Meiotic Scc2 recruits cohesin to the chromosome for two important functions: activation of gene expression and mediation of sister-chromatid cohesion. This study reveals that cohesin positively regulates transcription in a position-dependent manner. Therefore cohesin can also act as a transcriptional activator in budding yeast.
Abstract
To tether sister chromatids, a protein-loading complex, including Scc2, recruits cohesin to the chromosome at discrete loci. Cohesin facilitates the formation of a higher-order chromosome structure that could also influence gene expression. How cohesin directly regulates transcription remains to be further elucidated. We report that in budding yeast Scc2 is required for sister-chromatid cohesion during meiosis for two reasons. First, Scc2 is required for activating the expression of REC8, which encodes a meiosis-specific cohesin subunit; second, Scc2 is necessary for recruiting meiotic cohesin to the chromosome to generate sister-chromatid cohesion. Using a heterologous reporter assay, we have found that Scc2 increases the activity of its target promoters by recruiting cohesin to establish an upstream cohesin-associated region in a position-dependent manner. Rec8-associated meiotic cohesin is required for the full activation of the REC8 promoter, revealing that cohesin has a positive feedback on transcriptional regulation. Finally, we provide evidence that chromosomal binding of cohesin is sufficient for target-gene activation during meiosis. Our data support a noncanonical role for cohesin as a transcriptional activator during cell differentiation.
INTRODUCTION
Chromosome organization determines a wide array of cellular activities that include gene expression, replication, and DNA damage repair (Misteli, 2007). During cell differentiation, for example, modifications of the chromatin accompany epigenetic regulation of gene expression that is necessary for cell-fate change. Chromosome-associated factors, including nonhistone proteins, play pivotal roles in modifying chromatin and gene expression (Fraser and Bickmore, 2007; Phillips and Corces, 2009), yet their exact role and mode of action remain to be elucidated.
A highly conserved protein complex, cohesin, is recruited to the chromosome at late G1 phase to mediate cohesion between duplicated sister chromatids at S phase (Onn et al., 2008; Nasmyth and Haering, 2009). Originally identified in budding yeast, cohesin is composed of four subunits called Smc1, Smc3, Mcd1/Scc1, and Irr1/Scc3, which form the cohesin ring (Guacci et al., 1997; Michaelis et al., 1997). Rec8 largely replaces Mcd1 after cells commit to meiosis (Klein et al., 1999; Parisi et al., 1999; Watanabe and Nurse, 1999), demonstrating that cohesins are specific to cell type. Orthologues of cohesin subunits have been found in all eukaryotes investigated (Losada et al., 1998; Sumara et al., 2000; Hirano, 2006). A conserved protein complex, consisting of Scc2 and Scc4, is required for recruiting cohesin to the chromosome to tether sister chromatids together (Ciosk et al., 2000; Tomonaga et al., 2000). Cohesin is necessary for proper chromosome condensation and has been proposed to facilitate chromatin loop formation (Guacci et al., 1997; Novak et al., 2008).
Genome-wide and large-scale mapping shows that cohesin binds to the chromosome at discrete loci in yeast and vertebrate cells (Glynn et al., 2004; Lengronne et al., 2004; Parelho et al., 2008; Rubio et al., 2008; Wendt et al., 2008), although the exact mechanisms of cohesin recruitment in yeast and humans may differ. Mutational analysis in yeast demonstrates cohesin's primary role in generation of sister-chromatid cohesion, including S-phase cohesion and cohesion triggered by DNA double-strand break (DSB) repair (Onn et al., 2008; Nasmyth and Haering, 2009; Sjogren and Strom, 2010). In addition, cohesin association helps establish chromosomal boundaries. In both budding and fission yeasts, cohesin binds to the cryptic mating locus and helps restrict the spread of the gene-silencing information (Donze et al., 1999; Lau et al., 2002; Nonaka et al., 2002). On the other hand, vertebrate cohesins have been localized to the chromosome at numerous CTCF binding sites (Parelho et al., 2008; Rubio et al., 2008; Wendt et al., 2008), which can serve as insulators that block the interaction between an enhancer and the corresponding promoter (Phillips and Corces, 2009). The physical interaction between cohesin and CTCF implies a possible role for cohesin in transcriptional regulation, perhaps in mediation of long-range interchromatin or intrachromatin interactions that help organize the chromosome into distinctive functional domains (Hadjur et al., 2009; Nativio et al., 2009; Hou et al., 2010).
A few lines of evidence of cohesin function in development support a role for cohesin in gene expression. For example, axon pruning during mushroom-body neuron development requires cohesin activity in Drosophila (Pauli et al., 2008; Schuldiner et al., 2008). In flies, cohesin and the loading factor component Scc2 (called Nipped-B) have opposing effects on the expression of cut and other homeobox genes (Rollins et al., 1999, 2004), possibly through their separate roles in mediating enhancer and promoter interactions (Dorsett, 2009). Cleavage of cohesin subunit Rad21 causes transcriptional changes in fly salivary glands (Pauli et al., 2010). In zebrafish, cohesin acts as a positive regulator of the expression of the runx genes that are required for cell differentiation (Horsfield et al., 2007). Nonlethal mutations in genes that encode Smc1, Smc3, and Scc2 have been mapped in a human developmental disorder called Cornelia de Lange syndrome (Tonkin et al., 2004; Musio et al., 2006; Deardorff et al., 2007). It is intriguing that a yeast scc2 mutant that mimics the human mutation shows altered GAL gene expression and chromosome organization (Gard et al., 2009). Together, these observations suggest that, in addition to establishing chromosome structure for segregation during cell division, cohesin plays a noncanonical role in regulation of gene expression during cell differentiation and development, but direct evidence is lacking.
To explore the role of cohesin in cell differentiation, we took advantage of yeast meiosis, in which vegetative yeast cells can be easily induced to switch to the sexual reproductive program. Using a molecular-genetics approach, we depleted the cohesin loader Scc2 right before yeast cells were induced to undergo meiosis. We found that meiotic Scc2 is required for recruiting cohesin to the chromosome not only for generating sister-chromatid cohesion, but also for activating meiotic gene expression. Our work reveals that cohesin can act as a transcriptional regulator and provides insights into its epigenetic role in cell differentiation and development in higher eukaryotes, including humans.
RESULTS
Scc2 localizes to meiotic chromosomes, and its binding sites highly overlap with the cohesin-associated regions
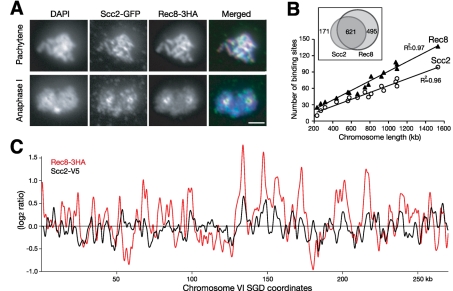
To investigate chromosome association of Scc2 during yeast meiosis, we localized Scc2 and Rec8 on surface-spread nuclei by indirect immunofluorescence (Figure 1A). At pachytene, where homologues are paired and synapsed, Scc2, like Rec8, was localized along the length of the chromosome (Figure 1A). The Scc2 signal appeared to be less continuous but overlapped with that of Rec8 (Figure 1A, upper right). This colocalization was also evident at anaphase I, when both Scc2 and Rec8 were enriched at the centromeres that were clustered around the poles (Figure 1A). These data suggest that Scc2 largely colocalizes with cohesin on meiotic yeast chromosomes.
