Abstract
Recent evidence suggests that the Myc and Mad1 proteins are implicated in the regulation of the gene encoding the human telomerase reverse transcriptase (hTERT), the catalytic subunit of telomerase. We have analyzed the in vivo interaction between endogenous c-Myc and Mad1 proteins and the hTERT promoter in HL60 cells with the use of the chromatin immunoprecipitation assay. The E-boxes at the hTERT proximal promoter were occupied in vivo by c-Myc in exponentially proliferating HL60 cells but not in cells induced to differentiate by DMSO. In contrast, Mad1 protein was induced and bound to the hTERT promoter in differentiated HL60 cells. Concomitantly, the acetylation of the histones at the promoter was significantly reduced. These data suggest that the reciprocal E-box occupancy by c-Myc and Mad1 is responsible for activation and repression of the hTERT gene in proliferating and differentiated HL60 cells, respectively. Furthermore, the histone deacetylase inhibitor trichostatin A inhibited deacetylation of histones at the hTERT promoter and attenuated the repression of hTERT transcription during HL60 cell differentiation. In addition, trichostatin A treatment activated hTERT transcription in resting human lymphocytes and fibroblasts. Taken together, these results indicate that acetylation/deacetylation of histones is operative in the regulation of hTERT expression.
Keywords: c-Myc
The Myc network proteins are transcription factors of the basic/helix-loop-helix/leucine zipper family that are involved in the control of proliferation, cell differentiation, and apoptosis (see ref. 1 for a review). Myc genes are deregulated in a large number of human tumors and affect both the development and progression of hyperproliferations. Myc stimulates the G1–S phase transition of the cell cycle, and, conversely, cell cycle exit and cellular differentiation are usually associated with down-regulation of Myc. Myc forms dimers with Max that bind to the 5′-CACGTG-3′ sequence (E-box) in promoters and transactivate target genes. The same E-box sequence can also be bound by a number of other transcription factors, including upstream stimulating factor (USF) (2). Transactivation by Myc may involve recruitment of histone acetyltransferases (3), which consequently would result in increased acetylation of the histones at the promoters. E-box sequences are found in the regulatory regions of a variety of genes, including ornithine decarboxylase, α-prothymosine, cdc25A, eIF4, carbamoyl-phosphatase/aspartate transferase/dihydroorotase (CAD), MrDb, cyclin D2, cdk4, and telomerase (4–7).
Max also forms dimers with the Mad family proteins, Mnt, and with itself, and all complexes bind to the same DNA sequence, 5′-CACGTG-3′ (8–10). In contrast to myc, the mad genes are expressed primarily in differentiated, nonproliferative tissues, and Mad proteins are negative regulators of cell growth. Mad1 inhibits cell cycle progression, represses transcription from E-box-containing reporter genes, inhibits apoptosis, represses c-Myc/Ha-Ras-mediated transformation, and inhibits transformation of certain tumor cells (see ref. 8 and references therein; refs. 11 and 12). Recruitment of mSin3 by Mad proteins is essential for their effects on cellular growth (13, 14). The mSin3 proteins serve as adapters for a number of proteins, including histone deacetylases (HDACs) and nuclear receptor corepressor, which in turn are thought to mediate Mad function (15–19). These findings have led to the suggestion that the repressive activity of Mad is mediated through remodeling of chromatin and modulation of gene expression (8, 20). The Myc/Max/Mad network may constitute a molecular switch where the abundance of Myc- versus Mad-containing heterodimers determines whether cells enter a differentiation pathway or remain in a proliferative, undifferentiated state.
Telomerase, a RNA-dependent DNA polymerase, extends chromosome ends with telomeric TTAGGG repeat sequences essential for the stability and integrity of linear chromosomes (21). Telomerase activity, although absent in most normal human somatic cells, is detected in a wide range of human tumors (21, 22). Compelling evidence suggests that the maintenance of telomere length by telomerase is required for sustained cell proliferation, and activation of telomerase thus is a critical step during cellular immortalization and malignant transformation. The catalytic subunit of the enzyme, the human telomerase reverse transcriptase (hTERT), has been shown to be the determinant for telomerase activity control (23–26). Recent studies indicate that Myc and Mad1 proteins are implicated in the regulation of hTERT/telomerase expression (7, 27–31). The hTERT proximal promoter harbors two E-boxes, which can be bound by Myc/Max or Mad1/Max complexes, as shown by in vitro experiments. Indeed, c-Myc induces whereas Mad1 represses the hTERT promoter activity and gene expression. However, almost all studies so far have been performed with the use of transfections in which c-Myc or Mad1 was expressed at levels significantly higher than physiological. With the chromatin immunoprecipitation (ChIP) assay we have analyzed the in vivo occupancy by endogenous c-Myc and Mad1 proteins, the acetylation status of the histones, and the effect of changes in histone acetylation at the hTERT promoter in proliferating and differentiated HL60 cells.
