RhoU, an atypical Rho family member with unique N- and C-terminal extensions, associates with activated EGFR in a GRB2-dependent manner via its N-terminal proline-rich motifs and is integrated into EGFR-mediated signaling, leading to AP1 transcriptional activity and cell migration.
Abstract
RhoU is an atypical Rho family member with high homology to CDC42 but containing unique N- and C-terminal extensions. The mechanisms regulating RhoU activation, as well as its downstream effectors, are not fully characterized. We show that after epidermal growth factor (EGF) stimulation RhoU colocalizes with EGF receptor (EGFR) on endosomes, which requires both its N- and C-terminal extension sequences. Moreover, RhoU physically associates with activated EGFR in a GRB2-dependent manner through specific proline-rich motifs within its N-terminus. Mutation of these proline-rich sequences or suppression of GRB2 by RNA interference abrogates the interaction of RhoU with activated EGFR, as well as EGF-stimulated RhoU GTP binding. In addition, RhoU is involved in EGFR-mediated signaling, leading to AP1 transcriptional activity and cell migration in pancreatic cancer cells, events that require its interaction with the Grb2–EGFR complex. Taken together, the data suggest a unique regulatory mechanism by which RhoU interaction with SH3 adaptor proteins might serve to integrate growth factor receptor signaling with RhoU activation.
INTRODUCTION
Rho-family GTP-binding proteins regulate many cellular functions, including cytoskeletal organization, cell polarity, cell cycle, survival, and gene transcription, all of which have been shown to be essential for the progression of human cancer (Jaffe and Hall, 2002; Fritz and Kaina, 2006). In general, Rho GTPases function as molecular switches and exist in active GTP-bound form, which allows them to interact with downstream effectors in a spatially and temporally controlled manner to regulate cellular functions, or an inactive GDP-bound state (Jaffe and Hall, 2005). The switch between the GDP- and GTP-bound forms of typical Rho GTPases is regulated by upstream guanine nucleotide exchange factors (Rossman et al. 2005) and GTPase-activating proteins (Tcherkezian and Lamarche-Vane, 2007).
RhoU, also known as Wrch-1, is an atypical member of the CDC42 subgroup of Rho GTPases and was first isolated as a gene transcriptionally up-regulated in wnt-1–transformed mouse mammary epithelial cells (Tao et al., 2001). Ectopic expression of a constitutively active RhoU mutant resulted in Wnt-like morphological transformation in mammary epithelial cells, as well as growth transformation in NIH3T3 cells (Tao et al., 2001; Shutes et al., 2004; Berzat et al., 2005). In addition to the well-established effects on the actin cytoskeleton such as dissolution of stress fibers, induction of filopodia, regulation of focal adhesion formation and cell motility in nonepithelial cells (Aspenström et al., 2004; Saras et al., 2004; Chuang et al., 2007; Ory et al., 2007; Ruusala and Aspenström, 2008), a recent study has also reported a role for RhoU in normal epithelial morphogenesis (Brady et al., 2009). These phenotypes, including loss of epithelial morphology and cell polarity, as well as F-actin reorganization, are also hallmarks of dedifferentiated cancer cells, suggesting that RhoU may be involved in cancer development and/or progression, although its exact functional and mechanistic roles remain poorly defined.
Although RhoU is a CDC42 homologue, there are striking differences regarding its GTP-loading properties, effector binding, and N- and C-terminal extension sequences. Although RhoU hydrolyzes GTP with similar kinetics to CDC42 in vitro, it has much higher intrinsic guanine nucleotide exchange activity, implying that it is constitutively GTP bound (Saras et al., 2004; Shutes et al., 2004). Compared to CDC42, human RhoU contains an N-terminal extension of 46 amino acid residues, which contains proline-rich modules capable of mediating interactions with SH3 domain–containing proteins (Saras et al., 2004; Shutes et al., 2004). Of interest, the N-terminal extension is not required for its GTPase activity in vitro, but a potential negative regulation of effector association via an intramolecular interaction has been suggested (Shutes et al., 2004). RhoU also contains a C-terminal extension of 21 residues terminated with an unusual CAAX sequence (CCFV). In contrast to most other Rho GTPases, which undergo isoprenylation on their CAAX sequence, RhoU uses palmitoylation as a membrane-targeting mechanism (Berzat et al., 2005). RhoU interacts with and activates PAK1 similar to CDC42 and RAC1, but it does not seem to bind the isolated CRIB domain of WASP (Aspenström et al., 2004). More recent studies have shown that RhoU interacts with PYK2 and PAR6, both of which bind RhoU in a GTP-dependent manner. Of interest, the N-terminal extension of RhoU is required for PYK2 but not PAR6 interaction (Ruusala and Aspenström, 2008; Brady et al., 2009). Taken together, these investigations suggest the presence of an additional set of effectors and unique mechanism(s) of regulation for RhoU.
The epidermal growth factor (EGF) receptor (EGFR) is a member of the ErbB receptor tyrosine kinase family. On ligand binding, EGFR dimerizes, autophosphorylates, and triggers downstream signaling cascades, including phosphatidylinositol-3-kinase/Akt, mitogen-activated protein kinase (MAPK), Jak/Stat, protein kinase C (PKC), and PLCγ1 activation (Nicholson et al., 2001). Ligand binding is also accompanied with accelerated internalization of EGFR through clathrin-coated pits, which is followed by lysosomal targeting of internalized receptors, resulting in receptor down-regulation (Sorkin and Goh, 2009). Many proteins are involved in mediating EGFR signaling and its internalization/degradation (Sorkin and Goh, 2009). In fact, the signaling adaptor protein GRB2, which contains an SH2 domain flanked by two SH3 domains, plays essential roles in both processes (Rozakis-Adcock et al., 1993; Yamazaki et al., 2002; Jiang et al., 2003). The SH2 domain of GRB2 recognizes the phosphotyrosine residues of activated EGFR (Lowenstein et al., 1992), and the SH3 domains can bind proline-rich sequences such as those found in the Ras GTP exchange factor, SOS. Thus GRB2 links the EGFR to distal signaling and degradation (Rozakis-Adcock et al. 1993).
Here we show that the N-terminal extension of RhoU, although not required for either GTP hydrolysis or GTP binding in vitro, provides a binding interface for the SH3 domains of GRB2, which physically links RhoU to EGFR on endosomes. We demonstrate that this interaction is essential in coupling RhoU to EGFR signaling, leading to the regulation of both EGF-induced JNK/AP1 transcriptional activity and cell motility.