FIGURE 1:
Chromosome association of Scc2 and Rec8 during yeast meiosis. (A) Immunofluorescence of Scc2-GFP and Rec8'3HA (strain HY2020). Yeast cells were induced to undergo synchronous meiosis, and nuclear surface spreads were prepared and stained with GFP and HA antibodies. Two representative stages, pachytene and anaphase I, are shown. Red, Rec8'3HA; green, Scc2-GFP; blue, DAPI. Bar, 2 μm. (B) The number of chromosome-associated regions of Scc2 and Rec8 was proportional to chromosome length. Chromatin immunoprecipitation combined with microarray was used to identify Scc2 and Rec8 chromosomal binding during meiosis (strain HY1644). Inset shows the overlap of the Scc2 and Rec8 binding sites. (C) Visual representation of chromosome association profile of Scc2 and Rec8 during yeast meiosis. The entire chromosome VI is shown as a representative. The scale of the y-axis is log2 ratio of immunoprecipitation to input. SGD coordinates of chromosome VI are shown at the bottom. Red, Rec8 ChIP; black, Scc2 ChIP.
Because cohesin binds to chromosomes at specific locations called cohesin-associated regions (CARs) (Blat and Kleckner, 1999; Laloraya et al., 2000), we asked whether meiotic Scc2 also binds to these chromosome addresses. We combined chromatin immunoprecipitation with a high-resolution tiling microarray to map both Scc2 and Rec8 binding sites throughout the yeast genome in cells that were staged at pachytene (Figure 1, B and C). A total of 792 Scc2 binding sites were identified, defined by a 1.8-fold or greater enrichment of Scc2 on the meiotic chromosome (Figure 1B). The number of Rec8 sites identified by the same threshold and statistical algorithm is 1116 (Figure 1B), a result consistent with our previous finding (Glynn et al., 2004). Like those of the CARs, the number of Scc2 binding sites on each chromosome had a linear correlation with chromosome length; intervals of ∼15 kb separate adjacent Scc2 binding sites (Figure 1B). More than 78% of the Scc2 sites identified from the yeast genome had corresponding Rec8 sites at the same chromosomal locations (Figure 1B). Taken together, these findings show that meiotic Scc2 binds to chromosomal addresses similar to those where cohesin binds, supporting a role for Scc2 as the cohesin loader.
Scc2 is required for nuclear division and sister-chromatid cohesion during meiosis
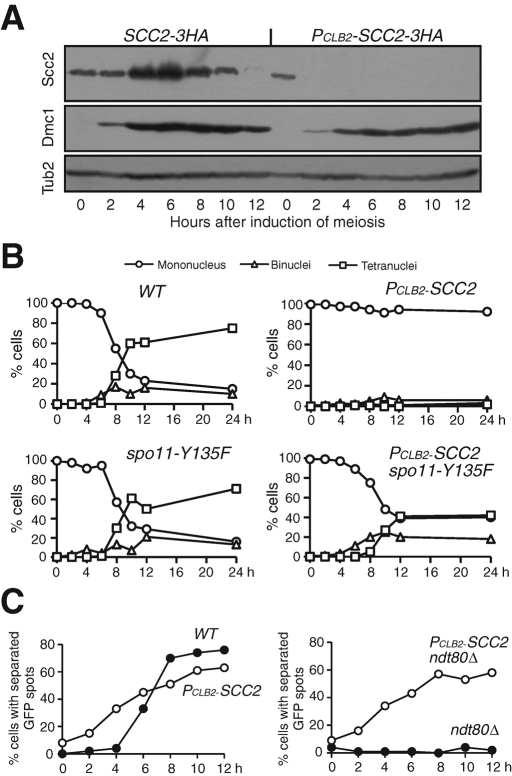
To address the meiotic function of Scc2, we generated a PCLB2-SCC2 allele, which specifically depleted the Scc2 protein in cells induced to undergo meiosis (Figure 2A). Because the meiosis-specific protein Dmc1 showed a similar level in wild-type and PCLB2-SCC2 cells after induction of meiosis (Figure 2A), we conclude that Scc2-depleted yeast cells were able to enter into the meiotic transcriptional program after conventional induction by nitrogen depletion. A 4’,6-diamidino-2-phenylindole (DAPI) staining assay of meiotic nuclear division revealed that ∼90% of the wild-type cells had completed meiosis I nuclear division within 12 h after induction (Figure 2B). In contrast, <5% of cells lacking Scc2 underwent nuclear division (Figure 2B). This lack of nuclear division is reminiscent of the phenotype of cohesin mutant rec8Δ, in which the recombination checkpoint is activated and arrests cells at prophase I (Klein et al., 1999). Indeed, Rad51 foci accumulate in Scc2-depleted meiotic cells (Supplemental Figure 1), suggesting that meiotic DSBs are formed but not repaired. We introduced a spo11 mutation to abolish the formation of meiotic DSBs, thus bypassing the recombination checkpoint in these cells. More than 70% of the PCLB2-SCC2 spo11-Y135F double-mutant cells could complete at least one nuclear division (Figure 2B), demonstrating that Scc2 is required for proper DSB repair and cell progression during meiosis.
FIGURE 2:
Scc2 is required for nuclear division and sister-chromatid cohesion during meiosis. (A) Depletion of Scc2 during yeast meiosis. Wild-type (3050) and PCLB2-SCC2 (HY1336) cells were induced to undergo synchronous meiosis, aliquots were withdrawn at indicated time points, and samples were prepared for immunoblot. Scc2-3HA was detected by an anti-HA antibody (16B12); Dmc1 was detected by a Dmc1-specific polyclonal antibody. Dmc1 is a meiosis-specific protein, indicating meiosis induction. The level of β-tubulin served as a loading control. (B) Nuclear division. Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at indicated time points and fixed in 1% formaldehyde for 1 h. Cells were stained with DAPI; nuclear division was determined by fluorescence microscopy. WT, NH144; PCLB2-SCC2, HY1336; spo11-Y135F, HY1499; PCLB2-SCC2 spo11-Y135F, HY1975. (C) Sister-chromatid cohesion assay. A tandem array of tetO was inserted at the URA3 locus of one homologue of chromosome V, and the separation of GFP dots was determined by fluorescence microscopy. WT, HY1294C; PCLB2-SCC2, HY2113; ndt80Δ, HY2130; PCLB2-SCC2 ndt80Δ, HY2115.
A role for meiotic Scc2 in regulating sister-chromatid cohesion was not known previously; we therefore performed a cohesion assay by marking the centromere of one homologue of chromosome V with tetO/tetR-GFP (Michaelis et al., 1997) (Figure 2C). In wild-type cells, duplicated sister chromatids were cohesive, forming a single green fluorescent protein (GFP) dot until sisters separated during meiosis I after ∼6 h after induction, forming two GFP dots (Figure 2C). In contrast, Scc2-depleted meiotic cells showed premature loss of sister chromatid cohesion; cells with two GFP dots appeared much earlier (Figure 2C). Furthermore, we arrested the cells at pachytene by deleting the NDT80 gene, which encodes a transcription factor required for the activation of middle and late meiotic genes (Xu et al., 1995). In ndt80Δ otherwise wild-type cells, a single GFP dot was observed, suggesting that sister chromatids remained cohesive when cells were blocked at pachytene (Figure 2C). More than 60% of PCLB2-SCC2 ndt80Δ cells showed two GFP dots 12 h after induction (Figure 2C). These observations establish experimentally that Scc2 is indeed required for sister-chromatid cohesion during meiosis.
