The distribution and function of aPKC are examined during sea urchin ciliogenesis. The kinase concentrates in a ring at the transition zone between the basal body and the elongating axoneme. Inhibition of aPKC results in mislocalization of the kinase and defective ciliogenesis. Thus aPKC controls the growth of motile cilia in invertebrate embryos.
Abstract
The atypical protein kinase C (aPKC) is part of the conserved aPKC/PAR6/PAR3 protein complex, which regulates many cell polarity events, including the formation of a primary cilium at the apical surface of epithelial cells. Cilia are highly organized, conserved, microtubule-based structures involved in motility, sensory processes, signaling, and cell polarity. We examined the distribution and function of aPKC in the sea urchin embryo, which forms a swimming blastula covered with motile cilia. We found that in the early embryo aPKC is uniformly cortical and becomes excluded from the vegetal pole during unequal cleavages at the 8- to 64-cell stages. During the blastula and gastrula stages the kinase localizes at the base of cilia, forming a ring at the transition zone between the basal body and the elongating axoneme. A dose-dependent and reversible inhibition of aPKC results in mislocalization of the kinase, defective ciliogenesis, and lack of swimming. Thus, as in the primary cilium of differentiated mammalian cells, aPKC controls the growth of motile cilia in invertebrate embryos. We suggest that aPKC might function to phosphorylate kinesin and so activate the transport of intraflagellar vesicles.
INTRODUCTION
Many motility or sensory processes rely on very conserved microtubule structures known as cilia or flagella (Gibbons 1981; Eggenschwiler and Anderson, 2007; Inaba, 2007; Pedersen and Rosenbaum, 2008; Nachury et al., 2010; Seeley and Nachury, 2010). Recent findings have revealed that cilia may also be involved in cell signaling and polarity establishment (Satir and Christensen, 2007; Anderson et al., 2008; Fischer and Pontoglio, 2009; Goetz and Anderson, 2010; Seeley and Nachury, 2010). Consequently, one expects that disorders affecting cilia structure, motility, or function may have important consequences for developmental events and cell physiology (Nigg and Raff, 2009; Konno et al., 2010). From studies of the single-celled alga Chlamydomonas, we have learned that virtually all cilia, including primary and sensory organ cilia, are constructed by a process called intraflagellar transport (IFT) (Snell et al., 2004; Satir, 2007), which assembles preexisting protein building blocks, such as tubulin and dynein, and a number of nine plus two architectural elements that are synthesized de novo during ciliogenesis (Pedersen and Rosenbaum, 2008).
Ciliogenesis occurs during sea urchin embryo development immediately prior to hatching into a swimming blastula and it ensures embryo motility up to the feeding pluteic larva stage. After fertilization, a series of rapid cleavages gives rise to a blastoderm consisting of several hundred cells arranged as a spherical monolayer of epithelial cells surrounding the blastocoel (Figure 1, A–G). At this stage, each blastomere synthesizes during interphase one single motile cilium (Figure 1H), itself composed of an axoneme made of a scaffold of nine doublet and two singlet microtubules and several hundred additional proteins attached to a basal body (Dutcher, 1995; Dawe et al., 2007; Silverman and Leroux, 2009). In the sea urchin swimming embryo, as in other multicellular models, the cilia emanate from basal bodies situated right under the apical surface of blastula epithelial cells (Stephens, 2008). During mitosis, each cell retracts and disassembles its axoneme, builds a mitotic spindle with the centrioles as spindle poles, divides, and then returns its centriole pair to the apical cell surface to grow a new cilium (Masuda and Sato, 1984; Dawe et al., 2007). Ciliogenesis is linked to the animal–vegetal cell polarity in the sea urchin embryo. While the blastula starts gastrulating (Figure 1I), the vegetal blastomeres grow short and very active cilia, which follow a metachronic beating movement coordinated along the animal–vegetal axis to provide directional motility to the blastula and propel it in seawater (Stephens, 1995). On the animal side a small group of epithelial cells, which appear thicker than the vegetal cells, form longer and poorly active cilia, which protrude anteriorly to the embryo in an entangled way (Figure 1, I–K). This “apical tuft” plays the role of rudder, preventing random swimming (Stephens, 2008).
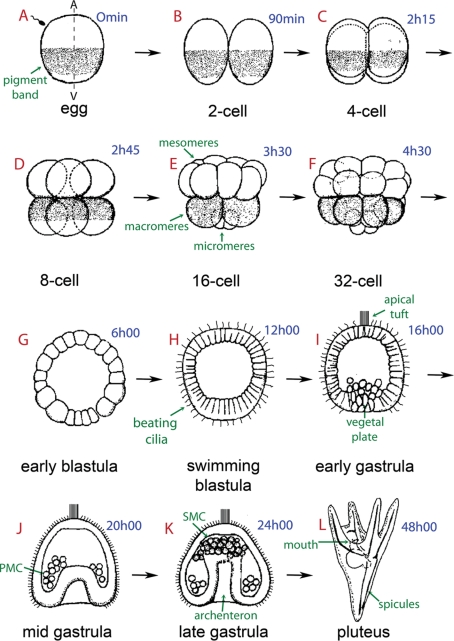
Figure 1:
Schematic representation of sea urchin embryonic development. The times in blue indicate hours postfertilization. The sea urchin oocyte is organized along an animal–vegetal (A/V) primary axis, which is observable in P. lividus due to the presence of a subequatorial cortical pigmented band visible under blue light (arrow in A). Fertilization is followed by first and second cleavages, which are meridional (parallel to the A/V axis) and perpendicular to each other (B, C). The third cleavage is equatorial, perpendicular to the first two cleavage planes, and separates the animal and vegetal hemispheres from one another (D). During the fourth cleavage, animal blastomeres divide equally to produce eight mesomeres, and the vegetal blastomeres divide unequally to produce large macromeres and small micromeres located at the vegetal pole of the embryo (E, F). Six hours after fertilization the sea urchin embryo enters the early blastula stage with an empty central cavity called a blastocoel (G). The cells start developing cilia on their outer surface to form a swimming blastula (H). Approximately 10–12 h after fertilization, the midblastula, composed of ∼600 cells, hatches out of the fertilization envelope (not shown). At the animal pole, the cilia are longer but do not beat. This “apical tuft” (I) provides directionality to swimming, as embryos almost always move with the apical tuft region forward. In the late blastula stage the embryo becomes thickened at the vegetal pole, forming the vegetal plate (I). This represents the gastrulation site where the primary mesenchyme cells (PMCs), which are derived from the micromeres and located in the center of the vegetal plate region, migrate into the blastocoel (I, J). The vegetal cells will continue to ingress in order to form the archenteron, led by filopodia extending from the secondary mesenchyme cells (SMCs), which eventually contact the animal pole at the future site of mouth formation (K). A prism (not shown) and finally a feeding pluteic larva will be formed 24 h later around an endoskeleton, which contains two spicules made of calcium carbonate secreted by the PMCs (L). A few days later, this pluteus will metamorphose into a tiny male or female adult urchin.