RESULTS
RhoU localizes to endosomes and interacts with activated EGFR
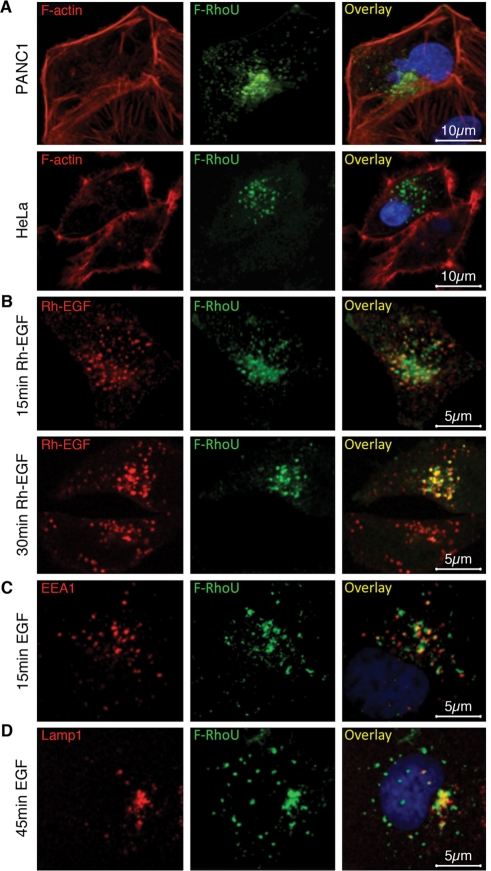
Localization of RhoU has been reported at several subcellular compartments, including endosomes, focal adhesions, plasma membrane, and cell–cell adhesion junctions depending on the cell type or the expression construct used. We therefore analyzed Flag-, Myc-, and green fluorescent protein (GFP)-tagged RhoU expression in a variety of epithelial cell lines in more detail by immunofluorescence staining and confocal microscopy. As shown in Figure 1, Flag-RhoU was consistently localized to endosome-like structures in pancreatic and cervical cancer cells (Figure 1), as well as in the prostate and breast cancer cell lines LNCaP and MDA-MB-231, respectively (Supplemental Figure S1A). In addition to endosomes, we also found RhoU at focal adhesions in a significant fraction of HeLa or PANC1 cells (unpublished data). However, localization at the plasma membrane was rarely observed in cells expressing low levels of RhoU and appeared predominantly with GFP-tagged RhoU (Supplemental Figure S1B). Neither disruption of microtubules by nocodazole nor F-actin depolymerization by cytochalasin D treatment significantly altered RhoU localization at endosomal structures (Supplemental Figure S1C). These analyses suggest that RhoU mainly resides on endosomes in these cancer cells when expressed at low levels.
FIGURE 1:
RhoU localizes to endosomes and colocalizes with EGFR on EGF stimulation. (A) Immunofluorescence staining of PANC1 and HeLa cells transfected with a low level of Flag–RhoU were stained with anti-Flag (green), phalloidin (red) for F-actin, and Hoechst 33342 (blue) for DNA. (B) Flag-RhoU–transfected and serum-starved HeLa cells were stimulated with Rh-EGF (10 ng/ml; red) and stained with anti-Flag (green). (C, D) HeLa cells were stimulated with EGF (20 ng/ml) and stained with anti-Flag (green), anti-EEA1 (red, C), or anti-LAMP1 (red, D), and Hoechst 33342 (blue) for DNA.
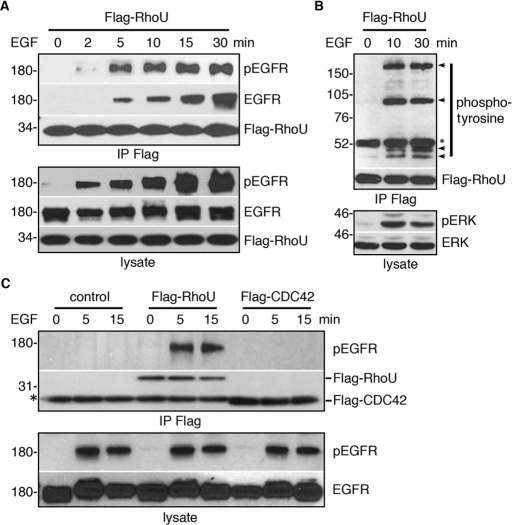
We next tested whether RhoU localization and/or its function is linked to growth factor–mediated signaling. On the basis of using rhodamine-conjugated EGF (Rh-EGF), it was evident that RhoU colocalized with internalized EGFR in a time-dependent manner (Figure 1B) at both EEA1-positive early endosomes (Figure 1C) and Lamp1-positive late endosome/lysosome (Figure 1D). Because RhoU colocalized with internalizing EGFR, we further determined whether RhoU physically interacts with EGFR. Indeed, phosphorylated EGFR coimmunoprecipitates with RhoU after EGF stimulation in a time-dependent manner (Figure 2A). Because activation of EGFR is accompanied with its autophosphorylation as well as tyrosine phosphorylation of other downstream substrates, we also examined whether RhoU coprecipitated with additional tyrosine phosphorylated protein(s). Several distinct bands, besides the apparent pEGFR (∼180 kDa), were detected (Figure 2B), suggesting that RhoU associates with the phosphorylated EGFR signaling complex on endosomes (Figure 1, B and C). However, despite high sequence homology between CDC42 and RhoU, we did not detect association of CDC42 with the EGFR under the same conditions (Figure 2C).
FIGURE 2:
RhoU, but not CDC42, physically associates with phosphorylated EGFR complex. (A, B) Flag-RhoU–transfected, serum-starved HeLa cells were stimulated with 20 ng/ml of EGF for the indicated time. Cells were lysed, immunoprecipitated, and immunoblotted with indicated antibodies. Arrows indicate tyrosine phosphorylated proteins coimmunoprecipitated with Flag–RhoU following EGF stimulation. Asterisk corresponds to nonspecific band from mouse immunoglobulin (Ig) G heavy chain. (C) HeLa cells were transfected with the empty vector, Flag–RhoU, and Flag–CDC42, respectively, immunoprecipitated, and immunoblotted with indicated antibodies. Asterisk denotes mouse IgG light chain.
Identification and validation of SH3-domain proteins as RhoU interactors
To identify potential interactors of RhoU that could link it to the EGFR, we performed yeast two-hybrid (YTH) screens. A putative constitutively active (CA) RhoU (Q110L; mouse) was used as bait to screen cDNA libraries from both human prostate and bone marrow (Supplemental Figure S2A). Of interest, among the >100 clones obtained and sequenced, a group of more than 10 clones all encoded SH3 domain–containing proteins (unpublished data). Each of these clones contains an insert encoding at least one intact SH3 domain, including two clones for the C-terminal SH3 of growth factor receptor–bound protein 2 (GRB2), suggesting that SH3 domain(s) from these proteins mediate interaction with RhoU. To validate the interaction of RhoU with these candidates as well as to further evaluate RhoU interaction with additional SH3 domains, we performed glutathione S-transferase (GST) pull-down assays using recombinant GST–SH3 fusion proteins with lysate from Flag-RhoU–transfected cells. The C-terminal GRB2 SH3, the N-terminal CRKL SH3 (but not the C-terminal SH3), and the LCK SH3 were confirmed to bind RhoU (Supplemental Figure S2B). Therefore at least one SH3 domain from each of the three tested candidates is able to bind RhoU, validating the YTH interactions. In addition, we found that SH3 domains from CIN85, CRKL-II, p85α, Abi-1, and Abi-2 were also able to bind RhoU (Supplemental Figure S2B). These results indicate that RhoU has the potential to interact with multiple SH3 domain–containing proteins and may be recruited into signaling pathways through these interactions.