Rec8 protein levels are dramatically lower in Scc2-depleted meiotic cells
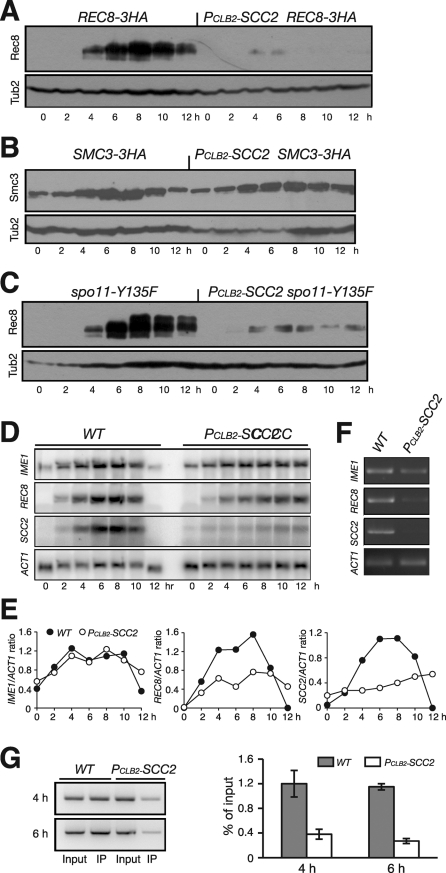
We next examined the effect of Scc2 depletion on the accumulation of the meiotic cohesin subunit Rec8, of which the encoding gene is induced for expression only after yeast cells commit to meiosis (Chu et al., 1998). Immunoblot analysis revealed that Rec8 protein levels were dramatically lower in Scc2-depleted cells than in cells showing the normal pattern of accumulation (Figure 3A). In wild-type cells, for comparison, Rec8 was detected 2 h after induction of meiosis, peaked around 6 h, and then was degraded before the completion of meiosis at ∼12 h (Figure 3A). In contrast, Rec8 was present at a much lower level in Scc2-depleted cells; only a trace amount could be detected by immunoblot (Figure 3A, right). Compared with those of the wild type, the protein levels of other cohesin subunits, for example Smc3, were not dramatically changed in Scc2-depleted meiotic cells (Figure 3B). Together, our data suggest that Scc2 is specifically required for maintaining normal Rec8 protein levels during meiosis.
FIGURE 3:
Scc2 is required for REC8 gene expression. (A) Rec8 protein level in wild-type (HY1503C) and PCLB2-SCC2 (HY1586) cells. Yeast cells were induced to undergo synchronous meiosis, then subjected to immunoblot as for Figure 2A. Rec8'3HA was detected by an anti-HA antibody (16B12). (B) Smc3 protein level during meiosis. Note that the levels of Smc3 remain normal in the absence of Scc2 in meiosis. SMC3-3HA, HY1750C; PCLB2-SCC2 SMC3-3HA, HY1750. (C) Rec8 protein level in spo11Δ cells during meiosis. Note that, in the double mutant PCLB2-SCC2 spo11Δ (HY1975), only residual Rec8-3HA was detected. (D) Northern blot showing the levels of IME1, REC8, SCC2, and ACT1 transcripts during meiosis. Wild-type (NH144) and PCLB2-SCC2 (HY1279) cells were induced to undergo synchronous meiosis; aliquots were withdrawn at indicated time points. Total RNA was extracted and prepared for Northern blots. Gene-specific probes sequentially probed the same blots. The level of ACT1 served as a loading control. (E) Quantification of Northern blots. The ratio of the gene of interest to ACT1 is shown. Wild type, filled circles; PCLB2-SCC2, open circles. (F) RT-PCR assay of transcripts. Aliquots were withdrawn 6 h after induction of meiosis. Total mRNA was extracted and reverse transcribed to cDNA. A semiquantitative PCR was used to amplify target cDNA with gene-specific primers. (G) ChIP of Pol II from WT (NH144) and PCLB2-SCC2 (HY1279) cells during meiosis. Yeast cells were induced to undergo synchronous meiosis, and ChIP was performed with an anti-Pol II CTD antibody (8WT16). Semiquantitative PCR was used to determine Pol II enrichment at the REC8 locus. Two representative time points are shown.
Scc2 is required for REC8 mRNA production
One interpretation of the foregoing result is that Rec8 is only fully produced after prophase I, at which PCLB2-SCC2 cells are arrested by the recombination checkpoint (Figure 2B). To test this hypothesis, we determined Rec8 protein levels in cells that bypass the recombination checkpoint because of the loss of Spo11 function (Figure 2B). We found that Rec8 remained low in Scc2-depleted cells, even though the majority of these cells could complete at least one meiotic nuclear division (Figures 2B and 3C).
Because sister-chromatid cohesion facilitates the formation of a higher-order chromosome structure that influences gene expression, an alternative explanation is that perhaps Scc2 exerts its effect on Rec8 at the transcriptional level. We performed Northern blots to determine the level of REC8 mRNA after inducing the cells to undergo meiosis (Figure 3, D and E). The meiosis-specific transcription factor IME1 is an early gene that governs meiotic entry (Kassir et al., 1988). The IME1 mRNA levels, which served as a positive control, were comparable in the wild type and the PCLB2-SCC2 mutant (Figure 3, D and E). This observation, along with the DMC1 expression pattern (Figure 2A), is consistent with the idea that Scc2-depleted cells are competent for induction of meiosis. In contrast, the level of REC8 mRNA decreased more than 50% in Scc2-depleted cells during meiosis (Figure 3E). In addition, we performed RT-PCR to confirm the decrease in REC8 transcripts in PCLB2-SCC2 cells (Figure 3F) and chromatin immunoprecipitation of the Pol II CTD to demonstrate lower Pol II binding to the REC8 gene during meiosis (Figure 3G). These data strongly suggest that Scc2 is required for a normal level of REC8 gene transcription during meiosis.
Our Northern blots showed that only a background level of SCC2 mRNA was present in PCLB2-SCC2 cells (Figure 3, D and E), demonstrating that the CLB2 promoter fully inactivated the expression of SCC2 during meiosis. On the other hand, although REC8 mRNA was significantly reduced, a basal level remained in PCLB2-SCC2 cells (Figure 3D), indicating that REC8 transcription was not completely abolished in Scc2-depleted meiotic cells.
Scc2 enhances REC8 promoter activity during meiosis
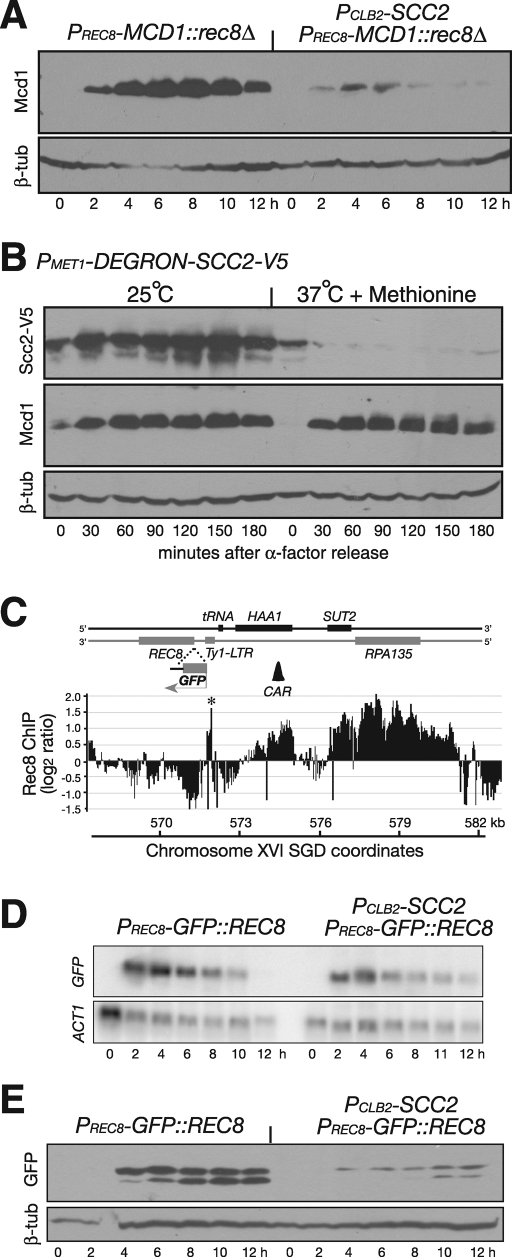
Having shown that Scc2 is required for both REC8 mRNA and protein accumulation during meiosis, we next determined whether Scc2 activates the REC8 promoter during meiosis. We replaced the REC8 open reading frame with that of MCD1, which encodes the mitotic kleisin subunit, the counterpart of Rec8 (Buonomo et al., 2000). When the MCD1 open reading frame was placed at the endogenous REC8 locus, the protein level of Mcd1 mirrored that of Rec8, such that only a trace amount of Mcd1 was detected in Scc2-depleted meiotic cells by immunoblot (Figure 4A). This result further suggests that Scc2 modulates the activity of the REC8 promoter. To address whether Scc2 has a similar role in transcriptional activation of the MCD1 gene that resides in its endogenous locus in vegetative cells, we generated a DEGRON allele of SCC2 to deplete the Scc2 protein in cells released from G1-phase arrest (Figure 4B). The Mcd1 protein was produced at similar levels in vegetative cells with and without Scc2 (Figure 4B). Therefore Scc2 is required for activation of REC8 gene expression in meiosis but not for MCD1 in mitosis. These results also indicate that the kleisin subunits (Mcd1 and Rec8) remain stable in Scc2-depleted cells if they are produced.