A set of key polarity regulators—Par3, Par6, and atypical protein kinase C (aPKC)—has been identified in all animal cells so far examined (Goldstein and Macara, 2007). These three proteins form a complex that is activated by the small G protein CDC42 (Munro, 2006; McCaffrey and Macara, 2009) and localizes asymmetrically along the cell periphery and regulates cell polarity. In mammalian epithelial cells the aPKC–PAR6–PAR3 complex associates with tight junctions, where its main function is to establish apical–basolateral polarity (Assémat et al., 2008). In general, PAR proteins were thought to control cortical actomyosin, thereby creating specialized cortical domains, which in turn would lead to specific spindle positioning and determinant localization (Cowan and Hyman, 2007; Munro and Bowerman, 2009). However, accumulating evidence supports the idea that the aPKC–PAR6–PAR3 complex also directly modulates several motility events commonly associated with the microtubule cytoskeleton. For instance, PAR6 associates with the spindle during meiosis in the mouse embryo (Vinot et al., 2004) and with the mitotic spindle together with PAR3 and aPKC in HeLa cells (Liu et al., 2006). The PAR complex also regulates polarized localization of the microtubule-organizing center (MTOC) during Drosophila melanogaster oogenesis (Goldstein and Macara, 2007) and polarized migration of wounded astrocytes (Etienne-Manneville and Hall, 2003). Moreover, the PAR complex, together with the Crumbs epithelial polarity complex (Bulgakova and Knust, 2009), has been shown to participate in primary cilium formation in cultured MDCK cells probably through its interaction with the microtubule motor KIF3A (Fan et al., 2004). Finally, aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during Drosophila development (Harris and Peifer, 2007).
The role of aPKC in early sea urchin development has been investigated in Strongylocentrotus purpuratus during early cleavages following fertilization (Alford et al., 2009). It was found that inhibition of aPKC at the two-cell stage leads to the formation of multipolar spindles, short asters, and improper spindle rotation. The inhibition of aPKC after the four-cell stage does not block cleavage but results in cleavage plane defects and aberrant subsequent unequal cleavages. It is only when inhibition occurs after the 16-cell stage that a blastula can form even though its epithelium does not look normal. The authors of this work concluded that cell polarity emerges at first cleavage in sea urchin embryos (Alford et al., 2009).
In this article we examine the distribution of aPKC during later embryonic development of the sea urchin Paracentrotus lividus embryo and analyze its function during ciliogenesis. We observe that this kinase, initially present in the whole cortex of the early embryo, is asymmetrically distributed starting from the 16-cell stage and is excluded from the vegetal pole, where asymmetric divisions occur and give rise to vegetal micromeres. We show that the most striking asymmetric distribution of aPKC appears during ciliogenesis, when the kinase is localized not only in the cortex and the membranes between cells, but also, remarkably, in the region of ciliary basal bodies. Inhibition of aPKC leads to altered cilium growth and defective swimming ability. Thus we conclude that in the sea urchin embryo, as in differentiated mammalian cells, a conserved function for aPKC is to control ciliogenesis.
RESULTS
Sea urchin aPKC has been addressed in two recent studies, which described its cloning and expression pattern in Hemicentrotus pulcherrimus (Shiomi and Yamaguchi, 2008) or its role in polarity establishment during first cleavage in Strongylocentrotus purpuratus and Lytechinus pictus (Alford et al., 2009). Here we resolve the puzzling finding (Shiomi and Yamaguchi, 2008) that the S. purpuratus genome predicts an aPKC with an unusual truncated amino terminus (Sp in Supplemental Figure S1), whereas the H. pulcherrimus cDNA encodes a normal full-length aPKC (Hp in Supplemental Figure S1). We go on examine the localization and function of aPKC in the sea urchin P. lividus.
Sea urchin embryos contain two aPKC isoforms
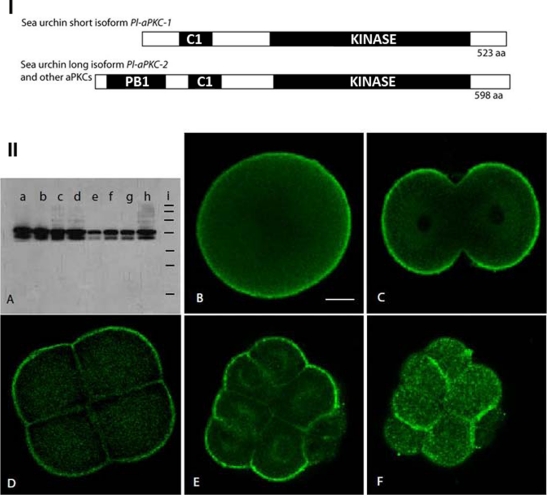
To clone the aPKC homologue from P. lividus, a DNA fragment encoding part of the well-conserved kinase domain was amplified by RT-PCR using degenerate oligonucleotide primers. The screen of a P. lividus egg cDNA library followed by 5′-RACE yielded two classes of clones, which differed in their 5′ termini and encoded proteins of 523 and 598 amino acids (Figure 2-I). The deduced protein sequence of the shorter P. lividus aPKC isoform (Pl-aPKC-1) is 98% identical to the predicted open reading frame encoding S. purpuratus Sp-aPKCι (National Center for Biotechnology Information [NCBI] Reference Sequence XP_780275.1) (Supplemental Figure S1). Pl-aPKC-1 and Sp-aPKC contain the characteristic cysteine-rich C1 and kinase domains, but, of interest, lack the Phox and Bem (PB1) protein interaction domain, which allows aPKC to bind PAR6 to form heterodimers (Hirano et al., 2005; Sumimoto et al., 2007). All other known aPKC proteins contain this PB1 domain, including those from nematodes, insects, and chordates. On the other hand, the long P. lividus aPKC isoform obtained in our screen (Pl-aPKC-2) is unlike Sp-aPKC at the amino-terminal region but is 95% identical to the H. pulcherrimus aPKC and exhibits 70% identity and 81% homology with its human counterpart (Supplemental Figure S1).
Figure 2:
I. Domain structure of sea urchin aPKCs. (A) The short isoform found so far in P. lividus and S. purpuratus encodes an aPKC that lacks the PB1 protein interaction domain. Kinase: serine threonine kinase domain; C1: cysteine-rich domain, which binds InsPtd(3,4,5)P3. The three urchin species examined so far (P. lividus, H. pulcherrimus, and S. purpuratus) also contain a long isoform that is similar to most other known aPKCs in its domain composition (see Supplemental Figure S1 for alignment). II. Two maternal aPKC proteins that localize in the cortex in early embryos of P. lividus. (A) Western blot with the SC216 anti-aPKC antibody on P. lividus extracts from (a) unfertilized eggs, (b) 2-cell stage, (c) 8-cell stage, (d) 16- to 32-cell stage, (e) swimming blastula, (f) early gastrula, (g) prism, (h) plutei. Equal amounts of protein were loaded in each lane. (i) Standard molecular weight markers from top to bottom: 200, 120, 100, 70, 50, 37, and 20 kDa. (B–F) Immunolocalization of aPKC in early sea urchin embryos observed by confocal microscopy. (B) Unfertilized egg, (C) 2-cell stage, (D) 8-cell stage, (E, F) two confocal sections of the same 16-cell embryo showing (E) the interior and (F) the surface. Scale bar, 20 μm.
As it was surprising that S. purpuratus aPKC would lack the conserved PB1 domain, especially since it was shown to interact with PAR6 (Alford et al., 2009), we searched the S. purpuratus genome for homology to the N-terminal sequence specific to our long isoform Pl-aPKC-2. Indeed, on the same scaffold 308/61100 (BAC clone accession numbers NW-001282827 and NW-001470470), we detected just 3 kb upstream of the 14 exons reported to encode Sp-aPKC (Shiomi and Yamaguchi, 2008) three additional exons that encode a strong PB1 domain but had been misannotated as Tanc-2, an ankyrin repeat protein (SPU_008179 in SpBase.org). The long and short transcripts, which are apparently produced by alternative splicing of a single aPKC gene, are both expressed in the S. purpuratus embryo according to Expressed Sequence Tag (NCBI) and microarray (http://urchin.nidcr.nih.gov/) databases.
