Abstract
The lymphoid tyrosine phosphatase LYP, encoded by the PTPN22 gene, recently emerged as a major player and candidate drug target for human autoimmunity. The enzyme includes a classical N-terminal protein tyrosine phosphatase catalytic domain and a C-terminal PEST-enriched domain, separated by an ∼300-amino acid interdomain. Little is known about the regulation of LYP. Herein, by analysis of serial truncation mutants of LYP, we show that the phosphatase activity is strongly inhibited by protein regions C-terminal to the catalytic domain. We mapped the minimal inhibitory region to the proximal portion of the interdomain. We show that the activity of LYP is inhibited by an intramolecular mechanism, whereby the proximal portion of the interdomain directly interacts with the catalytic domain and reduces its activity.
The lymphoid tyrosine phosphatase LYP,1 encoded by the PTPN22 gene, recently emerged as a key player and candidate drug target for human autoimmunity. LYP is an ∼105 kDa Class I protein tyrosine phosphatase (PTP) (1, 2) characterized by an ∼300-amino acid classical N-terminal PTP domain and an ∼200-amino acid C-terminal domain which includes four putative polyproline motifs [termed P1-P4 (Figure 1A)]. The catalytic domain and the C-terminal domain are separated by an ∼300-amino acid region called the “interdomain” (see Figure 1A) (3). In T cells, LYP and its mouse homologue called PEST-enriched phosphatase (PEP) are known to potently inhibit T cell receptor (TCR) signaling (4-8) through dephosphorylation of several substrates, including the Src family kinases Lck and Fyn, ZAP70, and TCRzeta (4, 9). A genetic variant of LYP, caused by a C1858T variation in PTPN22 and carrying Trp620 instead of Arg620 in the C-terminal domain, was first reported to increase the risk of type 1 diabetes (10-13) and rheumatoid arthritis (6, 12, 14, 15) and subsequently found to predispose humans to a whole range of autoimmune diseases, including systemic lupus erythematosus, Graves’ and Addison's disease, and others (reviewed in refs 16 and 17). LYP-W620 acts with a dominant mechanism and confers significant predisposition to autoimmunity even when present in a single copy. LYP-W620 was later shown to be a gain-of-function inhibitor of signaling and to have increased phosphatase activity (7, 18). The molecular mechanism of the gain-of-function phenotype of LYP-W620 is currently unknown. It has been proposed that small molecule inhibitors of LYP could reverse the effect of the pathogenic variation and have a therapeutic effect in the treatment of autoimmunity (19).
Figure 1.
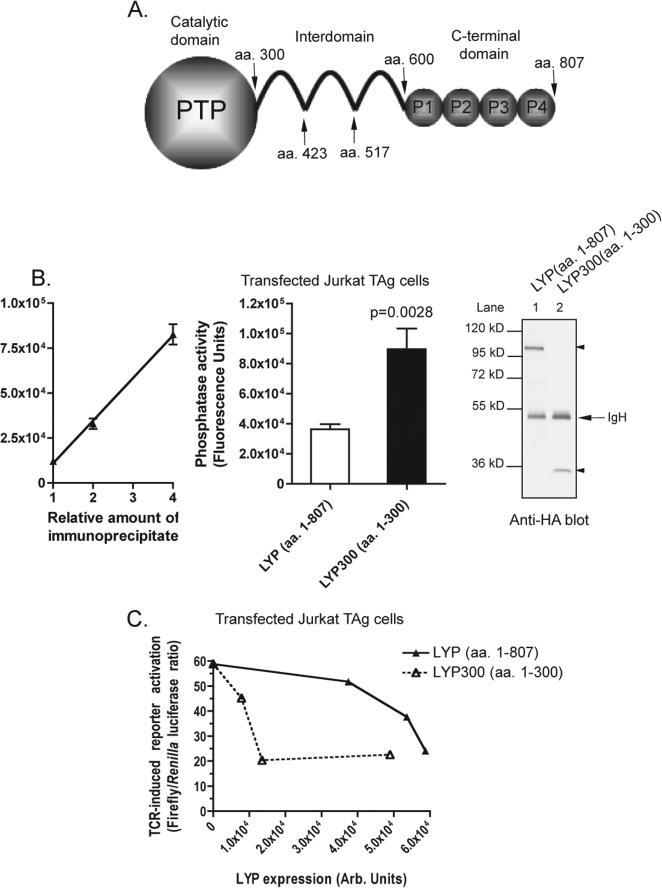
Truncation of LYP at amino acid 300 leads to a gain of function and increased phosphatase activity. (A) Schematic of the LYP structure showing the catalytic domain, the interdomain, and the C-terminal domain. Sites where truncations were performed for this study are denoted with arrows. (B) Immunoprecipitated LYP300 (amino acids 1−300) exhibits higher phosphatase activity than LYP. JTAg cells were transfected with HA-LYP or HA-LYP300. HA-LYP was immunoprecipitated from cell lysates, and its phosphatase activity was assessed under Vmax conditions using 0.25 mM DiFMUP as a substrate, as described in Materials and Methods. The left panel shows the linearity of the assay with respect to the amount of input immunoprecipitate in cells transfected with HA-LYP. The middle panel shows the average ± SD activity of immunoprecipitated HA-LYP (white column) or HA-LYP300 (black column) after reaction for 50 s, normalized for LYP expression as assessed by densitometry of anti-HA blots of fractions of the same immunoprecipitations. The right panel shows an anti-HA blot of an aliquot of immunoprecipitates from cells transfected with HA-LYP (lane 1) or HA-LYP300 (lane 2). IgH denotes heavy chains of immunoglobulins. The statistical significance of the difference between LYP and LYP300 was calculated with a Student's t test (p value shown above the LYP300 column). (C) Truncation of LYP at amino acid 300 leads to a gain of function. Activation of an NFAT/AP1 reporter in T cells transfected with HA-LYP or HA-LYP300. JTAg cells were cotransfected with a 3 × NFAT/AP1 luciferase reporter and LYP or LYP300. Cells were stimulated for 6 h with OKT3 and then lysed, and luciferase activity was measured on cell lysates. The average ± SD stimulation-induced increase in the ratio between firefly and Renilla luciferase activities of lysates of cells transfected with LYP (▲) or LYP300 (△), each measured in triplicate, was plotted vs the transfected protein expression in the same lysates as assessed by anti-HA WB.