FIGURE 4:
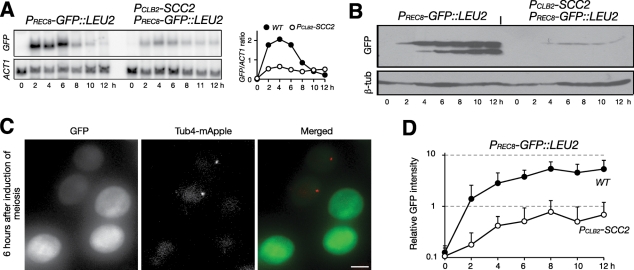
The specificity of Scc2 to REC8 promoter activation. (A) Ectopic expression of MCD1 in meiosis. The MCD1 open reading frame was inserted at the REC8 locus and was under the control of the endogenous REC8 promoter and its 5′ upstream sequence. Yeast cells were induced to undergo synchronous meiosis, and samples are prepared for immunoblot as for Figure 2A. An Mcd1-specific antibody was used to determine the Mcd1 protein level. Strains used: HY2500 and HY2502. (B) Depletion of Scc2 in vegetative cells. A DEGRON-SCC2 (HY1869) construct is under the control of the MET1 promoter. Yeast cells were arrested at G1 with α-factor; the culture was split into two and released to 25°C and 37°C simultaneously. Protein extracts were prepared for immunoblots. Note that the Mcd1 protein levels remained normal in Scc2-depleted vegetative cells. The level of β-tubulin served as a loading control. (C) A heterologous reporter assay of the REC8 promoter. The GFP open reading frame was inserted at the REC8 locus as in A. Black bars show cohesin enrichment on the chromosome as determined by ChIP on chip. The scale of the y-axis is log2 ratio of immunoprecipitation to input. SGD coordinates of chromosome XVI are shown at the bottom. Asterisks mark the long-terminal repeat (LTR) sequence as putative peaks because of its repetitive nature. The first CAR that is 5′ upstream of the REC8 promoter is depicted as a black triangle (∼3 kb away). (D) Meiotic expression of PREC8-GFP::REC8 (HY2203 and HY2107). Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at indicated time points. Samples were prepared for northern blots as for Figure 3D and probed with GFP- and ACT1-specific probes. (E) GFP protein level by immunoblot. Yeast cells were induced to undergo synchronous meiosis as in C. The GFP protein was detected by an anti-GFP antibody. This antibody recognizes a major GFP band and a minor one with a lower molecular weight.
To analyze further the interaction between Scc2 and the REC8 promoter, we developed a GFP-based heterologous reporter assay using the PREC8-GFP construct (Figure 4C; also see later discussion). When this construct was placed at the REC8 locus, at which the expression of GFP was under the control of the endogenous REC8 promoter and its 5′ upstream regulatory sequence, GFP was expressed after cells were induced to undergo meiosis (Figure 4, C–E). The levels of GFP mRNA and protein were lower when Scc2 was absent (Figure 4, D and E). These results confirm that Scc2 plays a role in the activation of the REC8 promoter during meiosis and demonstrate that GFP production can serve as a reliable indicator of promoter activation.
Transcriptional activation by Scc2 is specific to the REC8 promoter but is not restricted to the REC8 locus
The heterologous PREC8-GFP reporter permitted us to map the 5′ upstream DNA sequences required for activating the REC8 promoter during meiosis (Figure 5). We found that the 698–base pair DNA sequence upstream of the REC8 start codon was sufficient to promote the transcription of the GFP gene when the PRec8'698-GFP construct (Figure 5A) was inserted at the LEU2 locus on chromosome III (Figure 5). The REC8 promoter contains the URS1 sequence commonly seen in early-induced meiotic genes (Buckingham et al., 1990). Addition of the 766–base pair sequence, including the Ty-LTR, which is upstream of the REC8 promoter (Figure 5A, PRec8'1297-GFP), dramatically increased the transcription of the GFP reporter as assayed indirectly by fluorescence production (Figure 5, C and D), but further addition of upstream sequence, including the tRNAGly (tG(GCC)P1) gene, did not increase GFP expression; rather, it reduced it (Figure 5, A and D, PRec8'2158-GFP). Crucially, the expression of the PRec8'1297-GFP construct inserted at the LEU2 locus was subject to the same regulation by Scc2 (Figure 6). With Northern blots, we observed a more than 75% reduction of GFP transcription in Scc2-depleted cells (Figure 6A). A correspondingly dramatic reduction of the GFP protein was observed in mutant cells by immunoblot and fluorescence microscopy (Figure 6, B–D). Our data therefore show that Scc2 is required for the activation of the REC8 promoter and that its activation is not restricted to the REC8 locus.
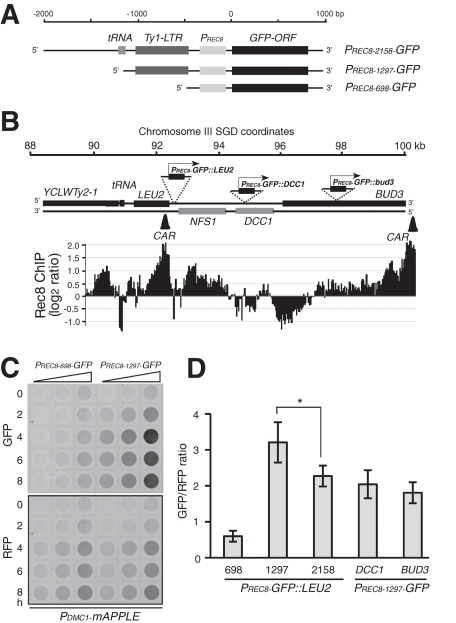
FIGURE 5:
Positional effect on REC8 promoter activation during meiosis. (A) A schematic diagram showing the three PREC8-GFP constructs. The only difference among them is the length of the 5′ upstream sequence. (B) Chromosomal location of the PREC8-GFP constructs. A schematic diagram showing the gene structure at the LEU2 locus on chromosome III. The location and orientation of the PREC8-GFP constructs are marked. Black bars show cohesin enrichment on the chromosome as determined by ChIP on chip. The scale of the y-axis is log2 ratio of immunoprecipitation to input. Two CARs in this region are depicted as black triangles. (C) GFP detected by a fluorescence scanner. Yeast cells were induced to undergo meiosis, and an aliquot was withdrawn at the indicated time, fixed in 1% formaldehyde, 1:2 serially diluted, and scanned with a fluorescence scanner. All strains harbored one copy of PDMC1-mAPPLE (RFP), which served as an internal control. Representative images are shown. (D) Quantitative analysis of PREC8-GFP gene expression. Three constructs were inserted at the LEU2 locus: PRec8'698-GFP (HY2666), PRec8'1297-GFP (HY2665), and PRec8'2158-GFP (HY2664) as shown in A. PREC8-GFP::DCC1 (HY2953) and PREC8-GFP::BUD3 (HY2952) were inserted at the DCC1 and BUD3 loci, respectively. GFP/RFP ratios were derived from samples at the 4-, 6-, and 8-h time points. Error bars show SD. *p value < 0.05, Student's t test.