RhoU N-terminus–mediated interaction with GRB2 is enhanced by EGF stimulation
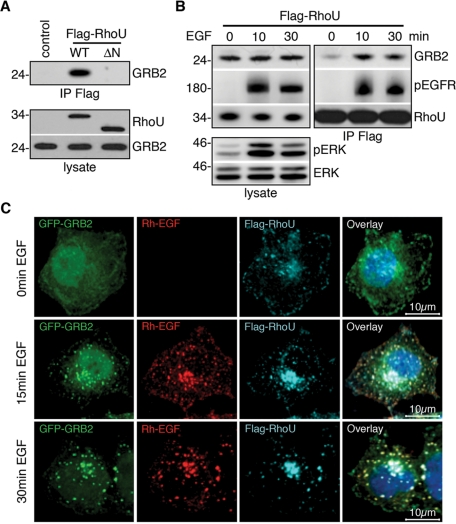
Among the potential RhoU-interacting proteins, GRB2 is of particular interest because of its critical role in transmitting EGFR signaling and in EGFR internalization. Although a direct interaction of RhoU with GRB2 has been reported (Shutes et al., 2004), whether this interaction is important in coupling RhoU to the activated EGFR is not known. Consistent with this previous report, GRB2 coimmunoprecipitated with wild-type (WT) RhoU but not the RhoU(ΔN) mutant lacking all potential SH3-binding sites (Figure 3A). Of significance, the RhoU–GRB2 interaction is markedly enhanced following EGF stimulation, which strongly correlates with EGF-triggered RhoU–EGFR interactions (Figure 3B). Confocal microscopy imaging revealed a strong colocalization of RhoU with GRB2 and internalized EGFR in response to EGF stimulation, as demonstrated by a punctuate localization pattern (Figure 3C), which is similar to the endosomal localization shown in Figure 1. Together, these data suggest that GRB2 interacts with RhoU via its N-terminal extension, which is enhanced by activated EGFR signaling.
FIGURE 3:
RhoU N-terminus–mediated interaction with GRB2 is enhanced by EGF stimulation. (A) HeLa cells were transiently transfected with empty vector, Flag–RhoU, and its N-terminal deletion mutant (ΔN) expression plasmids. Lysates were immunoprecipitated and immunoblotted as indicated. (B) HeLa cells transfected with Flag–RhoU were serum starved before treatment with 20 ng/ml of EGF for the indicated time. Lysates were immunoprecipitated and immunoblotted with indicated antibodies. Efficient EGF activation is documented by ERK phosphorylation in total lysate. (C) Immunofluorescence staining of HeLa cells cotransfected with Flag–RhoU and GRB2–eGFP expression plasmids. Serum-starved cells were stimulated with Rh-EGF (10 ng/ml; red) for the indicated time and stained with anti-Flag (cyan) and Hoechst 33342 (blue) for DNA.
GRB2 is essential in coupling RhoU to EGFR
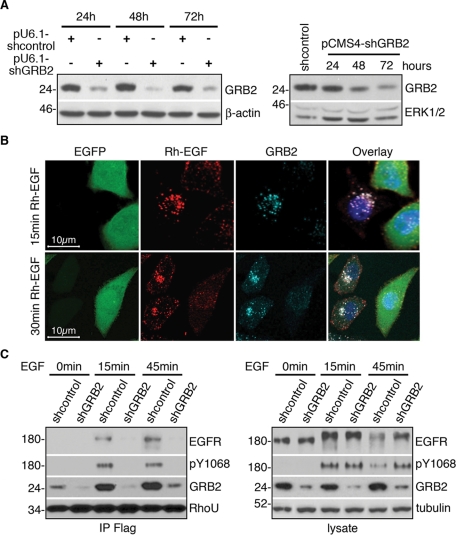
To determine the role of GRB2 in coupling RhoU to the EGFR, we generated GRB2 short hairpin RNA (shRNA) constructs in both pRNA-U6.1 and pCMS4 vectors that drive the shRNA expression by either U6 or H1 promoters, respectively. The pCMS4 vector also expresses EGFP from an independent promoter, allowing identification of the transfected cells. Efficient suppression of GRB2 was achieved with either vector system, as demonstrated by immunoblot analysis (Figure 4A). We next examined the effect of GRB2 suppression on EGFR internalization. In vector control transfected HeLa cells, EGFR was efficiently internalized and showed exclusive colocalization with GRB2 in endosomal compartments on EGF stimulation (Supplemental Figure S2C). In contrast, endogenous GRB2 is markedly reduced in shGRB2-transfected cells (GFP positive) compared with nontransfected neighboring cells (GFP negative) (Figure 4B), and consistent with previous data (Jiang et al., 2003), it was apparent that EGFR internalization was impaired in GRB2-suppressed cells, with a significant portion of EGFR remaining on the plasma membrane even after 30 min of EGF treatment (Figure 4B). Similar results were observed in cells with GRB2 suppression using the pU6-shGRB2 expression vector, which demonstrates more efficient GRB2 suppression (Figure S2D).
FIGURE 4:
GRB2 is essential in coupling RhoU to EGFR complex. (A) Western blot analysis of HeLa cells transfected with pU6-shGRB2, pCMS4-shGRB2, or respective scrambled control vectors for time-dependent GRB2 suppression as indicated. (B) The pCMS4-shGRB2–transfected cells were seeded on cover slips and allowed to grow for 48 h, followed by 16 h of serum starvation before stimulation with Rh-EGF (10 ng/ml; red) for the indicated time. Cells were stained for GRB2 (light blue) and Hoechst 33342 (blue) for DNA. (C) Western blot analysis of HeLa cells transfected with pU6-shGRB2 or scrambled control vector for 24 h, then sequentially transfected with Flag–RhoU for another 24 h, followed by serum starvation for 16 h before EGF stimulation (20 ng/ml). Lysates were precipitated with anti-Flag agarose. Immunoprecipitates and lysates were immunoblotted with indicated antibodies. Numbers on the left correspond to protein molecular weight marker in kilodaltons.
To determine the role of GRB2 in coupling RhoU to EGFR, we performed immunoprecipitation for Flag–RhoU in GRB2-suppressed and control cells. EGF stimulation resulted in a marked increase in association of GRB2 and phosphorylated EGFR with RhoU in vector-transfected cells (Figure 4C, left). In contrast, coimmunoprecipitation of EGFR with RhoU was abrogated in GRB2-suppressed cells as detected by both EGFR and phospho-EGFR (pY1068) antibodies, a primary site for GRB2 SH2 binding (Figure 4C, left). Note the decrease in EGFR protein level in the lysate at 45 min after EGF treatment in vector-transfected cells (Figure 4C, right), which is expected as a result of EGF-induced degradation. However, EGFR levels remained higher in GRB2-suppressed cells at this time point, which is in agreement with impaired degradation of EGFR in the absence of GRB2 (Jiang et al., 2003). Together, these data suggest that GRB2 is essential to couple RhoU to activated EGFR.