Elucidating how LYP activity is regulated is important in understanding the mechanism of action of LYP-W620, and it might lead to innovative approaches to modulating the phosphatase activity for therapeutic purposes. Recently, the Zhang group reported that the activity of LYP is regulated by phosphorylation of serine 35 in the catalytic domain (19). However, little else is known about the regulation of LYP activity, and in particular, there is no information available about possible regulatory effects of the interdomain. Here we report that truncation of human LYP at amino acid 300 immediately C-terminal to the catalytic domain causes a dramatic increase in phosphatase activity. We found that the 20 amino acids immediately proximal to the catalytic domain are responsible for most of the effects of the truncation. We show that the activity of LYP is inhibited by an intramolecular mechanism, whereby the proximal portion of the interdomain directly interacts with the catalytic domain and reduces its activity.
MATERIALS AND METHODS
Plasmids, Antibodies, and Other Reagents
Full-length LYP and its truncation mutants were cloned in the BamHI site of plasmid pEF5 (20) which allows the expression of constructs under control of the EFα promoter and in fusion with an N-terminal HA tag. Site-directed mutagenesis and deletions were performed by PCR. All mutants were assessed by full-length sequencing. The monoclonal anti-HA antibody (clone 16B12) was purchased from Covance (Berkeley, CA). The pTyr peptide (ARLIEDNEpYTAREG) and the peptide of amino acids 301−320 (HSGTESQAKHCIPEKNHTLQ) were ordered from Celtek Peptides (Nashville, TN). The control peptide of amino acids 604−643 (biotin-ATAPRIDDEIPPPLPVRTPESFIVVEEAGEFSPNVPKSLS) was purchased from Antagene (Mountain View, CA). The purity of all peptides was >95%. All peptides were purchased as lyophilized powder, and stock solutions were prepared in phosphatase buffer.
Purification of Recombinant Proteins
For purification of recombinant proteins from insect cell lysates, full-length LYP and its mutants and/or truncations were cloned in the BamHI site of pFastBac-HTa (Invitrogen, Carlsbad, CA) in frame with a FLAG tag, and recombinant bacmids and baculoviruses were produced using the Bac-to-Bac method (Invitrogen). Virus titers and times of incubation were optimized to yield high levels of expression of full-length recombinant proteins in Sf9 cells. The proteins were purified from lysates of insect cells using single-step affinity chromatography on M2-FLAG beads (Sigma, St. Louis, MO) and eluting with a combination of FLAG peptide and high concentrations of DTT. The final buffer consisted of 50 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, and 1 mM DTT. The purity of recombinant proteins was more than 80% as assessed by silver stain of polyacrylamide gels (Figure 2B). The yield of the isolation was around 1 μg of full-length protein/150 mm plate of infected Sf9 cells. For purification of proteins from lysates of Escherichia coli, the constructs were cloned in the BamHI and NotI sites of the pET28a vector (EMD Biosciences, San Diego, CA). Six-His-tagged proteins were purified from lysates of transformed BL21(DE3) cells by two-step affinity chromatography on a Ni2+-NTA column, followed by cleavage of the six-His tag by thrombin and further passage of the eluate on a benzamidine column for removal of the protease. The proteins were dialyzed against 50 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, and 1 mM DTT. The purity of eluted proteins was consistently more than 90% as assessed by Coomassie blue staining of polyacrylamide gels (data not shown).
Figure 2.