FIGURE 6:
A reporter assay showing that Scc2 modulates REC8 promoter activity. (A) Meiotic expression of PREC8-GFP::LEU2 (HY2157 and HY2228). Yeast cells were induced to undergo meiosis, and samples were prepared for Northern blots as in Figure 3D. Quantification is shown to the right. (B) GFP protein level by immunoblot. Note that only minimal GFP protein can be detected in Scc2-depleted cells. (C, D) A quantitative-microscopy method of determining GFP level in live meiotic cells. Wild-type (HY2157) and PCLB2-SCC2 (HY2301) cells were mixed and induced to undergo meiosis, and 2-μl aliquots were withdrawn at indicated time points and mounted on a microscope slide for fluorescence microscopy. The PCLB2-SCC2 cells harbor one copy of γ-tubulin–mApple, which distinguishes them from the wild-type cells in the same microscopy field. Projected images from 14 Z-stacks are shown. Exposure for each section is 80 ms. Quantification of GFP intensity from each strain is shown in D. Error bars show SD. At least 50 cells were analyzed at each time point. Wild type, filled circles; PCLB2-SCC2, open circles. Bar, 4 μm.
The specificity of Scc2 in activating the REC8 promoter was further demonstrated when a PDMC1-GFP construct was inserted at the same LEU2 locus (Supplemental Figure 2A). Unlike that of the PREC8-GFP::LEU2 construct, the expression of PDMC1-GFP::LEU2 appeared unaltered in Scc2-depleted cells during meiosis (Supplemental Figure 2B), consistent with our observation that the Dmc1 protein level remained normal in PCLB2-SCC2 cells (Figure 2A). Likewise, the PDMC1-REC8 construct produced similar levels of Rec8 with or without Scc2 in meiosis (Supplemental Figure 2C). Taken altogether, our data suggest that the activation of the REC8 promoter is controlled by a combination of cis-acting DNA elements and trans-acting meiosis-specific factors during meiosis.
Positional effect of a CAR on REC8 promoter activation
Scc2 and cohesin bind to the chromosome at CARs (Figure 1). We therefore inspected the DNA sequences at both the REC8 and the engineered LEU2 loci and found that at each locus an adjacent cohesin-associated region is positioned at the 5′ upstream of the REC8 promoter, with ∼2 kb at LEU2 and ∼3 kb at the endogenous REC8 locus (Figures 4C and 5B). Because the expression level of PREC8-GFP is about twofold higher at the LEU2 locus than at the REC8 locus (Figures 4E and 6A), we hypothesized that the distance from the REC8 promoter to its 5′ upstream CAR is inversely correlated with promoter activation. This reasoning is supported by the observation that PRec8'1297-GFP::LEU2 has a significantly higher expression level than PRec8'2158-GFP::LEU2; the two differ in that the REC8 promoter of the latter is positioned 861 base pairs further downstream of the cohesin-binding site at LEU2 (Figure 5D). In addition, we placed the GFP construct at two positions further downstream of the LEU2 CAR—at the DCC1 locus (PREC8-GFP::DCC1, ∼5 kb from the LEU2 CAR) and at the BUD3 locus (PREC8-GFP::BUD3, ∼8 kb from the LEU2 CAR) (Figure 5), and found a reduction of GFP production in both (Figure 5D). Together, these data suggest that the CAR at the LEU2 locus has a positional effect on REC8 promoter activation.
Positive feedback control of REC8 promoter by meiotic cohesin
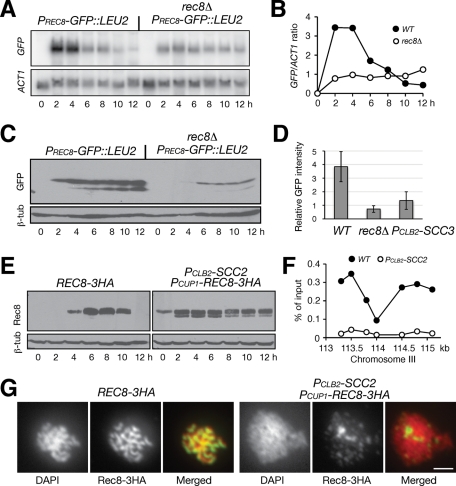
If a cohesin-associated region acts as a transcriptional regulatory sequence, Rec8-associated cohesin could be the trans-acting factor that activates the REC8 promoter. To test this hypothesis, we assayed the transcriptional activity of PREC8-GFP::LEU2 in rec8Δ cells (Figure 7A). As predicted, GFP transcription was decreased by 75% in the absence of Rec8 during meiosis (Figure 7, A and B). Accordingly, the level of GFP protein decreased to one-fifth of the level in the wild type (Figure 7, C and D), suggesting a positive feedback control by Rec8 of its own promoter. As in Scc2-depleted cells, a low basal level of GFP expression remained in rec8Δ cells (Figure 7, A and B), demonstrating that the REC8 promoter remained minimally active in the absence of meiotic cohesin. Because REC8 expression was also low in an scc3 mutant in meiosis (Lin et al., 2011), and because PREC8-MCD1 was expressed in the absence of Rec8 and suppressed rec8Δ phenotype in REC8 promoter activation (Figure 4A), our data suggest that cohesin acts as a transcriptional activator at the REC8 promoter.
FIGURE 7:
Feedback control of cohesin on REC8 promoter. (A) Rec8 activates its own promoter. Yeast cells were induced to undergo meiosis, and samples were prepared for Northern blot as for Figure 3D. PREC8-GFP::LEU2 (HY2157); PREC8-GFP::LEU2 rec8Δ (HY2229). (B) Quantification of Northern blots. Wild type, filled circles; rec8Δ, open circles. (C) GPF protein level by immunoblot. Cells were induced to undergo synchronous meiosis as for A, and protein extracts were prepared for immunoblot. (D) GFP production in live cells. Aliquots were withdrawn 6 h after induction of meiosis. GFP intensity was measured by fluorescence microscopy as for Figure 6D. PCLB2-SCC3, HY2285. (E) Ectopic production of Rec8 in Scc2-depleted cells. To induce PCUP1-REC8 (HY2122) expression, 100 μM CuSO4 was added to the culture medium after induction of meiosis. (F) ChIP of Rec8 at centromere III. Chromosome III SGD coordinates are shown on the x-axis. Ratio of immunoprecipitation to input is shown on the y-axis. Rec8'3HA, filled circles; PCUP1-Rec8'3HA, PCLB2-SCC2, open circles. (G) Rec8 localization on meiotic chromosomes. Yeast cells were induced for synchronous meiosis and nuclear spreads were prepared for immunofluorescence as for Figure 1A. Bar, 2 μm.
To determine whether additional genes are subjected to cohesin regulation during meiosis, we surveyed the global gene-expression pattern using the expression microarray (Supplemental Figure 3). A total of 204 genes are repressed by more than 50% and 85 genes by 75% in rec8Δ cells 6 h after induction of meiosis; no one gene showed a fourfold increase (Supplemental Figure 3A). Of the 85 genes, we confirmed that 82 were repressed by at least 50%, and 59 by 75%, in the spo11 rec8Δ double mutant (unpublished data). If the 85 genes were randomly positioned between two adjacent CARs, which are ∼11 kb apart on average throughout the yeast genome, we would expect a distribution of roughly 7.7 genes/kb. In contrast, 53 (62%) are positioned within 4 kb 5′ downstream of a corresponding CAR (Supplemental Figure 3B). Our survey of the 40 early meiotic genes (Chu et al., 1998) that were not subjected to cohesin regulation showed a random distribution of CARs (Supplemental Figure 3C). Therefore our data suggest an inverse relationship between the activation of the cohesin target promoter and its distance from the 5′ upstream CAR.
