Abstract
4-Hydroxynonenal (4-HNE) is an endogenous product of lipid peroxidation known to play a role in cellular signaling through protein modification and is a major precursor for protein carbonyl adducts found in alcoholic liver disease (ALD). In the present study, a greater than 2-fold increase in protein carbonylation of sirtuin 3 (SIRT3), a mitochondrial class III histone deacetylase, is reported in liver mitochondrial extracts of ethanol consuming mice. The consequence of this in vivo carbonylation on SIRT3 deacetylase activity is unknown. Interestingly, mitochondrial protein hyperacetylation was observed in a time-dependent increase in a model of chronic ethanol consumption; however, the underlying mechanisms for this remain unknown. Tandem mass spectrometry was used to identify and characterize the in vitro covalent modification of rSIRT3 by 4-HNE at Cys280, a critical zinc-binding residue, and the resulting inhibition of rSIRT3 activity via pathophysiologically relevant concentrations of 4-HNE. Computational-based molecular modeling simulations indicate that 4-HNE modification alters the conformation of the zinc-binding domain inducing minor changes within the active site, resulting in the allosteric inhibition of SIRT3 activity. These conformational data are supported by the calculated binding energies derived from molecular docking studies suggesting the substrate peptide of acetyl-CoA synthetase 2 (AceCS2-Kac), displays a greater affinity for native SIRT3 compared with the 4-HNE adducted protein. The results of this study characterize altered mitochondrial protein acetylation in a mouse model of chronic ethanol ingestion and thiol-specific allosteric inhibition of rSIRT3 resulting from 4-HNE adduction.
INTRODUCTION
Mitochondrial dysfunction is known to play a role in numerous pathologies, including neurodegenerative diseases, cancer, and alcoholic liver disease (ALD) (1–4). Components of the onset and progression of these pathologies are a consequence of metabolic disruption and/or increases in oxidative stress. In ALD microsomal and mitochondrial systems involved in ethanol metabolism contribute to hepatotoxicity through an increase in cytochrome P450-2E1 (CYP2E1), catalase and aldehyde dehydrogenase (ALDH) activities (5, 6). The oxidation of ethanol and acetaldehyde by alcohol dehydrogenase (ADH) and ALDH alters NADH/NAD+ redox status (7, 8). During prolonged exposure to biochemical stress, such as chronic ethanol metabolism, the generation of reactive oxygen species (ROS) overcomes cellular antioxidant systems, resulting in excesses of superoxide, hydrogen peroxide, and hydroxyl radicals (9). This enhanced ROS production alters the mitochondrial genome and proteome through a number of mechanisms, including the accumulation of the lipid peroxidation product 4-hydroxynonenal (4-HNE) in pathologic quantities near lipid rich cellular membranes (10–13). The cellular impact of 4-HNE generation is not fully understood; however, at basal levels it has been implicated in intracellular signaling and in excess it is known to interfere with proteomic activity via Michael-type and Schiff-base adduct formation with Cys>>EHis>Lys>>Arg amino acid residues of proteins (14).
Recently, increased mitochondrial protein acetylation in a model of chronic ethanol exposure was reported (15, 16); however, the underlying mechanism behind this increase was not determined and the physiological consequences of ethanol-induced mitochondrial protein hyperacetylation remain elusive. Sirtuins catalyze the deacetylation of the ε-amino group of lysine residues and are involved in regulating transcriptional activity and protein function (17). Only three mitochondrial sirtuins have been identified, SIRT3–5. Based on knockout studies, SIRT3 was determined to be the major mitochondrial protein deacetylase (18). Sirtuin activity is uniquely NAD+- and zinc-dependent and results in the generation of nicotinamide (NAM), O-acetyl-ADP-ribose, and the deacetylated substrate (19, 20). A recent proteomic survey reported that approximately 20% of mitochondrial proteins are acetylated, implicating SIRT3 as a key regulator of mitochondrial proteome function (21). Targets of SIRT3 deacetylation are widespread and are implicated in a host of cellular pathways, including, oxidative phosphorylation, fatty acid metabolism, oxidative stress response, and alcohol metabolism (21–24). It is therefore likely that alterations in protein acetylation due to metabolic and oxidative stress may negatively impact these pathways, disrupting normal mitochondrial processes. Although few studies have investigated the impact of chronic ethanol consumption on sirtuins, recent data suggests that SIRT1 is a target of protein carbonylation in a model of chronic obstructed pulmonary disease in human lung tissue (25–27). The sirtuin family of proteins contains a highly conserved enzymatic core with a large NAD+-binding domain and a smaller zinc-binding domain (26). Given the sequence homology among the sirtuin family of proteins, it is likely that SIRT3 is also susceptible to protein carbonylation. Furthermore, localization at the inner mitochondrial membrane exposes SIRT3 to elevated concentrations of 4-HNE (12, 28, 29).
The present study investigates the impact of the lipid peroxidation product 4-HNE on rSIRT3 deacetylase activity. An established ethanol feeding model in C57BL/6J mice allowed immunohistochemistry, lipid peroxidation, and NADH/NAD+ assays on liver tissue to investigate factors that may support mechanisms of mitochondrial protein hyperacetylation associated with alcohol consumption. It was found that the carbonylation of SIRT3 in vivo was significantly increased by over 2-fold. The in vitro exposure of rSIRT3 to 4-HNE uncovered significant inhibition at pathophysiologically relevant concentrations of aldehyde. A rSIRT3 protein adduct was identified on Cys280 using nano liquid chromatography tandem mass spectrometry (nLC-MS/MS) and computational-based molecular modeling demonstrates numerous 4-HNE induced conformational changes within the active site of rSIRT3. The results of this study suggest a potential mechanism for carbonyl-induced inhibition of SIRT3 in vivo arising from 4-HNE modification. As previously reported, numerous factors are likely to play a role in mitochondrial protein hyperacetylation; however, the results in this report demonstrate significant decreases in rSIRT3 activity resulting from thiol-specific modification by the lipid peroxidation product 4-HNE reflected in significant SIRT3 carbonylation in vivo.
MATERIALS AND METHODS
Materials
The reagents used in this study were purchased from Sigma-Aldrich (St. Louis, MO), unless stated otherwise. Purified human rSIRT3 protein was purchased from Cayman Chemical (Ann Arbor, MI). 4-HNE was synthesized in our laboratory as previously detailed (30). Protein content was determined by the Lowry-based Dc protein assay (Bio-Rad Labs, Hercules, CA).
Animals
The procedures and protocols used in this study were approved by the Institutional Animal Care and Use Committee of the University of Colorado and were performed in accordance with standard National Institute of Health guidelines. C57BL/6J mice were fed a modified Lieber-DeCarli liquid-ethanol diet over a period of six weeks as previously described (31). Control and ethanol diets were obtained from Bio-Serv (Frenchtown, NJ) and were prepared fresh weekly. The diet contains 44% fat-derived calories, 18% protein-derived calories and the remainder from maltodextrin and/or ethanol, with a maximum 31.8% ethanol-derived calories (EDC). In short, ethanol-fed mice began the study on a diet containing 10.8% EDC (2% v/v) and the ethanol content was increased weekly until the diet contained 31.8% EDC (6% v/v). Age-matched, pair-fed control mice received an identical caloric intake, supplemented with maltodextrin-derived calories rather than EDC. Drinking water was provided ad libitum. Upon completion of the one-, three-, and six-week feeding regimens, the animals were anesthetized by an intraperitoneal injection of sodium pentobarbital and euthanized by exsanguination. Livers were removed and either fixed in formalin for histochemical analysis, or homogenized and subjected to fractionation using differential centrifugation as previously described (32).
Immunohistochemistry
Following excision, livers were sectioned and placed in 10% neutral buffered formalin for 8 hours. Samples were then processed, embedded in paraffin and mounted to slides by Colorado HistoPrep (Fort Collins, CO). One pair of slides were stained with hematoxylin and eosin (H&E) for histological characterization, while the remaining slides were subjected to de-paraffinization and rehydration for immunohistochemical (IHC) characterization with antibodies directed against 4-HNE modified protein (Bethyl Labs, Montgomery, TX) and acetylated-lysine residues (Cell signaling, Boston, MA).
Lipid Peroxidation and Protein Carbonylation
The extent of lipid peroxidation was determined using the thiobarbituric acid reactive substances (TBARS) assay and were measured in whole liver homogenates (500 µg) by incubating samples in thiobarbituric acid reagent (15% w/v trichloroacetic acid, 0.375% w/v thiobarbituric acid and 0.25 N HCl). Samples were boiled at 100°C for 15 min, cooled to room temperature, and then centrifuged at 10,000 rpm. Absorbance of the cleared supernatant was monitored spectrophotometrically on a microplate reader at 535 nm, and the concentration of malondialdehyde (MDA) was calculated from the extinction coefficient (ε = 156,000 M-1 cm-1).
