Abstract
Clinical isolates (n = 63) of Pseudomonas aeruginosa obtained from various sites in 63 horses were compared using ERIC2 RAPD PCR to determine their genetic relatedness. Resulting banding patterns (n = 24 genotypes) showed a high degree of genetic heterogeneity amongst all isolates examined, indicating a relative non-clonal relationship between isolates from these patients, employing this genotyping technique. This study characterised 63 clinical isolates into 24 distinct genotypes, with the largest cluster (genotype E) accounting for 10/63 (15.9%) of the isolates. ERIC2 RAPD PCR proved to be a highly discriminatory molecular typing tool of P. aeruginosa in isolates recovered from horses. With the adoption of several controls to aid reproducibility, this technique may be useful as an alternative to PFGE, particularly in epidemiological investigations of outbreaks where speed may be a significant parameter. This is the first report of clonal heterogeneity amongst P. aeruginosa from horses and demonstrated that ERIC RAPD PCR is a rapid method for the examination of this species in horses, which may be useful in outbreak analysis.
Keywords: equine, ERIC2 RAPD, genotyping, horse, PCR, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa is an important Gram-negative bacterial pathogen in horses, particularly associated with the genital tract [9], although infection due to this causal agent may occur at virtually any anatomical site in the horse. Molecular genotyping of P. aeruginosa is of major importance in the elucidation of transmission routes. Genetic variability of clinical isolates from horses may be compared by examination of phylogenetic distances, which can give an important insight into the interrelationship of these bacterial isolates. Detailed genetic analysis at the subspecies (strain) level gives insights into the variability within a bacterial population and generates evidence on genome evolution, which in turn leads to bacterial adaptation to various environmental conditions. This information can be used in a clinical setting to help identify clinical isolates that occur frequently in animals, so that further work may be undertaken to identify common virulence characteristics in equine strains and help, for example, in directing choice of strains to use in vaccine development.
For the last two decades, PCR-based genotyping methods have played an important role in bacterial typing schemes. One of the PCR-based methods, namely random amplification of polymorphic DNA (RAPD)-PCR, also known as arbitrarily primed-polymerase chain reaction (AP-PCR), has been described to be useful on account of its simplicity and utility for analysis of large throughput samples [4]. This technique utilises a variable short length arbitrary primer, and is advantageous, as it does not require any previous knowledge of the target DNA sequence data. The primer is amplified arbitrarily at low stringency, where the oligonucleotide binds at complementary and partially mismatched sites and generates bands which differ in length and nucleotide composition. RAPD-PCR has been successfully applied for typing a wide variety of both Gram-positive and Gram-negative bacteria. Various genus specific and universal primers have been used, including the enterobacterial repetitive intergenic consensus (ERIC2) sequence and M13 primers, to successfully type both Gram-positive bacteria e.g. MRSA [5] and Gram-negative organisms e.g. P. aeruginosa [2], as a means of rapid strain differentiation and as an alternative to Pulsed-Field Gel Electrophoresis (PFGE). To date, there have been no reports in the literature examining the molecular epidemiology of P. aeruginosa from clinically significant isolates in horses. Therefore, the aim of this study was to examine the genetic relatedness of P. aeruginosa isolated from a variety of sites in Irish horses, over the five year period, 2003-2007, through employment of ERIC2-RAPD PCR.
Materials and methods
Description of isolates employed
P. aeruginosa isolates (n = 63) were obtained from the bacteriological culture archive of the Irish Equine Centre, Johnstown, Naas, Co. Kildare, Ireland. These strains were originally isolated from several sites in horses, including abscess, ear, eye, faecal, genito-urinary, guttural pouch, lung, nasal, pharynx, semen, skin scrapings, throat, tissue, urine and wound, during the five year period, 2003-2007, including 2007 (n = 19 isolates), 2006 (n = 15 isolates), 2005 (n = 17 isolates), 2004 (n = 8 isolates) and 2003 (n = 4 isolates). All isolates were revived from frozen storage and were identified phenotypically employing a combination of conventional identification methods (e.g. oxidase), as well as the API Identification scheme (API 20NE) (BioMérieux, Les Halles, France).
