We discovered that the relative durations of the phases of the yeast metabolic cycle change with the growth rate. These changes can explain mechanistically the transcriptional growth-rate responses of all yeast genes (25% of the genome) that we find to be the same across all studied nutrient limitations in either ethanol or glucose media.
Abstract
We studied the steady-state responses to changes in growth rate of yeast when ethanol is the sole source of carbon and energy. Analysis of these data, together with data from studies where glucose was the carbon source, allowed us to distinguish a “universal” growth rate response (GRR) common to all media studied from a GRR specific to the carbon source. Genes with positive universal GRR include ribosomal, translation, and mitochondrial genes, and those with negative GRR include autophagy, vacuolar, and stress response genes. The carbon source–specific GRR genes control mitochondrial function, peroxisomes, and synthesis of vitamins and cofactors, suggesting this response may reflect the intensity of oxidative metabolism. All genes with universal GRR, which comprise 25% of the genome, are expressed periodically in the yeast metabolic cycle (YMC). We propose that the universal GRR may be accounted for by changes in the relative durations of the YMC phases. This idea is supported by oxygen consumption data from metabolically synchronized cultures with doubling times ranging from 5 to 14 h. We found that the high oxygen consumption phase of the YMC can coincide exactly with the S phase of the cell division cycle, suggesting that oxidative metabolism and DNA replication are not incompatible.
INTRODUCTION
The response of gene expression to growth rate has been studied using chemostats to control steady-state growth rate and gene expression microarrays to study patterns of gene expression by several groups (Hayes et al., 2002; Pir et al., 2006; Regenberg et al., 2006; Castrillo et al., 2007; Brauer et al., 2008; Fazio et al. 2008). These studies demonstrated a common growth rate response (GRR) in steady-state, exponentially growing cultures limited by different natural nutrients, as well as by auxotrophic requirements, under anaerobic as well as aerobic conditions. However, in all these studies, the carbon source was glucose, which is highly preferred by Saccharomyces cerevisiae and unique in many ways (Zaman et al., 2008). Thus it is not clear how much of the identified GRR is specific to growth on glucose (Futcher, 2006; Zaman et al., 2009) as a sole carbon source and how much of the GRR is general to growth independent of the carbon source. Zaman et al. (2009) proposed that much of the observed common GRR might be triggered by sensing glucose, and thus be specific to growth on glucose. In this study, we set out to test this hypothesis directly.
A striking finding of previous studies was that many of the genes for which expression correlated with growth rate were also among the genes that oscillate in their expression in the metabolically synchronized cultures that are used to define the yeast metabolic cycle (YMC; Klevecz et al., 2004; Tu et al., 2005). It is significant that metabolic cycling as studied by these authors was induced by glucose starvation followed by refeeding at a constant rate. In glucose media, Brauer et al. (2008) observed that genes expressed during different phases of the YMC have GRRs that tend to be either positive or negative. This observation suggests that a cycle similar to the YMC might be present even in cells from nonsynchronized cultures such as the ones grown by Brauer et al. (2008). Consistent with this interpretation, Silverman et al. (2010) used gene–gene correlations to demonstrate that the YMC has a single cell origin. Futcher (2006) pointed out that some of the genes expressed periodically in the YMC of a glucose-limited culture are coexpressed in a cell division cycle (CDC) synchronized culture grown in ethanol media. However, it remained unclear whether a putative metabolic cycle in cells grown on ethanol as the sole carbon source (Keulers et al., 1996) is the same as the YMC induced in glucose media, and whether the GRR on ethanol as sole carbon source will correlate with the YMC as it does in cultures grown on glucose. We set out to explore these issues in the hope of finding a persuasive theory to relate GRR to yeast metabolic cycling.
In this paper, we show that most of the genes showing GRR found in glucose media also respond similarly to growth rate in ethanol media. This subset of ∼25% of all yeast genes thus shows a “universal” GRR. By comparing the GRR in glucose and ethanol, we could define an additional subset of genes with a GRR dependent on the carbon source. Remarkably, we found that all the genes with universal GRR are expressed periodically in the YMC as defined by gene expression patterns in metabolically synchronized cells (Klevecz et al., 2004; Tu et al., 2005). These results confirm and extend to all the genes with the universal GRR the proposition (Brauer et al., 2008) that genes whose expression levels increase with growth rate are expressed in the YMC phase with high oxygen consumption (HOC), while genes with expression levels that decrease with growth rate are expressed in the YMC phase with low oxygen consumption (LOC).
We hypothesize that it is the relative lengths of the HOC and LOC phases of the YMC that change with growth rate. Using metabolically synchronized cultures, we confirmed that the relative duration of the HOC phase increases with growth rate. We show that such a change in the distribution of cells in HOC and LOC phases can account, in a straightforward way, for the observed universal GRR. We also found that the HOC phase of the metabolic cycle can coincide exactly with the S phase of the CDC, suggesting that the origin of the metabolic cycle may not lie in the incompatibility of oxidative metabolism and DNA replication.
RESULTS
Physiological parameters of cultures grown on ethanol
To study the genetic and physiological responses to changes in the doubling time (growth rate) with a nonfermentable carbon source, we grew and characterized nine nonsynchronized cultures of a haploid prototrophic strain of S. cerevisiae (DBY11369) growing on ethanol as the only source of carbon and energy. Each culture was limited on one of three nutrients: ethanol, the carbon source (C); ammonium, the sole nitrogen source (N); and phosphate, the sole source of phosphorus (P). For each nutrient limitation, we grew three cultures with steady-state growth rates μ = 0.05, 0.10, and 0.14 h−1, corresponding to doubling times of 14, 7, and 5 h, respectively. For each steady-state culture, we measured cell density, distribution of cells sizes, residual ethanol, bud index, and gene expression.
In previous studies of this kind using glucose media, we also studied auxotrophic strains limited by their auxotrophic requirements. In this article, we concentrate only on the natural nutrients that limit the growth of prototrophs. Thus, in what follows, we have data for three conditions in ethanol (C, N, and P) and four in glucose (C, N, P, and also sulfate-limited [S]). Because the useful range of growth rates is less limited in glucose than in ethanol, we have data for five or six growth rates in glucose to compare with the three growth rates in ethanol.
In the ethanol cultures at steady-state, the cell density (Figure 1A) decreases monotonically with increasing growth rate, similar to the results of Brauer et al. (2008) for the analogous experiments with glucose as carbon source. This is entirely consistent with theoretical expectations, as described in the Supplementary Material. The concentrations of residual ethanol in the fermenter vessels also follow the expected trend. As the flux of medium is reduced, cells grow more slowly and spend more time in the reaction vessels. Both of these factors suggest that the concentration of residual ethanol should be inversely correlated to the growth rate of the cultures, as we observed (Figure 1B). The specific consumption of ethanol (i.e., ethanol consumed/steady-state biomass) is lower for the ethanol-limited cultures (C), suggesting that cultures limited on phosphate (P) or ammonium (N) might metabolize some fraction of the excess ethanol to acetate, possibly to generate reducing NADPH required for biosynthetic processes.