Materials and Methods
Cell Culture, Induction of Differentiation, and Trichostatin A (TSA) Treatment.
The human promyelocytic leukemic cell line HL60 was grown in RPMI 1640 medium (Life Technologies, Paisley, Scotland) containing 10% FCS, 100 units/ml penicillin, and 2 mM l-glutamine. To induce cell differentiation, cells at a density of 0.4 × 106 cells per ml were treated with 1.25% vol/vol DMSO (Merck) for 48 h. The differentiated cells exceeded 90%, as demonstrated by acquisition of the CD11b differentiation marker and morphological examination (data not shown). The specific HDAC inhibitor TSA (Sigma) was dissolved in 100% ethanol and added to HL60 cells in the presence or absence of DMSO overnight. Normal human T lymphocytes and fibroblasts, isolated from buffy coats of healthy individuals and derived from adult skin, respectively, were maintained in the same culture medium as above. The cells were incubated with TSA at different concentrations overnight. In parallel, part of the T lymphocytes was treated with 1 μg/ml of anti-human CD3 and 0.5 μg/ml of anti-CD28 antibodies (R & D Systems) overnight to activate the cells.
RNA Extraction, Competitive Reverse Transcription–PCR (RT-PCR), and Northern Blot.
Total cellular RNA was isolated with an ULTRASPEC-II RNA kit (Biotecx Laboratories, Houston). cDNA synthesis, RT, and competitive PCR were performed as described (32, 33). PCR for Mnt mRNA was carried out for 28 cycles (94°C for 30 s, 60°C for 45 s, and 72°C for 60 s) with the following specific primers: forward: 5′-CCA GTG AAC AGA AGA AGA-3′; reverse: 5′-AAT CTC CCA GTA CGT CCA T-3′. For Northern blot, 20 μg of total RNA derived from undifferentiated and differentiated HL60 cells was separated in 1% formaldehyde-agarose gels, transferred to a Hybond nylon membrane (Amersham Pharmacia), and hybridized with the Myc, Max, and Mad1 probes.
Immunofluorescence Staining and Western Blot for hTERT Protein.
Slides prepared by cytospin were fixed with paraformaldehyde, rehydrated, immersed in 10 mM citrate buffer (pH 6.0), and boiled in a microwave oven for 5 min two times, followed by incubation with a 1:300 dilution of a polyclonal goat antibody against hTERT protein (sc-7214; Santa Cruz Biotechnology) at 4°C overnight. Donkey anti-goat antibody conjugated with FITC was applied to identify antibody binding. The cells were evaluated under a Zeiss epifluorescence microscope. For Western blot, equal amounts of cellular proteins (100 μg) derived from control and DMSO-treated HL60 cells were separated by 10% SDS/PAGE and blotted to nitrocellulose membranes. The blots were first incubated with the anti-hTERT antibody described above (1:2,000) and then with horseradish peroxidase-conjugated anti-goat IgG (1:8,000) and visualized with the use of the Amersham Pharmacia ECL system.
Telomerase Activity Assay.
The telomeric repeat amplification protocol assay (22, 34) was used to determine telomerase activity. PCR was performed for 23 and 28 cycles for HL60 and T lymphocytes, respectively. Products were resolved on 12% polyacrylamide gels, stained with SYBR green I (Roche Diagnostics), and visualized under UV light.
Electrophoretic Mobility-Shift Assay (EMSA).
Whole-cell extracts and EMSA were carried out as described (11). For antibody supershifts, 0.05 μg of anti-Max (sc-197), 0.2 μg of anti-Mnt (sc-769), 0.2 μg of anti-c-Myc antibody (sc-764), or 0.2 μg of anti-Mad1 (sc-222) (all from Santa Cruz Biotechnology) were added to the binding reactions. Electrophoresis gels were dried and developed with the use of PhosphorImager technology (Molecular Dynamics). The following oligonucleotides were used as probes: hTERTGS1, 5′-GCG CTC CCC ACG TGG CGG AGG; and hTERTGS2, 5′-CCT CCG CCA CGT GGG GAG CGC.
ChIP Assay.