The spatial expression pattern of P. lividus aPKC mRNA was determined using a probe corresponding to the kinase domain, which should hybridize to both long and short transcripts. As in H. pulcherrimus (Shiomi and Yamaguchi, 2008), strong zygotic expression was observed at the gastrula stage in cells at the vegetal pole starting to invaginate, the primary mesenchyme, and at the tip of the archenteron (Supplemental Figure S2, F–H). No signal was evident in P. lividus eggs or in embryos prior to the blastula stage (Supplemental Figure S2, A–E), although in H. pulcherrimus there is a low level of maternal signal detectable by RT-PCR (Shiomi and Yamaguchi, 2008).
P. lividus aPKCs are maternal proteins that localize in the cortex in early embryos
The temporal and spatial distribution of aPKC proteins in P. lividus embryos was determined using an antibody targeted to the conserved C-terminal 20 amino acids (boxed in Supplemental Figure S1), which specifically recognizes aPKC in a wide variety of vertebrate and invertebrate species (Patalano et al., 2006). On immunoblots of P. lividus protein extracts (Figure 2-IIA), the aPKC antibody detected two bands in the 60- to 70-kDa weight range, corresponding to the expected sizes for the Pl-aPKC-1 and Pl-aPKC-2 as calculated from their sequences (60 and 68 kDa, respectively). Both positive signals disappeared when the antibody was preincubated with the immunogenic peptide but not with unrelated peptides (unpublished data). It is unlikely that the lower band represents a degradation product of the higher one, as a high concentration of proteolysis inhibitors was used during extract preparation. The aPKC proteins are abundant in unfertilized eggs and early embryonic stages until blastula, when their level decreased (Figure 2-IIA, lanes a–e). The amount of aPKC isoforms then increased at early gastrula and later stages (Figure 2-IIA, lanes f–h). However, the zygotic expression of the shorter isoform appears to be proportionally less than that of the longer form during late developmental stages. This has been confirmed by quantitative analysis of the signals corresponding to both isoforms (unpublished results).
Immunofluorescence localization of aPKC on fixed P. lividus embryos revealed a strong and uniform staining at the cell periphery of all blastomeres from unfertilized egg to 8-cell stage embryos (Figure 2-II, B–D, and Supplemental Figure S4). Starting at fourth cleavage (8- to 16-cell stage), micromeres are formed when one spindle pole moves close to the vegetal cortex (Schroeder, 1987). At the 16-cell stage, a clear asymmetric distribution of aPKC can be observed (Figure 2, E and F): there is a strong staining of the cortex in animal blastomeres, whereas no staining was apparent in the vegetal cortex of the four micromeres. This asymmetric distribution of aPKC persisted during the following two asymmetric divisions occurring between 16- and 64-cell stages as aPKC was continually excluded from the vegetal cortex (Supplemental Figure S3). These results are similar to what is observed in the Caenorhabditis elegans zygote, where the mitotic spindle migrates toward the posterior cortical region, which lacks the complex Par3–Par6–aPKC, causing unequal cleavage (Cowan and Hyman, 2007).
Sea urchin aPKC associates with microtubule structures during mitotic divisions
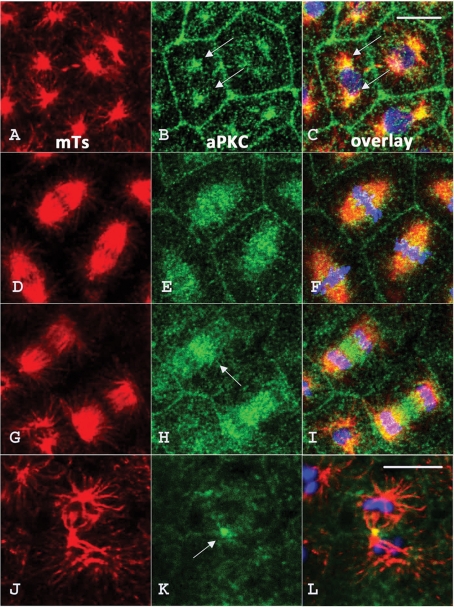
A careful examination of the staining of 16-cell embryos reveals that in addition to enrichment at the cortical layer, a significant amount of aPKC protein resides in the region of mitotic structures in the cytoplasm (Figure 2-IIE), suggesting that aPKC could also bind to microtubules. To further investigate this possibility, we double-stained embryos fixed at different stages of mitosis for microtubules and aPKC. Figure 3 shows the results obtained for a population of 64-cell stage embryos in which mitotic divisions are still fairly synchronous, except for the descendants of micromeres. During prophase (Figure 3, A–C), aPKC is found not only at the cell periphery but also in the two MTOCs (arrows in Figure 3, B and C) on both sides of the nucleus. During metaphase (Figure 3, D–F), aPKC is mostly present all along spindle microtubules, whereas at anaphase, aPKC becomes enriched in the region of the spindle midzone (Figures 3, G–I, arrow in H), suggesting that aPKC might travel toward the plus end of microtubules. The specific accumulation of aPKC in the midbody at the end of mitosis seems to confirm this possibility (Figure 3, J–L, arrow in K).
Figure 3:
aPKC is associated with microtubule structures during mitotic divisions in the early sea urchin embryo. Sixty-four–cell stage embryos were fixed and labeled with DM1A antitubulin antibody (red) and anti-aPKC antibody (green). aPKC associates with the MTOC (arrows in B and C) during prophase (A–C); decorates spindle microtubules during metaphase (D–F); starts to concentrate in the spindle midzone (arrow) during anaphase (G–I), and ends up in the midbody (arrow) during telophase (J–L). The nucleus (in C) and chromosomes (in F, I, and L) are stained in blue with Hoescht stain. Bar, 5 μm.
Sea urchin aPKC concentrates in a ring-shaped structure near the basal body of ciliated cells in swimming embryos
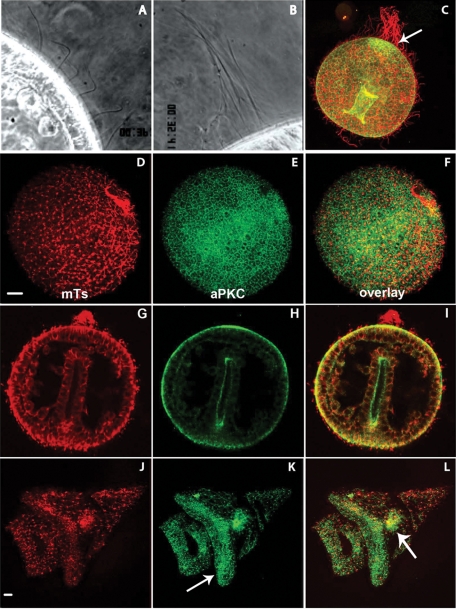
The localization of aPKC was examined in later stages of embryogenesis. Sea urchin embryos start swimming right after hatching due to the metachronic beating of hundreds of cilia, which lie all around the blastula (Figure 1H) and can be observed under dark-field or phase-contrast microscopy (Figure 4A). A handful of cilia located at the animal pole appear twice as long (∼40 μm) as beating cilia but are mostly immobile and often rolled up all around each other (Figure 4B). This “apical tuft” acts as a rudder to allow the linear swimming of the embryo. Labeling with antitubulin antibody shows that the cilia are preserved after methanol fixation in blastula-, gastrula-, and pluteus-stage embryos (Figure 4, D, G, and J). The immunofluorescence signal obtained for aPKC labeling was essentially the same in blastula and gastrula: the aPKC antibody stains the apical and lateral surface of all blastomeres (see Figure 5, H and I, for lateral view). The apical membrane of the internal archenteron is also labeled in gastrula-stage embryos (Figure 4C), closely correlating with a localized burst in zygotic expression (Supplemental Figure S2, F–H). Of interest, aPKC signal also concentrates at the animal pole, at the basis of the apical tuft (Figure 4, H and I, arrow in C). This strong enrichment of aPKC protein is likely due to accumulation of preexisting maternal protein because no zygotic expression of aPKC mRNA was observed in this region in P. lividus (Supplemental Figure S2) or in H. pulcherrimus (Shiomi and Yamaguchi, 2008). In late pluteus larva, aPKC protein is also detected at the basis of very long cilia nearby the mouth orifice, as well as in the ciliary band (Figure 4K, arrow), and in the stomach endoderm (Figure 4L, arrow).