SIRT3 protein carbonylation was monitored using biotin hydrazide (BH) treatment and avidin pull-down (31). Briefly, 200 µg aliquots of mitochondrial extract were treated in the dark for 2 hours at 25°C with 5 mM BH (Thermo Fisher Scientific Inc., Rockford, IL), from a stock of 50 mM BH in dimethyl sulfoxide (DMSO). To remove unreacted BH, the solution was then dialyzed 1:10,000 in 10 mM phosphate buffered saline (PBS) overnight at 4°C. NeutrAvidin agarose beads (Thermo Fisher Scientific) were added to the dialysand and the reaction mixture was gently rotated overnight at 4°C. The beads were washed five times in tris-buffered saline containing 0.2% tween 20 (TBST), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer was added and the carbonylated protein isolate was applied to SDS-PAGE and transferred as detailed below for immunodetection by anti-SIRT3 antibody.
Liver mitochondrial extracts from control and ethanol-fed mice were applied to SDS-PAGE and western blotting for analysis. In short, 12% SDS-PAGE gels were used to resolve mitochondrial extracts. The gels were then transferred to Hybond-P transfer membranes (GE Healthcare, Piscataway, NJ). The membranes were blocked in 5% (w/v) non-fat dry milk (NFDM) in TBST. These membranes were probed with anti-SIRT3 (a kind gift from Dr. Eric Verdin) and anti-Acetyl-Lys (Cell Signaling, Boston, MA) antibodies. VDAC (Abcam, Cambridge, MA) was used as the load control. A horseradish peroxidase-conjugated secondary antibody (Jackson Labs) was applied and the membranes were developed with ECL-Plus reagent (GE Health Sciences).
NADH/NAD+ Determination
Alterations in mouse liver NADH/NAD+ redox state were quantified using an EnzyChrom NADH/NAD+ assay according to the assay instructions (BioAssay Systems, Hayward, CA). Based on an alcohol dehydrogenase cycling reaction, the NADH generated in this assay reduces a formazan reagent, and the formation of the product, measured at 565 nm, was proportional to the NADH/NAD+ concentration in the samples.
Upon excision, liver sections were snap-frozen in 1 mL eppendorf tubes under liquid nitrogen and stored at −80°C. A 10 mg aliquot of tissue was prepared on ice and immediately placed into NAD or NADH assay buffer, each containing HCl or NaOH, respectively. The samples were homogenized, heated for 5 min at 60°C, buffer was added to neutralize pH, and finally the samples were centrifuged at 14,000 × g for 5 min. The supernatants were diluted 1:10 in assay buffer for analysis.
Covalent Modification of rSIRT3
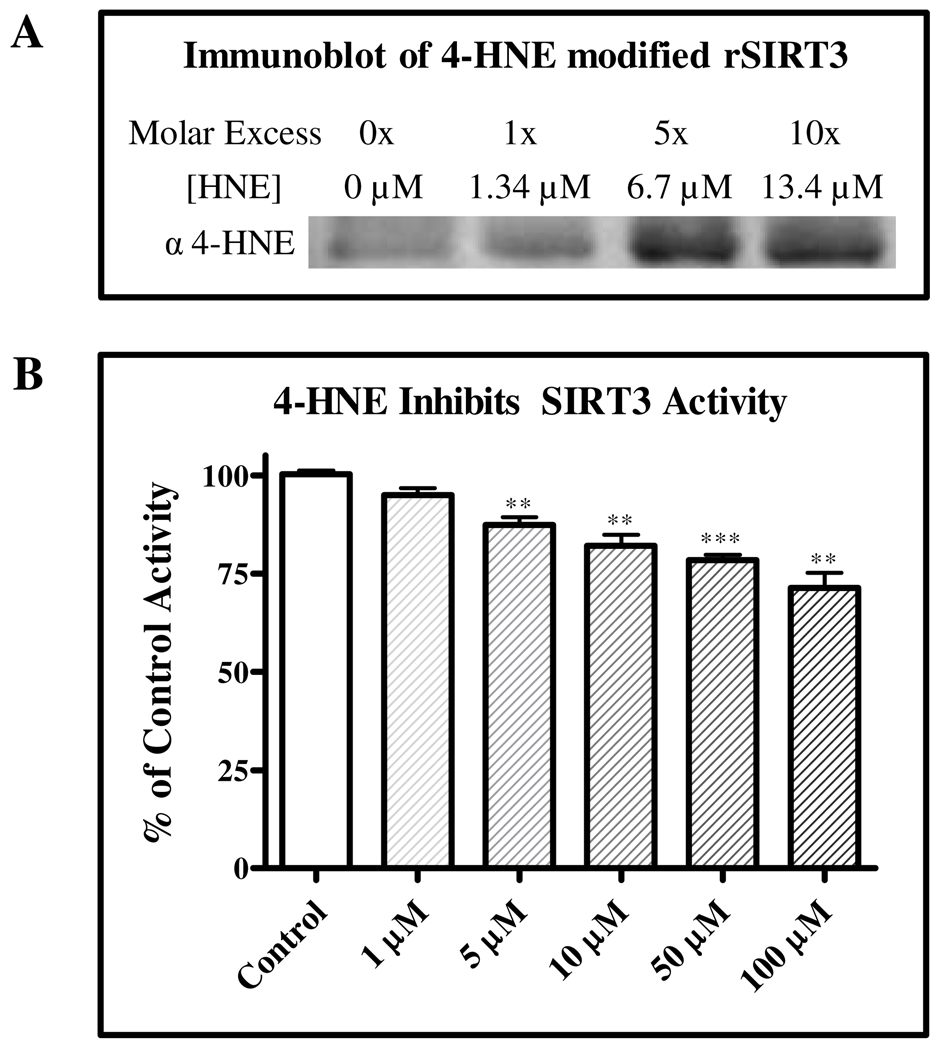
Adduction of human rSIRT3 was achieved by incubating 1 µg (27 pmoles, 1.34 µM) of protein with increasing concentrations of 4-HNE. Treatments included final concentrations of 1.34, 6.7, and 13.4 µM of 4-HNE for 30 min at 37°C. Upon completion, samples were applied to SDS-PAGE, transferred to Hybond-P transfer membranes and visualization of adduct formation was monitored by immunodetection with a custom polyclonal rabbit anti-4-HNE antibody, directed against 4-HNE modified keyhole limpet hemocyanin (Bethyl Labs).
rSIRT3 Inhibition
The rSIRT3 assay allows for the assessment of 4-HNE-mediated inhibition of rSIRT3 activity and uses the p53 sequence Gln-Pro-Lys-Lys(Acetyl)-aminomethylcoumarin, an acetylated substrate specific for sirtuins. The rSIRT3-mediated lysine deacetylation of the substrate liberates aminomethylcoumarin (AMC), which was quantified through fluorescence excitation at 350 nm and emission at 450 nm. rSIRT3 inhibition or activation was quantified and reported as a percentage of initial rSIRT3 activity. The concentration of 4-HNE treatment ranged from 1 µM to 100 µM. rSIRT3 was treated for 30 min at 25°C prior to assaying activity. To determine if 4-HNE inhibition occurred as a result of covalent modification, protein was dialyzed against 1X PBS for 5 hours at 4°C prior to assaying activity. Additionally, 10 mM O-phenanthroline was used as previously reported (20) to inhibit rSIRT3 activity and 0.3 to 30 mM N-Ethylmaleimide (NEM) was used as a cysteine-specific modifier to inhibit rSIRT3 activity (Supplemental Data).
Mass Spectrometry
Human rSIRT3 was treated with 4-HNE to identify modified residues using nLC-MS/MS. Briefly, 5 µg (0.13 nmoles, 2.6 µM) of rSIRT3 in 50µL of 50mM sodium phosphate buffer, pH 7.4, was treated with 25× 4-HNE (3.25 nmoles, 65 µM) molar excess for 30 min at 25°C. 4-HNE treatments were followed by NaBH4 (5 mM) reduction at 25°C for 30 min in order to stabilize protein adducts. The samples were resolved on a 12% SDS-PAGE gel and visualized by Coomassie blue staining. Protein bands were excised and subjected to proteolysis with trypsin (Promega, Madison, WI). The digested samples were analyzed using nano liquid chromatography (EASY-nLC, Proxeon) at a flow rate of 300 nL/min with a gradient of 5 to 40% acetonitrile (0.2% formic acid) over 40 min on C-18 Proxeon EASY-Column trapping (20 × 0.1 mm) and analytical columns (100 × 0.075 mm). The nLC was coupled to a nano-electrospray ionization source and esquire HCT ion trap mass spectrometer (Bruker Daltonics). The instrument was operated using data-dependent MS/MS with a threshold of 30,000 TIC. Data analysis was performed using Mascot (v 2.1.04, www.matrixscience.com) and Bruker Daltonics esquire v 5.2 data analysis package, and b/y ion fragmentation was predicted using ProteinProspector v 5.4 (http://prospector.ucsf.edu, UCSF Mass Spectrometry Facility). Secondary MS analysis was performed using an ABI 4800 Plus MALDI-TOF/TOF (Applied Biosystems) instrument to confirm the findings of the nLC-MS/MS analysis and to improve on overall peptide detection. The digested samples were prepared using C18 ZipTips (Millipore), mixed 1:1 with α-cyano-4-hydroxycinnamic acid (CHCA, 10 mg/mL) and spotted in 1 µL aliquots onto an Opti-TOF 384 well insert (Applied Biosystems).
