scholar]
- 32.Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG, Lu W. Proc Natl Acad Sci USA. 2009;106:4665. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popowicz GM, Czarna A, Siglinde W, Wang K, Wang W, Dömling A, Holak TA. Cell Cycle. 2010;9:1104. doi: 10.4161/cc.9.6.10956. [DOI] [PubMed] [Google Scholar]
- 34.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 35.Popowicz GM, Czarna A, Rothweiler U, Szwagierczak A, Krajewski M, Weber L, Holak TA. Cell Cycle. 2007;6:2386. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 36.Popowicz GM, Czarna A, Holak TA. Cell Cycle. 2008;7:2441. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 37.Joseph TL, Madhumalar A, Brown CJ, Lane DP, Verma C. Cell Cycle. 2010;9:1167. doi: 10.4161/cc.9.6.11067. [DOI] [PubMed] [Google Scholar]
- 38.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 39.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yang D, Wang S. Proc Natl Acad Sci USA. 2008;105:3933. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcíia-Echeverría C, Chene P, Blommers MJ, Furet P. J Med Chem. 2000;43:3205. doi: 10.1021/jm990966p. [DOI] [PubMed] [Google Scholar]
- 41.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, Maguire D, Lattanze J, Franks CF, Zhao S, Ramachandren K, Bylebyl GR, Zhang M, Manthey CL, Petrella EC, Pantoliano MW, Deckman IC, Spurlino JC, Maroney AC, Tomczuk BE, Molloy CJ, Bone RF. J Med Chem. 2005;48:909. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 42.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. J Biomol Screening. 2001;6:429. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 43.Koblish HK, Zhao S, Franks CF, Donatelli RR, Tominovich RM, LaFrance LV, Leonard KA, Gushue JM, Parks DJ, Calvo RR, Milkiewicz KL, Marugán JJ, Raboisson P, Cummings MD, Grasberger BL, Johnson DL, Lu T, Molloy CJ, Maroney AC. Mol Cancer Ther. 2006;5:160. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- 44.Parks DJ, LaFrance LV, Calvo RR, Milkiewicz KL, Marugán JJ, Raboisson P, Schubert C, Koblish HK, Zhao S, Franks CF, Lattanze J, Carver TE, Cummings MD, Maguire D, Grasberger BL, Maroney AC, Lu T. Bioorg Med Chem Lett. 2006;16:3310. doi: 10.1016/j.bmcl.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 45.Bissantz C, Kuhn B, Stahl M. J Med Chem. 2010;53:5061. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y, Parrish DA, Deschamps JR, Wang S. J Am Chem Soc. 2005;127:10130. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 47.Jacoby E, Boettcher A, Mayr LM, Brown N, Jenkins JL, Kallen J, Engeloch C, Schopfer U, Furet P, Masuya K, Lisztwan J. Methods Mol Biol. 2009;575:173. doi: 10.1007/978-1-60761-274-2_7. [DOI] [PubMed] [Google Scholar]
- 48.A. Boettcher, WO 2008119741, 2008.
- 49.A. Dömling, WO 2008130614, 2008.
- 50.Beck B, Leppert CA, Mueller BK, Dömling A. QSAR Comb Sci. 2006;25:527. [Google Scholar]
- 51.Allen JG, Bourbeau MP, Wohlhieter GE, Bartberger MD, Michelsen K, Hungate R, Gadwood RC, Gaston RD, Evans B, Mann LW, Matison ME, Schneider S, Huang X, Yu D, Andrews PS, Reichelt A, Long AM, Yakowec P, Yang EY, Lee TA, Oliner JD. J Med Chem. 2009;52:7044. doi: 10.1021/jm900681h. [DOI] [PubMed] [Google Scholar]
- 52.Phan J, Li Z, Kasprzak A, Li B, Sebti S, Guida W, Schönbrunn E, Chen J. J Biol Chem. 2010;285:2174. doi: 10.1074/jbc.M109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zondlo SC, Lee AE, Zondlo NJ. Biochemistry. 2006;45:11945. doi: 10.1021/bi060309g. [DOI] [PubMed] [Google Scholar]
- 54.Li C, Pazgier M, Liu M, Lu WY, Lu W. Angew Chem. 2009;121:8868. doi: 10.1002/anie.200904550. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:8712. [Google Scholar]
- 55.Madhumalar A, Jun LH, Brown CJ, Lane D, Verma CS. Cell Cycle. 2009;8:137. doi: 10.4161/cc.8.1.7530. [DOI] [PubMed] [Google Scholar]
- 56.Li C, Liu M, Monbo J, Zou G, Li C, Yuan W, Zella D, Lu WY, Lu W. J Am Chem Soc. 2008;130:13546. doi: 10.1021/ja8042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dastidar SG, Lane DP, Verma CS. J Am Chem Soc. 2008;130:13514. doi: 10.1021/ja804289g. [DOI] [PubMed] [Google Scholar]
- 58.Kerns EH, Di L. Drug-like Properties: Concepts, Strcuture Design and Methods. Academic; New York: 2008. [Google Scholar]
- 59.Dömling A. Curr Opin Chem Biol. 2008;12:281. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]