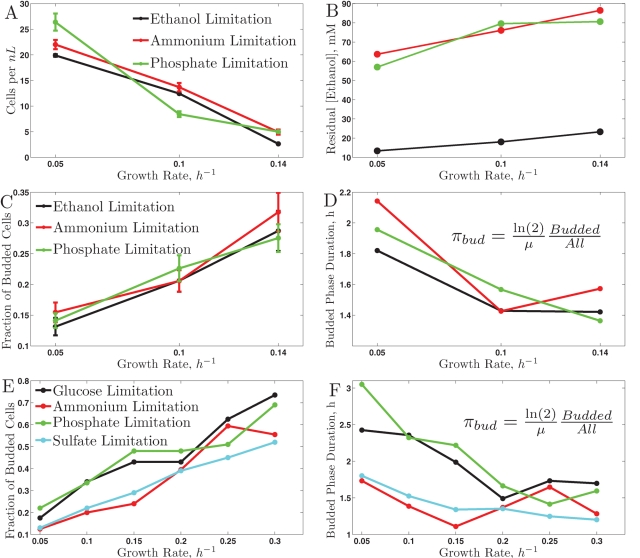
FIGURE 1:
Physiological responses to growth rate on ethanol (A–D) and on glucose (E–F), (Brauer et al., 2008). All responses are plotted as a function of the growth rate across the nutrient limitations indicated in the legends. (A) Biomass in ethanol carbon source; (B) residual ethanol concentration; (C) bud index in ethanol carbon source; (D) duration of the budded phase in ethanol carbon source; (E) bud index in glucose carbon source; (F) duration of the budded phase in glucose carbon source.
One of the advantages of budding yeast is that the cell morphology is indicative of the position of individual cells in their CDC (Hartwell, 1974; Hartwell et al., 1974). In particular, cells in G0/G1 are not budded, while cells in S, G2, or M are budded. In our ethanol cultures, the fraction of budded cells shows linear dependence (within experimental error) with respect to the growth rate, similar to the trend observed on glucose (Brauer et al., 2008). Since we set and thus know the doubling time of the cultures, we can use these data to compute the average duration of CDC phases during which the cells are budded, πbud:
The results shown in Figure 1D indicate that as the period of the CDC decreases (faster growth rates) the duration of the CDC phases during which the cells are budded (πbud) also decreases; the Spearman rank correlation between μ and πbud is −0.8 with a p value < 0.05. To investigate whether the observed change in the duration of CDC phases is limited only to nonfermentable carbon source or generalizes to a fermentable carbon source (glucose), we apply the same analysis to the bud index data (Figure 1E) collected by Brauer et al. (2008). The results indicate a similar trend in decreasing the duration of the budded period as the period of the CDC decreases, Figure 1F. This trend is similar to the one observed by Hartwell and Unger (1977) using mutations and drugs slowing the growth rate. Thanks to the chemostat, we can quantify this trend in more natural conditions without complications requiring normalization (Hartwell and Unger, 1977). Except for the phosphate limitation, πbud is shorter when ethanol is the carbon source compared with glucose, most likely reflecting the increased time cells have to spend in G0/G1 to build up the required biomass and energy when using ethanol as the only source of energy and carbon. The distribution of cell sizes also reflects the increased fraction of budded cells with growth rate (Supplementary Figure S5); as shown in the Supplementary Material, this distribution can be used to estimate the budded fraction (Figures S6 and S7).
Gene expression in cultures grown on ethanol
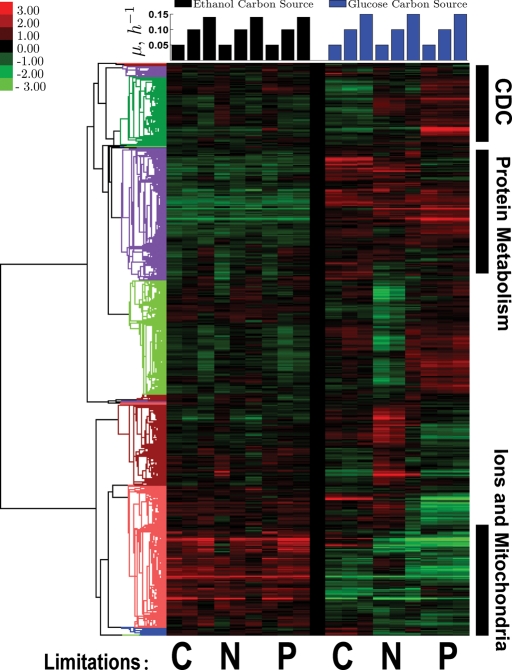
To visualize global trends, we first used hierarchical clustering of the data we obtained in ethanol media with the corresponding data in glucose obtained previously by Brauer et al., (2008). As can be seen in Figure 2, we found clusters with different mean expression in ethanol and glucose. Genes expressed at higher levels in ethanol are strongly (p <10−12) enriched for transitional metal transport, amine biosynthesis, oxidation-reduction processes, and mitochondrial functions. The higher expression of these genes is not unexpected, and likely reflects the increased respiratory requirements of cells growing on ethanol. Genes expressed more highly on glucose are enriched for protein complex biogenesis and proteolysis.
FIGURE 2:
A clustergram of the log2 gene expression data from continuous cultures on ethanol (left set of columns) and glucose (right set of columns; Brauer et al., 2008). Each column (carbon source) represents one limitation (carbon, nitrogen, or phosphorus) indicated below the columns as C, N, and P, respectively, and each limitation has three growth rates ordered from slowest to fastest growth (μ = 0.05, 0.10, 0.14/0.15 h−1). The similarity metric used for clustering (noncentered, variance-normalized correlations) is computed using all data shown in the clustergram.
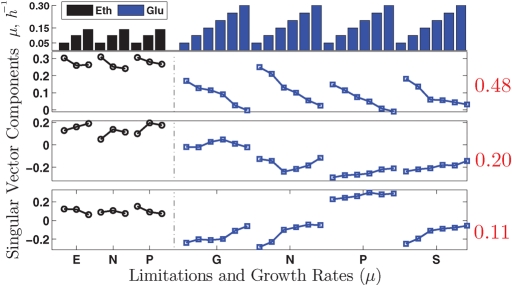
To explore the data further, we computed the singular value decomposition (SVD; Golub and Kahan, 1965; Alter et al., 2000) of the gene expression data for all conditions (carbon sources and limitations). Such decomposition allows one to summarize and visualize the changes in gene expression in terms of a set of orthogonal vectors, sometimes referred to as “eigengenes.” Eigengenes can be thought of as representative expression profiles that, together, represent all the variations in a set of experiments. The extent to which each vector represents the expression profiles of many genes is quantified by the magnitude of the corresponding singular value relative to all singular values. As shown in Figure 3, the results closely resemble those found by Brauer et al. (2008). The most prominent singular vector, which accounts for 48% of the variance (that is 48% of the change in gene expression), is strongly correlated 1 to the growth rate, which suggests that much of the glucose GRR is also present when the cells are grown on ethanol as the only carbon source. This means that 48% of the changes in gene expression, as a response to increase in growth rate, are monotonic increases or decreases in the expression of the same genes across all conditions. For this reason, we will refer to this as the universal GRR. The larger magnitudes of the elements corresponding to ethanol carbon source reflect the fact that the correlations between growth and gene expression changes in ethanol are stronger than the correlations previously found in glucose. The second most prominent singular vector also has a substantial growth rate component, but it is hard to interpret; this was also the case in glucose. The third most prominent vector is more interesting. It correlates strongly to growth rate, but in opposite directions for glucose and ethanol carbon sources, suggesting that this vector corresponds to genes having opposite GRRs in glucose and ethanol carbon sources. This vector provides strong evidence for a carbon source–specific GRR. In principle, one could identify all the genes corresponding to the universal and carbon source–specific GRRs from the loadings of the singular decomposition vectors. However, because both the universal and the carbon source–specific GRR are evident in the data more directly, we will follow the more intuitive approach of Brauer et al. (2008).