In an attempt to locate and identify SIRT3 in vivo, 500 µg of liver mitochondrial extract from control-and ethanol-fed mice were fractionated on a 12% SDS-PAGE gel and protein bands of 4 kDa were excised from the gel across a range of 24–36 kDa. Given that the molecular weight of murine SIRT3 is 28,822 Da, the excised range is sufficient to identify SIRT3 in vivo. Proteolysis and nLC-MS/MS analysis were performed as described above, using a gradient of 5 to 40% acetonitrile (0.2% formic acid) over 90 min.
Molecular Modeling
All simulations were performed using Discovery Studio software (Version 2.5.5; Accelrys Inc., San Diego, CA). The crystallographic coordinates of the 2.7Å human SIRT3 crystal structure (PDB code: 3GLS) (26) was obtained from the RCSB Protein Data Bank (http://www.rcsb.org). The 4-HNE adduct was built onto Cys280 and the adducted and unadducted SIRT3 structures were typed with the CHARMm force field, and subjected to minimization (1000 iterations) using the conjugate gradient method to a convergence of 0.001 kcal/mol (33). The substrate, an acetyl-CoA synthetase 2 acetylated peptide (AceCS2-Kac) containing residues 638–649 of human AceCS2 was constructed from the AceCS2-Kac peptide extracted from the 1.87Å human SIRT3 co-crystallized structure (PDB code: GLR), and NAD+ was constructed from ADP ribose bound to the AceCS2 peptide containing a thioacetyl lysine from the 2.2Å human SIRT3 co-crystallized structure (PDB code: 3GLT) (26). The AceCS2-Kac peptide and NAD+ were docked separately, using the flexible docking protocol (34), into the active site of the 4-HNE adducted and unadducted SIRT3 structures. Two separate 12Å binding spheres were selected for the AceCS2-Kac peptide and NAD+ cofactor, the AceCS2-Kac peptide binding sphere included the residues: Arg158, Gln228, Ala247, His248, Val292–Pro299, Phe302, Leu303, His305, and Leu322–Leu330; and the NAD+ cofactor binding sphere included: Met143–Ser159, Leu164, Glu177, Phe180, Leu195, Leu199, Asn207, Thr209, His210, Tyr226–Glu234, Ala247, His248, Ile317–Val324, Phe327, Ala328, Leu342, Ile343–Val348, and Val366. The AceCS2-Kac peptide was docked into the SIRT3 structures first and the highest ranked complex was used for NAD+ docking. The Generalized Born implicit solvent model was used to calculate binding energy (ΔGbind) of the docked complexes (35).
Data Analysis
Data, graphs and statistical analysis were performed using GraphPad Prism 4 (GraphPad Software, Inc., La Jolla, CA) and Excel (Microsoft Corp, Redmond, WA). Western blot quantification was achieved using an open source software application, ImageJ 1.42q (National Institutes of Health). All data is presented as mean +/− standard error of the mean (SEM) and each experiment was repeated for confirmation. Student’s t-test was used to determine statistical significance.
RESULTS
Chronic Ethanol Feeding Results in a Time-dependent Increase in Hepatic Protein Acetylation
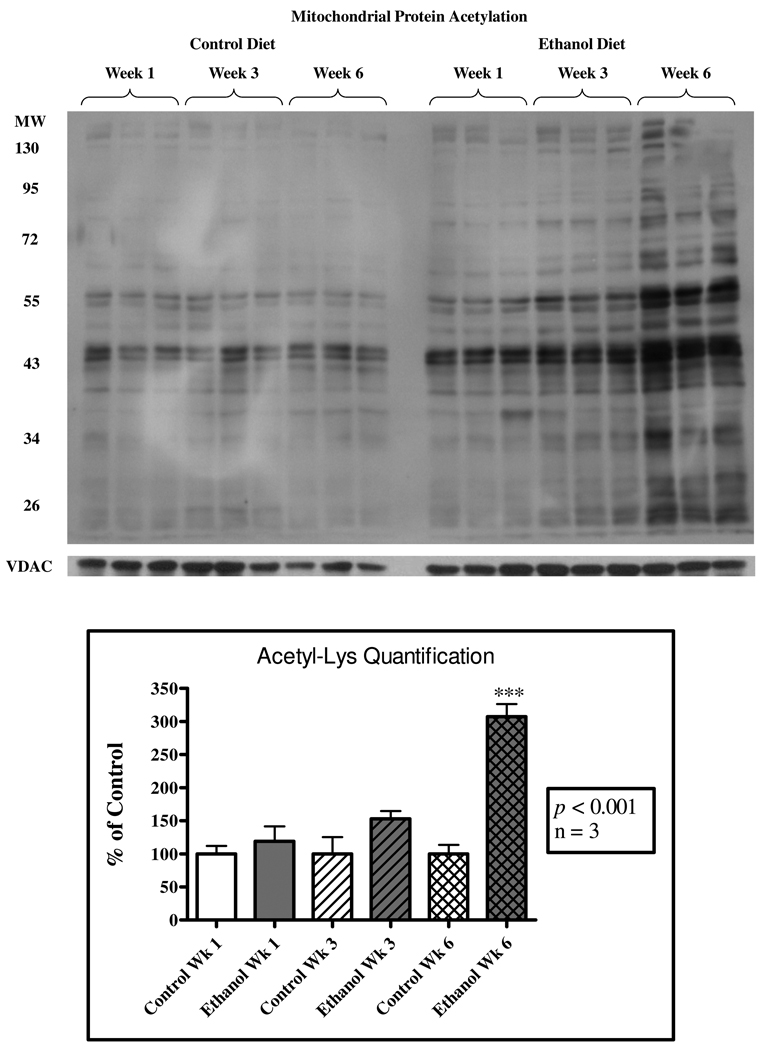
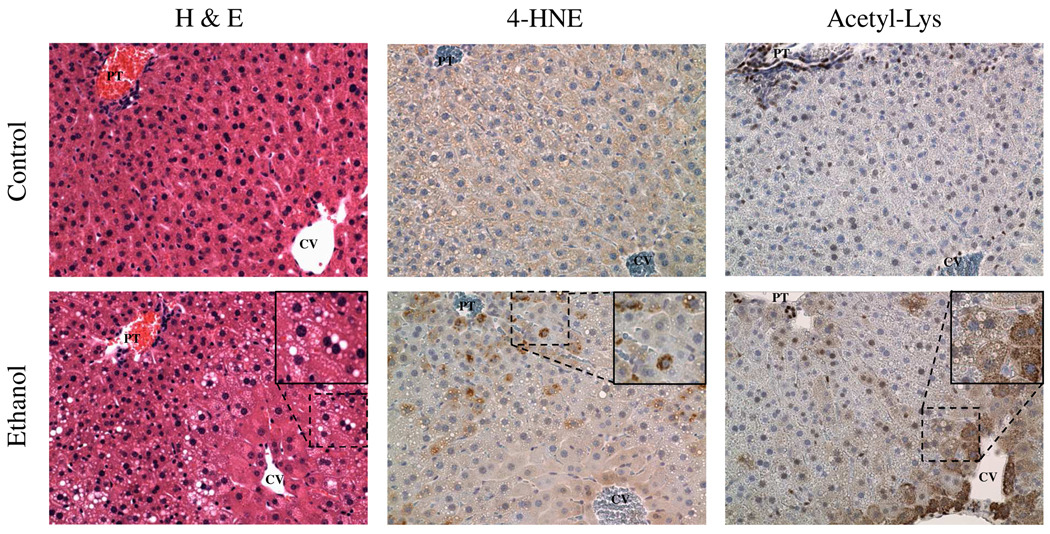
Control and ethanol diets were fed to age-matched mice over a 6-week “ethanol ramping” period. Standard markers for early-stage ALD reflect previously reported values and were statistically significant, such as elevated alanine aminotransferase activity, liver triglycerides, and increased liver to body weight ratio (31). As shown in Figure 1, dramatic increases in mitochondrial protein acetylation were observed, confirming previously reported alcohol-induced protein hyperacetylation (15, 16). Interestingly, protein hyperacetylation increased gradually, with no significant increase at one week of treatment, slight increases by week three, and in excess of 3-fold induction by week six. The presence of lipid, as shown by H&E staining in Figures 2A and 2B, also demonstrates disease progression in our ethanol model to steatosis. A marked increase in 4-HNE immunopositive proteins within the liver is readily visible in Figure 2C and 2D. Additionally, Figures 2E and 2F provide, for the first time, immunohistochemical analysis of acetylation in mouse liver tissue from control- and ethanol-fed groups at week 6 and demonstrate a lobular-specific up-regulation of this mitochondrial protein hyperacetylation in zone 3, with minor increases in zones 1 and 2.