The ChIP assay was performed as described by Orlando et al. (35). Briefly, logarithmically growing and differentiating HL60 cells (≈1 × 108 cells) were fixed with formaldehyde (final concentration 1% vol/vol) in serum-free RPMI 1640 medium at 4°C for 1 h. Glycine was added to a final concentration of 0.125 M to stop cross-linking. Fixed cells were pelleted by centrifugation and sequentially washed during rotation at room temperature for 15 min each in buffer A (10 mM Tris⋅HCl, pH 8.0/10 mM EDTA/0.5 mM EGTA/0.25% Triton X-100) and buffer B (10 mM Tris⋅HCl, pH 8.0/200 mM NaCl/1 mM EDTA/0.5 mM EGTA). The cells were then resuspended in RIPA (10 mM Tris⋅HCl, pH 8.0/140 mM NaCl/1 mM EDTA/1% Triton X-100/0.1% SDS/0.1% deoxycholate) and were sonicated (five times for 20 s each) to make soluble chromatin. Samples of total chromatin were taken at this point to use as a positive control in the PCRs (input chromatin). The cell lysates were precleared by incubation with protein A or G-Sepharose beads and then incubated with mAbs [6A10 (anti-c-Myc), 5E12 (anti-Mad1), D19 (anti-β-galactosidase; Roche Diagnostics] or with polyclonal antibodies from Santa Cruz Biotechnology [anti-Max (sc-197), anti-c-Myc (sc-764), anti-Mad1 (sc-222), anti-Mnt (sc-769), and anti-USF (sc-229)] overnight at 4°C. For analysis of acetylation status, antibodies against acetylated histone H3 (06–599) and histone H4 (06–866) (Upstate Biotechnology, Lake Placid, NY) were used for immunoprecipitations. DNA–protein complexes were collected with protein A or G-Sepharose beads followed by several rounds of washing. Bound DNA–protein complexes were eluted from the antibodies with two incubations in elution buffer (10 mM Tris, pH 8.0/1 mM DTT/0.5% SDS) at room temperature for 10 min. Iodacetamid was added to the combined eluates to neutralize DTT. A second immunoprecipitation was carried out to increase specificity with the use of the same protocol. Cross-links were reversed by the sequential addition of RNase A and proteinase K, followed by incubation at 65°C for 5 h. Samples were then extracted twice with phenol/chloroform and precipitated with ethanol overnight in the presence of 20 μg glycogen as a carrier. DNA fragments were recovered by centrifugation, resuspended in ddH2O, and used for PCR amplifications. The PCR products were fractionated on 2% agarose gels, stained with ethidium bromide, and quantified with the use of National Institutes of Health image software (version 1.61).
Results
Down-Regulation of hTERT Expression and Telomerase Activity in Differentiated HL60 cells.
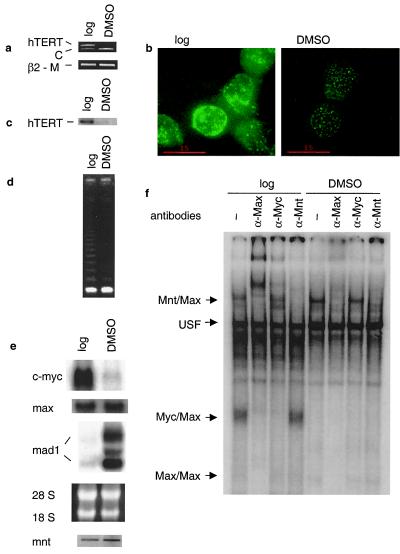
HL60 cells matured into granulocytes after exposure to DMSO. As demonstrated previously (23, 33), the amount of hTERT mRNA decreased rapidly and was undetectable after 48 h (Fig. 1a). Consistent with this observation, immunofluorescence and Western blot analyses revealed a significant reduction or disappearance of hTERT protein in the differentiated HL60 cells (Fig. 1b and c). As expected, the down-regulation of hTERT expression led to diminished telomerase activity in the DMSO-treated HL60 cells, which was found to be 10-fold lower compared with that in the control cells (Fig. 1d).
Figure 1.
Down-regulation of hTERT/telomerase activity is closely associated with changes in c-Myc and Mad1 expression during the differentiation of HL60 cells. HL60 cells were treated with DMSO for 48 h and analyzed for (a) hTERT mRNA expression by competitive RT-PCR; (b and c) expression of hTERT protein by immunofluorescence and Western blot; (d) telomerase activity by telomeric repeat amplification protocol assay; (e) c-Myc, Max, and Mad1 expression by Northern blot (Top), the ethidium bromide-stained gel visualizing the 18 and 28 S RNA (Middle), and Mnt expression by RT-PCR (Bottom); and (f) EMSA demonstrating DNA binding activities to the hTERT E-box in HL60 extracts. Antibodies used for supershifts are indicated at the top, and protein complexes binding to the hTERT oligo are indicated to the left. Log, logarithmically growing cells; DMSO, DMSO-treated (differentiated) cells; C, competitor for hTERT PCR product; β2-M, β2-microglobulin, internal control for the hTERT and Mnt RT-PCR.
Differential Expression of c-Myc and Mad1 and Their Binding Capacity for the hTERT E-Box-Containing Oligo in Protein Extracts of HL60 Cells.