Scc2 is required for loading meiotic cohesin to the chromosome to activate gene expression
To determine whether Scc2 is required for loading cohesin onto the chromosome during meiosis, we engineered a PCUP1-REC8 construct to produce Rec8 in Scc2-depleted cells (Figure 7E). On the addition of CuSO4 to the meiotic culture, PCUP1-REC8 was expressed and produced Rec8 at a level comparable to that in wild-type cells (Figure 7E). To determine Rec8's chromosome association, we performed Rec8 chromatin immunoprecipitation (ChIP) combined with a quantitative PCR method (Figure 7F). A representative experiment showed that ectopically produced Rec8 failed to bind to the centromere III region in Scc2-depleted meiotic cells (Figure 7F). In addition, we performed immunofluorescence to localize Rec8 on surface-spread yeast nuclei and found that Rec8 was only minimally localized to meiotic chromosomes without Scc2 (Figure 7G). We therefore conclude that Scc2 is required for loading meiotic cohesin to the chromosome. The observations also imply that Scc2 binds to the meiotic chromosome before cohesin. Indeed, meiotic Scc2 remained chromosome bound in rec8Δ cells, as revealed by indirect immunofluorescence (Supplemental Figure 4). Taken together, our data suggest that meiotic Scc2 loads cohesin onto the chromosome.
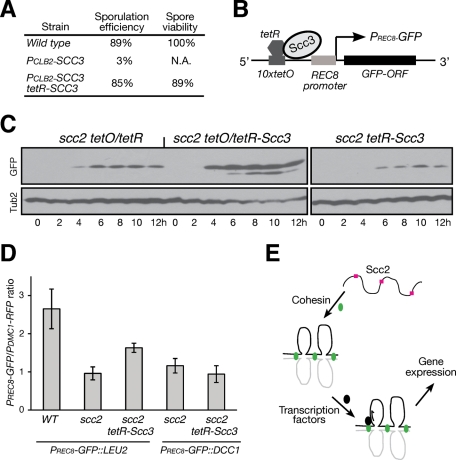
To determine whether chromosomal loading of cohesin is sufficient for gene activation, we reasoned that forced localization of cohesin to the chromosome would increase gene expression in Scc2-depleted cells. We constructed a functional tetR-SCC3 fusion allele because it suppressed a meiosis-specific Scc3-depletion allele (Figure 8A). Using the specific interaction between tetO and tetR, we tethered Scc3, and therefore cohesin, to the 10xtetO sequence, which was positioned <2 kb 5′ upstream of the REC8 promoter (Figure 8B). By assaying the production of Rec8-3HA, we found that forced localization of cohesin to the 5′ upstream sequence of the endogenous REC8 promoter increased Rec8 production (Supplemental Figure 5A). Similarly, forced localization of cohesin to the 5′ upstream sequence of the PREC8-GFP construct positioned at the LEU2 locus significantly increased GFP production in Scc2-depleted cells (Figure 8, C and D). Forced localization of Scc3 alone, however, was not sufficient to activate the REC8 promoter in Smc1-depleted cells (Supplemental Figure 5, B and C). In addition, when PREC8-GFP was positioned at the DCC1 locus that is ∼6 kb downstream of the 10xtetO sequence, its expression level did not increase in Scc2-depleted cells even in the presence of tetR-Scc3 (Figure 8D). Therefore activation of the REC8 promoter depends on a functional cohesin complex positioned adjacent to its 5′ upstream sequence. These results provide direct evidence that cohesin binding to the chromosome is both necessary and sufficient for activation of the REC8 promoter during meiosis.
FIGURE 8:
Activation of REC8 promoter by forced localization of cohesin. (A) Generating a functional tetR-SCC3 fusion allele. Yeast strains (NH144, 3200, and HY2636) were sporulated, and tetrads were dissected for determination of spore viability. We used the CLB2 promoter to replace the endogenous SCC3 promoter to generate PCLB2-SCC3. (B) A schematic diagram showing forced localization of Scc3, and therefore cohesin, to the 10× copies of tetO sequence located 5′ upstream of the REC8 promoter. The distance between the first tetO and the REC8 promoter is ∼2 kb. (C) Immunoblot showing the production of GFP. Cells were induced to undergo synchronous meiosis, and protein extracts were prepared for immunoblotting. Strains used: HY2685 (scc2 tetO), HY2684 (scc2 tetO/tetR-Scc3), and HY2692 (scc2 tetR-Scc3). (D) Quantification of PREC8-GFP production. Yeast cells were induced to undergo synchronous meiosis, and the fluorescence intensities of GFP and RFP were determined as for Figure 5C. Strains used: WT, HY3100; scc2 PREC8-GFP::LEU2, HY3044; scc2 tetR-SCC3 PREC8-GFP::LEU2, HY3045; scc2 PREC8-GFP::DCC1, HY3046; and scc2 tetR-SCC3 PREC8-GFP::DCC1, HY3047. All strains harbor one copy of PDMC1-mAPPLE (RFP) and 10 copies of tetO inserted at the LEU2 locus. (E) A model depicts Scc2 and cohesin action in transcriptional activation during meiosis. Scc2, red squares; cohesin, green ovals. Black ovals represent an unknown transcription factor. Chromosomes are shown as black and gray lines.
Scc2 regulates other cohesin target genes
Our expression microarray analysis of cohesin target genes (Supplemental Figure 3) permitted us to determine whether Scc2 plays a role in transcriptional activation of other meiotic genes. As an example, we show that Scc2 activates a second cohesin-regulated gene HIM1 in a manner similar to that of REC8 (Supplemental Figure S6). A cohesin-associated region was found <1 kb upstream of the 5′ sequence of the HIM1 promoter (Supplemental Figure 6A). Using a similar approach, we constructed a heterologous PHIM1-GFP reporter positioned at both the endogenous HIM1 locus and the LEU2 locus and found that Scc2 positively regulated PHIM1-GFP expression at both loci (Supplemental Figure 6, B and C). Taken together, our data suggest that Scc2 recruits cohesin to the chromosome to regulate the expression of cohesin target genes during yeast meiosis.
DISCUSSION
In the research reported here, we showed that meiotic Scc2 was required for the recruitment of cohesin to the chromosome to act as a transcriptional activator, a role not previously known in budding yeast. Our data provide direct evidence to support recent findings from other model organisms that, in addition to its canonical role in sister-chromatid cohesion, cohesin plays an important role in regulation of gene expression, in particular during cell differentiation and development.