Figure 1. Chronic ethanol consumption results in a marked increase in mitochondrial protein acetylation.
Liver mitochondrial extracts were obtained from control and ethanol-fed mice at 1-, 3- and 6-week intervals on a modified Lieber-DeCarli liquid diet. Western blotting using anti-acetyl-lysine antibody was used to probe for mitochondrial protein acetylation. Significant increases in protein acetylation were not observed at week 1. At week 3 a noticeable increase occurred in the ethanol-fed group and significant increases were observed by week 6. Quantification of these blots demonstrates an increase in over 300% protein acetylation in the week 6 ethanol-fed group versus the pair-fed control.
Figure 2. Histological characterization of control and ethanol-fed mouse liver tissue.
H&E staining of the 6-week pair-fed groups illustrates a marked increase in lipid accumulation, verifying the progression of our ethanol model to steatosis (A-Control, B-Ethanol). Comparative IHC analysis of 4-HNE accumulation demonstrates a sizeable increase in 4-HNE modified proteins in the ethanol-fed group (C-Control, D-Ethanol). Analysis of protein acetylation in the 6-week groups (E-Control, F-Ethanol) illustrates mitochondrial staining with a zonal distribution of increased mitochondrial protein acetylation in zone 3 and migrating into zones 2 and 1. Histologically, the liver is comprised of 3 zones, where zone 1 resides around the Portal Triad (PT) as shown in the top left of each slide, zone 3 encompasses the tissue surrounding the Central Vein (CV) as shown in the bottom right of each slide and zone 2 is centrally located between zones 1 and 3.
Enhanced Lipid Peroxidation and SIRT3 Protein Carbonylation
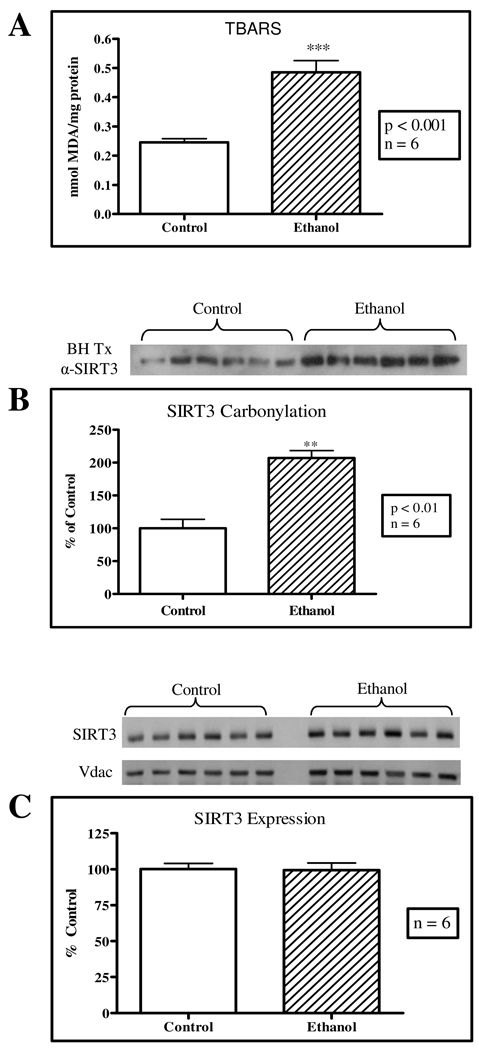
Studies have demonstrated that SIRT3 deacetylase activity alters the function of various proteins, including acetyl-CoA synthetase 2, glutamate dehydrogenase, isocitrate dehydrogenase 2 (ICDH2), electron transport chain Complex I and long-chain acyl-CoA dehydrogenase (22, 24, 36, 37). Therefore, the modification of SIRT3 provides a potential mechanism for altering these enzymatic pathways. Protein carbonylation is known to alter the function of many proteins and may play a role in regulating SIRT3 activity in vivo. As shown in Figure 3A, a 2-fold increase in lipid peroxidation was detected in ethanol-fed mice. Based on a standard method of determining protein carbonyl content, BH treatment of mitochondrial fractions was used to examine SIRT3 post-translational modification by reactive aldehydes. Figure 3B establishes that in our mouse model of chronic ethanol feeding, SIRT3 protein carbonylation was increased by over 2-fold in the ethanol-fed mice; however, SIRT3 expression levels remained unaltered (Figure 3C). This significant increase in SIRT3 carbonylation coincides with the observed 4-HNE accumulation and increase in lipid peroxidation as demonstrated in Figures 2 and 3.
Figure 3. Enhanced lipid peroxidation and SIRT3 protein carbonylation in a 6-week chronic ethanol model.
(A) A TBARS assay demonstrates a significant increase in lipid peroxidation in this chronic ethanol model, as quantified by detecting MDA levels. (B) Biotin hydrazide (BH) conjugation displays a significant increase in SIRT3 protein carbonylation in these week 6 mice (Control = 100%, SEM 13.7. Ethanol = 207%, SEM 11.3). (C) Chronic ethanol consumption does not alter SIRT3 protein expression. VDAC was used as the load control.
Chronic Ethanol Consumption Alters NADH/NAD+
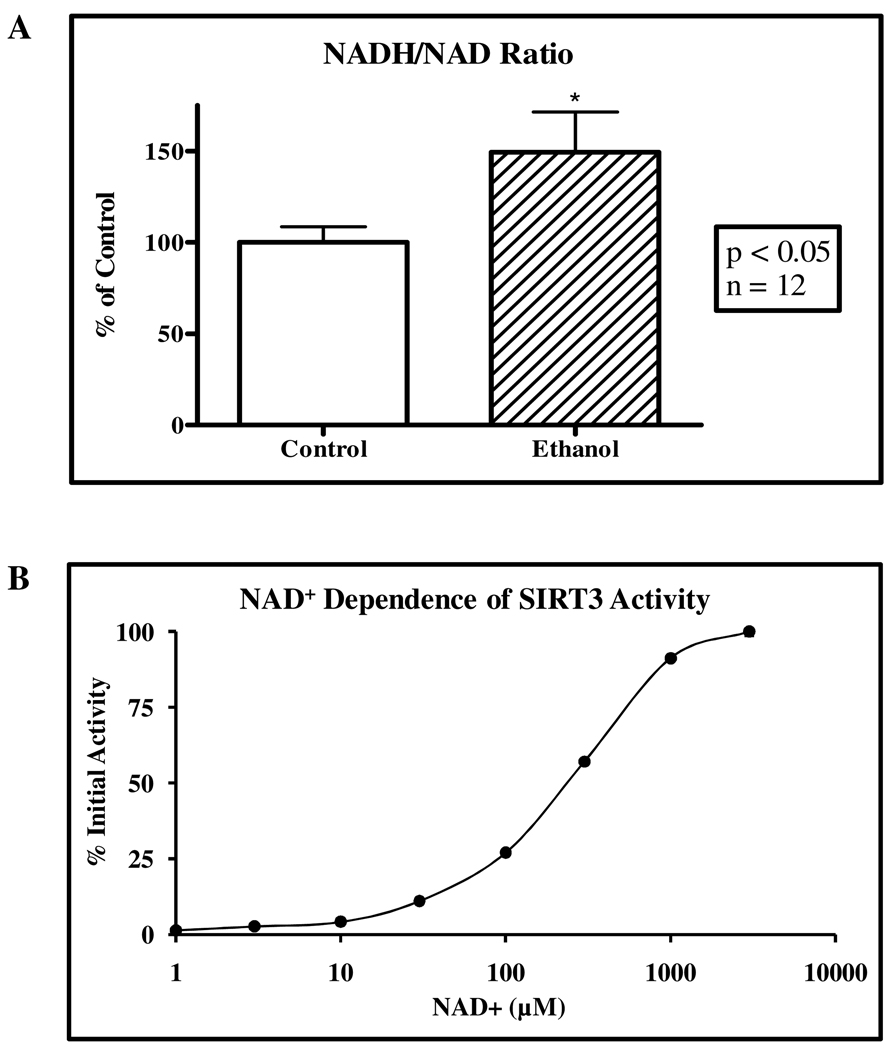
Sirtuins are classified as NAD+-dependent and require zinc binding for activity (26). This NAD+-dependence suggests that SIRT3 regulates mitochondrial metabolism and gene expression via nutrient sensitive signaling in response to altered NAD+ concentrations (22). Further characterization of this ethanol model was accomplished by measuring NADH/NAD+ ratios in whole liver extracts. The NADH/NAD+ redox status (Figure 4A) was significantly increased by over 50% in these ethanol-fed mice. Levels of mitochondrial NADH/NAD+ redox flux have been reported to be substantially higher, approaching a 5-fold increase under situations of ethanol exposure (38, 39). This significant alteration in NADH/NAD+ redox has the potential to negatively affect SIRT3 activity, however the physiological impact is yet to be determined (40, 41). As quantified by the rSIRT3 activity assay employed in Figure 4B, the measured Km value of rSIRT3 for NAD+ is 283 µM. The reported Km values of sirtuins for NAD+ are varied, ranging from 100 µM to 3.2 mM, thus displaying a low affinity for NAD+ (19) (Caliper LifeSciences, Application note 208). Evidence supports SIRT3 as a regulator of cellular metabolism via nutrient sensing during conditions of stress and toxicity through NAD+-dependence; however what role this plays in situations of chronic ethanol exposure remains unknown (19).