FIGURE 3:
Singular value decomposition (SVD) of the gene expression data. Top, first singular pair; middle, second singular pair; and bottom, third singular pair. The fraction of variance explained by each singular pair is indicated by the numbers in red to the right of the corresponding panels.
Identifying genes with universal GRR
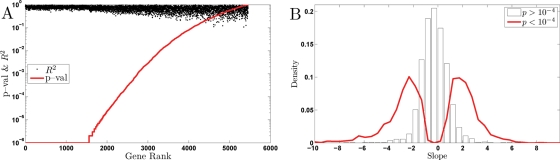
Given the large fraction of variance (48%) explained by the singular vector correlated to the growth rate, one may expect a large number of genes having universal GRR. To find such genes, we modeled the GRR (log2 expression values) of each gene as a linear function of the growth rate and a gene-specific constant similar to Airoldi et al. (2009) and Brauer et al. (2008). To specifically quantify the growth rate–related variance in the data without complications of carbon source or limitation of specific mean level of expression, we first normalized the expression of each gene for each combination of carbon source and nutrient limitation to mean zero. This normalization is particularly important in the presence of different carbon sources, because many genes have substantially different expression levels for glucose compared with ethanol as the carbon source, and the parameter of interest is the slope of gene expression as a function of growth rate, and not the mean value of gene expression (see Supplementary Material for more discussion on this point). Since we have data for more conditions from cultures grown on glucose, we gave higher weights to the ethanol carbon source conditions, so the model parameters are equally representative of both carbon sources. Figure 4 shows the results: At a very stringent p value cutoff of 10−6, the model explains a significant fraction of the variance in the expression for at least 1500 genes, that is, ∼25% of the genes in the genome. Figure 4A shows the number of genes with significant fit to the model at any level of statistical significance, and it is clear that a more lenient cutoff would result in a larger set of genes. For example, a threshold at 0.1% false discovery rate (FDR) results in 3019 genes whose variance is explained significantly by the model.
FIGURE 4:
Distribution of slopes. Rank-ordered p values and the corresponding R2 for all GRR gene expression data using a model accounting for the GRR only by a gene-specific slope and a constant. The p values are computed from 106 bootstrap resamplings of the data.
In this analysis, a low p value (and thus a significant fit to the model) for a gene indicates that a significant fraction of the variance in the expression levels of that gene can be explained by a single growth rate slope for all conditions; it does not mean that the gene has identical (or even statistically indistinguishable) slopes in all limitations and carbon sources. It does suggest, however, that the trends in the expression levels of genes with high R2 are likely to be similar across carbon sources and nutrient limitations, which can be seen to be the case when the patterns of gene expression for the 1500 genes with p < 10−6 selected by this analysis are displayed (Figure 5). We used the GO Term Finder (Boyle et al., 2004) to identify the biological processes overrepresented by this set of 1500 genes with universal GRR (Tables 1 and 2). The full list with the genes and the universal GRR corresponding to each GO term can be found at the GRR website (http://genomics-pubs.princeton.edu/grr).
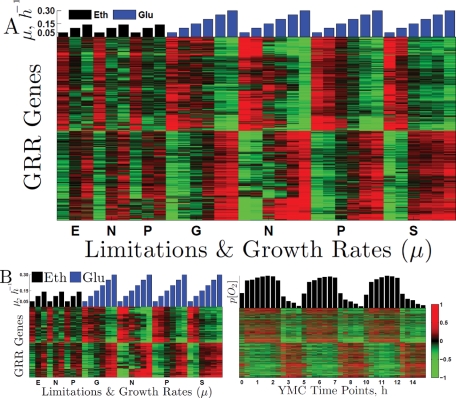
FIGURE 5:
Genes with universal GRR are periodic in the YMC. (A) Expression levels of the genes with best fits to the GRR model are normalized to mean zero for each limitation and clustered. The first nine columns (black bars) correspond to ethanol carbon source and limitations on ethanol (E), nitrogen (N), and phosphorus (P). The next columns (blue bars) correspond to glucose carbon source and limitations on glucose (G), nitrogen, phosphorus, and sulfur (S). The overrepresented GO terms for the genes with negative and positive slopes can be found in Tables 1 and 2, respectively. (B) The left panel shows the same mean-centered expression levels as Figure 5A. The right panel depicts the log2 expression levels (normalized to mean zero) of the same genes in the YMC (Tu et al., 2005). The genes in both panels are clustered based only on their expression levels in the growth rate experiments (not the YMC) and thus genes (rows) from the left panel correspond to the genes (rows) in the right panel. The dynamical range on all color heat maps is from −1 to 1 (twofold down- and up-regulation) as shown by the color bar.
TABLE 1:
Overrepresented GO terms for genes with negative UGRR.
| Gene ontology term | Cluster frequency | Genome frequency | Corrected p value | FDR (%) |
|---|---|---|---|---|
| Vacuolar protein catabolic process | 84/1148 | 118/7274 | 9 × 10−39 | 0.00 |
| Stress response | 269/1148 | 848/7274 | 1 × 10−33 | 0.00 |
| Autophagy | 84/1148 | 118/7274 | 9 × 10−28 | 0.00 |
| Cell differentiation | 79/1148 | 247/7274 | 1 × 10−7 | 0.00 |
TABLE 2:
Overrepresented GO terms for genes with positive UGRR.
| Gene ontology term | Cluster frequency | Genome frequency | Corrected p value | FDR (%) |
|---|---|---|---|---|
| Ribosome biogenesis | 174/1103 | 437/7274 | 1 × 10−33 | 0.00 |
| Cellular biosynthetic process | 510/1103 | 2203/7274 | 8 × 10−31 | 0.00 |
| Regulation of translation | 90/1103 | 190/7274 | 2 × 10−23 | 0.00 |
| Posttranscriptional regulation of gene expression | 510/1103 | 2203/7274 | 9 × 10−21 | 0.00 |
| Translation | 254/1103 | 962/7274 | 1 × 10−19 | 0.00 |
| Mitochondrial translation | 57/1103 | 110/7274 | 2 × 10−16 | 0.00 |
Connection between the universal GRR and the YMC
One of the most striking and puzzling results of previous studies was the connection between the GRR and the YMC. Brauer et al. (2008) suggested that the YMC might underlie the GRR and Silverman et al. (2010) provided further evidence supporting this idea. To explore this relationship, we examined whether genes with universal GRR are expressed periodically in the YMC, as seen in the data of Tu et al. (2005). The set of genes exhibiting the universal GRR, it should be recalled, show GRR in ethanol as well as in glucose. Figure 5B shows that genes expressed at higher levels during the HOC phase have positive slopes (expressed more highly at the higher growth rates) while genes expressed at higher levels during the LOC phase have negative slopes (expressed more highly at the slower growth rates).