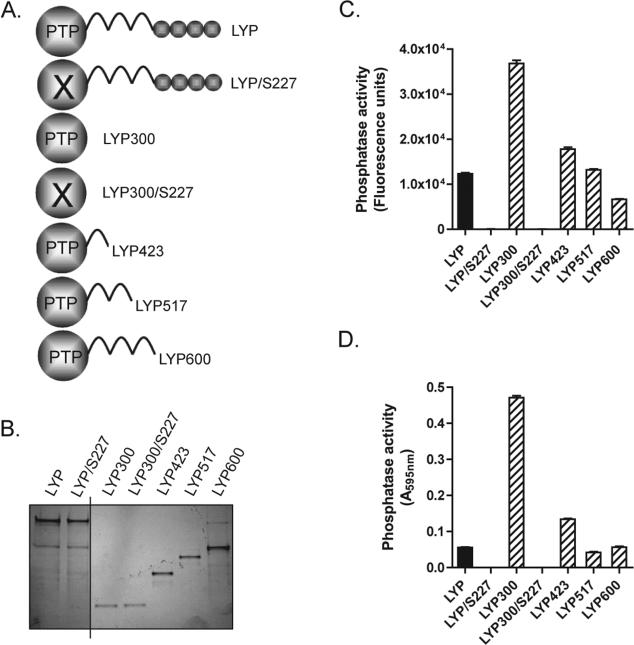
C-Terminal truncation of LYP within the interdomain shows that the N-terminal third of the interdomain inhibits the catalytic activity. Full-length recombinant LYP and its truncation mutants were isolated from lysates of baculovirus-infected insect cells as described in Materials and Methods. (A) Schematic of the C-terminal truncations of LYP used for the experiments shown in this figure. (B) Silver-stained gel showing 80 ng of LYP (lane 1), LYP/S227 (lane 2), LYP300 (amino acids 1−300, lane 3), LYP300/S227 (lane 4), LYP423 (amino acids 1−423, lane 5), LYP517 (amino acids 1−517, lane 6), and LYP600 (amino acids 1−600, lane 7). (C and D) Activity of LYP (black column), LYP600, LYP517, LYP423, and LYP300 on DiFMUP (C) or pTyr peptide (D). The histograms show that LYP/S227 and LYP300/S227 were completely inactive. The phosphatase assays were conducted under Vmax conditions. Panel C shows the average ± SD activity measured in triplicate, using 1 nM enzyme and 0.4 mM DiFMUP. Panel D shows the average ± SD activity measured in triplicate, using 1 nM enzyme and 0.16 mM peptide.
Cell Culture and Transfections
Jurkat cells expressing the SV-40 large T antigen (JTAg) (21) were kept at logarithmic growth in RPMI 1640 medium supplemented with 10% FCS, 1 mM sodium pyruvate, 10 mM HEPES (pH 7.3), 2.5 mg/mL d-glucose, 100 units/mL penicillin, and 100 μg/mL streptomycin. JTAg transfections were performed by electroporation at 240 V with a single 25 ms square pulse (7).
Immunoprecipitations/Pull-Downs and Western Blotting
For immunoprecipitations and pull-downs, cells were lysed in 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% NP-40 (TNE buffer), 1 mM PMSF, 10 μg/mL aprotinin/leupeptin, and 10 μg/mL soybean trypsin inhibitor. Lysates were precleared by centrifugation at 16000 rcf for 30 min. For immunoprecipitations, anti-HA antibody was added to the precleared lysates followed by PG-Sepharose beads. For pull-downs, recombinant proteins were added to the precleared lysates, followed by Ni2+NTA-Sepharose to precipitate the protein complex. Beads were washed four or five times in lysis buffer and resuspended in SDS sample buffer.
Luciferase Assays
Luciferase assays were performed as described in ref 7. The NFAT/AP-1 reporter plasmid (kindly provided by G. Crabtree, Stanford University, Stanford, CA) carries three tandem copies of the distal NFAT/AP-1 composite element of the human IL2 promoter (21) which are cloned upstream of the minimal IL2 promoter (from nucleotide −89 to +51) and the luciferase reporter gene. The NFAT/AP-1 reporter plasmid and the control pGL3 plasmid, driving constitutive expression of Renilla luciferase, were cotransfected in JTAg cells with variable amounts of the appropriate HA-tagged constructs. Cells were subjected to TCR stimulation with 1.5 μg/mL OKT3 (22) for 6 h, and then luciferase assays were performed in triplicate on cell lysates using the dual luciferase kit from Promega (Madison, WI) according to the manufacturer's instructions, and reading luminescence on a plate reader. The difference in the ratio between firefly and Renilla luciferase activity in stimulated versus unstimulated cells (TCR-induced increase in the level of activation of reporter) was then plotted against the level of expression of constructs assessed by densitometric scanning of anti-HA blots of total lysates.
Phosphatase Assays
Phosphatase assays were conducted using 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) or a 14-amino acid phosphotyrosine (pTyr) peptide (ARLIEDNEpYTAREG) based on the Lck Y394 autophosphorylation site as a substrate (23). The buffer consisted of 50−100 mM Bis-Tris (pH 6.0) and 1−5 mM DTT buffer (phosphatase buffer). When DiFMUP was used as a substrate, phosphatase activity was detected by continuously monitoring the increase in fluorescence (λex = 360 nm, and λem = 460 nm). When the pTyr peptide was used as a substrate, reactions were stopped by the addition of BIOMOL GREEN REAGENT (BIOMOL International, Plymouth Meeting, PA), and the absorbance of the solution was measured at 595 nm. The activity measured in triplicate was corrected for the nonspecific signal of identical reactions performed also in triplicate without addition of enzyme. For detection of phosphatase activity of LYP immunoprecipitated from transfected cells using DiFMUP as a substrate, the immunoprecipitates were washed three times in Bis-Tris (pH 6.0) and then resuspended in phosphatase buffer. The activity corrected for the background fluorescence of the substrate alone was then normalized for LYP expression as assessed by anti-HA blot of fractions of immunoprecipitates taken before resuspension in the final phosphatase buffer. This assay showed a good range where the signal varied linearly with the amount of immunoprecipitate added to the reaction (Figure 1B). Kinetic parameters were calculated by plotting enzyme activity versus substrate concentration and fitting the points to the Michaelis–Menten equation. For all phosphatase assays, the time of reaction and the amount of enzyme were optimized to keep the activity in initial rate.