Figure 4. Ethanol metabolism alters NADH/NAD+.
(A) Chronic ethanol consumption results in a significant shift in NADH/NAD+ redox status in mouse liver whole cell extract. NAD+ concentrations significantly decreased from 1.05 mM/g (0.1) in control mice to 0.89 mM/g (0.07) in ethanol-fed mice (p < 0.05) while NADH significantly increased from 0.67 mM/g (0.09) in control mice to 0.84 mM/g (0.12) in ethanol-fed mice (p < 0.05). (B) rSIRT3 was incubated with increasing NAD+ concentrations in an in vitro activity assay, displaying an apparent Km of ~283 µM for NAD+. Error bars represent SEM.
4-HNE Modification Inhibits rSIRT3 Deacetylase Activity
Accumulation of the lipid peroxidation product 4-HNE is a hallmark of ALD, consequently, a significant portion of protein carbonyl adducts found in vivo include 4-HNE (10, 13, 14, 42). Furthermore, recent publications linking cigarette smoke to decreased rSIRT1 activity by 4-HNE modification provide evidence that the sirtuin family of proteins may be susceptible to 4-HNE adduction (25, 27). Therefore, the chemical reactivity of 4-HNE towards human rSIRT3 was determined using immunodetection. As illustrated in Figure 5A, increasing levels of 4-HNE (0–13.4 µM) were applied to characterize a concentration dependent increase in rSIRT3 modification. Consequently, 4-HNE inhibition of rSIRT3 activity was verified using an activity assay based on human rSIRT3 and a sirtuin specific peptide, comprising amino acids 317–320 of human p53 conjugated to AMC. The results in Figure 5B demonstrate a 13% decrease (p < 0.01) in rSIRT3 activity at 5 µM 4-HNE and a 20% decrease (p < 0.01) in activity upon exposure to a pathophysiologically relevant 4-HNE concentration of 10 µM. The levels of 4-HNE applied remain over an order of magnitude below the reported concentrations (5 µM to 5 mM) associated with situations of sustained oxidative stress (13, 14, 43). Both positive controls of O-phenanthroline and NEM (Supplemental Data) inhibited SIRT3 activity and demonstrate the impact of zinc-dependent and thiol-specific inhibition on SIRT3 activity. To further verify these findings, Supplemental Figure 3 demonstrates 4-HNE inhibition of rSIRT3 using an independent activity assay (MBL Intl.)
Figure 5. 4-HNE covalently modifies and inhibits rSIRT3.
(A) Covalent modification of rSIRT3 by 4-HNE increases in a concentration-dependent manner, as determined by western blot. (B) rSIRT3 activity is inhibited in a concentration-dependent manner, showing a 13% decrease (p < 0.01) in activity at 5 µM 4-HNE and a 20% decrease (p < 0.01) in activity upon exposure to 10 µM 4-HNE. Error bars represent SEM.
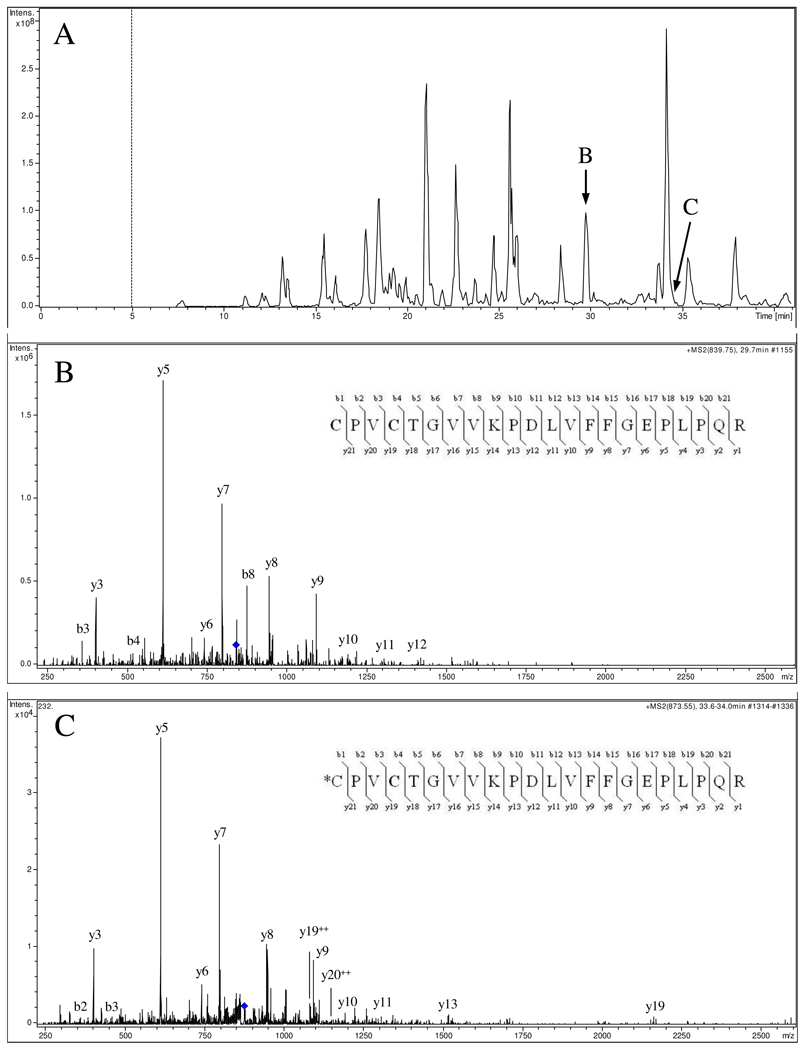
MS Identification of 4-HNE Modified rSIRT3
In vitro adduction of rSIRT3 by 4-HNE was performed to determine the location and number of protein modifications. Chemical reduction was used to stabilize and improve the detection of labile 4-HNE adducts. Analysis of tryptic digests of modified rSIRT3 resulted in MS peptide sequence coverage of 78%, which included all four zinc-binding cysteine residues (Cys256, Cys259, Cys280, and Cys283) and the histidine active site residue (His248). The resulting mass spectra identified two amino acid sequences with 4-HNE adducts; the first contained His354 and the second consisted of two cysteine residues, Cys280 and Cys283 (Table 1). The His354 residue is located distal to the zinc-binding pocket and the active site and is not known to play a significant role in protein function. However, the nLC-MS identified the triply charged parent ion 873.55 m/z, which corresponds to the peptide containing two cysteine residues, Cys280 and Cys283, and is part of the zinc-binding motif on the protein small domain, where four cysteine residues coordinate one zinc atom. In Figure 6, the identified peptide included one cysteine containing a reduced 4-HNE adduct, while the other cysteine contained a carbamidomethyl derivative resulting from iodoacetamide treatment. Using MS/MS fragmentation and the resulting b/y ion distribution, we were able to confirm that Cys280 was the residue susceptible to modification by 4-HNE. Additionally, NEM modification resulted in an adduct specific to Cys280 and demonstrates this residue is susceptible to electrophilic modification (Supplemental Data). A recent publication examined the structural basis for sirtuin function and reported that zinc binding results in subtle allosteric shifts that occur within the active site binding cleft and is necessary for deacetylase activity (44). Previous work has also demonstrated that point mutations of the four zinc-binding cysteine residues prevents SIRT3 deacetylase activity (20). Therefore, 4-HNE adduction of Cys280 impedes zinc binding and alters the structural integrity of the active site, providing a mechanism of rSIRT3 inhibition via thiol-specific electrophilic adduction. Interestingly, increasing the concentration of 4-HNE applied to rSIRT3 did not result in different sites of modification, as only Cys280 and His354 were modified.