Proline-rich motifs in the N-terminus of RhoU mediate its interaction with GRB2 and the EGFR
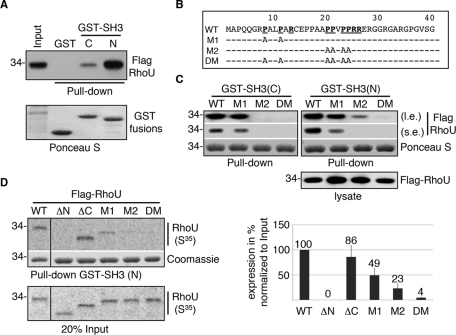
Having established that GRB2 is essential in coupling RhoU to the activated EGFR, we were interested in delineating which proline-rich motif within the N-terminus of RhoU is involved in mediating RhoU/GRB2 interactions and whether one or both GRB2 SH3 domains are involved. Although both GRB2 clones from YTH screening contained only the C-terminal SH3, we tested whether the N-terminal SH3 could also interact with RhoU. In fact, the N-terminal SH3 domain of GRB2 pulled down significantly more RhoU compared with the C-terminal domain (Figure 5A). We next examined the putative SH3-binding proline-rich sequences within the RhoU N-terminus. Mouse RhoU harbors two class II SH3-binding consensus (PXXPXR) motifs that are fully conserved in human RhoU (Figure 5B). We generated mutations in each of the potential binding sites designated as mutant 1 (M1), mutant 2 (M2), and a double mutant (DM) as shown in Figure 5B. To determine the effect of these mutations on RhoU interaction with GRB2 SH3 domains, we used recombinant GST fusion proteins of GRB2 N- and C-terminal SH3 domains for pull-down assays with lysate from cells transfected with the indicated RhoU constructs. For the N-terminal SH3, mutation of either site resulted in reduced binding, although the M2 had a more dramatic effect (Figure 5C, right). For the C-terminal SH3, M1 had only a moderate effect, whereas M2 largely abolished this interaction (Figure 5C, left). Of significance, neither SH3 domain bound to the DM, suggesting that both domains likely contribute to the interaction with GRB2 (Figure 5C). To further characterize the interactions between RhoU SH3-binding consensus and the SH3 domain, as well as to explore the potential effect of GTP-loading status on this interaction, we performed GST pull-down assays with in vitro–translated [35S]methionine-labeled Flag–RhoU with desired deletions/mutations. As shown in Figure 5D, mutation of either SH3 binding site markedly reduced the binding, whereas deletion of the N-terminal extension or mutation of both SH3-binding consensus sites within the N-terminus abolished the interaction (Figure 5D). However, deletion of the C-terminal extension did not reduce binding in this assay. These results suggest that both proline-rich sequences within the RhoU N-terminus and both GRB2 SH3 domains are involved in maintaining the optimal interaction between these two proteins.
FIGURE 5:
Proline-rich motifs in the N-terminus of RhoU mediate its interaction with GRB2 and the EGFR. (A) Lysates from Flag-RhoU–transfected HeLa cells were used for GST pull-down assays with either the N- or C-terminal SH3 domain of GRB2 (top). The same blot stained with Ponceau S documents the fusion protein input (bottom). (B). The amino acid sequence of mouse RhoU N-terminal extension. The two class II SH3-binding consensuses (PXXPXR) are underlined in bold. The mutations by substitution of prolines with alanines in each mutant are specified. (C) Lysates from WT and mutant Flag-RhoU–transfected cells were used for GST pull-down assays with GRB2 N- and C-terminal SH3 fusion proteins, designated GST–SH3(N) and GST–SH3(C), respectively. Top two panels show Flag–RhoU bound to GST–SH3 fusion proteins as detected by anti-Flag antibodies (l.e. and s.e. indicate longer and shorter exposure, respectively). Ponceau S staining documents the input of recombinant GST fusion proteins. The expression level of Flag–RhoU constructs is shown in the bottom panel. (D) GST SH3(N) was used in pull-down assay with in vitro–translated [35S]methionine-labeled RhoU containing indicated deletion/mutations. A representative result of RhoU proteins bound to GST SH3(N) is shown in the top panel followed by the Coomassie staining of the same gel to the indicated input of GST–SH3 fusion proteins. The input (20%) of in vitro–translated RhoU proteins is shown at the bottom. The average band intensity for GST-SH3–precipitated RhoU from three independent experiments along with SD is shown in the histogram (right).
The N- and C-terminal extensions, but not GTP loading, are required for RhoU–GRB2/EGFR interactions
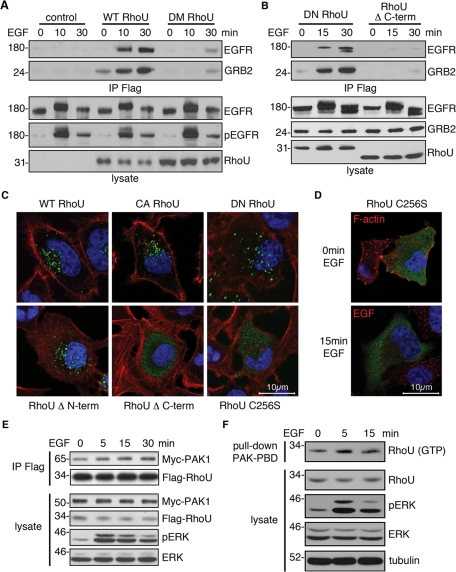
To determine the necessity of the RhoU SH3 binding sites in coupling to the GRB2/EGFR complex in vivo, we coimmunoprecipitated endogenous EGFR/GRB2 with Flag–RhoU. In agreement with the in vitro pull-down assay results, the DM RhoU largely lost its ability to interact with GRB2/EGFR in EGF-stimulated cells (Figure 6A). Of interest, although dominant negative (DN) RhoU (T63N) interacted with EGFR/GRB2 complexes, deletion of the RhoU C-terminal extension abolished this interaction (Figure 6B). Because the C-terminus of RhoU is involved in its posttranslational modification critical for proper membrane interactions, we analyzed the subcellular localization of this mutant. Of significance, WT, CA, DN, and N-terminal–deleted mutants of RhoU remain associated with endosomal structures (Figure 6C). However, deletion of the C-terminal extension or mutation of the specific palmitoylation site (C256S) near the C-terminus resulted in a largely diffuse cytoplasmic distribution of RhoU (Figure 6C). Consistent with this result, the C256S mutant was no longer able to colocalize with activated EGFR (Figure 6D). Of interest, phosphorylation of RhoU by Src has been predicted to result in its inactivation and redistribution from the plasma membrane to endosomes (Alan et al., 2010). Strikingly, we find that this mutant, RhoU(Y254F), localizes to endosomes in the absence of phosphorylation by Src and interacts with the EGFR on endosomes (Figure S3A). These data suggest that the N-terminal extension of RhoU is required for the physical association of RhoU with the activated EGFR but not endosomal localization. On the other hand, palmitoylation within the C-terminal extension of RhoU is critical for endosomal localization and, thus, interaction with activated EGFR at this membranous structure.
FIGURE 6:
The N- and C-terminal extensions, but not GTP loading, are required for RhoU–GRB2/EGFR interactions. (A, B) Western blot analysis of HeLa transfected with indicated Flag–RhoU expression vectors for 24 h, followed by serum starvation for 16 h before EGF stimulation (20 ng/ml). Equal amount of lysate was precipitated with anti-Flag agarose. Immunoprecipitates and lysates were immunoblotted with indicated antibodies. (C, D) Immunofluorescence staining of HeLa cells expressing the indicated RhoU constructs (green). Transfected cells were maintained in normal medium for 24 h (C) or serum starved for additional 16 h and stimulated with EGF stimulation (20 ng/ml) (D) before fixing and staining with anti-Flag (green), phalloidin (red) for F-actin, and Hoechst 33342 (blue) for DNA. (E) HeLa cells were cotransfected with Flag–RhoU and Myc–PAK1 for 24 h, followed by 16 h of serum starvation. Lysates were prepared after stimulation with EGF (50 ng/ml) for the indicated time, immunoprecipitated, and blotted with RhoU, myc, and additional antibodies as indicated. (F) HeLa cells were transfected with Flag–RhoU and treated with EGF as in E. Cell lysates were prepared for pull-down assay with GST–PBD. The pull-down samples and lysate were immunoblotted with indicated antibodies.