Figure 4:
aPKC localization in ciliated swimming sea urchin embryos. (A, B) Dark-field videomicroscopy showing (A) short, active cilia and (B) the “apical tuft” made of a few long and immobile cilia at the animal pole. (C–L) Confocal sections of swimming embryos fixed and labeled for microtubules (red) and aPKC (green). (C) Early gastrula showing the longer cilia (in red) and the concentration of aPKC at the animal pole (arrow). (D–F) Swimming blastula. (G–I) Late gastrula. (J–L) Pluteus larva; arrows: ciliary band in K and stomach in L.
Figure 5:
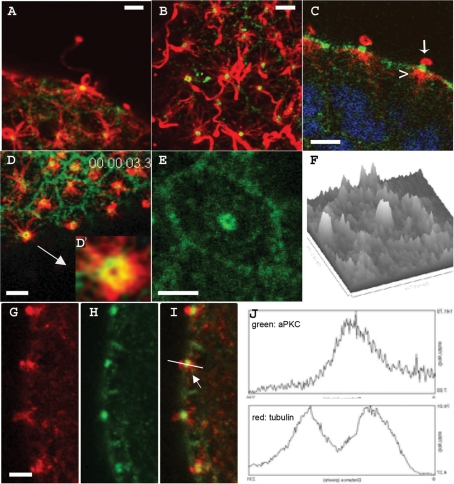
Sea urchin aPKC localizes to a disk situated between the basal body and the elongating axoneme of motile cilia. (A–E, G–I) Confocal sections of cells on the surface of swimming blastulas stained for microtubules (red) and aPKC (green). (A, B, D, E) Surface views; (C, G–I) sagittal views. The arrowhead and arrow in C show the absence of aPKC in the basal body and in the elongating axoneme, respectively. (D′, E) Higher-magnification views of aPKC ring-shaped labeling at the bases of two different cilia. (F) Relative intensity of the aPKC signal observed in (E) using the surface plot quantitation function of the ImageJ program. (J) Relative quantitation of intensity of the fluorescence signals (y-axis) over distance (x-axis) obtained in the green (top) and red (bottom) channels along the line drawn in I using the plot profile function of the ImageJ image processing program. All bars, 2 μm.
Detailed analysis at higher magnification shows that aPKC protein concentrates around the base of cilia in swimming sea urchin embryos (Figure 5, A and B). The core of each cilium is composed of an axoneme elongating from and anchored within the cell basal body. A careful examination of sagittal confocal sections through the middle of ciliary structures showed that aPKC signal was located neither at the level of the basal body within the cell (Figure 5C, arrowhead) nor at the level of the elongating cilium outside the cell (Figure 5C, arrow) but rather between them in a zone defined as the cilium transition zone (Eggenschwiler and Anderson, 2007). This was confirmed with a γ-tubulin antibody, which labels the basal body within the cell at a position distinct from and adjacent to the aPKC label (Supplemental Figure S5). Quantitative analysis of fluorescence signal intensities (Figure 5J) showed clearly that the peak intensity corresponding to aPKC (green) occurred at a location different from that of the maxima of the tubulin signals (red). Visual and quantitative analysis of higher-magnification surface views (Figures 5, D, D′, and F) showed that aPKC is associated with a structure forming a ring around the axoneme in the transition zone below the plasma membrane. Thus we conclude that aPKC is enriched in the cilium transition zone, which is where IFT particles are loaded onto kinesins before being transported toward the tip of the cilium (Silverman and Leroux, 2009).
aPKC is required for ciliogenesis during sea urchin development
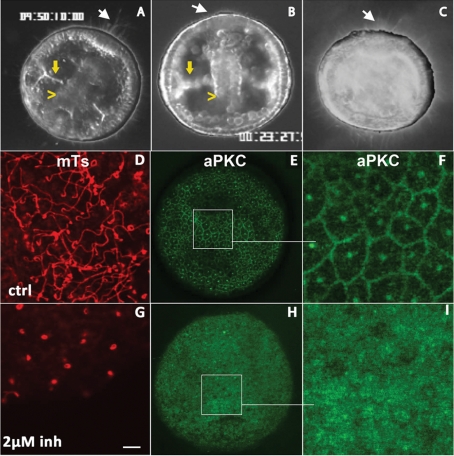
The localization of aPKC in the transition zone of the cilium suggests that it plays a role in sea urchin embryo ciliogenesis. To test this possibility, we analyzed the effect of aPKC inhibitors on the growth process of cilia. First we recorded by videomicroscopy under dark-field illumination the development and behavior of 24-h swimming gastrulas (Figure 1K), which were cultured for 2 h either in the absence (Figure 6A) or presence (Figure 6B) of the inhibitor GF109203X, which has been successfully used to block aPKC function in astrocytes (Etienne-Manneville and Hall, 2003). The overall morphology of the embryos treated with the inhibitor was similar to that of control embryos, with apparent normal archenteron ingression and spicule formation (compare Figure 6A and B, yellow arrows and yellow arrowheads). The only difference between control and treated embryos was the short size of the cilia and the absence of an apical tuft at the animal pole (Figure 6B, white arrow) when aPKC inhibitor is present. These treated embryos all displayed defective swimming. To rule out the possibility that cilia shortening was due to a nonspecific effect of GF109203X on other PKCs, we repeated the experiment using Gö 6976, a potent inhibitor of Ca2+-activated PKC shown to have no effect on aPKCs (Rosse et al., 2009). In this case, the morphology of the treated embryos differs significantly from that of the control (Figure 6C): the archenteron does not form, and the skeletal spicules cannot be detected. However, ciliogenesis did not seem to be perturbed in these Gö 6976–treated embryos, as they were able to swim properly and displayed a regular apical tuft at their animal pole (Figure 6C, arrow). The requirement of aPKC for ciliogenesis was confirmed using a second aPKC inhibitor—the very specific aPKC pseudosubstrate peptide. In embryos treated with 2 μM pseudosubstrate peptide, cilia appear much shorter compared with control (Figure 6, G and D, respectively). Because inhibitor was added to swimming gastrula embryos whose cilia were already formed, this indicates that aPKC is required for maintenance of cilia length. Cell division during the incubation period may also contribute to the apparent shortening of cilia, as reflecting impaired ciliogenesis in the new daughter cells. In addition, aPKC localization was perturbed: the normal enrichment at cell membranes and the base of cilia seen in control embryos (Figure 6, E and F) was not observed in embryos treated with pseudosubstrate (Figure 6, H and I). Thus inhibition of aPKC by its pseudosubstrate peptide resulted in the mislocalization of the kinase.
Figure 6:
Sea urchin embryos treated with aPKC inhibitors do not swim and exhibit short cilia. (A–C) Dark-field videomicroscopy images taken from time-lapse sequences of late gastrulas control (A) or treated with the PKC inhibitors GF109203X, 10 μM (B) and Gö 6976, 20 μM (C). White arrows indicate the presence (A, C) or the absence (B) of the apical tuft at the animal pole. Yellow arrowheads and yellow arrows indicate the archenteron and the forming spicules, respectively. (D–I) Confocal surface views of gastrula-stage embryos either (D–F) untreated or (G–I) after treatment with the myristoylated aPKC pseudosubstrate inhibitor. Fixed embryos were labeled for tubulin (D, G) or aPKC (E, F, H, I). F and I are higher-magnification views of the regions boxed in E and H.