Table 1.
nLC-MS/MS and MALDI-TOF/TOF MASCOT results from rSIRT3 analysis: Untreated and modified by 4-HNE (NaBH4 reduced).
| AA | Exp | Calc | Sequence | MS/MS |
|---|---|---|---|---|
| 100–120 | 1970.41 | 1969.97 | RSISFSVGASSVVGSGGSSDK | * |
| 123–133 | 1255.71 | 1255.71 | LSLQDVAELIR | * |
| 140–158 | 1901.00 | 1902.01 | VVVMVGAGISTPSGIPDFR | * |
| 140–158 | 1916.98 | 1917.00 | VVVMVGAGISTPSGIPDFR (1 Met Oxidation) | * |
| 198–214 | 2110.06 | 2110.06 | ELYPGNYKPNVTHYFLR | * |
| 215–219 | 624.33 | 624.36 | LLHDK | |
| 215–224 | 1176.74 | 1176.73 | LLHDKGLLLR | |
| 225–235 | 1320.67 | 1320.67 | LYTQNIDGLER | * |
| 236–243 | 757.43 | 757.43 | VSGIPASK | |
| 244–269 | 2861.40 | 2861.36 | LVEAHGTFASATCTVCQRPFPGEDIR (1 Cys Carb) | * |
| 244–269 | 2918.38 | 2918.38 | LVEAHGTFASATCTVCQRPFPGEDIR (2 Cys Carb) | * |
| 280–301 | 2514.29 | 2514.28 | CPVCTGVVKPDIVFFGEPLPQR (2 Cys Carb) | * |
| 280–301 | 2615.61 | 2615.39 | CPVCTGVVKPDIVFFGEPLPQR (1 Cys 4-HNE [+158], 1 Cys Carb) | * |
| 341–345 | 627.38 | 627.41 | LLINR | |
| 341–356 | 1868.99 | 1869.07 | LLINRDLVGPLAWHPR | |
| 346–356 | 1259.68 | 1259.80 | DLVGPLAWHPR | * |
| 346–356 | 1418.04 | 1417.81 | DLVGPLAWHPR (1 His 4-HNE [+158]) | * |
| 359–384 | 2896.45 | 2896.43 | DVAQLGDVVHGVESLVELLGWTEEMR (1 Met Oxidation) | |
| 385–389 | 629.34 | 629.35 | DLVQR | |
| 385–393 | 1044.54 | 1044.54 | DLVQRETGK | |
| 390–399 | 1058.74 | 1058.52 | ETGKLDGPDK | * |
Amino acid number (AA), experimental (Exp.) and calculated (Calc.) nominal masses for each peptide are shown.
MS/MS analysis confirmation was obtained. (Carb = Carbamidomethyl)
Figure 6. 4-HNE modified rSIRT3 characterized by nLC-MS/MS analysis.
(A) Base peak chromatogram of trypsin-digested human rSIRT3 following 4-HNE exposure. (B) The native peptide containing Cys280 and Cys283 was identified with 2 Cys carbamidomethyl derivatives (CPVCTGVVKPDIVFFGEPLPQR). (C) The 4-HNE (+158) modified peptide was also identified and MS/MS analysis determined the exact residue of adduction, Cys280 (*CPVCTGVVKPDIVFFGEPLPQR).
Figure 7 provides a UniProt sequence comparison between the active forms of human and murine SIRT3 (www.uniprot.org). This alignment suggests a shared susceptibility to electrophilic adduction at that specific cysteine residue for both human and murine forms of SIRT3. Unfortunately, our attempts to identify murine SIRT3 in vivo were unsuccessful. We were able to identify 112 unique proteins from the excised bands spanning 24–36 kDa from a 500 µg aliquot of mitochondrial extract using our highly sensitive nLC-MS/MS instrumentation; however, the expression levels of SIRT3 were not at MS detectable concentrations and SIRT3 was not identified.
Figure 7. Sequence alignment of human and murine SIRT3.
The active forms of human and murine SIRT3 share a highly conserved amino acid sequence, as only 37 of the 257 amino acid residues differ. Specifically, the peptide microenvironment surrounding Cys280 (highlighted in bold rectangle) is identically matched for 10 residues towards the N terminus and 16 residues towards the C terminus. This similarity suggests a shared susceptibility to thiol-specific 4-HNE modification in situations of increased lipid peroxidation. (human Cys280 = murine Cys138)
In Silico Modeling of 4-HNE Adducted SIRT3
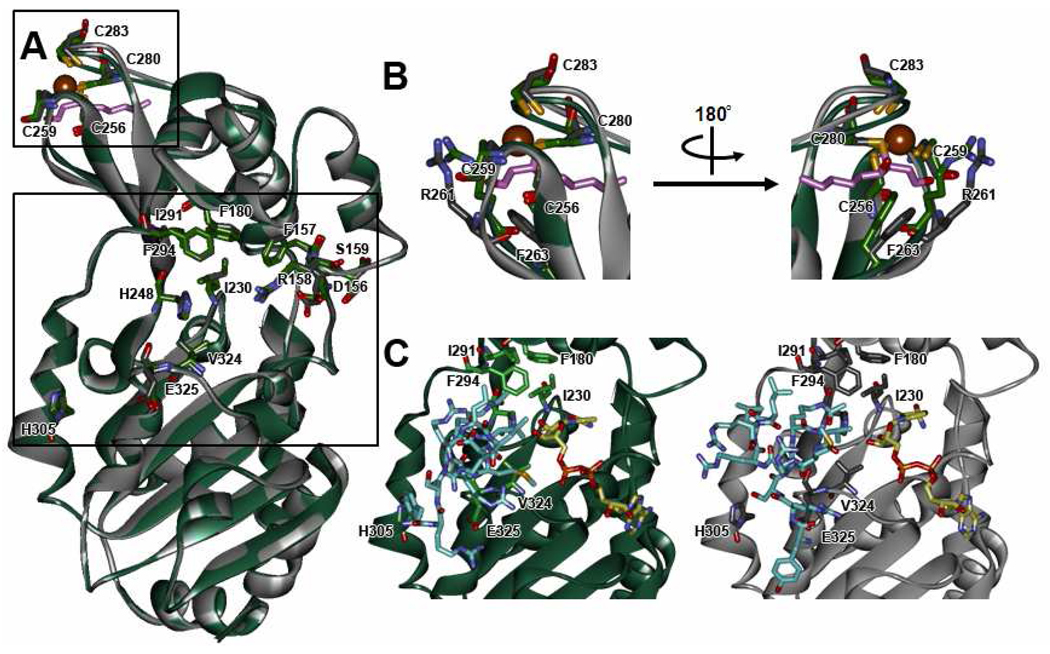
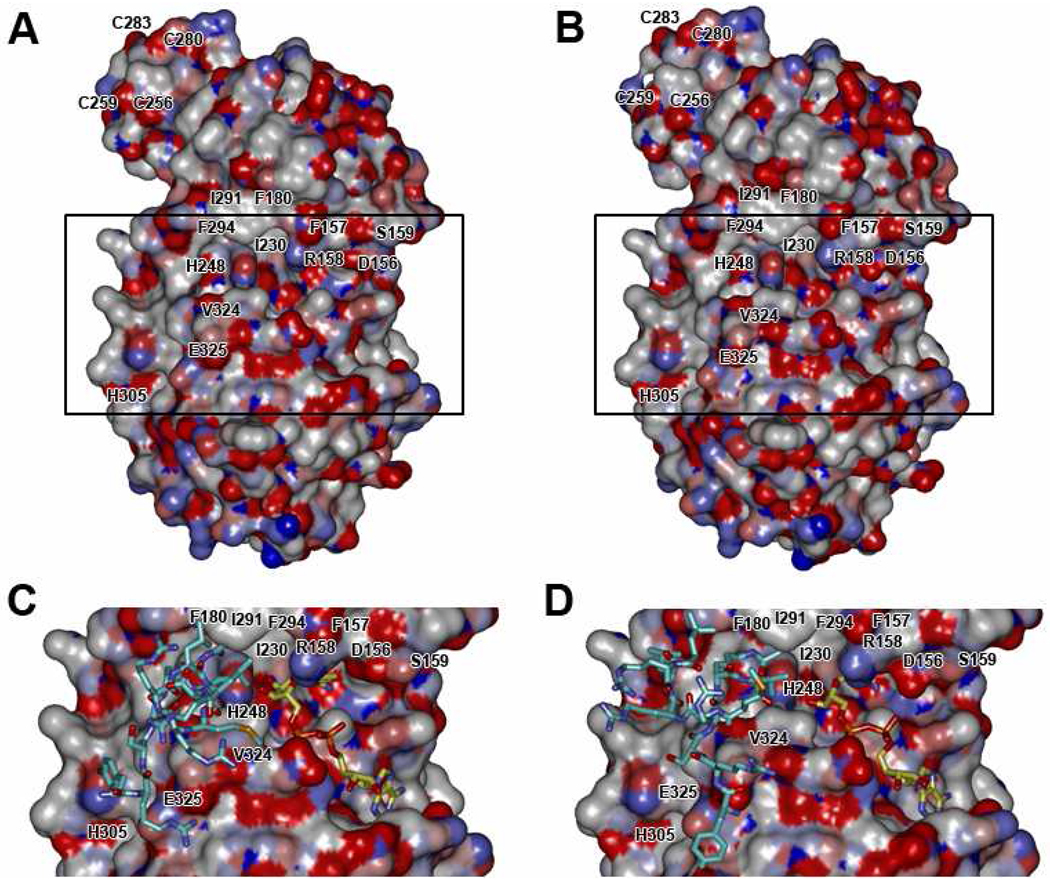
Computational-based molecular modeling studies were performed to determine the impact of Cys280 adduction by 4-HNE on the conformation of the SIRT3 zinc-binding pocket and accessibility for zinc binding. The minimization of the 4-HNE adducted SIRT3 structure resulted in a number of conformational changes in the zinc-binding site and in regions on the periphery of the active site that may affect the catalytic activity of SIRT3, as illustrated in Figure 8A. The 4-HNE adducted to Cys280 occupies the zinc-binding pocket and could sterically prevent zinc from binding to the other cysteine residues. Furthermore, Figure 8B illustrates the adduction of Cys280 by 4-HNE alters the conformation of the zinc-binding site; the Cα of the four cysteine residues are shifted away from the binding site by 0.22Å (Cys256), 0.45Å (Cys259), 1.02Å (Cys280), and 0.76Å (Cys283) relative to the unadducted protein, and shifts of 0.71Å and 1.45Å were also observed for Phe263 and Arg261, respectively. Although distortions in the protein backbone were observed in the zinc-binding site, Cα shifts of a similar magnitude were not observed in the active site of the protein. The ribbon overlay in Figure 8A and molecular surface representations in Figures 9A and 9B clearly show differences in the conformation of the Val324 and Glu325 residues on the periphery of the substrate binding site and between the Asp156, Phe157, Arg158, and Ser159 residues near the NAD+ binding site, which may affect the binding of the substrate and NAD+ cofactor.