We performed Northern blot and RT-PCR analyses to determine the expression of c-Myc, Max, Mnt, and Mad1 mRNA in HL60 cells. As expected, the untreated cells expressed high levels of c-Myc mRNA, whereas DMSO exposure led to its rapid down-regulation (Fig. 1e). In contrast, Mad1 mRNA was undetectable in the untreated cells but was dramatically induced after differentiation. One major and one minor Mad1 mRNA species, around 3.8 kb and 6.5 kb, respectively, were detected in the differentiated cells (Fig. 1e). These results are in agreement with those obtained by Larsson et al. (36). The DMSO treatment resulted in slight increases in expression of Max and Mnt mRNA (Fig. 1e). To evaluate the potential influence of the change in c-myc expression on the DNA binding capacity of the c-Myc protein, EMSA was performed by incubating protein extracts of HL60 cells with an E-box-containing oligo derived from the hTERT promoter region. The c-Myc/Max complex was readily detected in logarithmically growing cells and was greatly diminished in differentiated HL60 cells (Fig. 1f), suggesting that the level of c-Myc expression correlates with Myc activity in binding to the hTERT E-box in HL60 extracts in vitro. Supershift experiments with Myc and Max antibodies confirmed the identity of the c-Myc/Max complex. The Mad1/Max complex in the DMSO-treated cells could be visualized upon longer exposure of the gel (data not shown). Mnt/Max, Max/Max, and USF complexes were readily observed, and their identity was confirmed with the use of specific antibodies in supershift analysis (Fig. 1f and data not shown). The E-box binding activity of Mnt increased slightly upon HL60 differentiation.
In Vivo Interaction Between c-Myc, Mad1, and the hTERT Promoter in HL60 Cells.
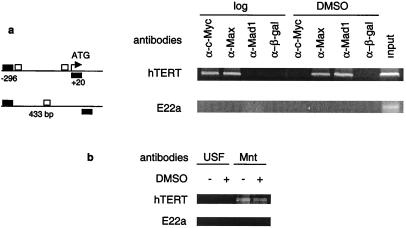
To obtain direct evidence of in vivo interaction of Myc and Mad1 with the E-boxes in the hTERT promoter in HL60 cells, we used ChIP analysis on both logarithmically growing and differentiated cells. After formaldehyde cross-linking and precipitation of the chromatin with c-Myc, Max, Mad1, or β-galactosidase antibodies, the precipitated DNA was subjected to PCR amplification with the use of specific primers for the hTERT proximal promoter region. A strong hTERT signal was observed in the c-Myc and Max immunoprecipitations, whereas no amplification of hTERT promoter sequences was detected in the Mad1 immunoprecipitation (Fig. 2a). This demonstrates that c-Myc/Max but not Mad1/Max binds in vivo to the E-boxes in the hTERT proximal promoter in exponentially growing HL60 cells. Upon differentiation of the cells, the binding of c-Myc to the promoter became undetectable, and instead Mad1 binding was observed (Fig. 2a). The amount of Max bound to the promoter was fairly constant. These data show a switch in binding at the hTERT promoter from Myc/Max to Mad1/Max during differentiation of HL60 cells. As a control, the chromatin was immunoprecipitated with β-galactosidase antibodies. In these immunoprecipitates no hTERT sequences could be detected, demonstrating the specificity of the procedure (Fig. 2a). The same changes occurred at other c-Myc targets analyzed (N.P., D.X., and M.H., unpublished observations). However, Myc and Mad1 do not bind to every E-box in the genome, inasmuch as Myc, Max, and Mad1 proteins were absent at the E22a and E22b fragments harboring E-boxes but located in regions of chromosome 22 that are not transcribed (Fig. 2a and data not shown).
Figure 2.
In vivo binding of c-Myc/Max, Mad1/Max, Mnt/Max, and USF to the hTERT proximal promoter in HL60 cells. A ChIP assay was performed on logarithmically growing and DMSO-treated HL60 cells, and the precipitated chromatin was PCR-amplified with the use of specific primers. (a) (Left) Schematic presentation of E-boxes (□) in the hTERT promoter and in the E22a locus, and the location of the respective primer sequences (■) for PCR analysis. The numbers below the hTERT promoter indicate the position of the PCR primers relative to ATG. The size of the E22a PCR product is shown. (Right) In vivo identification of reciprocal E-box occupancy by c-Myc, Max, and Mad1 at the hTERT promoter in logarithmically growing (log) and differentiated (DMSO) HL60 cells. β-Galactosidase antibodies were used as controls. Input: PCRs performed on total chromatin from differentiated HL60 cells. There is an absence of c-Myc, Max, and Mad1 binding to the control E22a locus that contains E-boxes but is not transcribed. (b) Absence of USF and presence of Mnt at the hTERT promoter in vivo in undifferentiated (−) and DMSO-differentiated (+) HL60 cells. Neither Mnt nor USF was present at the control E22a locus.
We also examined the in vivo binding of USF and Mnt to the E-box at the hTERT promoter, with the use of the ChIP assay. Surprisingly, the hTERT promoter sequence was undetectable in the USF antibody-precipitated chromatin DNA derived from either actively proliferating or differentiated HL60 cells (Fig. 2b). These data indicate that the in vitro binding activity of USF as assessed with EMSA does not necessarily reflect its E-box occupancy at specific promoters in vivo. The ChIP result showed the presence of Mnt at the hTERT promoter in vivo, and there was no significant difference in its binding between logarithmically growing and differentiated HL60 cells (Fig. 2b). Taken together, our data indicate that c-Myc/Max complexes were replaced by Mad1/Max at the hTERT promoter after HL60 cell differentiation.