The simplest hypothesis that can explain this remarkable correlation is to suppose that fast-growing cultures have a large fraction of their cells in a HOC-like phase of a cell-intrinsic metabolic cycle (Silverman et al., 2010). Such a change in the fraction of HOC cells is a likely outcome of changing the relative durations of HOC and LOC with growth rate, since the fraction of cells in HOC (ΨHOC) is directly proportional to the ratio of durations of HOC and LOC, ΨHOC ∝ THOC/TLOC, in nonsynchronized cultures.
To test this prediction, we measured how the YMC changes with growth rate. Following a protocol similar to that used by Klevecz et al. (2004) and Tu et al. (2005), we starved and refed chemostat cultures with glucose-limited media to induce metabolic cycling at three different growth rates varying from 0.05 to 0.14 h−1 (corresponding to doubling times ranging from 14 to 5 h). We followed the metabolic cycles by measuring dissolved oxygen as a function of time.
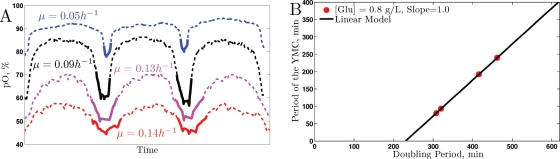
Figure 6 displays the dissolved oxygen traces we obtained, rescaled on the time axis so that the oscillations have the same period (Figure 6A) for better visualization of the relative shapes of the dissolved oxygen traces. The relationship between the YMC period and the growth rate is plotted on a linear scale in Figure 6B. It is clear from the shapes of the traces in Figure 6A that the relative duration of the HOC phase increases with the growth rate. This result is entirely consistent with the hypothesis that the universal GRR and YMC are actually reflections of the same underlying metabolic arrangements.
FIGURE 6:
Changes in the YMC with growth rate. (A) Three cycles from four metabolically synchronized cultures at growth rates μ = 0.05, 0.09, 0.13, 0.14 h−1 are scaled to have the same period for emphasizing the change in relative (rather than absolute) durations of the YMC phases. The absolute durations (YMC periods) are shown as a function of the doubling time of the cultures (B).
The linear dependence shown in Figure 6 is reminiscent of the linear dependence of the bud index on growth rate Figure 1. As the growth rate decreases, the budded phases of the CDC become a smaller fraction of the total CDC. Similarly, it appears that many of the functions in metabolism (notably those that require HOC) become a smaller fraction of the total duration of the metabolic cycle.
Connection between the metabolic cycle and the CDC
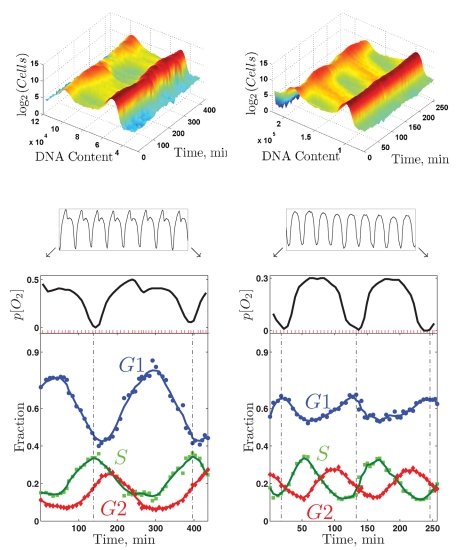
The ability to metabolically synchronize cells at various growth rates offered the possibility of a direct test of a hypothesis offered by many workers in the metabolic cycle field, including our own group (Klevecz et al., 2004; Tu et al., 2005; Chen et al., 2007; Chen and McKnight, 2007; Murray et al., 2007; Silverman et al., 2010). The idea was that the metabolic cycle might serve to separate in time DNA replication from the HOC characteristic of slow growth on glucose and, of course, on carbon sources like ethanol. We therefore synchronized cultures at several growth rates and followed both oxygen consumption and DNA content (by labeling DNA with a fluorescent dye [Cytox Green] and determining DNA content of 100,000 cells per time-point sample in a fluorescence-activated cell sorter).
Figure 7 shows these data for two wild-type prototrophic cultures (DBY12007): one growing at 0.133 h−1 fed with media containing 800 mg/l glucose, and another growing at 0.100 h−1 fed with media containing 400 mg/l glucose. It is clear from the figure that the S phase of the CDC coincides almost exactly with the HOC phase of the metabolic cycle in the more slowly growing culture, while the two phases are almost exactly opposite in the rapidly growing culture.
FIGURE 7:
Coupling between the CDC and the YMC. DNA content and oxygen consumption of two metabolically synchronized cultures of wild-type diploid cells at μ = 0.100 h−1 (left set of panels) and at μ = 0.133 h−1 (right set of panels). Each culture cycled steadily (as indicated by the oxygen trains) and was sampled at the positions indicated by the red tick marks during about two metabolic cycles (enlarged oxygen traces). Top, the raw DNA content data are shown as two-dimensional distributions. Bottom, the fraction of cells in different phases of the CDC as inferred from the two-dimensional distributions of DNA content.
Growth rate prediction
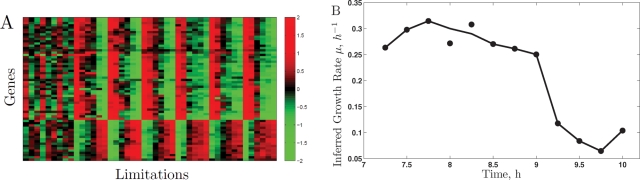
Our results show that there is a universal GRR that extends to cells growing on ethanol as the sole carbon source. We examined whether the growth rate prediction model used by Airoldi et al. (2009) could be improved and extended by using genes with expression that best correlates with the universal GRR. Each of the genes selected by Airoldi et al. (2009) for their growth rate model has very similar slopes across all limitations on glucose (Figure 8). On ethanol as the carbon source, many of those genes have expression profiles similar to those in glucose and still correlate well to growth rate, while the expression of other genes does not show good correlation to growth rate in ethanol. This is interesting, because the analysis of the diauxic shift experiments by Brauer et al. (2005) and Airoldi et al. (2009) predicted an unrealistically high growth rate when growth resumes in ethanol: higher than the growth rate inferred on glucose at the beginning of the experiment and significantly higher than the highest growth rate we have measured in 100 mM ethanol even for DBY11369, a strain selected for good growth on ethanol. We sought to improve the predictive power of the model on ethanol carbon source. To improve the model, we selected a predictive set of genes on the basis of large slopes and high R2 (across all conditions) as Airoldi et al. (2009) did. The predicted growth rate by such a model for the batch culture grown by Brauer et al. (2005) is shown in Figure 8B. The results indicate that a model based on the genes with universal GRR results in realistic predictions for all time points. We also selected a smaller subset of genes by sparse l1 (first norm) (1) regularized regression (Malioutov et al., 2005). Details of the models are given in the Supplementary Material.
FIGURE 8:
Predicting growth rate. (A) Genes used in predicting growth rate by Airoldi et al. (2009). The expression levels are zero-centered and depicted using the same notation as in Figure 5. (B) Predicted growth rate for a batch culture (Brauer et al., 2005) using genes with universal GRR.