Given that RhoU is physically coupled to phosphorylated EGFR, we sought to determine whether RhoU activation is functionally integrated into EGFR signaling as well. PAK1 is among the best-characterized RhoU effectors and, like CDC42, the p21-binding domain (PBD) of PAK1 interacted with GTP-loaded CA and WT RhoU, but not DN RhoU (Figure S3B, C). Thus we chose PAK1 as a proxy effector molecule to test whether RhoU GTP binding is regulated by EGFR signaling. We first examined RhoU association with exogenously expressed PAK1 following EGF stimulation. It was apparent that RhoU associated with PAK1 even in serum-starved cells, consistent with the basal level of high GTP loading (Saras et al., 2004; Shutes et al., 2004). However, EGF stimulation further enhanced the association of RhoU and Pak in a time-dependent manner, suggesting that RhoU GTP binding is significantly enhanced by EGF signaling (Figure 6E, Supplemental Figure S3D). Of interest, we were also able to observe a significant increase in active RhoU after EGFR stimulation in a pull-down assay using the PAK1 PBD (Figure 6F, Supplemental Figure S3E), which largely correlated with increased PAK1 binding. These results suggest that EGFR activation can stimulate RhoU GTP binding and increase its interactions with effector molecules.
RhoU enhances EGF-induced JNK activation
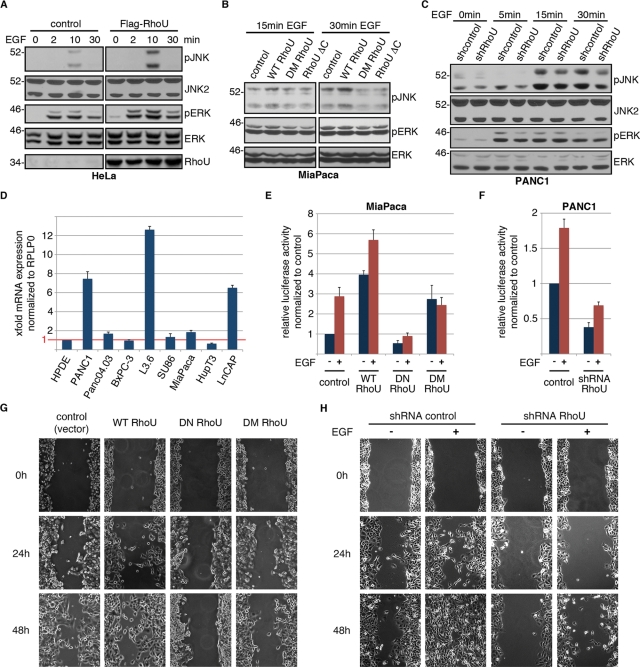
To determine whether the EGF-augmented interaction of RhoU affects downstream signaling cascades, we evaluated the phosphorylation of several major components downstream of activated EGFR in cells overexpressing WT RhoU. Of interest, we found that overexpression of RhoU correlates with an increased phosphorylation of JNK and ERK (Figure 7A) but not PLCγ or AKT on EGF stimulation (Supplemental Figure S3F). Increased JNK phosphorylation was also observed in MiaPaca2 cells overexpressing WT RhoU but not in cells expressing mutants deficient in GRB2 binding (DM RhoU) or deletion of C-terminal extension (Figure 7B). This role of RhoU in EGF-stimulated JNK phosphorylation was further evaluated in PANC1 cells, which express high levels of endogenous RhoU compared with the immortalized normal human pancreatic ductal epithelial cells (Figure 7D). Although PANC1 cells revealed different EGF-induced JNK phosphorylation kinetics from HeLa cells (Figure 7A), depletion of RhoU by lentivirus-mediated shRNA expression (Figure S4, B, C) significantly reduced EGF-induced JNK phosphorylation (Figure 7C, Supplemental Figure S3G). It was surprising that EGF-stimulated ERK phosphorylation kinetics appeared unaltered in the absence of RhoU (Figure 7C).
FIGURE 7:
RhoU synergize with EGF in JNK/AP1 activation and cell migration. (A) HeLa cells transfected with control vector or Flag–RhoU were serum starved for 16 h before EGF stimulation (20 ng/ml) for the indicated period of time. Cell lysates were prepared and immunoblotted for activating phosphorylation of the major EGF-dependent signaling pathways with indicated antibodies. (B) Miapaca2 cells transfected with WT or mutant RhoU expression constructs were serum starved, treated with EGF (100 ng/ml). and immunoblotted with the indicated antibodies. (C) PANC1 cells with stable RhoU suppression or scramble control were analyzed for JNK phosphorylation following EGF treatment (100 ng/ml). Phosphorylated ERK documents successful EGF stimulation, and ERK staining reflects relatively even sample loading in A–C. (D) Quantitative RT-PCR for RhoU mRNA expression in immortalized human ductal epithelial cells (HPDEs), different pancreatic cancer cell lines, and the prostate cancer cell line LNCaP cells. Results are normalized to RPLP0 and HPDE cells. (E, F) Luciferase reporter assay for AP1 activity in low-level (Miapaca2, E) and high-level (PANC1, F) RhoU-expressing cells. Miapaca2 cells were transfected with the 6xAP1 luciferase reporter and indicated constructs for 24 h and subsequently treated with EGF (20 ng/ml) or DMSO for 24 h. PANC1 cells were transfected with shRNA against RhoU or control shRNA for 36 h and subsequently treated with EGF (20 ng/ml) or DMSO for 24 h. Results are mean of three independent experiments ±SD. (G) For wound-healing analysis PANC1 cells were transfected with indicated constructs for 12 h and serum starved for another 12 h. At a transfection efficiency of >70% a cell-free space was made by scraping through the monolayer using a pipette tip. (H) PANC1 cells stably expressing RhoU shRNA or scrambled control were seeded at a high density and serum starved overnight. Scratches were made as in G, and EGF 20 ng/ml or DMSO was supplemented as indicated. Wound closure was documented every 12 h. Results from one representative experiment of three independent studies are shown.
RhoU and EGF synergize in JNK/AP1 activation and cell migration
We next examined whether RhoU expression or alteration of the EGFR/GRB2–RhoU signaling axis affects EGF-mediated cellular functions. A well-established target of activated JNK is c-Jun, a major component of the AP1 transcription factor complex. We therefore determined the effect of RhoU on AP1 activation using a luciferase reporter containing six tandem AP1 consensus sites. In MiaPaca2 cells, AP1-driven reporter activity is induced nearly threefold by EGF treatment alone, which is nearly doubled by simultaneous expression of WT RhoU. On the other hand, although WT RhoU enhances basal AP1 reporter activity about fourfold, expression of DN RhoU abrogated basal AP1 reporter activity and prevented EGF-induced activation, indicating that GTP loading is essential for this function (Figure 7E). Of interest, the DM RhoU mutant sustained basal AP1 activity but was unable to promote EGF-stimulated AP1 transcriptional activity (Figure 7E). Of significance, suppression of RhoU in PANC1 cells resulted in a reduction of both basal and EGF-induced AP1 reporter activity (Figure 7F). Consistent with the idea that RhoU membrane localization is critical for coupling EGFR signaling to AP-1, we find that expression of WT RhoU but not the C-terminal deletion or palmitoylation-site mutant (C256S) could enhance EGF-stimulated AP-1 transcriptional activity (Supplemental Figure S3H). These results are consistent with a previous report that RhoU expression leads to activation of JNK and AP1 (Tao et al., 2001). Of importance, our result further identifies that physical coupling of RhoU to EGFR signaling can further enhance AP1 transcriptional activity.