DNA extraction
Genomic DNA was extracted from confirmed isolates of P. aeruginosa. Isolates were cultured on Columbia Blood Agar (Oxoid CM0331) supplemented with 5% (v/v) defibrinated horse blood, for 24 hours at 37°C. All DNA isolation procedures were carried out in a Class II Biological Safety Cabinet (MicroFlow, England) in a room physically separated from that used to set up nucleic acid amplification reaction mixes and also from the 'post-PCR' room. This was performed in accordance with the Good Molecular Diagnostic Procedures (GMDP) guidelines of Millar et al. [6], in order to minimise contamination and hence the possibility of false positive results. Bacterial genomic DNA was extracted from a single colony of P. aeruginosa from each isolate, under standard conditions, by employment of the Roche High Purity PCR Template Preparation Kit (Roche, England), in accordance with the manufacturer's instructions. Extracted DNA was stored at -80°C prior to PCR amplification. For each batch of extractions, a negative extraction control containing all reagents minus organism, was performed, as well as an extraction positive control with P. aeruginosa.
ERIC2 RAPD PCR amplification
All reaction mixes were set up in a PCR hood in a room separate from that used to extract DNA and the amplification and post-PCR room in order to minimise contamination. Initially, PCR amplification conditions were optimised by separately varying magnesium chloride concentration, annealing temperature, primer concentration and DNA template concentration. Following optimisation, reaction mixes (25 μl) were set up as follows: -10 mM Tris- HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 200 μM (each) dATP, dCTP, dGTP and dTTP; 1.25U of Taq DNA polymerase (Amplitaq; Perkin Elmer), 0.1 μM of the RAPD primer, ERIC2 (enterobacterial repetitive intergenic consensus sequence) (5' - AAG TAA GTG ACT GGG GTG AGC G - 3') and 1 μl of DNA template. The reaction mixtures following a 'hot start' were subjected to the following empirically optimised thermal cycling parameters in a Perkin Elmer 2400 thermocycler: 94°C for five minutes, followed by 40 cycles of 94°C for one minute, 50°C for one minute, 72°C for one minute, followed by a final extension at 72°C for 10 minutes. Multiple negative (water) amplification controls were included in every set of PCR reactions. Following amplification, aliquots (10 μl) were removed from each reaction mixture and examined by electrophoresis (80 V, 45 min) in gels composed of 2% (w/v) agarose (Gibco, UK) in TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, pH 8.3), stained with ethidium bromide (5 μg/100 ml). In order to improve resolution of images, resulting PCR amplicons were resolved on 2% (w/v) metaphor agarose (Flowgen Ltd., UK), as described above. Ethidium bromide stained agarose gels were visualised under UV illumination using a gel image analysis system (UVP Products, England) and all images archived as digital graphic files (*.bmp). The genetic relationship between banding patterns was interpreted in accordance with the criteria as defined previously by Tenover et al. [11] and individual ERIC2 genotypes were assigned an arbitrary code based on ascending alphabetic order. Genotypes were assigned a novel status when differing by a one band shift. RAPD-PCR reproducibility was examined with P. aeruginosa isolates on at least two further occasions.
Results
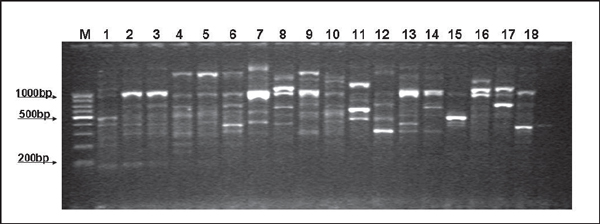
All P. aeruginosa isolates examined in this study generated an ERIC2 RAPD banding pattern ranging in size, from approximately 200 bp to >1,000 bp, with between three and eight bands per isolate, with a mean band number of 5.3 bands per isolate examined (Figure 1). Overall, there was a high degree of clonal heterogeneity between all isolates examined, as 24 distinct RAPD genotypes were recorded (Genotype A-Genotype X) from the 63 isolates examined. Clustering of isolates was observed, where Genotype E was the most frequent cluster with 10 (10/63; 15.9% of total isolates) members, followed by Genotype R (8/63; 12.7%), Genotype C (6/63; 9.5%), Genotype B (5/63; 7.9%) and Genotype V (5/63; 7.0%). These five largest clusters accounted for 34/63 isolates (69.8%) examined in the study. Of the remaining genotypes observed, there were three genotypes each containing three isolates, five genotypes each containing two members and 11 genotypes represented by a single isolate. Certain genotypes with larger numbers of isolates had isolates from several years, i.e., for genotype E with 10 members, these isolates were representative from 2005-2007, whilst for genotype R, isolates here represented 2003-2006. Thus, this study demonstrated the persistence and recurrence of certain genotypes over a four year period within the equine population.