Gel Filtration Chromatography
Gel filtration chromatography was performed using the AKTA FPLC system (GE Healthcare). Standards and experimental samples were separated using Superdex 75 10/300 GL (GE Healthcare) gel filtration columns. Standards [conalbumin (75 kDa), carbonic anhydrase (29 kDa), and ovalbumin (43 kDa)] were run through the column to determine the elution volume of each. The samples were centrifuged at 35000g for 10 min using an ultracentrifuge followed by fractionation at a flow rate of 0.6 mL/min. A sample volume of 100 μL corresponding to 100 μg was loaded for each run. The elution volume of samples was compared to the standards. Both standards and samples were fractionated using TNE or phosphatase buffer.
Graphs and Statistics
Proteins were aligned using ClustalW (24, 25). Graphs, curve fittings, and kinetic parameter calculations were performed using Graphpad Prism (Graphpad, San Diego, CA). Statistics were calculated also using the same software or using Excel (Microsoft, Redmond, WA). All SDs of differences and ratios were calculated according to the error propagation rules described by Taylor (26).
RESULTS
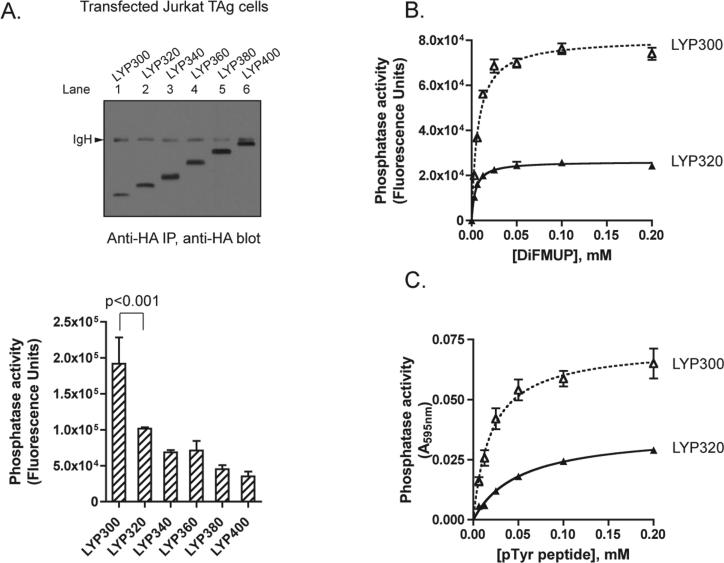
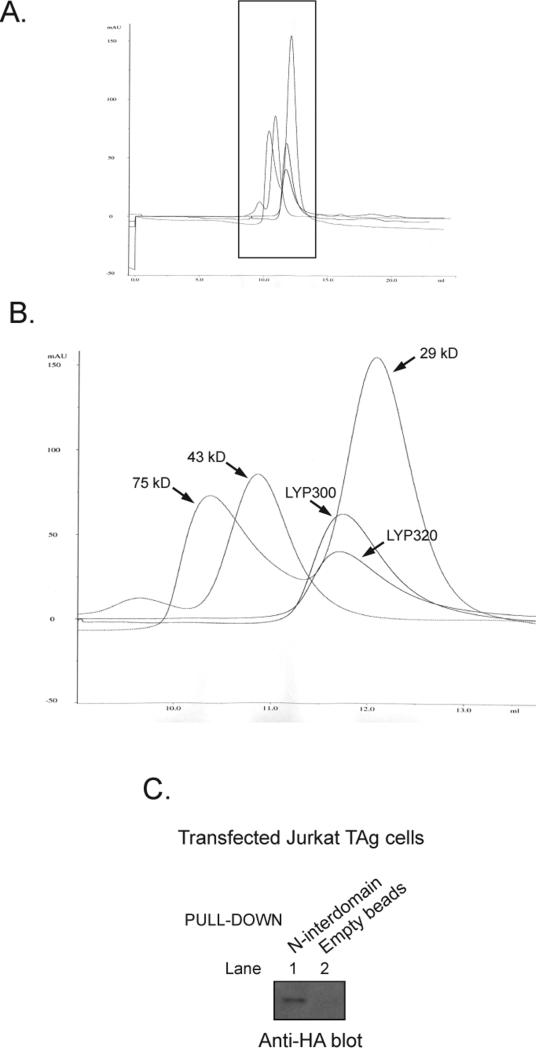
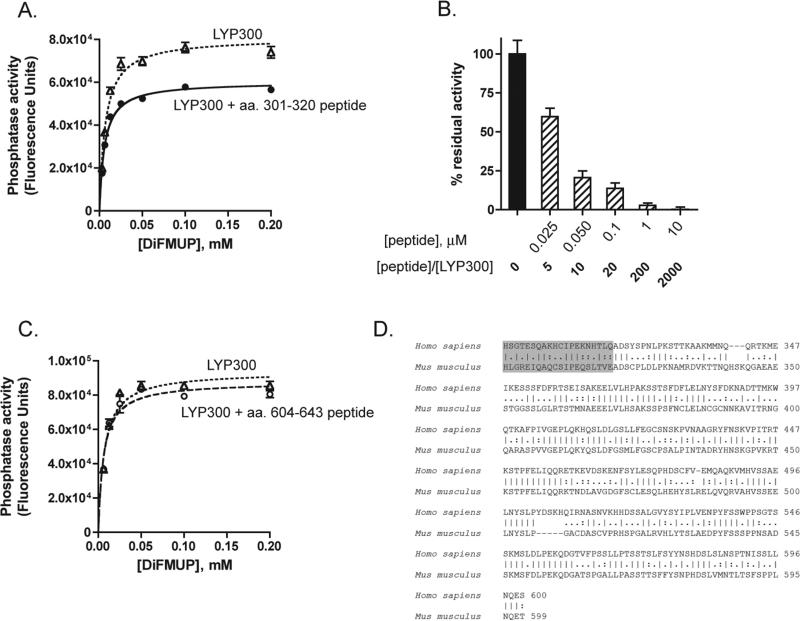
Gjorloff-Wingren et al. reported that the catalytic activity of immunoprecipitated full-length PEP is much lower than the activity of the catalytic domain alone (amino acids 1−290) and suggested that the activity of PEP might be regulated by an intramolecular mechanism (5). Since the degree of conservation between human LYP and mouse PEP is relatively low outside the catalytic domain (the identity between amino acids 1−300 of LYP and amino acids 1−300 of PEP is 87.4%, while the identity between amino acids 301−807 of LYP and amino acids 301−802 of PEP is 60.7%), we first assessed whether truncation of LYP immediately C-terminal to the catalytic domain (LYP300, amino acids 1−300) leads to a similar gain of function in the human phosphatase. Figure 1B shows that immunoprecipitated LYP300 has approximately double the activity of immunoprecipitated LYP in a Vmax phosphatase assay. Next we assessed whether truncation at amino acid 300 also leads to a gain of function in TCR signaling modulation. Figure 1C shows that LYP300 was a several-fold more potent inhibitor of TCR signaling in an NFAT-AP1 reporter assay, suggesting that the inhibition of the catalytic domain of LYP by the C-terminal portion is physiologically relevant in reducing the activity of the phosphatase. The increased gain of function observed in the signaling assay compared to the effect on the phosphatase activity is possibly due to amplification of signaling downstream of the phosphatase or to the amplifying effect of TCR-induced post-translational modifications of the phosphatase. Next, we purified full-length recombinant LYP, LYP300, and a series of LYP truncation mutants as recombinant proteins from lysates of insect cells. An analysis of the interdomain with DISOPRED (Bioinformatics Group, University College London, London, U.K.) did not reveal any predictable secondary structure. However, to minimize the chance of disrupting secondary structure elements, truncation sites were placed in regions which the software predicted to be less likely to be ordered. The mutants included truncations immediately C-terminal to the interdomain at amino acid 600 (LYP600), at two-thirds of the interdomain at amino acid 517 (LYP517), and at one-third of the interdomain at amino acid 423 (LYP423) (Figures 1A and 2A). When the activity of these proteins was compared in Vmax phosphatase assays on two different substrates, LYP300 consistently exhibited greatly increased phosphatase activity, while LYP423 was the only other one to exhibit slightly increased