EGF stimulation has been shown to increase cell motility in multiple cell lines (Giehl et al., 2000). Using pancreatic cancer cell lines expressing high (PANC1) or low (HupT3) levels of endogenous RhoU as a model, we examined the effect of RhoU on cell motility in a wound-healing assay. We found that overexpression of either DN or DM RhoU inhibits spontaneous migration of PANC1 cells compared with control transfected cells (Figure 7G). However, the WT RhoU did not seem to simulate cell migration. Of interest, in HupT3 cells, which contain very low amounts of endogenous RhoU and are poorly migratory (Figure 7D), overexpression of WT RhoU induced cell migration, which was further increased by simultaneous treatment with EGF (Supplemental Figure S4A). It was surprising that expression of DM RhoU alone also slightly increased migration, although EGF treatment of DM cells had no synergistic effect on migration as observed in WT RhoU-transfected cells (Supplemental Figure S4A). In line with results from other groups, EGF treatment of PANC1 cells in the scrambled control group leads to increased cellular motility, whereas stable depletion of RhoU markedly inhibits spontaneous, as well as EGF-induced, cellular motility (Figure 7H). This is likely not the consequence of RhoU depletion on cell morphology or cell proliferation, as these phenotypes are not markedly different between the two cell populations (Supplemental Figure S4, D–F). Taken together, these data demonstrate a role for RhoU in spontaneous cell migration, as well as a role for the RhoU–GRB2/EGFR signaling axis in regulating EGF-stimulated migration in pancreatic cancer cell lines.
DISCUSSION
Recent studies have described an important role for RhoU in a variety of cellular functions, including actin cytoskeleton reorganization, focal adhesion formation, and cell motility. Of importance, RhoU activity has been shown to correlate with cellular transformation. However, the regulatory mechanisms underlying RhoU activation, as well as the signaling pathways in which it functions, remain poorly understood. Here we show that RhoU is physically and functionally coupled to activated EGFR complex via GRB2. Of importance, the interaction of RhoU with the activated EGFR occurs on endosomes and requires an interaction with GRB2 via its N-terminal extension and palmitoylation within its C-terminus. This EGF-induced RhoU–GRB2 interaction leads to increased GTP loading on RhoU and increased PAK1 association. In addition, we show that RhoU activity augments EGFR signaling specifically through the JNK pathway and enhances EGFR-induced JNK/AP1 activation and cell motility. These data not only provide a unique mechanism of signaling-regulated activation of RhoU via N-terminal–mediated SH3 interaction, but also identify a previously unappreciated role for RhoU in regulating EGF signaling and provide an additional potential mechanism by which RhoU can contribute to cancer phenotypes.
The 46–amino acid extension in the N-terminus of RhoU is one of the major features that distinguish it from CDC42. It was postulated in the original cloning paper that the proline-rich N-terminal extension might mediate interaction with SH3 domains (Tao et al., 2001). Subsequent work from both the Aspenström and Der groups demonstrated that the N-terminus could interact with several SH3 domain–containing proteins, including GRB2, NCKβ, and PLCγ, and a GRB2–RhoU interaction was further confirmed in vivo (Saras et al., 2004; Shutes et al., 2004). In addition, removal of the N-terminal extension enhances its ability to interact with and activate PAK to cause growth transformation (Shutes et al., 2004). These studies suggest a potentially important role of the N-terminal extension in the regulation of RhoU activity/function. However, whether or how RhoU–SH3 protein interactions are regulated and their functional significance are not known. To address these questions, we performed a large-scale YTH screening and isolated a number of SH3 domain–containing proteins as RhoU interactors in addition to those previous reported. Thus our data suggest an even broader spectrum of SH3 proteins capable of interacting with RhoU, which further strengthens the notion that the N-terminal extension may function as a dedicated SH3-binding module and raises a pivotal question as to how the interactions are regulated in vivo to achieve specificity.
Our extensive in vitro as well as in vivo analysis of RhoU and GRB2 mutants suggest that the second PXXPXR SH3-binding consensus and the N-terminal SH3 domain serve as an important docking interface linking RhoU to the activated EGFR on endosomes. Of most significance, our results demonstrated that the RhoU–GRB2 (SH3) interactions functionally couple RhoU into specific EGFR-mediated signaling and cellular activities. This result is consistent with previous reports showing that RhoU activation alone is sufficient to phosphorylate JNK/cJun via phosphorylation/activation of PAK1 (Tao et al., 2001; Chuang et al., 2007). Of greater importance, we further show that RhoU is also capable of synergizing with activated EGFR in JNK/AP1-mediated transcriptional activity in a manner dependent on SH3 binding, as the DM RhoU, which is physically uncoupled from GRB2/EGFR complex, is unable to synergize with activated EGFR in either JNK/AP1 activation or increased cell motility.
The mechanism responsible for enhanced RhoU activity by EGF signaling as reflected by both increased GTP loading and PAK1 association is not clear. Because of its rapid guanine nucleotide exchange rate when compared with Cdc42, it has been postulated that RhoU is constitutively GTP bound and therefore highly active (Saras et al., 2004; Shutes et al., 2004). It is surprising that, although our results confirm high intrinsic basal RhoU–GTP levels, RhoU–GTP binding can be enhanced downstream of EGF stimulation. These data are consistent with previous studies demonstrating that the putative GTP-bound active mutant (Q107L) demonstrates a stronger phenotype in its transforming activity in NIH3T3 cells and more efficient phosphorylation of c-Jun, PAK1, and PYK2 than WT RhoU (Tao et al., 2001; Shutes et al., 2004; Ruusala and Aspenström, 2008; Brazier et al., 2009). Of interest, the RhoU/GRB2 (SH3) association is markedly enhanced by EGF stimulation. We suggest that in unstimulated cells, the distinct subcellular localization of GRB2 and RhoU is responsible for the low basal interaction. In fact GRB2 localizes in the cytosol in complex with SOS (Figure 3C, top), whereas RhoU mainly resides on endosomal membranes. However, on EGF stimulation, GRB2 is recruited to phosphorylated EGFR via binding to pY1068/1086 and internalized with EGFR via the endosomal pathway where RhoU is accumulated. The dynamic localization of GRB2/EGFR along the endosomal membrane is likely responsible for increased association of RhoU–GRB2/EGFR. In addition, EGF stimulation may cause a conformational change in GRB2 leading to increased binding affinity to RhoU. Because It has been shown that the N-terminus of RhoU may act as a negative regulator of RhoU interaction with downstream effectors (Shutes et al., 2004), our data are consistent with a model in which EGF simulation enhances RhoU–GRB2 (SH3) interactions, which in turn relieves the N-terminus–mediated autoinhibition of effector binding, resulting in overall increased RhoU activity.
In addition to GTP loading, the ability of Rho proteins to regulate specific pathways is also dictated by their localization to specific subcellular compartments via lipid modification of their C-terminal CAAX domain. Compared to CDC42, RhoU also has a C-terminal extension of 21 residues, which is critical for both its subcellular localization and transforming activity. In contrast to CDC42, it has been shown that RhoU uses palmitoylation as a membrane-targeting mechanism instead of prenylation (Berzat et al., 2005). Of significance, RhoU endosomal localization appears to be largely independent of serum level in the culture medium, GTP loading status, or the presence of the N-terminal extension. However, the C-terminal extension, and specifically the palmitoylation site, is essential for proper localization. In fact our data demonstrate that deletion or mutation of the C-terminus results in a loss of RhoU endosomal localization. In addition, although the C-terminus is not required for RhoU–GRB2 binding in vitro, deletion or mutation of the palmitoylation site abolished RhoU–GRB2 interaction in cells and its coupling to EGFR-mediated JNK/AP1 transcriptional activity and cell motility.