Cilia growth limitation by aPKC pseudosubstrate inhibitor is dose dependent and reversible
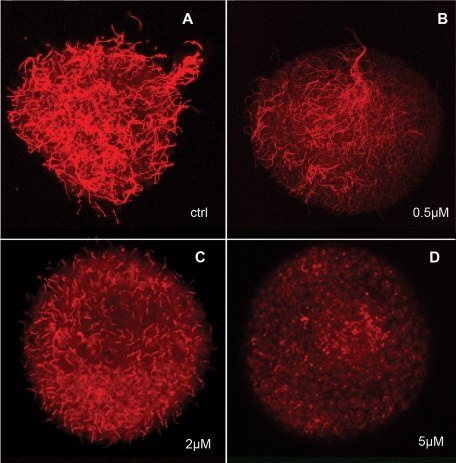
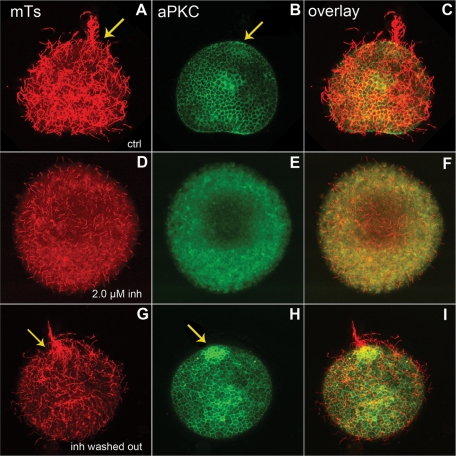
To quantify the effect of aPKC inhibitor on cilia growth, a “deciliation–reciliation” assay was used (Auclair and Siegel, 1966). This method generates a homogeneous, cilia-less starting point from which cilia growth measurements can be compared. Twenty-four-hour–old swimming gastrulas were deciliated using a high osmotic stress in hypersaline (∼1.3 M NaCl) seawater for 5 min. This treatment causes the clear cut of the axoneme from the basal body without affecting cell contacts (Stephens, 1995). Embryos were allowed to regrow their cilia in MFSW supplemented with different concentrations of pseudosubstrate inhibitor and then observed for swimming behavior or fixed for immunofluorescence and cilia length measurements (Figure 7). Embryos treated with a low concentration of aPKC inhibitor (0.5 μM) were able to regrow cilia slightly shorter than those of controls (Figure 7B and Table 1) and were able to swim linearly because of the presence of an adequate apical tuft at the animal cap. Such was not the case at higher inhibitor concentrations: average ciliary length dropped rather rapidly, reaching 0 for 5.0 μM inhibitor (Figure 7D and Table 1). This effective pseudosubstrate inhibitory concentration is lower than that generally used to inhibit aPKC in mammalian cells (10–50 μM) (Sun et al., 2005) but similar to the 4 μM used to inhibit aPKC during S. purpuratus early development (Alford et al., 2009). The effect of aPKC inhibition on ciliogenesis is almost fully reversible, as 85% of embryos washed after treatment with 2 μM pseudosubstrate inhibitor and left to reciliate showed a ciliary pattern similar to that of control embryos (compare Figure 8, A–C and G–I) and aPKC localization was restored (Figure 8I); note the normal apical aPKC enrichment at the animal pole (Figure 8I, arrow).
Figure 7:
Cilia length is proportional to level of aPKC inhibition. Confocal surface views of embryos fixed and labeled for tubulin. Embryos were deciliated by osmotic stress and then allowed to reciliate in the absence (A) or in the presence (B–D) of myristoylated aPKC pseudosubstrate inhibitor. Inhibitor final concentrations used are indicated.
TABLE 1:
Average length of cilia in control and in embryos treated with increasing amounts of myristoylated aPKC pseudosubstrate inhibitor. The given values were obtained by measuring at least 30 cilia on four or five different embryos for each concentration of the inhibitor.
| Inhibitor concentration, μM | Average cilium length, μm | Apical tuft | Swimming |
|---|---|---|---|
| 0 | 21,7 ± 1.7 | Present | Linear |
| 0.5 | 18.3 ± 1.2 | Present | Linear |
| 1.0 | 12.1 ± 2.1 | Absent | Circular |
| 2.0 | 7.4 ± 1.3 | Absent | No |
| 5.0 | No cilia | Absent | No |
Figure 8:
The inhibition of aPKC and its effect on ciliogenesis can be reversed. (A–C) Control embryos were deciliated and left to reciliate for 2 h before fixation and staining for microtubules (red) and aPKC (green). The arrows show the apical tuft and the accumulation of aPKC at the animal pole. (D–F) Embryo deciliated and left to reciliate in the presence of 2.0 μM pseudosubstrate, fixed, and stained like the control. (G–I) Embryo treated as in D, then washed to remove the inhibitor and allowed to reciliate for 2 h. The arrows show the reformation of the apical tuft and apical accumulation of aPKC when the kinase is no longer inhibited.
DISCUSSION
Two aPKC isoforms in sea urchins present in eggs and early embryos
This study reveals that in the sea urchins P. lividus and S. purpuratus two isoforms of the ser/thr kinase aPKC are expressed: the complete aPKC, which is closest to the aPKC iota of vertebrates, and a shorter form that lacks the N-terminal 228 nucleotides encoding the PB1 domain (Figure 2 and Supplemental Figure S1). In the case of H. pulcherrimus, a directed search by 5′-RACE failed to find the alternative short form (Shiomi and Yamaguchi, 2008), but it may have escaped detection due to a low level of transcripts or because it is provided as a maternal protein as in P. lividus. The detection of the two forms in S. purpuratus emphasizes the importance of manually verifying gene model predictions made by automated genome annotation. Frequently, incomplete or erroneous annotation leads to mistaken conclusions about coding sequences, as has been uncovered for ascidian PAR3 (Patalano et al., 2006) and numerous myosin genes (Odronitz and Kollmar, 2007).
The novel sea urchin aPKC isoform must have special roles during development, but given the strong maternal stock of the two aPKC proteins (Figure 2-IIA), it will be difficult to analyze their functions separately. An aPKC protein lacking the PB1 domain would be unable to interact with a variety of partners, notably PAR6, which inhibits aPKC in a CDC42- or phosphorylation-dependent manner (Wirtz-Peitz et al., 2008). Thus this truncated form of aPKC may respond to some other signaling pathway or may be less sequestered on membranes and hence free to localize elsewhere. Along these lines, possibly the two aPKC isoforms could correspond to the two subcellular localizations we observed—cortical or microtubule structures (Figures 2-II and 3, respectively). Quantitative analysis of aPKC signals on developmental Western blots (unpublished data) reveals that the longer isoform is proportionally more abundant than its shorter counterpart in swimming embryos. This might suggest that the longer isoform is responsible for the control of ciliogenesis. However, the fact that zygotic expression occurs only in inner cells during late development indicates that this new aPKC protein is not produced in the ciliated cells. Thus it will be necessary to raise antibodies specific for the unique N termini of both isoforms to determine whether the two maternal aPKC proteins are partitioned into distinct cell lineages and whether one of them is specifically allocated to cilia. It will also be interesting to determine whether the unusual urchin aPKC lacking the PB1 domain is present in any other animal groups. To our knowledge, the only other case of shortened aPKC isoforms is found in ascidians, in which all examined species contain the PB1 domain but lack the cysteine-rich domain C1 (Patalano et al., 2006).