activity (Figure 2C,D). Importantly, in these assays, the inactive mutants of LYP and LYP300 (LYP/S227 and LYP300/S227) did not exhibit any activity (Figure 2C,D). These data suggest that the N-terminal one-third of the interdomain between amino acids 300 and 423 (here called the N-interdomain) is responsible for the inhibition of the activity of LYP300. To assess whether we could further map the region of the protein responsible for the inhibition of the activity, we generated smaller truncation mutants of LYP within the N-interdomain and measured their activity in Vmax assays. Figure 3A shows that truncation of the 20 amino acids immediately C-terminal to LYP300 had the strongest effect on the activity of the phosphatase, suggesting that the region between amino acids 301 and 320 is responsible for most of the inhibitory effect of the N-interdomain on LYP300. In line with this hypothesis, when LYP300 and LYP320 were purified from bacterial lysates and their activity was compared, LYP320 was consistently less active than LYP300 on both DiFMUP and the pTyr peptide. The kcat was 9.4 ± 0.2 (standard error) s−1 on DiFMUP and 11.0 ± 0.4 (standard error) s−1 on pTyr peptide for LYP300 versus 2.8 ± 0.0 (standard error) s−1 on DiFMUP and 5.5 ± 0.2 (standard error) s−1 on pTyr peptide for LYP320, while the KM was 6.1 ± 0.5 (standard error) μM on DiFMUP and 19.9 ± 2.4 (standard error) μM on pTyr peptide for LYP300 versus 3.9 ± 0.3 (standard error) μM on DiFMUP and 51.1 ± 4.4 (standard error) μM on pTyr peptide for LYP320 (Figure 3B,C). Next we performed gel filtration chromatography of recombinant LYP300 and LYP320 in TNE and phosphatase buffers and found that both proteins elute as monomers with an elution volume of 12.0 mL, in a molecular mass range between the 43 kDa (elution volume of 11.0 mL) and 29 kDa (elution volume of 12.2 mL) standards (Figure 4A,B and data not shown). The chromatogram excluded the fact that the difference in activity between LYP300 and LYP320 is due to dimerization or multi-merization of LYP320. Overall, the data suggest that the proximal portion of the interdomain inhibits the activity of LYP through an intramolecular mechanism. Figure 4C shows that the recombinant N-interdomain could pull down LYP300 from lysates of JTAg cells, which suggests that the intramolecular inhibition of the phosphatase activity depends on a physical interaction between the N-interdomain and LYP300. We then assessed whether a peptide encompassing the minimal inhibitory region between amino acids 301 and 320 was able to inhibit the phosphatase activity when incubated with LYP300 in trans. As a control, we also included a peptide of amino acids 604−643, which spans the P1 and P2 motifs of LYP. Figure 5A shows that incubation of LYP300 with the peptide of amino acids 301−320 led to substantial inhibition of the phosphatase activity. The kcat was 9.0 ± 0.2 (standard error) s−1 in the absence of peptide versus 5.2 ± 0.1 (standard error) s−1 in the presence of the peptide of amino acids 301−320 at 25 nM, while the KM was 5.2 ± 0.4 (standard error) μM in the absence of peptide versus 4.1 ± 0.2 (standard error) μM in the presence of the peptide of amino acids 301−320. Figure 5B shows that inhibition of the activity of LYP300 by the peptide of 25 nM 301−320 was dose-dependent. Figure 5C shows that incubation of LYP300 with a control peptide of 25 nM 604−643 did not lead to any significant inhibition of the phosphatase activity. The kcat was 8.8 ± 0.2 (standard error) s−1 in the absence of peptide versus 8.2 ± 0.3 (standard error) s−1 in the presence of the peptide of amino acids 604−643 at 50 nM, while the KM was 7.0 ± 0.9 (standard error) μM in the absence of peptide versus 5.9 ± 0.9 (standard error) μM in the presence of the peptide of amino acids 604−643.