In Vivo Association Between Acetylated Histones and the hTERT Promoter in HL60 Cells.
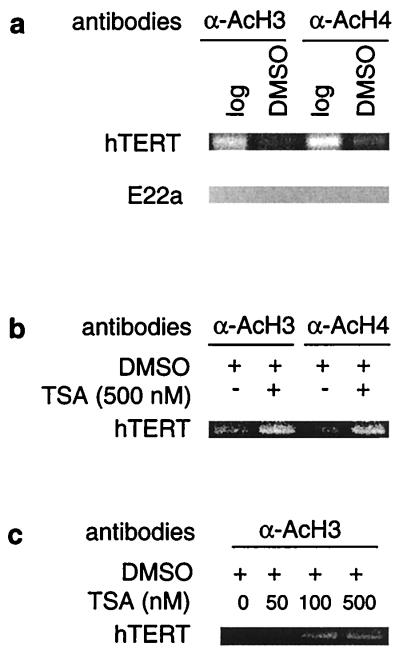
Histone acetylation has been implicated in Myc-mediated transactivation of target genes, whereas deacetylation has been shown to be important for Mad1 repression. We therefore wanted to determine whether there was a change in histone acetylation at the hTERT promoter upon differentiation of HL60 cells. Antibodies to acetylated histones were used to precipitate the cross-linked chromatin derived from logarithmically growing and DMSO-treated HL60 cells. Abundant levels of acetylated H3 and H4 histones were found to be present at the hTERT promoter in exponentially proliferating HL60 cells, and there was a significant reduction in the acetylation status after cellular differentiation (Fig. 3a). This reduction correlated with a decreased hTERT expression and telomerase activity (Fig. 1 a–d). We also analyzed the acetylation status of histones interacting with the nontranscribed E-box-containing E22a and E22b fragments. No acetylation was detected at either H3 or H4 histones independently of differentiation status at these loci (Fig. 3a and data not shown). Thus histone deacetylation seems to selectively occur at specific promoters during certain stages of differentiation of HL60 cells.
Figure 3.
Changes in histone acetylation at the hTERT promoter and TSA-mediated hyperacetylation of histones during HL60 differentiation. A ChIP assay was performed on logarithmically growing and DMSO-treated HL60 cells as described in Materials and Methods. (a) A decrease in acetylation of H3 and H4 histones at the hTERT promoter in differentiated HL60 cells. The absence of acetylated histones at the E22a fragment that contains an E-box but is not transcribed. Log, logarithmically growing cells; DMSO, DMSO-treated (differentiated) cells. (b) Abolishment of the differentiation-associated deacetylation of histones H3 and H4 at the hTERT promoter by TSA treatment in HL60 cells. Cells were induced to differentiate by DMSO overnight in the absence or presence of TSA as indicated. (c) TSA-mediated dose-dependent accumulation of histone H3 at the hTERT promoter in differentiating HL60 cells. Cells were treated with DMSO overnight in the absence or presence of various concentrations of TSA as indicated.
Partial Abolishment of Differentiation-Mediated hTERT Repression in HL60 Cells and Activation of hTERT Transcription in Normal Resting T Lymphocytes by TSA.
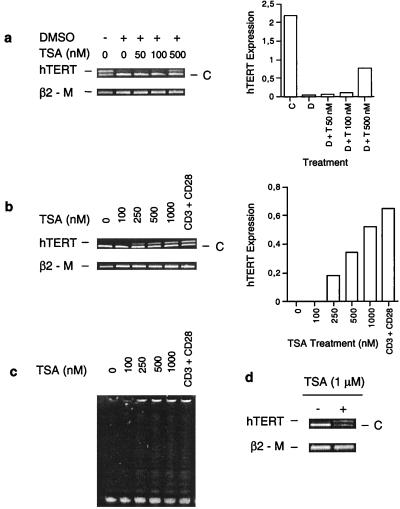
The amount of acetylated histones interacting with the hTERT proximal promoter was significantly reduced upon HL60 cell differentiation and correlated with repression of hTERT gene transcription. We therefore asked whether inhibition of histone deacetylation was capable of maintaining active transcription of the hTERT gene. For this purpose, HL60 cells were treated with TSA, a potent inhibitor of HDACs, followed by an overnight incubation with DMSO and analysis of hTERT mRNA expression. In the absence of TSA, DMSO treatment led to a reduction in the level of hTERT mRNA (Fig. 4a). However, in the presence of TSA the down-regulation of hTERT mRNA triggered by DMSO was partially blocked in a dose-dependent manner. When exponentially proliferating HL60 cells were incubated with TSA alone at a low concentration (<100 nM), a slight increase in hTERT transcripts was observed (data not shown). It has been well established that TSA itself induces differentiation and inhibits the growth of various cell types, including HL60 cells (37). Therefore, it is unlikely that the observed prevention of hTERT mRNA reduction by TSA results from an inhibition of HL60 cell differentiation. To determine whether this effect of TSA resulted from an increased accumulation of acetylated histones at the hTERT promoter, we performed ChIP analysis on the DMSO-treated HL60 cells in the presence and absence of TSA. As shown in Fig. 3b, a significantly higher level of acetylated histones was found at the hTERT promoter in TSA/DMSO-treated cells compared with cells incubated with DMSO alone. Consistent with the hTERT mRNA expression profile, a dose-dependent TSA-mediated