Figure 8. Structural changes in SIRT3 upon 4-HNE adduction.
(A) Ribbon representation of the superimposition of SIRT3 (green) and 4-HNE adducted SIRT3 (gray). (B) Superimposition of the zinc-binding domain of SIRT3 (green) with zinc (colored brown) and 4-HNE (carbon atoms colored salmon) adducted at Cys280 of SIRT3 (gray). (C) AceCS2-Kac peptide (carbon atoms colored cyan) and NAD+ (carbon atoms colored yellow) docked into the active site of SIRT3 (ribbon and carbon atoms colored green) and 4-HNE adducted SIRT3 (ribbon and carbon atoms colored gray).
Figure 9. Global structural changes in SIRT3 upon 4-HNE adduction.
(A) Molecular surface of SIRT3 protein (colored by atom charge). (B) Molecular surface of SIRT3 protein with 4-HNE adducted at Cys280 (colored by atom charge). A comparison of the AceCS2-Kac peptide and NAD+ in the active site of (C) unadducted and (D) adducted SIRT3 illustrates subtle changes throughout the binding cleft, consequently impacting both substrate and NAD+ docking into their respective binding sites.
Molecular docking simulations were performed to examine the binding of the AceCS2-Kac peptide and NAD+ cofactor in the respective binding sites of the 4-HNE adducted and unadducted SIRT3 protein. The complexes were ranked based on binding mode and the calculated ΔGbind. In the highest ranked docked complex of the AceCS2-Kac peptide in the active site of the unadducted SIRT3 protein shown in Figure 8C, the carbonyl carbon atom of the acetyl group was 2.93Å from the nitrogen of His248, with the remainder of the molecule oriented outside of the active site. However, in the 4-HNE adducted SIRT3 protein, the AceCS2-Kac peptide adopts a conformation that has rotated 180° from that observed in the unadducted complex, the acetyl group of the AceCS2-Kac peptide was still directed towards His248 in the active site of 4-HNE adducted SIRT3; however, a distance of 3.56Å was recorded between the carbonyl carbon atom of the acetyl group and the nitrogen of His248. This change in binding conformation clearly affects the interaction of the AceCS2-Kac peptide with SIRT3 active site residues, most notably between the Tyr of the AceCS2-Kac peptide and His305, which may be required to stabilize the SIRT3-AceCS2-Kac complex for subsequent NAD+ binding. Furthermore, the average calculated ΔGbind values indicate that the AceCS2-Kac peptide has a greater affinity for unadducted SIRT3 compared with the 4-HNE adducted protein (−17.1kcal/mol versus −12.3kcal/mol, respectively). Interestingly, although some differences in the position of adenine ring were noted, the docking of the NAD+ cofactor was only slightly affected by 4-HNE adduction, with ΔGbind values of −25.4 kcal/mol and −23.5 kcal/mol for the unadducted and 4-HNE adducted protein, respectively.
DISCUSSION
Oxidative stress and the accumulation of 4-HNE are known factors contributing to the progression of ALD, impacting numerous cellular processes (5, 12, 45–48). The inhibition of rSIRT3 by 4-HNE modification at Cys280 is another example of altered protein function through electrophilic adduction. The biological impact of oxidative stress is substantial and was recently detailed in a review which examined the consequences of altered thiol redox on cellular systems (49). The review highlights cysteine-rich zinc fingers and zinc-binding domains of proteins, as these regions provide a mechanism for redox regulated control of enzymatic activity, either directly at the active site or through allosteric activation/inhibition. The rSIRT3 activity assay and mass spectral data detailed in the present study supports this allosteric control mechanism for SIRT3 activity through 4-HNE post-translational modification of a zinc-binding residue distal to the active site. The characterization of this post-translational modification by 4-HNE using MS and in silico molecular modeling provides insight into the biochemical mechanism of this thiol-specific regulation of rSIRT3 activity.
Our molecular modeling simulations identified sizeable shifts in the zinc-binding motif of the small subunit of SIRT3 as a result of 4-HNE modification. Furthermore, the active site binding cleft located between the large and small protein subunits was altered enough to prevent the substrate from binding in the correct orientation, which is normally stabilized through zinc binding at the small domain. These data demonstrate that 4-HNE adduction of Cys280 inhibits rSIRT3 activity through covalent modification of a residue distal to the active site, rather than directly modifying the active-site histidine (His248) or other residues within the active site. Interestingly, the ΔGbind values derived from molecular docking studies provide evidence for altered substrate binding while NAD+ binding appears unaffected. Our data agrees with the recently reported isothermal titration calorimetry (ITC) and crystal structure studies which used the identical substrate and cofactor, AceCS2-Kac and NAD+, confirming that the peptide substrate binds first to SIRT3 followed by NAD+ binding (26). Collectively, these results indicate 4-HNE modification of rSIRT3 results in altered substrate binding by inducing subtle conformational changes in the active site as a result of this thiol-specific modification. Therefore, substrate binding is the “rate-limiting” step for SIRT3 deacetylase activity and inhibition by 4-HNE adduction is independent of NAD+ concentrations. SIRT3 activity assay data reported by assay manufacturers indicates that substrate binding may determine the affinity of SIRT3 for NAD+ binding, as two separate SIRT3 activity assays report a Km of 241 µM (p53 substrate peptide, Cayman Chem) and 3.2 mM (Histone H3 substrate peptide, Caliper LifeSciences) for NAD+ binding. This difference is likely due to substrate-induced conformational changes within the cofactor binding site, where a set of highly conserved catalytic residues modulate NAD+ binding. Therefore, the subtle shifts which occur within the active site as a result of 4-HNE modification may impact SIRT3 substrate binding to varying degrees. The numerous factors involved in substrate, cofactor and intermediate binding kinetics are detailed extensively elsewhere (19, 26, 41, 44, 50). In aggregate, these data suggest an induced specificity of SIRT3 for varied substrate deacetylation in a nutrient sensing, NAD+-dependent manner, which maintains the potential for alteration by post-translational modifications such as 4-HNE, as demonstrated by our inhibition assay, MS data and in silico modeling studies. Taking into account the numerous known and unknown factors that have the potential to impact SIRT3 activity in vivo, such as NAD+ concentrations, substrate induced alterations in the Km of SIRT3 for NAD+, and protein adduction by lipid electrophiles, among others, it remains difficult to draw any specific conclusions relating to the underlying mechanism of ethanol-induced protein hyperacetylation.