Of interest, a recent study identified a tyrosine residue (Y254) near the C-terminal palmitoylation site, which can be phosphorylated by Src and regulate its localization as well as its activity in a non–small cell lung cancer cell line (Alan et al., 2010). In serum-starved cells, N-terminal GFP-tagged RhoU localization was mainly observed on the plasma membrane, whereas serum stimulation resulted in cytoplasmic redistribution (Alan et al., 2010). In contrast to this report, we find that the RhoU(Y254F) mutant, which cannot be phosphorylated by Src, is still capable of localizing to endosomes and associating with the internalizing EGFR. Taken together, our data are consistent with the idea that RhoU localizes to several different subcellular compartments, including endosomes, focal adhesions, and the plasma membrane, and its localization appears to be dynamically regulated in a signaling- or cell type–dependent manner.
RhoU is widely expressed in human tissues and possesses transforming potential in both mouse mammary epithelial and mesenchymal cells (Shutes et al., 2004; Berzat et al., 2005; Tao et al., 2001). RhoU is also reported to stimulate cell migration and reduces focal adhesion formation, and RhoU activation also leads to disrupted epithelial tight junction and integrity, resembling the phenotype of dedifferentiated cancer cells (Saras et al., 2004; Chuang et al., 2007; Ory et al., 2007; Brady et al., 2009). Thus RhoU activation may contribute to both tumorigenesis and progression. In fact, in addition to being a transcriptional target of wnt-1, RANKL, or GP130/STAT3 as has been reported (Tao et al., 2001; Brazier et al., 2006; Schiavone et al., 2009), our study suggests that RhoU activity can also be further modulated by the EGFR, another important regulator of cancer progression. Conversely, active RhoU synergizes with EGFR in driving JNK/AP1 activation and cell motility. This GRB2-dependent RhoU–EGFR cross-talk is of particular importance considering that overexpression/activation of EGFR is observed in majority of human epithelial tumors and is associated with metastasis, poor prognosis, and resistance to chemotherapy (Nicholson et al., 2001). In this regard, our findings not only provide a physiologically relevant mechanism for the regulation of RhoU activity, but also link RhoU to a well-known oncogenic pathway, which may have significant implication in cancer development and progression.
MATERIALS AND METHODS
Expression plasmids
Both mouse and human RhoU were used in this study as specified. To generate mouse RhoU cDNA expression plasmids, we amplified the full-length RhoU coding sequence by PCR from an expressed sequence tag clone obtained from American Type Culture Collection (Manassas, VA; GenBank Accession No. AW475926) and cloned into pCMV-Tag2 (Stratagene, Santa Clara, CA) in frame with the N-terminal Flag. The cDNA fragments encoding N- and C-terminal deletion mutants encoding amino acid residues 48–261 and 1–240 were cloned into the same vector, respectively. Mutagenesis for the SH3-binding consensus and the CA RhoU (Q110L) and DN RhoU (T66N) mutants were generated using QuikChange Site-Directed Mutagenesis Kit (Stratagene). For YTH screening, the mouse RhoU Q110L mutant was cloned into pGKB-T7 vector in frame with N-terminal GAL4 DNA-binding domain. The human RhoU coding sequence was amplified from a RhoU cDNA plasmid obtained from Open Biosystems (Thermo Biosystems, Huntsville, AL; GenBank Accession No. CA392555) and cloned into pCI vector (Promega, Madison, WI) with an N-terminal Flag tag. The G58V, T63N, Y254F, and C256S mutations were generated with Site-Directed Mutagenesis kit as mentioned. The N- and C-terminal deletion mutants encoding amino acids 46–258 and 1–237 were cloned by PCR in the same vector. The WT RhoU cDNA sequence is also subcloned into pCMV-Tag2 vector with an N-terminal Myc tag or GFP tag in pEGFP N1 (Clontech, Mountain View, CA), respectively. To generate shRNA expression constructs for RhoU, the human RhoU cDNA coding fragments 568–586 (5′-CTGTGCGCCGAGGAAATCA-3′) and 656–674 (5′-TCGTCGCTGGCATTCAATA-3′) were used as targets and cloned as hairpins into the pCMS4 vector. The pCMS4-eGFP-H1P vector has been previously reported (Gomez et al., 2006).
The pCDNA3-PAK1 expression vector was kindly provided by Mengwei Zang and Zhijun Luo (Boston University, Boston, MA). To generate GRB2–EGFP fusion expression vector, GRB2 cDNA encoding amino acids 1–216 was PCR amplified with the following primers: 5′-CCGCTCGAGCCACCATGGAAGCCATCGCCAAATATG (forward) and 5′-CCCAAGCTTTCCACCTCCCCCTCCGACGTTCCGGTTCACGGGGGTG-3′ (reverse), digested with XhoI/HindIII, and cloned into pEGFP N1 vector at the same sites. This vector expresses GRB2 fused with C-terminal EGFP with four glycine residues as spacer replacing the stop codon of GRB2. To generate shRNA expression constructs for GRB2, the human GRB2 cDNA coding fragment 609–627 (5′-CATGTTTCCCCGCAATTAT-3′) was used as target and cloned into the pRNA-U6.1/Hygro vector (GenScript, Piscataway, NJ) and pCMS4 vector. For GST fusion protein expression, cDNA fragments encoding PBD (p21-binding domain of PAK1) and various SH3 domains were amplified by PCR and cloned into pGEX-5x expression vector (Amersham Pharmacia Biotech, Piscataway, NJ). All cDNA and shRNA expression plasmids were verified by direct sequencing at the Mayo Molecular Biology Core Facility.
Reagents
RhoU antiserum was generated with a synthetic peptide corresponding to amino acid residues 232–248 of human RhoU and purified as previously described (Zhang et al., 2005). Monoclonal antibody specific to early endosomal antigen (EEA1) was obtained from BD Transduction Laboratories (Lexington, KY). Rabbit polyclonal antibodies to ERK1/2, GRB2, SAPK/JNK, phosphorylated EGFR (pY1068), pAKT (S473), and pPLCγ (Y783), monoclonal antibodies to pERK1/2 (T202/Y204), pJNK (T183, Y185), and phosphotyrosine (4G10), and Myc tag (9B11) were purchased from Cell Signaling Technology (Beverly, MA). Monoclonal antibodies to lysosomal-associated membrane protein 1 (Lamp1) and polyclonal anti-EGFR were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-GRB2 (2GB04) for immunofluorescence analysis of endogenous GRB2 was obtained from Thermo Scientific (Waltham, MA). Recombinant EGF, monoclonal antibodies to β-actin, and α-tubulin (including its fluorescein conjugate), anti-FLAG M2 agarose, as well as rabbit polyclonal anti-Flag, were from Sigma-Aldrich (St. Louis, MO). EGF conjugated with rhodamine (Rh-EGF), rhodamine and Alexa-Fluor 633–labeled phalloidin, and fluorescently conjugated secondary reagents (goat anti-mouse and goat anti-rabbit) were all purchased from Invitrogen (Carlsbad, CA). The TNT coupled in vitro transcription/translation system was obtained from Promega. The Matchmaker GAL4 Two-Hybrid System 3 and related regents were from Clontech.
Cell culture, transfection, and treatment
HeLa cells were passaged in RPMI 1640 medium supplemented with 5% fetal bovine serum (FBS) and 5% fetal calf serum (FCS). LNCaP and HupT3 cells were maintained in RPMI 1640 with 7 and 10% FBS, respectively. The MDA MB231 and PANC1 were maintained in DMEM with 10% FBS. All culture media were supplemented with 2 mM l-glutamine. HeLa cells were transfected using electroporation (315–350 V, one pulse, 10 ms). LNCaP, MDA MB231, and PANC1 cells were transfected with Lipofectamine 2000 (Invitrogen). Transfected cells were used after 48–72 h for suppression or 16–24 h for expression unless specified. Cells were serum starved for 16–24 h for experiment involving EGF stimulation. In some cases cells were pretreated with nocodazole (5 μg/ml) or cytochalasin D (10 μM) for 30–40 min.