aPKC localization during early sea urchin development
We observed two subcellular localizations for aPKC in the early P. lividus embryo: on the cortex, especially apical surfaces (Figure 2-II), and on microtubule-based structures during mitosis (Figure 3). The cortical enrichment quite resembles the localization recently reported for polarity proteins PAR6 and CDC42 in the S. purpuratus embryo (Alford et al., 2009). However, in that study, aPKC protein was found to be enriched at the cleavage furrow of the two-cell stage, a pattern we did not observe (Supplemental Figure S4). Initially we thought that the discrepancy was due to the different fixation conditions used in the two studies; however, repeating our analysis using a formaldehyde fixation (Foe and von Dassow, 2008) instead of cold methanol did not alter aPKC localization in early P. lividus embryos (see Supplemental Figure S4). Thus it may be a species difference, possibly related to the fact that, unlike other sea urchins, the eggs of P. lividus exhibit visible cortical polarity along the animal–vegetal axis before fertilization (Schroeder, 1980). Finally, it may be that distinct aPKC isoforms are preferentially recognized by aPKC antibody in fixed embryos of the two species.
In P. lividus, the cortical distribution of aPKC is uniform up until the asymmetric fourth cleavage, when aPKC becomes depleted from the vegetal cortex of micromeres (Figure 2-II, E and F, and Supplemental Figure S3). This is reminiscent of asymmetric divisions in the C. elegans zygote and in Drosophila neuroblasts, where aPKC, PAR6, and PAR3 are excluded from the cortex of the smaller daughter cell (Goldstein and Macara, 2007; Chia et al., 2008). It is likely that in the sea urchin as well, the aPKC polarity complex plays an important role in the dramatic unequal cleavages of the vegetal pole. Multiple localizations and functions for aPKC in the early sea urchin embryo could explain the complex phenotypes caused by early inhibition of aPKC, which included a variety of mitotic defects and improper spindle positioning as well as aberrant apical–basal polarity (Alford et al., 2009).
The pattern of aPKC distribution on microtubule-based structures changes during the mitotic cycle in a manner that suggests it travels toward the plus end of microtubules (Figure 3). Similarly, in HeLa cells (Liu et al., 2006) aPKC was found on the spindle poles and on spindle microtubules during prometaphase and metaphase, respectively, and then in the spindle midzone to end up in the midbody during telophase. Experiments using an anti–phospho-aPKC antibody led the authors to propose that the active phosphorylated form of aPKC migrates on spindle microtubules until anaphase, when it is inactivated and accumulates in the spindle midzone. It would be interesting to compare the distribution of active and inactive aPKC in sea urchin, although our experience is that most antibodies raised against vertebrate antigens do not recognize orthologues in marine invertebrate models, thus rendering this kind of approach difficult.
Affinity for microtubules could account for the exclusion of aPKC from the cortex of sea urchin micromeres (Figure 2-II, Supplemental Figure S3). During these unequal cleavages, one spindle pole moves snug against the vegetal cortex, and all of its microtubule plus ends point toward the center of the cell (Schroeder, 1987). Thus migration toward the plus ends of microtubules would cause aPKC to decrease at the vegetal cortex and to accumulate at the spindle midzone and finally at the membrane forming the boundary between the micromere and its sister cell.
Role of aPKC in ciliogenesis and association with microtubule motors
The localization and inhibitor phenotypes reported here suggest that the function of aPKC in sea urchin ciliogenesis might be regulation of IFT. The cilium transition zone where urchin aPKC concentrates (Figure 5 and Supplemental Figure S5) is the region where IFT particles are formed and loaded onto the axoneme before their transport (Eggenschwiler and Anderson, 2007). Like IFT in other systems, aPKC is required both for cilia formation, as shown by experiments on deciliated embryos (Figures 7 and 8), and cilia maintenance, as found when inhibitor is added to embryos with already formed cilia (Figure 6). It is known that the maintenance of the cilium structure requires a constant intraflagellar transport in order to recycle and regenerate inactive molecules such as receptors (for review, see Silverman and Leroux, 2009). Of interest, we noted that the localization of aPKC itself is disrupted by inhibitor treatment (Figures 6 and 8). This could be a direct effect if phosphorylation of some target protein by aPKC is necessary to maintain its localization, or instead an indirect effect due to a general disruption of polarity, as the lack of aPKC signal on membranes seems to suggest.
Our results are consistent with a mechanism whereby aPKC could regulate intraflagellar transport and ciliogenesis via association with kinesin, as previously suggested by Fan et al. (2004), who showed that the aPKC polarity complex can cosediment with microtubules in the presence of adenylyl-imidophosphate. In MDCK cells, the polarity complex exhibits a localization pattern like that of KIF3a, the primary anterograde motor involved in intraflagellar transport (Lin et al., 2003). A similar association of polarity complex components with KIF3 is found in neurons (Nishimura et al., 2004), where disruption of PAR3–KIF3A interaction inhibited the accumulation of PAR3 and aPKC at the tip of the neurites and abolished neuronal polarity. The phenotype of short, nonmotile cilia observed on aPKC inhibition (Figures 7 and 8) is similar to that seen in swimming embryos of the sea urchin L. pictus that had been injected with a specific anti–kinesin II antibody (Morris and Scholey, 1997). In addition, we sometimes observed punctate accumulations of aPKC along the cilia of gastrula or plutei (unpublished data), which strongly resemble the distribution of the kinesin II component KAP in sea urchin embryo cilia (Morris et al., 2004), as well as the distribution of aPKC in ciliated layers of MDCK cells (Fan et al., 2004). Furthermore, as described earlier, the dynamic localization pattern of aPKC during the cell cycle indicates that the kinase associates with a plus end–directed motor. Thus we propose that sea urchin aPKC could associate directly or via PAR3 (Nishimura et al., 2004) with the heterotrimeric kinesin II in intraflagellar transport vesicles. aPKC could then phosphorylate and activate the heterotrimeric kinesin. Although there is no evidence for a phosphorylation-dependent activation of KIF3A/KIFB yet, a specific mitotic phosphorylation of KAP3 in HeLa cells (Haraguchi et al., 2006) has been proposed to regulate its interaction with cargo proteins that are involved in spindle formation and chromosome segregation.
The sea urchin embryo, in which one can repeatedly deciliate and induce cilia regrowth, should prove a simple and favorable model in which to dissect the molecular mechanisms linking polarity and ciliogenesis.
MATERIALS AND METHODS
Culture of sea urchin embryos
All experiments used the Mediterranean sea urchin P. lividus, which was collected by divers from the bay of Villefranche-sur-Mer. To collect gametes, adults were opened using scissors rinsed with tap water. Female gonads were manually removed and left to spawn their eggs in a beaker containing 0.22-μm of filtered seawater (MFSW). Eggs were separated from gonad tissues by filtration through a 100-μm nylon mesh. The sperm was collected dry in Falcon tubes and kept on ice until use. For fertilization the sperm was added to the sea water containing the eggs (final ratio sperm/oocyte solution 1/20,000 vol/vol) in the presence of 1 mM 3-amino-1,2,4-triazole, 0.5 mM EDTA, and 20 mM Tris-HCl, pH 8.2, to block hardening of the fertilization membrane. Weakened fertilization membranes were removed by gentle filtration through a 60-μm nylon mesh. Eggs were left to settle down and were then washed several times with MFSW to remove excess of sperm, 3 amino-(1,2,4)-triazole, and EDTA. Cultures of fertilized eggs were incubated at 18°C under gentle stirring for the required amount of time for embryonic development.