Figure 3.
Cis inhibition of LYP300 phosphatase activity by the segment of amino acids 301−320. (A) Mapping of the minimal inhibitory interdomain segment. JTAg cells were transfected with HA-LYP300 (amino acids 1−300), HA-LYP320 (amino acids 1−320), HA-LYP340 (amino acids 1−340), HA-LYP360 (amino acids 1−360), HA-LYP380 (amino acids 1−380), or HA-LYP400 (amino acids 1−400). HA-tagged proteins were immunoprecipitated from cell lysates, and their phosphatase activity was assessed under Vmax conditions using DiFMUP as a substrate, as in Figure 1B. The top panel shows the anti-HA WB of an aliquot of immunoprecipitates from cells transfected with HA-LYP300 (lane 1), HA-LYP320 (lane 2), HA-LYP340 (lane 3), HA-LYP360 (lane 4), HA-LYP380 (lane 5), or HA-LYP400 (lane 6). IgH denotes heavy chains of immunoglobulins. The bottom panel shows the average ± SD activity of immunoprecipitated HA-tagged proteins, normalized for LYP expression as assessed by densitometry of anti-HA blots of fractions of the same immunoprecipitations. Data were analyzed using ANOVA, followed by post-test analysis of the difference between each truncation and the immediately preceding one in the series by the Bonferroni test. Only significant results are shown. (B and C) LYP300 (amino acids 1−300) and LYP320 (amino acids 1−320) were isolated from lysates of E. coli as described in Materials and Methods. (B) Activity of 5 nM LYP300 ( ) and LYP320 (
) and LYP320 ( ) on DiFMUP. Points are the average ± SD activity of LYP300 and LYP320 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation. (C) Activity of 1 nM LYP300 (
) on DiFMUP. Points are the average ± SD activity of LYP300 and LYP320 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation. (C) Activity of 1 nM LYP300 ( ) and LYP320 (
) and LYP320 ( ) on the pTyr peptide. Points are the average ± SD activity of LYP300 and LYP320 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation.
) on the pTyr peptide. Points are the average ± SD activity of LYP300 and LYP320 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation.
Figure 4.
Inhibition of LYP300 by the interdomain is due to an intramolecular mechanism. (A and B) Gel filtration chromatograms of protein standards, LYP300 (amino acids 1−300), and LYP320 (amino acids 1−320). Panel A shows the full chromatogram. Panel B shows a magnification of the protein peaks. The proteins were eluted in phosphatase buffer [100 mM Bis-Tris (pH 6.0) and 1 mM DTT]. Similar results were obtained when the same proteins were eluted in TNE buffer (data not shown). The standards shown are conalbumin (75 kDa), ovalbumin (43 kDa), and carbonic anhydrase (29 kDa). (C) Interaction between the N-terminal one-third of the interdomain (amino acids 301−423) and LYP300. JTAg cells were transfected with HA-tagged LYP300, and total lysates were subjected to pull-down using Ni2+-NTA bead-bound six-His-bound N-interdomain (lane 1) or beads alone (lane 2). Panel C shows the anti-HA WB of pull-downs.
Figure 5.
Dose-dependent trans inhibition of LYP300 phosphatase activity by a peptide of amino acids 301−320. (A) Activity of 5 nM LYP300 (amino acids 1−300) on DiFMUP in the absence ( ) or presence of a peptide of amino acids 301−320 at 25 nM (
) or presence of a peptide of amino acids 301−320 at 25 nM ( ). Points are the average ± SD activity of LYP300 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation. (B) Dose–response inhibition of LYP300 activity in the presence of increasing amounts of the peptide of amino acids 301−320. The peptide was added to the buffer at the concentrations and peptide:enzyme molar ratios indicated on the X-axis. The phosphatase assays were conducted under Vmax conditions as described in Materials and Methods using 5 nM enzyme and 0.4 mM DiFMUP and following the activity continuously. Graphs show the average ± SD percent residual activity at 2 min, measured in triplicate. (C) Activity of 5 nM LYP300 (amino acids 1−300) on DiFMUP in the absence (
). Points are the average ± SD activity of LYP300 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation. (B) Dose–response inhibition of LYP300 activity in the presence of increasing amounts of the peptide of amino acids 301−320. The peptide was added to the buffer at the concentrations and peptide:enzyme molar ratios indicated on the X-axis. The phosphatase assays were conducted under Vmax conditions as described in Materials and Methods using 5 nM enzyme and 0.4 mM DiFMUP and following the activity continuously. Graphs show the average ± SD percent residual activity at 2 min, measured in triplicate. (C) Activity of 5 nM LYP300 (amino acids 1−300) on DiFMUP in the absence ( ) or presence of the control peptide of amino acids 604−643 at 50 nM (
) or presence of the control peptide of amino acids 604−643 at 50 nM ( ). Points are the average ± SD activity of LYP300 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation. (D) Alignment of the interdomains of LYP from Homo sapiens (gi:48928054) and PEP from Mus musculus (gi:6679555). The fragment of amino acids 301−320 is highlighted in gray. Lines indicate identities; colons indicate conservative substitutions, and periods indicate semiconservative substitutions.