Identifying genes with carbon source–specific GRR
In the analysis of the gene expression data (Figure 2) we identified a set of genes (corresponding to the third eigenvector in Figure 3) with GRR specific to the carbon source. Many of the genes in this set are readily interpretable in the light of well-understood biochemistry of carbon and energy metabolism. As expected, genes encoding enzymes of each of the steps in ethanol utilization and many enzymes involved in gluconeogenesis are included in the set of genes showing the carbon source–specific GRR. For many of these genes, it is interesting to explore the isoenzymes for which levels change significantly, as these isoenzymes are likely to mediate the expected changes in metabolic fluxes.
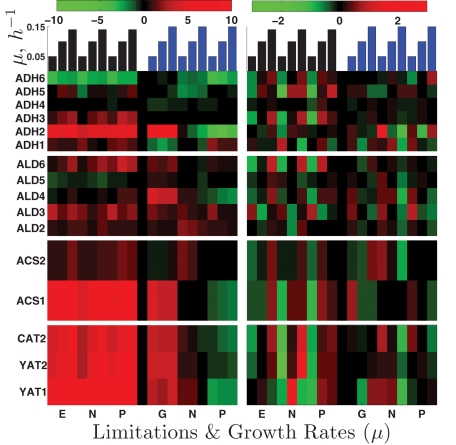
Alcohol dehydrogenases.
The first reaction in ethanol utilization is its oxidation to acetaldehyde. The expression levels of the six isoenzymes (Adh1p to Adh6p) that can catalyze this reaction are shown in the top panels of Figure 9. The isoenzyme showing strongest induction (∼30-fold) across all nutrient limitations when ethanol is the carbon source is Adh2p. Interestingly, ADH2 gene expression is induced equally strongly in the glucose-limited cultures, whereas it is repressed in phosphate- and ammonia-limited cultures using glucose as a carbon source. This expression pattern suggests that ADH2 expression is more likely repressed by glucose than induced by ethanol. ADH3 is also induced in ethanol carbon source (approximately two- to fourfold) with positive slopes in all nutrient limitations, which could reflect the shuttling of NADH from the mitochondria to the cytoplasm that increases with the growth rate.
FIGURE 9:
Enzymes participating in ethanol utilization and catabolism. Each row of panels shows the expression levels of a set of enzymes catalyzing a reaction from ethanol catabolism. The left panels show the log2 expression levels relative to the reference and the right panels show zero-transformed data to enhance the viability of growth rate trends.
Aldehyde dehydrogenases.
The second reaction in ethanol utilization is oxidation of acetaldehyde to acetate. There are five isoenzymes, Ald2p to Ald6p (second set of panels in Figure 9). The isoenzymes induced most strongly are Ald2p, Ald3p, and Ald6p. The first two are known to be repressed by glucose and induced by ethanol and stress (Navarro-Avino et al., 1999). Ald6p is unique among the aldehyde dehydrogenases in using NADP instead of NAD as a cofactor, (Saint-Prix et al., 2004). This cofactor specificity is crucially important in the context of growth, as reduced NADP (NADPH) is required in many biosynthetic reactions but produced by only a few reactions. In growth on ethanol, the only other reaction generating NADPH-reducing power is catalyzed by isocitrate dehydrogenase (Adp2p), the mRNA of which is also very strongly induced across all limitations on ethanol as carbon source. Consistent with expectations for increasing demand for NADPH with increasing growth rate, the expression of ALD6 (and ADP2) has positive slopes on ethanol as carbon source (Figure 9).
Acetyl-CoA synthetases.
The third reaction in ethanol utilization is the transfer of acetate to CoA (third set of panels in Figure 9). This transfer may be catalyzed by acetyl-CoA synthetases (Acs1p and Acs2p), which use acetate, CoA, and ATP as substrates, or by CoA transferase (Ach1p), which transfers CoA from succinyl-CoA to acetate. Expression of ACS1 has a positive slope, and is induced very highly (up to 120-fold) across all limitations on ethanol as carbon source and, only in the glucose limitation (to a slightly smaller extent), on glucose as carbon source in a manner analogous to ADH2. ACS2 shows only modest induction (up to two- to threefold) with a small positive slope (Figure 9). The gene encoding acetyl-CoA transferase ACH1 is also induced highly, up to 30-fold.
Acetyl-CoA transport.
The next step in ethanol catabolism is transporting acetyl-CoA across the mitochondrial and peroxisomal membranes. Major players in this process are encoded by the carnitine acetyl-CoA transferase genes (CAT2, YAT1, and YAT2; fourth set of panels in Figure 9). All three genes show very strong up-regulation (up to ∼250-fold) across all limitations when ethanol is the sole carbon source and very large positive slopes (up to 40) except for YAT1, which is induced highly even at slow growth.
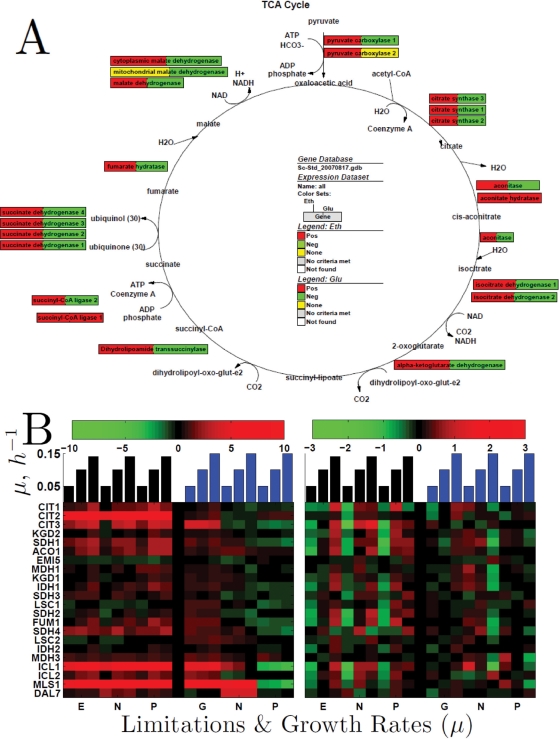
Tricarboxylic acid cycle.
Since the tricarboxylic acid (TCA) cycle is a major hub through which the carbon from ethanol has to pass before it can be used for energy or in anabolic precursors, the flux through the TCA cycle is expected to increase with growth rate on the ethanol carbon source. Consistent with this expectation, the mean level of expression (relative to the glucose carbon source reference) and the slopes of mRNAs corresponding to TCA cycle enzymes are overwhelmingly positive (Figure 10).
FIGURE 10:
Growth rate slopes and expression levels (log2) of enzymes from the Krebs and glyoxylate cycles. (A) Growth rate slopes of the mRNAs encoding enzymes catalyzing reactions from the Krebs cycle. Each box corresponds to an enzyme and the colors to the growth rate slopes of the corresponding mRNA in ethanol (left) and glucose (right) as indicated in the legend. (B) The left panels show the log2 expression levels relative to the reference and the right panels show zero-transformed data to enhance the visibility of growth rate trends.
Gluconeogenesis.