Cloning of P. lividus aPKCs
A sea urchin aPKC DNA probe was generated by PCR on P. lividus cDNA using degenerate primers corresponding to conserved sequences in the ser/thr kinase domain. This fragment was labeled with P32 and used to probe a cDNA library synthesized from P. lividus egg mRNA in the ZAP vector (Stratagene, Santa Clara, CA). The screen yielded several identical long cDNA clones encoding the 3′ portion of sea urchin aPKC. The complete open reading frames of P. lividus aPKC were obtained by 5′-RACE-PCR with the BD SMART RACE Kit (BD Biosciences Clontech, Mountain View, CA) using specific primers 5′-TCGGTCTGAACCCAGTCTATATCCTCATCGTCGGTC and 5′-CTCGTCGCCTTCAGTTCCACAAGGAGCACCTTGGC followed by cloning in the RN3 vector.
Whole-mount in situ hybridization
Whole-mount in situ hybridization of sea urchin was performed following Harland (1991) and Lepage et al. (1992). Digoxigenin (DIG)-labeled probes were detected using an alkaline phosphatase (ALP)–conjugated anti-DIG antibody (Roche Applied Science, Indianapolis, IN) with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-1-phosphate as chromogenic substrates. The probe for aPKC mRNA was a 820–base pair fragment corresponding to amino acids 320–593 (kinase domain) of Pl-aPKC2 long isoform and amino acids 245–518 of Pl-aPKC2 short isoform.
Treatment with aPKC inhibitors
Myristoylated-aPKC pseudosubstrate peptide inhibitor (Sigma-Aldrich, St. Louis, MO) was dissolved in distilled water at the concentration of 500 μM. GÖ 6976 PKC inhibitor (Sigma) and bisindolylimaleimide broad PKC inhibitor (GF 109203X; Calbiochem, Nottingham, United Kingdom) were dissolved in dimethyl sulfoxide at 10 mM. Embryos were allowed to reach the swimming gastrula stage (24 h postfertilization; see Figure 1K) and then treated with inhibitors at the desired final concentration (10 μM for GF109203X, 20 μM for Gö 6976) and incubated for 2 h before observation or fixation (Figure 6). To assay reversibility, gastrula embryos were treated for 2 h with 2 μM pseudosubstrate inhibitor and then washed several times in MFSW and left to reciliate for an additional 2 h (Figure 8, G–I).
Deciliation–reciliation of swimming embryos
Deciliation of swimming embryos was induced by osmotic stress according to Auclair and Siegel (1966). A total of 160 ml of 5 M NaCl was added per liter of culture of embryos at swimming gastrula stage in seawater, to give a final NaCl concentration of ∼1.3 M. After 5 min at room temperature, the loss of cilia was checked by dark-field microscopy, and embryos were then left to settle for 2 min and rinsed twice in normal MFSW. Deciliated washed embryos were then incubated for 2 h in the absence or in the presence of increasing concentrations of pseudosubstrate inhibitor and then either observed live under dark-field videomicroscopy or fixed for immunolabeling (Figure 7).
Antibodies and immunolabeling
The following primary antibodies were used: Anti-aPKC: rabbit polyclonal C-20 (Santa Cruz Biotechnology SC216) dilution 1:200 for immunofluorescence and 1:1000 for Western blotting. Anti-tubulin: mouse monoclonal DM1A (Sigma-Aldrich), dilution 1:1000. Anti–γ-tubulin: mouse monoclonal GTU 88 (Sigma-Aldrich) dilution 1:200. Anti–mouse and anti–rabbit secondary antibodies coupled to fluorescein isothiocyanate or to tetramethyl rhodamine isothiocyanate (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at a dilution 1:200. Anti–rabbit secondary antibodies coupled to horseradish peroxidase (Amersham, GE Healthcare Bio-Sciences, Piscataway, NJ) were used at a dilution of 1:7500 for Western blotting. For all immunolabeling studies except Supplemental Figure S4, embryos were fixed in cold methanol (−20°C) containing 50 mM ethylene glycol tetraacetic acid (EGTA) pH 7.5. Fixed samples were rehydrated three times for 5 min each in phosphate-buffered saline (PBS) in plastic 96-well microtiter plates, blocked three times for 15 min each with PBT (PBS, pH 7.4, containing 3% bovine serum albumin and 0.05% Triton X-100), and incubated with primary antibodies diluted in PBT for either 2 h at room temperature or overnight at 4°C. Embryos were then washed three times for 15 min each in PBT, incubated with secondary antibodies supplemented with 1 μg/ml Hoescht 33342 (Sigma-Aldrich) to label DNA, washed again in PBT, and mounted in Citifluor (Chem Lab, Canterbury, United Kingdom). For analysis of cilia length (Table 1, Figure 7), embryos were fixed in 2% paraformaldehyde dissolved in 20 mM 1,4-piperazinediethanesulfonic acid, pH 6.5, 0.5 mM EDTA, 20 mM MgCl2, and 0.1% Triton X-100 and labeled with DM1A antitubulin antibody. Fluorescent images of fixed immunolabeled embryos were acquired with a Leica SP2 confocal microscope and processed using ImageJ (National Institutes of Health, Bethesda, MD) or Photoshop (Adobe, San Jose, CA) software.
Western immunoblotting
Protein extracts of P. lividus prepared by lysis of packed eggs or embryos in 10 volumes of low–ionic strength buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 0.5 mM EGTA, 5 mM dithiothreitol, 0.1% NP40) containing antiprotease inhibitors cocktail, followed by centrifugation (40,000 rpm, 1 h, 4°C). The supernatant was then mixed with 1/3 volume of 4X Laemmli sample buffer and heated for 2 min at 95°C. Immunoblots were prepared by standard SDS–PAGE followed by transfer to nitrocellulose membranes (Schleicher & Schuell BioScience, Dassel, Germany). The nitrocellulose was blocked for 2 h with 3% low-fat dry milk dissolved in PBS containing 0.1% Tween-20 before incubation with primary antibody SC216. Membranes were then washed three times for 15 min in PBS–0.1%Tween-20, incubated with horseradish peroxidase–conjugated anti–rabbit secondary antibody (Amersham), washed three times for 15 min in PBS Tween-20, and detected on ECL Hyperfilm (Kodak, Rochester, NY) using chemiluminescent substrates (Amersham).
Videomicroscopy to analyze swimming behavior
Embryos were observed by dark field microscopy with an Olympus BH2 microscope equipped with dry (Zeiss 25×) or oil immersion lenses. In the latter case, the objective lens used was an Olympus 40× D-Apo UV-oil 1.30 with diaphragm combined with an Olympus Dark Field oil condenser DWC 1.2–1.4. Recordings were made using stroboscopic illumination by a Strobex (Chadwick-Helmuth, El Monte, CA). Video images were recorded with a Panasonic WV-F 15 E S-VHS video camera (constant frame rate of the European standard, 50 Hz) synchronized to the stroboscopic illumination with a fiber optic “video-sync” (module 9630; Chadwick-Helmuth, El Monte, CA), connected to a Hamamatsu video image processor (DVS 3000) and to a Panasonic AG 7330 S-VHS video recorder. Still images of embryos were obtained by digitalization of individual frames recorded on S-VHS with a Formac FS10 digitizer and were further processed using Adobe Photoshop 7.0 software for Macintosh.
Supplementary Material
Acknowledgments
This work was supported by grants from the Agence Nationale pour la Recherche, Centre National de la Recherche Scientifique–Action Concertée Incitative, Association pour la Recherche sur le Cancer, and Association Française contre les Myopathies to C.S. and by Centre National de la Recherche Scientifique and Université Pierre et Marie Curie grants to the UMR7009. We thank Christian Rouviere for assistance with image processing, Olivier Dijoux and Faisal Bekkouche for computer assistance, and Sebastien Motreuil, David Luquet, and Laurent Giletta for collecting sea urchins.