). Points are the average ± SD activity of LYP300 measured in triplicate plotted vs substrate concentration. Lines are fits of data to the Michaelis–Menten equation. (D) Alignment of the interdomains of LYP from Homo sapiens (gi:48928054) and PEP from Mus musculus (gi:6679555). The fragment of amino acids 301−320 is highlighted in gray. Lines indicate identities; colons indicate conservative substitutions, and periods indicate semiconservative substitutions.
DISCUSSION
The interdomain of LYP is known to harbor putative phosphorylation and protein–protein interaction motifs, but little is known about its function. Our data suggest that the interdomain plays an important role in regulating the catalytic activity through a direct intramolecular interaction and inhibition of the catalytic domain. Several cytosolic PTPs reportedly are physiologically regulated by intramolecular inhibition (27, 28). For example, SHP-1 is inhibited by a constitutive interaction between the catalytic and N-terminal SH2 domain, which is released following recruitment of the domain to phosphorylated targets (28).
The inhibition of the catalytic domain by the interdomain might play an important physiological role. For example, alterations of the interaction between these two domains could indirectly mediate the functional effects of post-translational modifications and/or protein interactions located in other portions of the protein.
We showed that the interaction between the proximal 20 amino acids of the interdomain and LYP300 is responsible for a substantial portion of the inhibitory effect of the interdomain on the activity of the phosphatase. As shown in Figures 2C,D and 3A, segments of the interdomain distal to amino acid 320 also contribute to the inhibition of LYP activity. Despite their relatively weak effect in our assays, distal portions of the interdomain could still play a critical physiological role in the cellular context. Also, all truncations distal to amino acid 320 were either purified or precipitated from eukaryotic cells, where post-translational modifications might be further complicating the picture. Further analysis is needed to refine our knowledge of the role of distal segments of the interdomain in the activity of the catalytic domain.
Our data suggest that the proximal 20 amino acids of the interdomain are able to inhibit the phosphatase activity in cis and in trans phosphatase assays by a direct interaction with LYP300. Some speculation about the structural basis of this phenomenon can be undertaken. The crystal structure of the catalytic domain of LYP has been determined by the Zhang (19) and Knapp (29) groups (Protein Data Bank entries 2P6X and 2QCT, respectively) and recently by us (manuscript submitted for publication). Each of these structures shows that the last helix of the catalytic domain (α6 helix, amino acids 275−300) lies in the proximity of the D-loop and shows extensive interactions with the α3 helix (which directly follows the D-loop on the primary structure). Thus, it is possible that the interaction between the segment of amino acids 301−320 of the interdomain and the catalytic domain somehow affects the position and range of movement of the D-loop. Structural analysis of larger truncations of LYP would be extremely valuable in understanding the mechanism of intramolecular inhibition.
The results of our study are in line with the one reported for mouse PEP showing that truncation at amino acid 290 leads to increased phosphatase activity (5). Conservation throughout evolution suggests that inhibition of the catalytic domain by the proximal interdomain is physiologically relevant. However, little can be concluded at the moment about the mechanism of inhibition by looking at conservation of primary structure. The critical segment of amino acids 301−320 does not show a higher level of conservation between human LYP and mouse PEP than the rest of the interdomain: in the segment of amino acids 301−320, nine amino acids are identical (45% identity) and 14 are similar (70% similarity) (Figure 5D), while the whole interdomain (amino acids 301−600 of LYP and amino acids 301−599 of PEP) showed 52% identity and 65.1% similarity between human LYP and mouse PEP.
In conclusion, we showed that there is an intramolecular regulation mechanism of LYP phosphatase activity, which is mediated by an interaction between the interdomain of the molecule and the catalytic domain. Further dissection of the interaction between the interdomain and LYP300 is desirable to identify the structural basis of the activity inhibition.
ACKNOWLEDGMENT
We thank Sophia Tsai for help with the structure prediction software and Tobias Ulmer for useful discussion. We declare no conflict of interest in relation to the data reported in this paper.
Footnotes
This work was supported by a grant from the National Institutes of Health (AI070544), a grant from the Juvenile Diabetes Research Foundation (1-2005-342), and a grant from the Arthritis National Research Foundation (ANRF) to N.B. S.M.S. was supported by National Institutes of Health Predoctoral Training Grant GM067587 in Cellular, Biochemical and Molecular Biology at the University of Southern California. V.O. and E.F. were supported by Master&Back fellowships from the Sardinian government.