Yeast growing on ethanol must synthesize glucose for the essential carbohydrates and glycosylated proteins and as an intermediate in the synthesis of ribose and deoxyribose for nucleic acids. The only known pathway for making glucose from acetyl-CoA is the glyoxylate cycle. The first biochemical reaction separating the glyoxylate cycle from TCA is the formation of succinate and glyoxylate from isocitrate; this pathway is catalyzed by isocitrate lyase (encoded by ICL1 and ICL2). Consistent with the expectation for increased flux, Figure 10 shows that ICL1 is strongly induced when ethanol is the carbon source and ICL2 is induced to a smaller extent. Remarkably, both isocitrate lyases are also induced in the glucose limitation, suggesting that it is the lack of glucose, rather than the presence of ethanol, that results in the increased expression levels. This mechanism of glucose repression, rather than ethanol induction, is possible, as a single repression mechanism can accomplish the regulation that otherwise might require many induction mechanisms (e.g., ethanol, glycerol, acetate, and pyruvate induction mechanisms). The next pair of isoenzymes in the glyoxylate cycle (Msl1p) and (Dal7p/Msl2p) catalyzes the synthesis of malate from the acetyl-CoA and the glyoxylate produced in the first reaction. Those two malate synthase genes have elevated levels with ethanol carbon source, especially MSL1, the expression pattern of which is similar to ICL1. Both malate synthases and ICL1 show nitrogen derepression in the ammonium-limited cultures on glucose carbon source, which most likely reflects the role of those enzymes in nitrogen salvage from purine catabolism.
Hierarchical clustering of GO terms
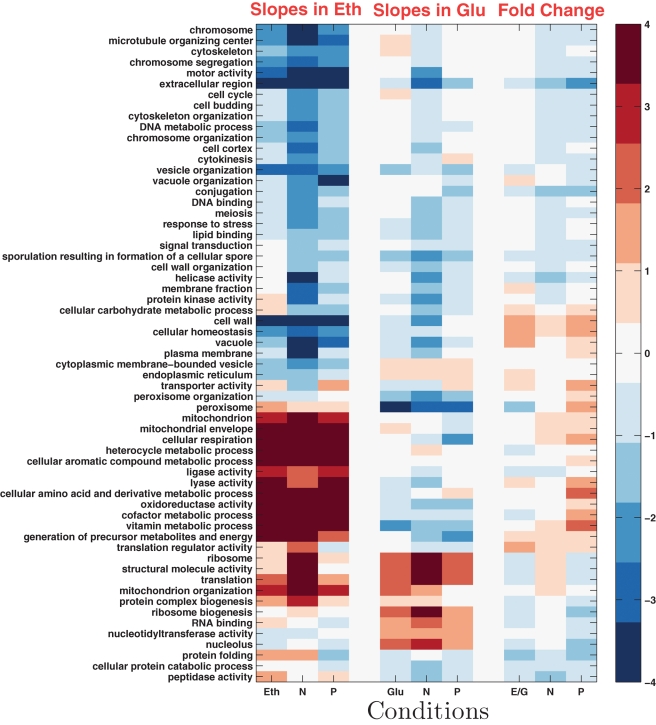
To get a system-level overview of nutrient-specific GRRs, we can summarize the mean slopes and fold changes between ethanol and glucose carbon source with a clustergram of gene ontology(GO) terms. Computing a single slope for each gene across all conditions is useful for identifying growth rate trends common to most conditions (Brauer et al., 2008). It fails, however, to capture nutrient and carbon source–specific responses (Boer et al., 2008). To characterize such specific responses, we can compute slopes specific to each condition (limitation and carbon source) for each gene. This makes it possible to compare and quantify (using the Wilcoxon rank-sum text) the differences between the distributions of nutrient-specific slopes for predefined sets of genes (such as GO groups) and the distribution of slopes for all genes. Figure 11 presents the results of this analysis. Consistent with the requirement for increased respiration at fast growth on ethanol carbon source, many mitochondrial and respiration-related genes have positive slopes. The genes with positive slopes are the same across all limitations on ethanol as carbon source, and a subset of the genes with positive slopes in ethanol also have positive slopes in glucose as carbon source (see Supplementary Material). Less expected is the difference in slopes (positive in ethanol and negative in glucose carbon source) for the genes involved in the generation of precursor metabolites and energy (Figure 11). Some of the genes in this set are mitochondrial, such as the TCA cycle (Figure 10), which explains at least in part the significantly positive slopes of this set of genes. Distributions of slopes and statistical analysis for all groups of genes represented on Figure 11 can be found on the supporting website (http://genomics-pubs.princeton.edu/grr).
FIGURE 11:
Growth rate slopes and fold changes by GO terms. Each row corresponds to a GO term. The first and second sets of three columns correspond to slopes in ethanol and glucose carbon source, respectively. The third set of three columns corresponds to fold change differences. The columns within each set correspond to limitations on carbon (C; ethanol or glucose), nitrogen (N), and phosphorus (P). The similarity metric (noncentered correlations) is computed using all data shown in the clustergram.
DISCUSSION
Carbon source–independent GRR
We obtained genome-wide patterns of gene expression obtained from yeast growing at different steady-state growth rates and under diverse nutrient limitations, but all with ethanol as the only source of carbon and energy. These results have served to clarify the connection between GRR and YMC first noted in previous studies with glucose as sole carbon and energy source (Brauer at al., 2008). In ethanol, most of the genes identified by Brauer et al. (2008) have very similar GRRs (first eigenvector in Figure 3), but a subset of them have significantly different GRRs (third eigenvector). Thus the growth rate data from cultures grown on ethanol allow us to separate out the glucose-dependent part of the GRR described by Brauer et al. (2008) and to define a core set of growth rate–responsive genes independent of the source of carbon and energy and the nature of the limiting nutrient in steady-state growth.
Whenever the growth rate changes in response to any natural nutrient, each gene that displays a universal GRR either increases or decreases its level of expression. The large number of genes included in the universal GRR is a remarkable finding. It fulfills the expectation that making protein (see Table 2) is a major bottleneck for rapidly proliferating cells (Maaløe, 1979) and that slowly growing cells need to recycle proteins and organelles (see Table 1). Consistently, genes involved in mitochondrial translation are strongly overrepresented in the universal positive GRR, underscoring that under steady-state experimental conditions (slow growth in chemostats) all cultures respire even when there is substantial residual glucose and that growth of cells requires growth of the mitochondria.
Carbon source–dependent GRR
The genes that comprise the carbon source–specific GRR are defined by their different slopes of gene expression relative to growth rate in ethanol and glucose media. Many of these genes are genes involved in respiratory functions, including all the steps of ethanol metabolism. These genes have a positive slope with respect to growth rate in ethanol but a negative slope in glucose. This is clarifying, because it means that in glucose, yeast respire more as the steady-state growth rate decreases, whereas in ethanol, they respire less as the growth rate decreases. An additional inference we can make is that both of these responses, in glucose or in ethanol, happen even when the growth rate is limited by nutrients (phosphate or ammonium) other than carbon. This means, in turn, that the control of respiration at the systems level is by growth rate and carbon and energy metabolites.
It is worth noting that these results are consistent with what is known about respiratory control during the diauxic shift between exponential fermentative growth on glucose and subsequent exponential respiratory growth on the evolved ethanol. During the fermentative phase, oxidative metabolic genes increase in their expression level as the instantaneous growth rate slows (Brauer et al., 2005). After a period of nearly zero growth, when growth resumes on ethanol, the level of expression of these genes is higher still.