accumulation of acetylated histone H3 at the hTERT promoter was observed in the DMSO-treated HL60 cells (Fig. 3c). These results suggest the involvement of histone deacetylation in the repression of hTERT transcription during HL60 cell differentiation. We further tested the effect of TSA on human T lymphocytes. hTERT expression and telomerase activity are usually very low or undetectable in resting human lymphocytes (38). TSA treatment of T cells induced hTERT mRNA expression in a dose-dependent manner to a level comparable to that observed in activated lymphocytes stimulated with anti-CD3 and CD28 antibodies (Fig. 4b). Concomitantly, telomerase was activated in the TSA-treated T cells (Fig. 4c). In addition, hTERT mRNA was also induced in human fibroblasts exposed to TSA (Fig. 4d). Taken together, these results indicate that histone deacetylation may be responsible for the repression of the hTERT gene observed in human somatic cells.
Figure 4.
Inhibition of HDACs by TSA attenuates down-regulation of hTERT mRNA expression in differentiated HL60 cells and activates hTERT transcription and telomerase in resting human T cells and fibroblasts. C, Competitor for hTERT fragment; β2-M, β2-microglobulin, internal control for the RT-PCR. (a) (Left) Competitive RT-PCR for hTERT mRNA in HL60 cells treated with TSA and/or DMSO. HL60 cells were incubated with TSA at various concentrations for 30 min, followed by overnight culture together with DMSO and analysis for hTERT mRNA expression. (Right) Quantitative expression of hTERT mRNA levels based on the signals to the left. D, DMSO; T, TSA. (b) (Left) Competitive RT-PCR for hTERT mRNA in T cells treated with different concentrations of TSA or anti-CD3 and anti-CD28 antibodies. (Right) Quantitative expression of hTERT mRNA levels based on the signals to the left. (c) Telomerase activity in T cells treated with different concentrations of TSA or anti-CD3 and anti-CD28 antibodies. (d) Competitive RT-PCR for hTERT mRNA in normal human fibroblasts treated with TSA.
Discussion
A number of recent observations support the identification of the hTERT gene as a c-Myc target: (i) ectopic expression of c-Myc induced hTERT expression and activated telomerase activity in human cells (27); (ii) the specific Myc binding motifs, E-boxes, are present in the hTERT promoter, and the activity of the promoter is significantly increased by transient expression of c-Myc (7, 28, 29, 39); (iii) ectopic expression of the Mad1 protein results in repression of hTERT expression (30, 31); and (iv) in vivo interaction between N-Myc and the hTERT promoter was recently observed in a neuroblastoma cell line transfected with N-myc (40). However, the effects obtained from artificially expressed Myc and Mad1 proteins at high levels do not necessarily demonstrate a physiological role, inasmuch as several more abundant proteins present in mammalian cells, such as USF, are capable of binding to E-boxes as well. To determine whether the hTERT promoter is regulated by endogenous c-Myc and Mad1 proteins, we used the ChIP assay in HL60 cells. These cells express different proteins of the Myc/Max/Mad network, depending on their state of differentiation. c-Myc is expressed during logarithmic growth and is down-regulated upon differentiation. At the same time Mad1 is induced, making HL60 a suitable system for the study of Myc and Mad1 target genes. We found that c-Myc binds in vivo to the hTERT promoter in logarithmically growing HL60 cells and that the binding activity decreased upon differentiation. In contrast, Mad1 expression and its binding to the hTERT promoter were observed only in differentiated cells. The presence of Max and Mnt at the hTERT promoter was fairly unchanged upon HL60 differentiation, whereas no binding of USF was observed. Thus, Myc/Max binding to the hTERT promoter is predominantly replaced by Mad1/Max, and this reciprocal hTERT promoter occupancy by c-Myc and Mad1 in vivo is compatible with activation and repression of hTERT transcription in logarithmically growing and differentiated HL60 cells, respectively. Taken together, these data suggest that hTERT is a physiological target of c-Myc and Mad1.