Because cohesin and the defined cohesin-associated regions influenced its target promoter in a position-dependent manner and because forced localization of cohesin increases cohesin target gene expression, our data fit the enhancer and transcriptional-activation model shown in Figure 8E. We propose that the cohesin-associated region acts as an upstream activating sequence for downstream gene activation. On the binding of cohesin, cohesin recruits a yet unknown transcriptional factor(s) to activate the downstream promoter (Figure 8E). This model is consistent with a recent finding in mouse embryonic stem cells, where cohesin directs the mediator to the chromosome for gene regulation (Kagey et al., 2010). Alternatively, cohesin could mediate chromatin-loop formation, which modifies local chromatin structure, making it favorable for gene expression. Of note, the location of a majority of CARs in the 3′ intergenic regions (Blat and Kleckner, 1999; Glynn et al., 2004; Lengronne et al., 2004; our unpublished results) might account for only a small number of genes subjected to cohesin regulation in budding yeast. Indeed, a CAR that is located to the 3′ downstream sequences of our reporter construct appears to have little effect on target promoter activation (Figure 5). In addition to its requirement for activating zebrafish runx genes during cell differentiation (Horsfield et al., 2007), cohesin also physically interacts with the transcriptional activator NF-κB to increase the activity of the HIV-LTR promoter (Lara-Pezzi et al., 2004); in contrast, cohesin primarily represses gene expression in flies (Dorsett, 2009). It is intriguing that in flies the Smc1 subunit is localized to the actively transcribed genes in the nonrepetitive regions that are surveyed by chromatin immunoprecipitation (Misulovin et al., 2008), implying that cohesin can also influence transcriptional elongation and/or termination. Cohesin regulation of gene expression therefore appears to be organism specific and involves a complex interaction between cis and trans factors. Our microarray analysis of gene expression in yeast meiosis shows that a few cohesin target genes are located directly downstream of the Ty-LTR or the full-length Ty1 transposable element, and insertion of the Ty-LTR sequence in front of the DMC1 promoter can increase DMC1 expression in a cohesin-dependent manner (unpublished data), suggesting that cohesin can interact with other cis-DNA elements, including the Ty-LTR, for gene regulation. The biological significance of this cohesin interaction remains to be elucidated.
Loading of cohesin onto the meiotic chromosome in budding yeast depends on Scc2 and, presumably, Scc4. We have found that the majority of Scc2's binding sites overlap with cohesin-associated regions during meiosis, supporting Scc2's role as a cohesin loader (Ciosk et al., 2000). In vertebrates, the Scc2/Scc4 complex is required for cohesin chromosome association (Watrin et al., 2006), but the transcriptional factor CTCF can also bind to cohesin and appears to be responsible for directing cohesin binding to the many CTCF sites during interphase (Parelho et al., 2008; Wendt et al., 2008). This discrepancy suggests that in vertebrates multiple pathways can lead to cohesin association with the chromosome and might have differentiated cohesin's role in sister-chromatid cohesion from that in gene expression. An equivalent of CTCF has not been found in budding yeast, but the role of meiotic Scc2 is twofold. First, it is required for sister-chromatid cohesion and nuclear division. Second, it acts as a transcriptional regulator by recruiting cohesin to the chromosome to regulate gene expression, so Scc2 may play a role similar to that of CTCF in yeast meiosis. Determining whether these two meiotic Scc2 functions are separable in yeast will be important. Though cohesin acts downstream of Scc2 in gene regulation, our data do not exclude the possibility that Scc2 can activate gene expression through a cohesin-independent pathway. In contrast to the Scc2/Scc4 complex, the vertebrate CTCF transcription factor binds to the DNA motif CCCTC (Lobanenkov et al., 1990), which presumably also determines the sequence specificity for cohesin association in vertebrates. We notice that a previous study in yeast vegetative cells identified Scc2/Scc4 as being localized preferentially to the tRNA genes and other locations bound by the TFIII C factor (D'Ambrosio et al., 2008), but a recent report showed that mitotic Scc2/Scc4 is predominantly associated with the cohesin-associated regions (Kogut et al., 2009). Our genome-wide mapping of meiotic Scc2 chromosome binding is more consistent with this later result that Scc2-associated regions largely overlap with CARs throughout the yeast genome.
Meiosis in yeast is triggered by a combination of internal and external cues, including the MAT locus information and nitrogen depletion from the environment (Mitchell, 1994; Kassir et al., 2003). On meiotic induction, transcriptional reprogramming accompanied by chromosome remodeling regulates the expression of many early meiotic genes and is followed by the expression of middle and late genes when cells progress further into meiosis (Vershon and Pierce, 2000). Chromosomal association of cohesin is thought to facilitate loop formation and other structural modifications of the chromatin (Guacci et al., 1997; Novak et al., 2008; Hadjur et al., 2009; Nativio et al., 2009). On the formation of a DSB, cohesin is recruited to the break site, where a localized modification of histone phosphorylation takes place (Unal et al., 2004). Cohesin also interacts with chromatin-remodeling factors to mediate sister-chromatid cohesion (Chai et al. 2005), although the exact mechanism remains to be elucidated. We found that minimizing the activity of the histone deacetylase Sir2 can partially suppress the scc2 phenotype of reduced REC8 promoter activity (Lin W, Jin H, Yu H-G, unpublished data), suggesting that chromosome structural modification mediated by cohesin and its loader protein Scc2 can also trigger an epigenetic response during yeast meiosis. The histone acetyltransferase Gcn5 is required for the activation of an array of meiotic genes when a vegetative yeast cell switches to the sexually reproductive program (Kassir et al., 2003). We speculate that chromosome reorganization during meiosis is necessary for this epigenetic regulation of cell differentiation and that cohesin facilitates it. Conceivably, when the yeast cell returns to vegetative growth during spore germination, corresponding chromosomal reorganization by cohesin and epigenetic regulation of gene expression will reverse its course.
Nonlethal mutations in cohesin and cohesin-associated factors lead to human developmental disorders, including Cornelia de Lange syndrome and Roberts syndrome, called cohesinopathies (Liu and Krantz, 2008; Bose and Gerton, 2010). The molecular mechanisms of these diseases remain to be elucidated. Our finding that cohesin and its loader protein Scc2 modulate meiotic gene expression further confirms that, in addition to its primary function in sister-chromatid cohesion, cohesin plays an important role in gene expression when the cell switches to a different developmental program. Future work with model organisms, including budding yeast, could provide further insights into the causes of human cohesinopathies.
MATERIALS AND METHODS
Yeast strains and plasmids
All yeast strains are diploid isogenic to SK1, except that the DEGRON-SCC2 strain is a haploid from the S288C background (Supplemental Table 1). To construct the PCLB2-SCC2 allele, we used a PCR-based approach to replace the endogenous SCC2 promoter with that from the CLB2 gene (Lee and Amon, 2003). The same PCR-based method was used to construct the DEGRON-SCC2 allele with p378 and the PCUP1-REC8 allele with pHG40. PREC8-GFP was inserted at the BUD3 locus by PCR with pHG115 and at the DCC1 locus by plasmid integration using pHG123. The spo11-Y135F has been described previously (Cha et al. 2000). We previously reported the PCLB2-SMC1 and PCLB2-SCC3 alleles (Lin et al., 2011). Integration of tetO/tetR-GFP at the URA3 locus was performed as previously described (Michaelis et al., 1997). PREC8-MCD1 was constructed as previously described (Buonomo et al., 2000). C-terminal protein tags (Scc2-GFP, Scc2-V5, Smc3–3HA, and Rec8'3HA) were performed as previously described (Jin et al., 2009). Primer information is listed in Supplemental Table 2.
To clone the REC8 promoter, we amplified a 2158–base pair DNA sequence upstream of the REC8 start codon from the SK1 background genomic DNA by PCR and cloned it into pRS305 to generate plasmid pHG88. The GFP open reading frame followed by a 283–base pair 3′ UTR from SMC1 was cloned into the PacI and SacI sites of pHG88 to form pHG105. Deletions of the 5′ upstream sequence of the REC8 promoter generated plasmids pHG107 and pHG108. For integration of PREC8-GFP at the LEU2 locus, pHG105, pHG107, and pHG108 were digested with AflII, then subjected to a standard yeast transformation protocol. For integration of the GFP open reading frame at the endogenous REC8 locus, pHG105 was digested with MluI before transformation. The DMC1 promoter was cloned previously (Yu and Koshland, 2005) and was fused with the GFP open reading frame at the PacI and SacI sites of pHG88 to form pHG112. The GFP open reading frame in pHG112 was replaced with the mApple open reading frame to form pHG140. The SCC3 open reading frame was cloned by PCR and fused with tetR to generate plasmid pHG141. Expression of tetR-SCC3 was under the control of the URA3 promoter. Ten copies of the tetO sequences were inserted at the SalI site of pHG106 to form pHG173. Similarly, we cloned the HIM1 promoter (∼1 kb upstream of the open reading frame), which was fused with the GFP to form plasmid pHG188. Integration of PHIM1-GFP at the LEU2 and HIM1 loci was achieved by transform of pHG188, which was digested with AflII and PacI, respectively.