A related component of the carbon source–specific growth response is that the peroxisomal genes, which constituted a major part of the negative GRR in glucose, have positive GRR in ethanol, and thus are not part of the universal GRR. This switch in the GRR of peroxisomal genes likely reflects increased oxidation in peroxisomes (such as fatty acid beta-oxidation) at slow growth on glucose, which may not occur to the same level during fully respiratory growth on ethanol, which involves more mitochondrial oxidation. Similar to peroxisomal genes, genes related to oxidoreductase activity and coenzyme metabolism and to generation of precursors and energy have negative GRR in glucose but positive in ethanol. This change likely reflects different biosynthetic pathways depending on the carbon source (for example gluconeogenesis) as well as increased respiration and oxidative phosphorylation during fast growth on ethanol. In contrast, as the rate of growth on glucose increases, respiration and oxidative phosphorylation decrease and fermentation increases. Other biological processes overrepresented by the set of genes with negative GRR on glucose but not on ethanol include monosaccharide, carbohydrate, and hexose metabolic processes, which, for obvious reasons, are specific to growth on glucose.
Given the central role of precursor metabolites and energy in growth, it is surprising that this set of genes does not have significantly positive slopes on the glucose carbon source. A diverse group of genes involved in the synthesis of cofactors and vitamins or using cofactors have significantly positive slopes on the ethanol carbon source across all limitations. A subset of those genes also have modestly positive slopes in glucose across all limitations but, as a whole, the slopes in glucose of this gene set are not significantly different from the slopes for all genes. Many of the vitamin-related genes in this group are cofactors for mitochondrial enzymes that show transcriptional induction themselves, such as the enzymes catalyzing reactions from the TCA cycle. Thus up-regulation of genes from this set is another indicator of growth rate–induced increase in the TCA cycle and related biochemical reactions involved in the production of energy and intermediate metabolites. Many of the genes with oxidoreductase activity have positive slopes, and some of them are related to mitochondria, which reinforces the observation that most mitochondria-related genes have positive GRR on the ethanol carbon source. More importantly, the strong GRR of those genes indicates the challenges posed by respiratory growth and the physiological response mounted to meet them.
GRR and the YMC
The definition of a universal GRR also helps to clarify the connection between the GRR and the YMC. Whereas in the past the correlation between the two was strong, the removal of the glucose-specific features of the GRR reduced the relationship to one of identity. All the genes that are in the universal GRR set oscillate with the metabolic cycle in synchronized cultures. Furthermore, the relationship between growth rate and metabolic oscillations can be understood as variation in the relative durations of the HOC and LOC phase of the metabolic cycle. As growth rate decreases, the length of the LOC phase increases, and thus the expression levels of genes peaking during the LOC phase increase as well, while the reverse holds for the HOC phase and the expression of HOC genes.
These findings raise important questions. How is the growth rate dependence of oxygen consumption during the YMC reflected in gene expression? What is the exact relationship between the YMC measured in synchronized populations (Klevecz et al., 2004; Tu et al., 2005) and the single cell intrinsic cycle (Silverman et al., 2010)? Genes with universal GRR tend to be coexpressed in higher eukaryotes (Slavov and Dawson, 2009; Janes et al., 2010; Ozbudak et al., 2010), suggesting that the cell intrinsic cycle underlying the GRR may be conserved in higher eukaryotes. The relative prominence of the YMC in cells growing on ethanol and slowly on glucose suggests that there must be an underlying connection between the YMC and respiration. The possibility that the YMC is absent in purely fermentative conditions (i.e., fast growth in glucose) is an important question currently under investigation.
DNA synthesis, CDC, and the metabolic cycle
As noted above, experiments with metabolically synchronized cultures provided clear instances in which the HOC phase of the metabolic cycle coincides exactly with the S phase (DNA synthesis) of the CDC. This result casts some shadow over the nearly universal suggestion made by all groups studying the YMC (Klevecz et al., 2004; Tu et al. 2005; Chen et al., 2007; Chen and McKnight, 2007; Murray et al., 2007), including ourselves (Chen et al., 2007; Chen and McKnight, 2007; Silverman et al., 2010), that the logic and the evolutionary driver of metabolic cycling might have been the incompatibility of DNA replication with the generation of reactive oxygen species during respiration. Our data do not address these ideas directly, but they do suggest that further examination of this question is warranted. Our bud index data indicate that in fast-growing cultures a larger fraction of the cells is in the budded phases (S/G2/M) of the CDC when compared with slowly growing cultures. Based on this observation, fast-growing cultures should have higher population average expression for genes expressed during S/G2/M. Surprisingly, however, this is not what we observed or what Brauer et al. (2008) reported. We observed that the distributions of growth rate slopes for CDC genes are centered at 0 or even slightly negative values, indicating that the same mechanism that explains so well the connection between the genes with universal GRR and the YMC fails to explain the GRR of CDC genes. Yet, examining the slopes of individual genes indicated that the majority of CDC genes have the same-sign growth rate slopes across all conditions. Some CDC genes have large and consistently positive growth rate slopes, while other genes have large and consistently negative growth rate slopes. We currently cannot convincingly explain what distinguishes the CDC genes with positive and negative growth rate slopes. It is possible that many genes with functions in the CDC are expressed periodically in the YMC, and thus their GRRs cannot be explained only in terms of the growth rate changes in the CDC without accounting for the growth rate changes in the YMC.
On ethanol carbon source, the distribution of growth rate slopes for CDC genes is shifted toward more negative values compared with glucose carbon source. Many CDC genes still have growth rate slopes similar to those in glucose, and the major difference is that negative slopes in ethanol tend to be larger by absolute value and positive slopes in ethanol tend to be smaller in absolute value. This difference may be due to a difference in the growth rate dependence of the YMC on ethanol carbon source. In particular, growing on ethanol is likely to result in extended HOC phase and large changes in the LOC/HOC periods due to the additional burden of gluconeogenesis and completely respiratory growth.
MATERIALS AND METHODS
Cultures and physiological measurements
We used a haploid prototrophic strain (DBY11369) for all growth rate experiments on ethanol carbon source. The strain was selected for its good growth on ethanol, reaching μ = 0.15 h−1 in exponential phase of a batch culture with excess of all nutrients and at 30°C. All nonlimiting nutrient concentrations were the same as the ones used by Saldanha et al. (2004) and Brauer et al. (2005, 2008). The only carbon source in all cases was ethanol 30 mM for the ethanol limitation and 100 mM for the nitrogen and phosphorus limitations. All limiting concentrations were first determined as limiting for final biomass in batch growth and then confirmed to be limiting and within the linear response range in continuous cultures. The nitrogen-limited media contained [(NH4)2SO4] = 100 mg/l as the only source of nitrogen, and the phosphorous-limited media contained [KH2P O4] = 20 mg/l as the only source of phosphorous. Chemostats were established in 500-ml fermenter vessels (Sixfors, Infors AG, Bottmingen, Switzerland) containing 250 ml of culture volume, stirred at 400 rpm, and aerated with humidified and filtered air. Chemostat cultures were inoculated, monitored, and grown to steady state as described previously (Brauer et al., 2005). All cultures were monitored for changes in cell density and dissolved oxygen and grown until these parameters remained steady for at least 48h. The fraction of budded cells, cell density, cell sizes, and ethanol concentrations were measured as previously described by Brauer et al. (2005, 2008).
We synchronized metabolically a diploid prototrophic strain (DBY12007) with S288c background and wild-type HAP1 using starvation followed by refeeding. The media was limited on glucose 800 mg/l with composition described by Saldanha et al. (2004) and Brauer et al. (2005, 2008). Chemostats were established in 500-ml fermenter vessels (Sixfors) containing 300 ml of culture volume, stirred at 400 rpm, and aerated with humidified and filtered air.