Abbreviations: AP1, activator protein 1; DiFMUP, 6,8-difluoro-4-methylumbelliferyl phosphate; DTT, dithiothreitol; EDTA, ethylene-diaminetetraacetic acid; EFα, elongation factor α; Fyn, oncogene related to SRC, FGR, and YES; FPLC, fast performance liquid chromatography; HA, hemagglutinin; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; JTAg; Jurkat large T antigen; Lck, leukocyte-specific protein tyrosine kinase; LYP, lymphoid phosphatase; NFAT, nuclear factor of activated T cells; NTA, nitrilotriacetic acid; PCR, polymerase chain reaction; PEP, PEST-enriched phosphatase; PMSF, phenylmethanesulfonyl fluoride; PTP, protein tyrosine phosphatase; pTyr, phosphotyrosine; RPMI, Roswell Park Memorial Institute; SD, standard deviation; SDS, sodium dodecyl sulfate; TCR, T cell receptor; WB, Western blotting; ZAP70, ζ-chain-associated protein kinase 70.
REFERENCES
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Mustelin T, Alonso A, Bottini N, Huynh H, Rahmouni S, Nika K, Louis-dit-Sully C, Tautz L, Togo SH, Bruckner S, Mena-Duran AV, al-Khouri AM. Protein tyrosine phosphatases in T cell physiology. Mol. Immunol. 2004;41:687–700. doi: 10.1016/j.molimm.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93:2013–2024. [PubMed] [Google Scholar]
- 4.Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J. Exp. Med. 1999;189:111–121. doi: 10.1084/jem.189.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur. J. Immunol. 1999;29:3845–3854. doi: 10.1002/(SICI)1521-4141(199912)29:12<3845::AID-IMMU3845>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 6.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SY, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, Mustelin T, Bottini N. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 8.Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, Jallal B. The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp. Hematol. 2002;30:237–244. doi: 10.1016/s0301-472x(01)00794-9. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Katrekar A, Honigberg LA, Smith AM, Conn MT, Tang J, Jeffery D, Mortara K, Sampang J, Williams SR, Buggy J, Clark JM. Identification of substrates of human protein-tyrosine phosphatase PTPN22. J. Biol. Chem. 2006;281:11002–11010. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 10.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 11.Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, Barratt BJ, Guja C, Ionescu-Tirgoviste C, Savage DA, Dunger DB, Widmer B, Strachan DP, Ring SM, Walker N, Clayton DG, Twells RC, Gough SC, Todd JA. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53:3020–3023. doi: 10.2337/diabetes.53.11.3020. [DOI] [PubMed] [Google Scholar]
- 12.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoledziewska M, Perra C, Orru V, Moi L, Frongia P, Congia M, Bottini N, Cucca F. Further evidence of a primary, causal association of the PTPN22 620W variant with type 1 diabetes. Diabetes. 2008;57:229–234. doi: 10.2337/db07-0289. [DOI] [PubMed] [Google Scholar]
- 14.Gregersen PK. Pathways to gene identification in rheumatoid arthritis: PTPN22 and beyond. Immunol. Rev. 2005;204:74–86. doi: 10.1111/j.0105-2896.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- 15.Orozco G, Sanchez E, Gonzalez-Gay MA, Lopez-Nevot MA, Torres B, Caliz R, Ortego-Centeno N, Jimenez-Alonso J, Pascual-Salcedo D, Balsa A, de Pablo R, Nunez-Roldan A, Gonzalez-Escribano MF, Martin J. Arthritis Rheum. 2005;52:219–224. doi: 10.1002/art.20771. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. [DOI] [PubMed] [Google Scholar]
- 16.Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin. Immunol. 2006;18:207–213. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: Ssetting thresholds for autoimmunity. Semin. Immunol. 2006;18:214–223. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J. Immunol. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Sun JP, He Y, Guo X, Liu S, Zhou B, Hudmon A, Zhang ZY. Structure, inhibitor, and regulatory mechanism of Lyp, a lymphoid-specific tyrosine phosphatase implicated in autoimmune diseases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19767–19772. doi: 10.1073/pnas.0706233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito K, Williams S, Bulankina A, Honing S, Mustelin T. Association of protein-tyrosine phosphatase MEG2 via its Sec14p homology domain with vesicle-trafficking proteins. J. Biol. Chem. 2007;282:15170–15178. doi: 10.1074/jbc.M608682200. [DOI] [PubMed] [Google Scholar]
- 21.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 22.Kung P, Goldstein G, Reinherz EL, Schlossman SF. Monoclonal antibodies defining distinctive human T cell surface antigens. Science. 1979;206:347–349. doi: 10.1126/science.314668. [DOI] [PubMed] [Google Scholar]
- 23.Montalibet J, Skorey KI, Kennedy BP. Protein tyrosine phosphatase: Enzymatic assays. Methods. 2005;35:2–8. doi: 10.1016/j.ymeth.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor JR. An introduction to error analysis. 2nd ed. University Science Books; Sausalito, CA: 1997. [Google Scholar]
- 27.Hao L, Tiganis T, Tonks NK, Charbonneau H. The noncatalytic C-terminal segment of the T cell protein tyrosine phosphatase regulates activity via an intramolecular mechanism. J. Biol. Chem. 1997;272:29322–29329. doi: 10.1074/jbc.272.46.29322. [DOI] [PubMed] [Google Scholar]
- 28.Pei D, Lorenz U, Klingmuller U, Neel BG, Walsh CT. Intramolecular regulation of protein tyrosine phosphatase SH-PTP1: A new function for Src homology 2 domains. Biochemistry. 1994;33:15483–15493. doi: 10.1021/bi00255a030. [DOI] [PubMed] [Google Scholar]
- 29.Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]