Histone acetylation and deacetylation have been suggested to play important roles in the regulation of gene transcription (41). In general, transcriptionally repressive chromatin is characterized by the presence of nonacetylated histones, whereas acetylation of histones leads to chromatin remodeling, allowing unfolding of the associated DNA, access by transcription factors, and gene expression. Acetylation and deacetylation of histones are catalyzed by histone acetyltransferases and HDACs, respectively, and a number of transcription factors, including Myc, are believed to regulate gene expression via recruitment of histone acetyltransferases or HDACs (3, 41). Substantial evidence suggests that the Mad1/Max dimer recruits mSin3A, mSin3B, HDACs, and corepressors, which in turn results in increased deacetylation of histones and repression of target gene transcription. McMahon et al. (3) have shown that the cofactor TRRAP associated with c-Myc is part of the SPT-ADA-GCN5 acetyltransferase (SAGA) complex, which among other factors, contains hGCN5, a human histone acetyltransferase, which contributes to increased histone acetylation. As a result, chromatin remodeling events occur that consequently initiate and/or accelerate transcription of c-Myc target genes. We observed an association between the switch from Myc/Max to Mad1/Max binding and a decrease in histone acetylation at the hTERT proximal promoter. These alterations were consistent with changes in the levels of hTERT expression. Similar changes were found at other c-Myc target genes during HL60 differentiation (N.P., D.X., and M.H., unpublished observations). These data thus suggest that c-Myc is responsible for the accumulation of acetylated histones at these promoters and Mad1 for their deacetylation. In addition, our findings that inhibition of HDACs by TSA resulted in abolishment of deacetylation at the hTERT promoter and blocked the repression of hTERT expression occurring during differentiation of HL60 cells demonstrate a functional role for histone acetylation at the hTERT promoter in the activation of transcription. This role is further supported by our observation that TSA induced hTERT expression in resting human T lymphocytes and fibroblasts. All of these data support a central role for regulation of the hTERT gene by changes in histone acetylation.
Interestingly, in a recent study, neither binding of c-Myc to the CAD promoter nor of N-Myc binding to the hTERT promoter was found to be correlated with the acetylation status at these promoters (40). The authors therefore draw the conclusion that Myc binding is not necessarily associated with changes in histone acetylation. One explanation for the discrepancy between our results is that the experiments were carried out in different ways and in different cells. Eberhardy et al. (40) attempted to determine whether the amount of acetylated histones correlated with the amount of bound N-Myc in SHEP neuroblastoma cells with inducible N-Myc expression. In this respect it is interesting to note that these cells harbor hTERT amplification (42) and therefore might have saturated acetylation status at the promoter independently of Myc expression. We analyzed HL60 cells that carry the normal hTERT locus (42) for changes in histone acetylation during differentiation. The decrease in histone acetylation that we observe at the hTERT promoter during HL60 differentiation is probably a combination of decreased Myc and increased Mad1 binding, resulting in active deacetylation of the histones. We have not been able to address whether there are changes in acetylation status at the CAD promoter during HL60 differentiation, because we only detected CAD promoter products in the total chromatin but not in any of our immunoprecipitations (data not shown).
Telomerase activity and hTERT expression are widespread in many human tissues throughout fetal development but become repressed in most somatic cells before or shortly after birth. The stringent repression of hTERT and telomerase activity in somatic tissues is a potent blockade to uncontrolled cellular proliferation (21). The mechanism of hTERT repression in somatic cells remains unknown. Our observations that inhibition of HDACs abolishes down-regulation of hTERT expression during cellular differentiation and induces telomerase in human resting T cells and fibroblasts suggest a potential repressive pathway: the down-regulation of hTERT transcription by deacetylation of histones may be a universal mechanism that leads to telomerase inactivation during differentiation and development of human normal somatic tissues.
Given that activation of telomerase is critical to oncogenesis, transactivation of the hTERT gene might be one essential mechanism by which c-Myc exerts its potent transforming effect: on one hand, it promotes cell proliferation by inducing cell cycle entry and progression, leading to accelerated telomere loss, and, on the other hand, it activates telomerase to stabilize telomere sizes and to maintain chromosomal integrity required for sustained cellular replication. Interestingly, a recent study demonstrates that normal human fibroblasts immortalized by hTERT transfection exhibit a significant up-regulation c-Myc expression, further highlighting an intimate interaction between Myc and hTERT during cellular immortalization (43). Our finding that endogenous c-Myc binds in vivo to the hTERT promoter suggests that the deregulated c-Myc expression, commonly observed in a wide range of human tumors, is most likely responsible for the abnormal growth and telomerase activation occurring in these tumors. Therefore, targeting c-Myc regulatory pathways is probably one approach to inhibiting telomerase activity and cell proliferation in human malignancies.
Acknowledgments
We thank B. Lüscher for discussions and mAbs, I. Chernov for help with the initial ChIP assay, and K. Pokrovskaja for critical reading of the manuscript. This study was supported by the Swedish Cancer Society, the Royal Swedish Academy of Sciences, Karolinska Institutet, and the Stockholm County Council. D.X. is supported by the David and Astrid Hagelens Foundation (Sweden). A.R.M. was a recipient of stipends from the Federation of European Biochemical Societies and from the Cancer Research Institute and Concern Foundation (USA).