Figure 1.
Representative banding profiles associated with ERIC2 RAPD PCR in P. aeruginosa isolated from horses. Lane M: Molecular weight marker (100 bp); lane 1: genotype A; lanes 2 and 3, genotype B; lanes 4, 5 and 9, genotype C; lane 6, genotype D; lanes 7 and 13, genotype E; lane 8, genotype F; lane 10, genotype G; lane 11, genotype H; lane 12, genotype H; lane 14, genotype J; lane 15, genotype K; lane 16, genotype L; lane 17, genotype M; lane 18, genotype N.
Discussion
In this study, we attempted to examine the genetic relatedness of a collection of 63 clinical isolates of P. aeruginosa, obtained from an equal number of horses throughout Ireland during the five year period, 2003-2007, employing an ERIC RAPD PCR molecular technique. Performing molecular epidemiological studies with such isolate collections is important to help understand acquisition of risk factors for P. aeruginosa and to help trace epidemic strains within such patient populations, as well as to help identify common virulent and/or transmissible strains. Previously, several molecular typing schemes have been applied to help elucidate the molecular epidemiology of populations of P. aeruginosa in human clinical medicine and these methodologies have been reviewed comprehensively by Speert [10], including pulsed-field gel electrophoresis (PFGE). In the current study, we employed a random amplification of polymorphic DNA (RAPD) method for the sub-species differentiation of a population of equine P. aeruginosa, with ERIC2 as the arbitrary primer used, as previously described by Clarke et al. [2], who applied this method to help differentiate P. aeruginosa, which originated from a human clinical population of patients with cystic fibrosis (CF). Our choice of method and primer was largely driven by the previous publication by Clarke et al. [2], as well as Renders et al. [8], who reported positively on the value of this method against pulsed-field gel electrophoresis (PFGE) in a CF study of clinical P. aeruginosa. In their study, these workers demonstrated that RAPD, using the ERIC2 primer, could expect full concordance with typing data obtained by PFGE, when single band differences are neglected. In addition, these workers showed a higher degree of genetic relatedness amongst the 19 CF patients examined, as demonstrated by highly similar banding profiles.
In this study, we have employed the Tenover criteria [11] to help interpret the banding/clustering patterns and hence the interstrain comparison. Although originally designed to help interprete banding patterns associated with PFGE analysis, many workers have adopted the clustering algorithms associated with the Tenover criteria, with other molecular typing methods, such as RAPD analysis. This may not be the most ideal scenario, but at present, RAPD techniques have not got robust clustering algorithms to help with strain comparison. With such an amount of development of methods to aid in the characterisation of P. aeruginosa, it is surprising that similar reports, applying these highly discriminatory molecular methods to collections of P. aeruginosain horses, are absent from the literature. The description of previous typing studies on equine P. aeruginosa is limited to the relatively old report by Atherton and Pitt [1] from 1982, who employed phenotypic serotyping and phage-typing. This study demonstrated that somatic type O3 accounted for 5% of isolates and that all serotypes had the potential to be pathogenic, given favourable culture conditions. Our current manuscript attempts to redress this lack of application of molecular techniques to collections of P. aeruginosa from horses, by applying a simple molecular PCR technique, as a suggested method to other equine microbiologists and others in the field, who wish to examine the genetic relatedness within their equine populations. Consequently, further epidemiological studies are needed from equine centres throughout different continents to help elucidate the molecular epidemiology of P. aeruginosa and its disease association with horses, so that common virulent strains may be identified and standardised protocols for the molecular typing of P. aeruginosa adopted internationally, resulting in a common technique, with added-value, thus aiding in the ability to interpret such typing data on a global basis.