We thank especially Philippe Huitorel, Alexandre Paix, Kazuo Inaba, and Alain Debec for helpful discussions and suggestions and Philippe Dru, bioinformatician in our lab, for his expertise concerning the search for aPKC isoforms in the S. purpuratus genome. We thank Guy Lhomond for the gift of P. lividus cDNA libraries used for the cloning of aPKC. We also thank Evelyn Houliston, Janine Beisson, Ronald Melki, and Alain Debec for the generous gift of anti–γ-tubulin antibodies. We thank the reviewers and editor whose thoughtful suggestions helped us to improve the manuscript.
Abbreviations used:
- aPKC
atypical protein kinase C
- Par
partition defective
- Pl
Paracentrotus lividus
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0844) on April 20, 2011.
REFERENCES
- Alford LM, Ng MM, Burgess DR. Cell polarity emerges at first cleavage in sea urchin embryos. Dev Biol. 2009;330:12–20. doi: 10.1016/j.ydbio.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Castillo AB, Brugmann SA, Helms JA, Jacobs CR, Stearns T. Primary cilia: cellular sensors for the skeleton. Anat Rec. 2008;291:1074–1078. doi: 10.1002/ar.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assémat E, Bazellières E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta Biomembr. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Auclair W, Siegel BW. Cilia regeneration in the sea urchin embryo: evidence for a pool of ciliary proteins. Science. 1966;154:913–915. doi: 10.1126/science.154.3751.913. [DOI] [PubMed] [Google Scholar]
- Bulgakova NA, Knust E. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J Cell Sci. 2009;122:2587–2596. doi: 10.1242/jcs.023648. [DOI] [PubMed] [Google Scholar]
- Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 2007;134:1035–1043. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3[beta] and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Fan S, Hurd TW, Liu C, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14:1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Fischer E, Pontoglio M. Planar cell polarity and cilia. Semin Cell Dev Biol. 2009;20:998–1005. doi: 10.1016/j.semcdb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Foe VE, von Dassow G. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J Cell Biol. 2008;183:457–470. doi: 10.1083/jcb.200807128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ. Cilia and flagella of eukaryotes. J Cell Biol. 1981;91:107s–124s. doi: 10.1083/jcb.91.3.107s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T. Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem. 2006;281:4094–4099. doi: 10.1074/jbc.M507028200. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev Cell. 2007;12:727–738. doi: 10.1016/j.devcel.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Yoshinaga S, Takeya R, Suzuki NN, Horiuchi M, Kohjima M, Sumimoto H, Inagaki F. Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. J Biol Chem. 2005;280:9653–9661. doi: 10.1074/jbc.M409823200. [DOI] [PubMed] [Google Scholar]
- Inaba K. Molecular basis of sperm flagellar axonemes. Ann NY Acad Sci. 2007;1101:506–526. doi: 10.1196/annals.1389.017. [DOI] [PubMed] [Google Scholar]
- Konno A, Kaizu M, Hotta K, Horie T, Sasakura Y, Ikeo K, Inaba K. Distribution and structural diversity of cilia in tadpole larvae of the ascidian Ciona intestinalis. Dev Biol. 2010;337:42–62. doi: 10.1016/j.ydbio.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Lepage T, Ghiglione C, Gache C. Spatial and temporal expression pattern during sea urchin embryogenesis of a gene coding for a protease homologous to the human protein BMP-1 and to the product of the Drosophila dorsal-ventral patterning gene tolloid. Development. 1992;114:147–163. doi: 10.1242/dev.114.1.147. [DOI] [PubMed] [Google Scholar]
- Lin F, Hiesberger T, Cordes K, Sinclair AM, Goldstein LSB, Somlo S, Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Xie X, Miki T. Inhibition of protein kinase C zeta blocks the attachment of stable microtubules to kinetochores leading to abnormal chromosome alignment. Cell Signal. 2006;18:2314–2323. doi: 10.1016/j.cellsig.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Masuda M, Sato H. Reversible resorption of cilia and the centriole cycle in dividing cells of sea urchin blastulae. Zool Sci. 1984;1:445–462. [Google Scholar]
- McCaffrey LM, Macara IG. Widely conserved signaling pathways in the establishment of cell polarity. Cold Spring Harbor Perspect Biol. 2009;1:a001370–a001370. doi: 10.1101/cshperspect.a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, English CN, Lou JE, Dufort FJ, Nordberg J, Terasaki M, Hinkle B. Redistribution of the kinesin-II subunit KAP from cilia to nuclei during the mitotic and ciliogenic cycles in sea urchin embryos. Dev Biol. 2004;274:56–69. doi: 10.1016/j.ydbio.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Munro E, Bowerman B. Cellular symmetry breaking during Caenorhabditis elegans development. Cold Spring Harb Perspect Biol. 2009;1:a003400–a003400. doi: 10.1101/cshperspect.a003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro EM. PAR proteins and the cytoskeleton: a marriage of equals. Curr Opin Cell Biol. 2006;18:86–94. doi: 10.1016/j.ceb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328–334. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2269 manually annotated myosins from 328 species. Additional data file 1: Problems and pitfalls of automatic gene annotation, gene collection, domain prediction, and sequence alignment. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patalano S, Pruliere G, Prodon F, Paix A, Dru P, Sardet C, Chenevert J. The aPKC-PAR-6-PAR-3 cell polarity complex localizes to the centrosome attracting body, a macroscopic cortical structure responsible for asymmetric divisions in the early ascidian embryo. J Cell Sci. 2006;119:1592–1603. doi: 10.1242/jcs.02873. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH, Parker PJ. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol. 2009;7:e1000235. doi: 10.1371/journal.pbio.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. Cilia biology: stop overeating now! Curr Biol. 2007;17:R963–R965. doi: 10.1016/j.cub.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. Expressions of the prefertilization polar axis in sea urchin eggs. Dev Biol. 1980;79:428–443. doi: 10.1016/0012-1606(80)90128-1. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. Fourth cleavage of sea urchin blastomeres: microtubule patterns and myosin localization in equal and unequal cell divisions. Dev Biol. 1987;124:9–22. doi: 10.1016/0012-1606(87)90454-4. [DOI] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi K, Yamaguchi M. Expression patterns of three Par-related genes in sea urchin embryos. Gene Expr Patterns. 2008;8:323–330. doi: 10.1016/j.gep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Snell WJ, Pan J, Wang Q. Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell. 2004;117:693–697. doi: 10.1016/j.cell.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Stephens RE. Ciliogenesis in sea urchin embryos—a subroutine in the program of development. Bioessays. 1995;17:331–340. doi: 10.1002/bies.950170409. [DOI] [PubMed] [Google Scholar]
- Stephens RE. Ciliogenesis, ciliary function, and selective isolation. ACS Chem Biol. 2008;3:84–86. doi: 10.1021/cb8000217. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007:re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- Sun R, Gao P, Chen L, Ma D, Wang J, Oppenheim JJ, Zhang N. Protein kinase C zeta is required for epidermal growth factor-induced chemotaxis of human breast cancer cells. Cancer Res. 2005;65:1433–41. doi: 10.1158/0008-5472.CAN-04-1163. [DOI] [PubMed] [Google Scholar]
- Vinot S, Le T, Maro B, Louvet-Vallée S. Two PAR6 proteins become asymmetrically localized during establishment of polarity in mouse oocytes. Curr Biol. 2004;14:520–525. doi: 10.1016/j.cub.2004.02.061. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblish J. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.