Abbreviations
- CAD
carbamoyl-phosphatase/aspartate transferase/dihydroorotase
- ChIP
chromatin immuno-precipitation
- EMSA
electrophoretic mobility-shift assay
- HDAC
histone deacetylase
- hTERT
human telomerase reverse transcriptase
- TSA
trichostatin
- USF
upstream stimulating factor
- RT-PCR
reverse transcription–PCR
Note Added in Proof.
During the submission of this manuscript, Cong and Bacchetti reported that TSA induced hTERT expression and telomerase activity in human fibroblasts (44).
References
- 1.Henriksson M, Lüscher B. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 2.Gregor P D, Sawadogo M, Roeder R G. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 3.McMahon S, Wood M A, Cole M D. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole M D, McMahon S B. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermeking H, Rago C, Schumacher M, Li Q, Barrett J F, Obaya A J, O'Conell B C, Mateyak M K, Tam W, Kohlhuber F, et al. Proc Natl Acad Sci USA. 1999;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu K-J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R D. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber-Agus N, DePinho R A. BioEssays. 1998;20:808–818. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 9.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro C L, Messali S, Zollo M, Ledbetter D H, et al. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurlin P J, Quéva C, Eisenman R N. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 11.Bejarano M T, Albihn A, Cornvik T, Osterdahl Brijker S, Asker C, Osorio L, Henriksson M. Exp Cell Res. 2000;260:61–72. doi: 10.1006/excr.2000.4996. [DOI] [PubMed] [Google Scholar]
- 12.Gehring S, Rottmann S, Menkel A R, Mertsching J, Krippner-Heidenreich A, Lüscher B. J Biol Chem. 2000;275:10413–10420. doi: 10.1074/jbc.275.14.10413. [DOI] [PubMed] [Google Scholar]
- 13.Ayer D E, Lawrence Q A, Eisenman R N. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 14.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 15.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 16.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 17.Heinzel T, Lavinsky R M, Mullen T-M, Söderström M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 18.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 19.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Lüscher B. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 20.Wolffe A P. Nature (London) 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 21.Harley C B, Kim N W, Prowse K R, Weinreich S L, Hirsch K S, West M D, Bacchetti S, Hirte H W, Counter C M, Greider C W, et al. Cold Spring Harbor Symp Quant Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 23.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 25.Bodnar A, Ouellette M, Frolkis M, Holt S, Chiu C, Morin G, Harley C, Shay J, Lichtsteiner S, Wright W. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 26.Nugent C I, Lundblad V. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Xie L Y, Allan S, Beach D, Hannon G J. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cong Y-S, Wen J, Bacchetti S. Hum Mol Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg R A, O'Hagan R C, Deng H, Xiao Q, Hann S R, Adams R R, Lichtsteiner S, Chin L, Morin G B, DePinho R A. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 30.Gunes C, Lichtsteiner S, Vasserot A P, Englert C. Cancer Res. 2000;60:2116–2124. [PubMed] [Google Scholar]
- 31.Oh S, Song Y H, Yim J, Kim T K. Oncogene. 2000;19:1485–1490. doi: 10.1038/sj.onc.1203439. [DOI] [PubMed] [Google Scholar]
- 32.Xu D, Gruber A, Peterson C, Pisa P. Br J Haematol. 1998;102:1367–1375. doi: 10.1046/j.1365-2141.1998.00969.x. [DOI] [PubMed] [Google Scholar]
- 33.Xu D, Gruber A, Björkholm M, Peterson C, Pisa P. Br J Cancer. 1999;80:453–457. doi: 10.1038/sj.bjc.6690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klapper W, Singh K K, Heidorn K, Parwaresch R, Krupp G. Biochim Biophys Acta. 1998;1442:120–126. doi: 10.1016/s0167-4781(98)00155-9. [DOI] [PubMed] [Google Scholar]
- 35.Orlando V, Strutt H, Paro R. Methods Enzymol. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 36.Larsson L-G, Pettersson M, Öberg F, Nilsson K, Lüscher B. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 37.Marks P A, Richon V M, Rifkind R A. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 38.Norrback K F, Roos G. Eur J Cancer. 1997;33:774–780. doi: 10.1016/S0959-8049(97)00059-2. [DOI] [PubMed] [Google Scholar]
- 39.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 40.Eberhardy S R, D'Cunha C A, Farnham P J. J Biol Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- 41.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 42.Zhang A, Zheng C, Lindvall C, Hou M, Ekedah J, Lewensohn R, Yan Z, Yang X, Henriksson M, Blennow E, et al. Cancer Res. 2000;60:6230–6235. [PubMed] [Google Scholar]
- 43.Wang J, Hannon G J, Beach D H. Nature (London) 2000;405:755–756. doi: 10.1038/35015674. [DOI] [PubMed] [Google Scholar]
- 44.Cong Y S, Bacchetti S. J Biol Chem. 2000;275:35665–35668. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]