Importantly, the observed mitochondrial protein hyperacetylation in our model of chronic ethanol consumption further demonstrates mitochondrial dysfunction in ALD, even though factors contributing to this altered acetylproteome and its downstream effects remain unknown. Acetylation is known to play an important role in metabolic regulation and antioxidant response pathways within the liver (15–18, 21, 22, 41, 51, 52). Recent reports indicate that SIRT3 alters the expression of superoxide dismutase 2 (SOD2) and the activity of ICDH2, both of which are intimately involved in metabolizing oxidative species and the maintenance of antioxidants within the mitochondria (24, 53). Unfortunately, the in vivo consequences of SIRT3 inhibition are poorly understood, with only a handful of direct interactions currently reported. The IHC characterization of our ethanol model presents lipid accumulation and oxidative stress, with significant induction of lipid peroxidation and 4-HNE generation. This increase in 4-HNE generation is likely to react readily with proteins, as observed by an over 2-fold increase in SIRT3 carbonylation. Since we were unable to detect SIRT3 in vivo, conclusions regarding the nature and consequence of these modifications remain unknown.
The characterized 4-HNE adduction of rSIRT3 at the zinc-binding domain may provide insights into potential mechanisms of allosteric inhibition applicable to the entire sirtuin family of proteins. Recent reviews have detailed efforts into the development of sirtuin inhibitors and activators (54, 55). Sirtuin inhibition/activation has the potential to impact numerous pathologies, including, ALD, cancer, neurodegenerative diseases, inflammation, cardiovascular diseases, diabetes, and metabolic syndrome (54, 55). While the majority of research has focused on the nuclear sirtuin, SIRT1, continued efforts to examine other sirtuins, such as SIRT3, remain critical to understanding mechanisms of sirtuin function throughout subcellular components. Further studies are needed to fully elucidate the metabolic consequences of SIRT3 carbonylation in vivo and its impact on structure and function. Understanding the role of mitochondrial protein acetylation in regulating enzyme activity, protein-protein interaction and gene expression will provide further insight into the tight regulation of cellular processes and the pathological implications of an altered acetyl-proteome and involvement in the pathogenesis of ALD.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Dr. Eric Verdin and Dr. Matthew Hirschey for the generous gift of anti-SIRT3 antibody and Dr. David Orlicky for assistance in immunohistochemical data interpretation.
FUNDING SUPPORT
This work was supported, in part by Grants NIH/NIAAA F31AA018606-01 (J.J.G.), NIH/NIAAA F31AA18898-01 (R.L.S.), NIH/NIAAA 5R37AA009300-15 and NIH/NIDDK 5R01DK074487-03 (D.R.P.).
REFERENCES
- 1.Reeve AK, Krishnan KJ, Turnbull DM. Age related mitochondrial degenerative disorders in humans. Biotechnol J. 2008;3:750–756. doi: 10.1002/biot.200800066. [DOI] [PubMed] [Google Scholar]
- 2.Mantena SK, King AL, Andringa KK, Eccleston HB, Bailey SM. Mitochondrial dysfunction and oxidative stress in the pathogenesis of alcohol- and obesity-induced fatty liver diseases. Free Radic Biol Med. 2008;44:1259–1272. doi: 10.1016/j.freeradbiomed.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee R, Starkov AA, Beal MF, Thomas B. Mitochondrial dysfunction in the limelight of Parkinson's disease pathogenesis. Biochim Biophys Acta. 2009;1792:651–663. doi: 10.1016/j.bbadis.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albano E. Free radicals and alcohol-induced liver injury. In: Sherman VPaRW., editor. In Ethanol and the Liver. London: Taylor and Francis; 2002. pp. 153–190. [Google Scholar]
- 6.Das SK, Vasudevan DM. Alcohol-induced oxidative stress. Life Sci. 2007;81:177–187. doi: 10.1016/j.lfs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Cederbaum AI. Microsomal generation of reactive oxygen species and their possible role in alcohol hepatotoxicity. Alcohol Alcohol Suppl. 1991;1:291–296. [PubMed] [Google Scholar]
- 8.Mantle D, Preedy VR. Free radicals as mediators of alcohol toxicity. Adverse Drug React Toxicol Rev. 1999;18:235–252. [PubMed] [Google Scholar]
- 9.Albano E. Alcohol, oxidative stress and free radical damage. Proc Nutr Soc. 2006;65:278–290. doi: 10.1079/pns2006496. [DOI] [PubMed] [Google Scholar]
- 10.Niemela O. Aldehyde-protein adducts in the liver as a result of ethanol-induced oxidative stress. Front Biosci. 1999;4:506–513. doi: 10.2741/niemela. [DOI] [PubMed] [Google Scholar]
- 11.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 12.Roede JR, Jones DP. Reactive species and mitochondrial dysfunction: mechanistic significance of 4-hydroxynonenal. Environ Mol Mutagen. 2010;51:380–390. doi: 10.1002/em.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 14.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 15.Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376:615–619. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 16.Shepard BD, Tuma DJ, Tuma PL. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol Clin Exp Res. 34:280–291. doi: 10.1111/j.1530-0277.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci. 2009;1173 Suppl 1:E10–E19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauve AA. Sirtuin chemical mechanisms. Biochim Biophys Acta. 2010;8:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 21.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. 2010;1804:1645–1651. doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 24.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 25.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, Milne JC, Westphal CH, Perni RB. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009 doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caito S, Rajendrasozhan S, Cook S, Chung S, Yao H, Friedman AE, Brookes PS, Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 29.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 31.Roede JR, Stewart BJ, Petersen DR. Decreased expression of peroxiredoxin 6 in a mouse model of ethanol consumption. Free Radic Biol Med. 2008;45:1551–1558. doi: 10.1016/j.freeradbiomed.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 32.Carbone DL, Doorn JA, Kiebler Z, Sampey BP, Petersen DR. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem Res Toxicol. 2004;17:1459–1467. doi: 10.1021/tx049838g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koska J, Spassov VZ, Maynard AJ, Yan L, Austin N, Flook PK, Venkatachalam CM. Fully automated molecular mechanics based induced fit protein-ligand docking method. J Chem Inf Model. 2008;48:1965–1973. doi: 10.1021/ci800081s. [DOI] [PubMed] [Google Scholar]
- 35.Zou XQ, Sun YX, Kuntz ID. Inclusion of solvation in ligand binding free energy calculations using the generalized-born model. J Am Chem Soc. 1999;121:8033–8043. [Google Scholar]
- 36.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieber CS. [Clinical biochemistry of alcohol and its metabolic and hepatic effects] Journ Annu Diabetol Hotel Dieu. 1992:183–210. [PubMed] [Google Scholar]
- 39.Veech RL, Guynn R, Veloso D. The time-course of the effects of ethanol on the redox and phosphorylation states of rat liver. Biochem J. 1972;127:387–397. doi: 10.1042/bj1270387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T, Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 2006;8:E632–E643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuma DJ, Casey CA. Dangerous byproducts of alcohol breakdown--focus on adducts. Alcohol Res Health. 2003;27:285–290. [PMC free article] [PubMed] [Google Scholar]
- 43.Usatyuk PV, Parinandi NL, Natarajan V. Redox regulation of 4-hydroxy-2-nonenal-mediated endothelial barrier dysfunction by focal adhesion, adherens, and tight junction proteins. J Biol Chem. 2006;281:35554–35566. doi: 10.1074/jbc.M607305200. [DOI] [PubMed] [Google Scholar]
- 44.Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: What we know and what we don't. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37:937–945. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Doorn JA, Hurley TD, Petersen DR. Inhibition of human mitochondrial aldehyde dehydrogenase by 4-hydroxynon-2-enal and 4-oxonon-2-enal. Chem Res Toxicol. 2006;19:102–110. doi: 10.1021/tx0501839. [DOI] [PubMed] [Google Scholar]
- 47.Carbone DL, Doorn JA, Petersen DR. 4-Hydroxynonenal regulates 26S proteasomal degradation of alcohol dehydrogenase. Free Radic Biol Med. 2004;37:1430–1439. doi: 10.1016/j.freeradbiomed.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J Pharmacol Exp Ther. 2005;315:8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 49.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirschey MD, Shimazu T, Huang JY, Verdin E. Acetylation of mitochondrial proteins. Methods Enzymol. 2009;457:137–147. doi: 10.1016/S0076-6879(09)05008-3. [DOI] [PubMed] [Google Scholar]
- 52.Shepard BD, Tuma PL. Alcohol-induced protein hyperacetylation: mechanisms and consequences. World J Gastroenterol. 2009;15:1219–1230. doi: 10.3748/wjg.15.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, Fini M, Russo MA. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem. 2009;106:643–650. doi: 10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- 54.Alcain FJ, Villalba JM. Sirtuin inhibitors. Expert Opin Ther Pat. 2009;19:283–294. doi: 10.1517/13543770902755111. [DOI] [PubMed] [Google Scholar]
- 55.Alcain FJ, Villalba JM. Sirtuin activators. Expert Opin Ther Pat. 2009;19:403–414. doi: 10.1517/13543770902762893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.