Employment of this primer allowed for the generation of banding patterns containing multiple amplicons, which aided in the differentiation of the isolates. In addition, there was good reproducibility when examining isolates with ERIC2, in that similar banding patterns and hence clusters were obtained for the same isolates examined on different days. This consistency in reproducibility was due to the adoption of several control interventions throughout the study, including: (i) standardisation of genomic DNA template; (ii) employment of single batches of reagents and thermal cycler; (iii) optimisation (iv) standardisation of the RAPD protocol; and, (v) standardisation of image-capture protocol.
Although it has been reported by various groups that RAPD typing is reproducible, historically there have been several reports of problems associated with its reproducibility. Grif et al. [3] reported that RAPD suffered from the lack of standardisation using different methods in different laboratories and this may affect the quality of results. RAPD-PCR can also suffer from the same factors that affect ordinary PCR, including magnesium concentration, primer and reagent 'batch-to-batch' variation and quality of the thermal cycler. Power [7] suggested that for RAPD to be a definitive typing technique, the reproducible generation and interpretation of RAPD fingerprints needs to be developed. To achieve this criterion, the use of an automated system for DNA preparation combined with the use of manufactured RAPD master mixes, as well as use of the same thermal cycler and standard procedures for visualisation of fingerprints, may aid reproducibility. Furthermore, RAPD offers a nucleic acid (DNA) amplification-based molecular alternative to PFGE genotyping.
In conclusion, our study demonstrated that there was a high degree of clonal diversity between isolates within this collection, characterising 63 clinical isolates into 24 distinct genotypes, with the largest cluster (genotype E) accounting for 10/63 (15.9%) of the isolates. ERIC2 RAPD PCR proved to be a highly discriminatory molecular typing tool of P. aeruginosa in isolates recovered in horses. With the adoption of several controls to aid reproducibility, this technique may be useful as an alternative to PFGE, particularly in epidemiological investigations of outbreaks where speed may be a significant parameter.
Acknowledgements
This work was partially funded by the Research and Development Office, HPSS(NI) through ID-RRG Grant 9.3.
References
- Atherton JG, Pitt TL. Types of Pseudomonas aeruginosa isolated from horses. Eq Vet J. 1982;14(4):329–332. doi: 10.1111/j.2042-3306.1982.tb02446.x. [DOI] [PubMed] [Google Scholar]
- Clarke L, Moore JE, Millar BC. et al. Molecular epidemiology of Pseudomonas aeruginosa in adult patients with cystic fibrosis in Northern Ireland. Brit J Biomed Sci. 2008;65(1):18–21. doi: 10.1080/09674845.2008.11732789. [DOI] [PubMed] [Google Scholar]
- Grif K, Karch H, Schneider C. et al. Comparative study of five different techniques for epidemiological typing of Escherichia coli O157. Diag Microbiol Infect Dis. 1998;32(3):165–176. doi: 10.1016/S0732-8893(98)00103-5. [DOI] [PubMed] [Google Scholar]
- Gürtler V, Mayall BC. Genomic approaches to typing, taxonomy and evolution of bacterial isolates. Int J Sys Evol Microbiol. 2001;51(1):3–16. doi: 10.1099/00207713-51-1-3. [DOI] [PubMed] [Google Scholar]
- Kurlenda J, Grinholc M, Jasek K. et al. RAPD typing of methicillin-resistant Staphylococcus aureus: a seven-year experience in a Polish hospital. Med Sci Monitor. 2007;13(6):MT13–18. [PubMed] [Google Scholar]
- Millar BC, Xu J, Moore JE. Risk assessment models and contamination management: implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol. 2002;40(5):1575–1580. doi: 10.1128/JCM.40.5.1575-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power EG. RAPD typing in microbiology - a technical review. J Hosp Infect. 1996;34(4):247–265. doi: 10.1016/S0195-6701(96)90106-1. [DOI] [PubMed] [Google Scholar]
- Renders N, Romling Y, Verbrugh H. et al. Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsedfield gel electrophoresis of DNA macrorestriction fragments. J Clin Microbiol. 1996;34(12):3190–3195. doi: 10.1128/jcm.34.12.3190-3195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper JC, Tibary A. Disease transmission in horses. Theriogenology. 2006;66(3):551–559. doi: 10.1016/j.theriogenology.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Speert DP. Molecular epidemiology of Pseudomonas aeruginosa. Front Biosci. 2002;7:e354–361. doi: 10.2741/speert. [DOI] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV. et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]