Measuring mRNA
To measure RNA levels we sampled 10–30 ml of steady-state culture and vacuum filtered the cells, which were immediately frozen in liquid nitrogen and stored in a freezer at −80°C. RNA for microarray analysis was extracted by the acid–phenol–chloroform method. RNA was amplified and labeled using the Agilent low RNA input fluorescent linear amplification kit (P/N 5184–3523, Agilent Technologies, Palo Alto, CA). This method involves initial synthesis of cDNA by using a poly(T) primer attached to a T7 promoter. Labeled cRNA is subsequently synthesized using T7 RNA polymerase and either Cy3 or Cy5 UTP. Each Cy5-labeled experimental cRNA sample was mixed with the Cy3-labeled reference cRNA and hybridized for 17 h at 65°C to custom Agilent Yeast oligo microarrays 8 × 15 K having 8 microarrays per glass slide. Microarrays were washed and scanned with an Agilent DNA microarray scanner (Agilent Technologies), and the resulting TIF files were processed using Agilent Feature Extraction Software, version 7.5. Resulting microarray intensity data were submitted to the PUMA Database for archiving.
Models
Similar to Brauer et al. (2008) and Airoldi (2009), we use an exponential phenomenological model to quantify the dependence between the mRNA level and the growth rate, mRNAi = bieaiμ. In semi-log space, such an exponential model “appears” linear log(mRNAi) = log(bi) + aiμ and, assuming Gaussian noise, can be solved by least-squares linear regression. All slopes throughout this paper are computed based on regression models in the l2 sense, minimizing the sum of squared residuals. As a measure of goodness of fit, we use the fraction of variance explained by the model which for the jth gene is quantified by Rj:
 |
In (2), yj is a vector of expression levels of the jth gene, yj is its mean expression level, i is an index enumerating the set of conditions α used in the model and fij is the model predication for the ith condition and jth gene. To assess statistical significance, we use bootstrapped p values equal to the fraction of randomly permuted data sets having R2 equal or greater than the R2 of the data. More details are available in the Supplementary Material and at http://genomics-pubs.princeton.edu/grr.
Supplementary Material
Acknowledgments
We thank Tomas Fox for sharing with us the strain used in the growth rate experiments (DBY11369) and Benjamin Tu for sharing with us the oxygen consumption data during the YMC. We also thank Sanford J. Silverman, Amy Caudy, Viktor Boer, and David Gresham for technical advice and stimulating discussions, as well as Sanford J. Silverman, Patrick A. Gibney, and Edoardo Airoldi for feedback on the manuscript. Research was funded by a grant from the National Institutes of Health (GM046406) and the NIGMS Center for Quantitative Biology (GM071508).
Abbreviations used:
- CDC
cell division cycle
- FDR
false discovery rate
- GO
gene ontology
- GRR
growth rate response
- HOC
high oxygen consumption
- LOC
low oxygen consumption
- SVD
singular value decomposition
- TCA
tricarboxylic acid cycle
- YMC
yeast metabolic cycle
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0132) on April 27, 2011.
The sign of the correlation does not matter since positive and negative correlation mean the same in this context.
REFERENCES
- Airoldi EM, Huttenhower C, Gresham D, Lu C, Caudy AA, Dunham MJ, Broach JR, Botstein D, Troyanskaya OG. Predicting cellular growth from gene expression signatures. PLoS Comput Biol. 2009;5:e1000257. doi: 10.1371/journal.pcbi.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci USA. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder–open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo J, et al. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, McKnight SL. A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle. 2007;6:2906–2912. doi: 10.4161/cc.6.23.5041. [DOI] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- Fazio A, Jewett M, Daran-Lapujade P, Mustacchi R, Usaite R, Pronk J, Workman C, Nielsen J. Transcription factor control of growth rate dependent genes in Saccharomyces cerevisiae: a three factor design. BMC Genomics. 2008;9:341. doi: 10.1186/1471-2164-9-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B. Metabolic cycle, cell cycle, and the finishing kick to start. Genome Biol. 2006;7:107. doi: 10.1186/gb-2006-7-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub G, Kahan W. Calculating the singular values and pseudo-inverse of a matrix. J SIAM Numer Anal, Ser B. 1965;2:205–224. [Google Scholar]
- Hartwell LH. Saccharomyces cerevisiae cell cycle. Microbiol Mol Biol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:4651. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol. 1977;75:422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes A, Zhang N, Wu J, Butler PR, Hauser NC, Hoheisel JD, Lim FL, Sharrocks AD, Oliver SG. Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods. 2002;26:281–290. doi: 10.1016/S1046-2023(02)00032-4. [DOI] [PubMed] [Google Scholar]
- Janes KA, Wang C-C, Holmberg KJ, Cabral K, Brugge JS. Identifying single-cell molecular programs by stochastic profiling. Nat Methods. 2010;7:311–317. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keulers M, Suzuki T, Satroutdinov AD, Kuriyama H. Autonomous metabolic oscillation in continuous culture of Saccharomyces cerevisiae grown on ethanol. FEMS Microbiol Lett. 1996;142:253–258. doi: 10.1111/j.1574-6968.1996.tb08439.x. [DOI] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaløe O. The regulation of the protein-synthesizing machinery ribosomes, tRNA, factors, and so on. In: Goldberger RF, editor. Biological Regulation and Development. New York: Plenum; 1979. [Google Scholar]
- Malioutov D, Çetin M, Willsky AS. Homotopy continuation for sparse signal representation. Proceedings of the IEEE Proceedings of the IEEE International Conference on Acoustics, Speech, and Signal Processing; Hoboken, NJ. Wiley-IEEE Press; 2005. pp. 733–736. [Google Scholar]
- Murray DB, Beckmann M, Kitano H. Regulation of yeast oscillatory dynamics. Proc Natl Acad Sci USA. 2007;104:2241–2246. doi: 10.1073/pnas.0606677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Avino JP, et al. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast. 1999;15:829–842. doi: 10.1002/(SICI)1097-0061(199907)15:10A<829::AID-YEA423>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Tassy O, Pourqui O. Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc Natl Acad Sci USA. 2010;107:4224–4229. doi: 10.1073/pnas.0909375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pir P, Kirdar B, Hayes A, Onsan ZI, Ulgen K, Oliver S. Integrative investigation of metabolic and transcriptomic data. BMC Bioinformatics. 2006;7:203. doi: 10.1186/1471-2105-7-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Grotkjaer T, Winther O, Fausboll A, Akesson M, Bro C, Hansen L, Brunak S, Nielsen J. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Prix F, et al. Functional analysis of the and gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+ -dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology. 2004;150:2209–2220. doi: 10.1099/mic.0.26999-0. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman SJ, et al. Metabolic cycling in single yeast cells from unsynchronized steady-state populations limited on glucose or phosphate. Proc Natl Acad Sci USA. 2010:6946–6951. doi: 10.1073/pnas.1002422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Dawson KA. Correlation signature of the macroscopic states of the gene regulatory network in cancer. Proc Natl Acad Sci USA. 2009;106:4079–4084. doi: 10.1073/pnas.0810803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 2009;5:245–245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.