Synchronous meiotic yeast culture was performed as previously described (Jin et al., 2009). We used α-factor (10 ng/ml) to arrest the DEGRON-SCC2 cells at G1 for 2 h at 25°C. Cells were washed twice with H2O, then were split into two cultures, one incubated at 25°C and the other at 37°C.
Northern blot, RT-PCR, and gene-expression microarray
Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at the time points indicated. Samples were immediately frozen at –80°C. Total RNA was extracted and then subjected to a standard Northern blot protocol (Carlile and Amon, 2008). For RT-PCR, we used the RNeasy kit (Qiagen, Valencia, CA) to extract and purify total RNA from yeast cells. Purified mRNA was reverse transcribed to cDNA (Invitrogen, Carlsbad, CA), and a semiquantitative PCR method was used to determine the concentration of target cDNA. Reverse-transcribed cDNA was labeled and hybridized to the 4×72K yeast expression array (Roche NimbleGen, Madison, WI). Scanned signals were analyzed by ArrayStar (DNAStar, Madison, WI).
Immunoblot
Protein extraction and immunoblot were performed essentially as previously described (Jin et al., 2009). Scc2-3HA, Smc3-3HA, and Rec8-3HA were detected by an anti-HA antibody (16B12, Santa Cruz Biotechnology, Santa Cruz, CA). GFP was detected by an anti-GFP polyclonal antibody (ab209; Abcam, Cambridge, MA). Mcd1 was detected by a Mcd1-specific polyclonal antibody (a gift from Vincent Guacci, Carnegie Institution of Washington, Baltimore, MD). The meiosis-specific protein Dmc1 was detected by a Dmc1 polyclonal antibody (a gift from Douglas Bishop, University of Chicago, Chicago, IL).
Chromatin immunoprecipitation and microarray
Yeast cells were fixed in 1% formaldehyde for 2 h at room temperature, then subjected to a chromatin immunoprecipitation procedure as described previously (Yu and Koshland, 2005). A semiquantitative method was used to detect the enrichment of Rec8 at the centromere III (Figure 7F). To map genome-wide Scc2 and Rec8 chromosome association, we amplified total DNA and ChIP samples using the WGA kit (Sigma-Aldrich, St. Louis, MO) and hybridized them to a 385K ChIP array (Roche NimbleGen). Signal profile was displayed by the SignalMap software (Roche NimbleGen). To determine the Scc2- and Rec8-associated regions throughout the yeast genome, we identified peak values above the 1.8-fold threshold and extracted active regions from the SignalMap gff files. Active regions <500 base pairs in size were ignored. A small gap of 50 base pairs allowed within an active region minimized internal noise. From each peak, we extended the region to the left to include all positive measurements until encountering a gap of negative measurement larger than the gap threshold. Similarly, we extended the region to the right. An identified active region of Scc2 was defined to overlap with that of Rec8 if those regions shared base pairs. For image display (Figure 1C), Rec8 and Scc2 peaks were smoothed with a nine-point Gaussian-weighted moving average and filtered with a left and right rise <0.1 and a height <0.5 (log2 space) (Glynn et al., 2004). To determine the chromosome association of RNA polymerase II (Pol II), we performed ChIP using an anti-Pol II CTD antibody (clone 8WG16; Millipore, Billerica, MA).
Fluorescence microscopy
Live-cell microscopy was carried out on a DeltaVision imaging system (Applied Precision, Issaquah, WA), at 30°C with a 60× (numerical aperture [NA] = 1.41) objective lens. A total of 14 Z-stacks was collected at each time point. Each optical section was 0.5 μm thick. Exposure time for each optical section was set at 80 ms. Acquired images were projected and quantified with Softworx (Applied Precision). For determination of nuclear division and assay of sister-chromatid cohesion (Figure 3, B and C), cells were induced to undergo synchronous meiosis, fixed in 1% formaldehyde, stained with DAPI, and visualized under a fluorescence microscope.
Yeast surface nuclear spreads were performed as previously described (Jin et al., 2009). Scc2-GFP was detected by a GFP antibody (ab209, Abcam); Rec8-3HA was detected by an anti-HA antibody (12CA5; Roche, Mannheim, Germany). Immunofluorescence images were acquired with a 100× objective lens (NA = 1.40) mounted on a motorized epifluorescence microscope (Axio Imager; Zeiss, Jena, Germany).
Fluorescence from cells in the 96-well format was scanned with a Typhoon phosphorimager/fluorescence imager (GE Healthcare Bio-Sciences, Piscataway, NJ). The ratio of signal intensity of GFP over red fluorescent protein (mApple) was determined (Figure 5) with IPLab (BD Biosciences, Franklin Lakes, NJ).
Supplementary Material
Acknowledgments
We thank H. W. Bass, Y. Wang, W. Deng, and the members of H.-G.Y.’s lab for discussions and comments. A. Amon, V. Guacci, D. Bishop, and M. Davidson provided strains and antibodies. S. Miller and K. Shirk provided technical assistance. A. B. Thistle assisted with text editing. This work was supported in part by the National Science Foundation (MCB#0718384) and the Florida Biomedical Research Program (08BN-08).
Abbreviations used:
- CAR
cohesin-associated region
- DSB
double-strand break
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0545) on April 20, 2011.
REFERENCES
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bose T, Gerton JL. Cohesinopathies, gene expression, and chromatin organization. J Cell Biol. 2010;189:201–210. doi: 10.1083/jcb.200912129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham LE, Wang HT, Elder RT, McCarroll RM, Slater MR, Esposito RE. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:9406–9410. doi: 10.1073/pnas.87.23.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol. 2009;187:455–462. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Guacci V, Yu HG. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–725. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi E, Pezzi N, Prieto I, Barthelemy I, Carreiro C, Martinez A, Maldonado-Rodriguez A, Lopez-Cabrera M, Barbero JL. Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J Biol Chem. 2004;279:6553–6559. doi: 10.1074/jbc.M307663200. [DOI] [PubMed] [Google Scholar]
- Lau A, Blitzblau H, Bell SP. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 2002;16:2935–2945. doi: 10.1101/gad.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Jin H, Yu H-G. Cohesin plays a dual role in gene regulation and sister-chromatid cohesion during meiosis in Saccharomyces cerevisiae. Genetics. 2011;187:1041–1051. doi: 10.1534/genetics.110.122358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5’-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misulovin Z, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Hoog C. Cohesin Smc1β determines meiotic chromatin axis loop organization. J Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Parisi S, McKay MJ, Molnar M, Thompson MA, Van Der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JH, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, van Bemmel JG, Oliveira RA, Itoh T, Shirahige K, van Steensel B, Nasmyth K. A direct role for cohesin in gene regulation and ecdysone response in Drosophila salivary glands. Curr Biol. 2010;20:1787–1798. doi: 10.1016/j.cub.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Weicsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren C, Strom L. S-phase and DNA damage activated establishment of sister chromatid cohesion—importance for DNA repair. Exp Cell Res. 2010;316:1445–1453. doi: 10.1016/j.yexcr.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break–specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Vershon AK, Pierce M. Transcriptional regulation of meiosis in yeast. Curr Opin Cell Biol. 2000;12:334–339. doi: 10.1016/s0955-0674(00)00104-6. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-G, Koshland DE. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell. 2005;123:397–407. doi: 10.1016/j.cell.2005.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.