GST pull-down and GTPase activation assays
GST-fusion protein expression/purification and GST–SH3 pull-down assays with either whole-cell extracts or in vitro–translated [35S]methionine-labeled proteins were performed as previously described (Zhang et al., 2001). For GTPase activation assay, the GTP-bound form of transfected Flag–CDC42 and Flag–RhoU and their mutants were detected using the modified GST–PBD affinity precipitation assay as previously reported (Hamann et al., 2007). To test the effect of EGF on RhoU GTP loading, HeLa cells were transfected for 24 h and serum starved for an additional 16–20 h before the assay. To determine the effect of GRB2 suppression on EGF-stimulated RhoU GTP loading, HeLa cells were sequentially transfected first with shRNA expression vector for 24 h, then with RhoU expression vectors for another 24 h, followed by a serum starvation of additional 16 h before EGF stimulation and GST–PBD pull-down assay.
Immunoprecipitation and Western blot analysis
To test the effect of EGF stimulation and/or GRB2 suppression on RhoU-mediated interaction with either GRB2 or EGFR, transfected cells were serum starved for 16–24 h before EGF stimulation. Immunoprecipitation (from 500 to 1000 μg of protein) and lysates (50–100 μg of protein) were prepared and analyzed by immunoblot.
Immunofluorescence and confocal microscopy
To determine the subcellular localization of RhoU in HeLa, electroporated cells were seeded directly onto cover slips and grown 16–24 h before being fixed with 4% paraformaldehyde and prepared for immunofluorescence as described (Zhang et al., 2007). For LNCaP, MDA MB231, and PANC1 cells, they were transfected with Lipofectamine 2000 in tissue culture dishes. At 24 h posttransfection, cells were split and grown onto cover slips (precoated with polylysine for LNCaP cells) for an additional 24 h before fixing and staining. To study the effect of EGF on RhoU localization and colocalization with EGFR or GRB2, electroporated HeLa cells were allowed to grow on cover slips for 24 h, then serum starved for another 16 h before EGF stimulation (20 ng/ml) for indicated times. Cells were fixed, permeabilized, blocked, stained with primary and secondary reagents (indicated previously), and mounted in SlowFade Gold antifade reagent (Invitrogen) as previously described (Nolz et al., 2008). Images were obtained with either an LSM-510 or LSM-710 laser scanning confocal microscope (Carl Zeiss, Jena, Germany) using the 100x/1.46 numerical aperture (NA) oil objective (Carl Zeiss) and analyzed using the LSM software packages.
YTH screening
The Matchmaker GAL4 Two-Hybrid System 3 was used for screening with the full-length mouse RhoU (Q110L) as bait. The integrity and expression of the fusion construct pGBK-T7-mRhoU in the host AH109 cells were confirmed with anti-Myc antibody. No autoactivation of the reporters was associated with this bait construct, as determined by cotransformation of pGBK-T7-mRhoU with prey library vector pGADT7 in AH109 cells. Sequential library scale transformations were performed for both normal prostate and borrow marrow cDNA libraries. A total of 2.5 million clones were screened and selected on high-stringency plates (SD/–Ade/–His/–Leu/–Trp) coated with X-β-Gal. The cDNA inserts from yeast clones were amplified by PCR using primers 5′-CTATTCGATGATGAAGATACCCCACCA (forward) and 5′-GTGAACTTGCGGGGTTTTTCAGTATCTACGA-3′ and sequenced at the Mayo Molecular Biology Core Facility.
RNA isolation and quantitative RT-PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA). To produce cDNA, 1 μg of total RNA was processed with the Superscript III Kit (Invitrogen) according to the manufacturer's instructions. Quantitative PCR was performed using the comparative CT method and specific primers for human RhoU (5′- TTTATGCGTGTGACAGTGTAT-3′; 5′ AGCCGCCTCCTACATC 3′) and human RPLP0 (5′-AGATCCGCATGTCCCTTC-3′; 5′-CCTTGCGCATCATGGTGTT-3′) as reference gene primers. The quantitative PCR was performed with the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) by using the ABI Prism 7900TM Sequence Detection System (Perkin Elmer-Applied Biosystems, Foster City, CA) at a annealing temperature of 60°C. Experiments were performed in triplicate using three independent cDNAs.
Reporter assay
A pGL3-based luciferase reporter construct containing six tandem AP1 consensus sites was used for the reporter assays as previously described (Zhang et al., 2001). Transfected cells were serum starved and treated with 50 ng/ml of EGF or dimethyl sulfoxide (DMSO) for 24 h. Luciferase activity was determined by using the Lumat LB 9501 luminometer (Berthold Technologies, Oak Ridge, TN). Firefly luciferase activity was normalized to Renilla luciferase activity. The results were expressed as mean “fold induction” normalized to DMSO-treated vector control transfected cells. Mean values of at least three independent experiments are displayed ±SD.
Migration assay
Cell migration was determined by wounding assay as previously described (Stähle et al., 2003). Briefly, stable RhoU-depleted PANC1 cells or control PANC1 were seeded at high density in 35-mm cell culture dishes. Stable depleted cells were directly used, and control PANC1 were transfected at a confluence of ∼90% with indicated constructs and GFP. Transfection efficiency was determined 24 h after transfection, and cells showing more than 75% GFP-positive cells were used for the assay. Wounding was performed by scraping through the confluent cell monolayer with a pipette tip, and cells were washed twice with serum-free medium and incubated with EGF or serum free medium as indicated. For kinetic analysis, two individual fields within the dish were photographed at room temperature using an Axiovert 200 microscope (Carl Zeiss) with a digital camera (AxioCam HRC; Carl Zeiss) using a 10x/0.25 NA objective at the indicated times postwounding. Images were captured with the AxioVision software package.
Cell proliferation assays
Cell proliferation was measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) assay (Promega) and cell cycle analysis. For the MTS assay, 5000 cells/well were seeded in a 96-well culture plates and incubated in DMEM medium containing 10% FCS or serum-free medium for 46 h. Medium was removed, and fresh medium was added to each well along with 1:10 dilution of MTS solution. After 2 h of incubation, the plates were analyzed with a microplate reader at a wavelength of 490 nm (Molecular Devices, Sunnyvale, CA). Cell cycle analysis was performed by using propidium iodide staining and flow cytometry as previously described (Singh et al., 2010). Briefly, stable PANC1 cells maintained under indicated conditions were trypsinized, washed with phosphate-buffered saline (PBS), and fixed in 70% ethanol for 25 min. After washing with PBS, cells were incubated with PBS containing 20 μg/ml of RNase and 50 μg/ml of propidium iodide for 1 h at room temperature under light protection. The DNA content of 50,000 cells was analyzed on a Becton Dickinson FACS Caliber flow cytometer (Beckman Coulter, Brea, CA). The fractions of cells in the G0/G1, S, and G2/M phases were calculated using ModFit (Verity Software House, Topsham, ME).
Lentivirus-mediated RhoU suppression and PANC1 stable clones
LKO.1 vector carrying selective RhoU shRNA (Sigma-Aldrich MISSION shRNA library, NM_021205.4–1105s1c1) and scrambled control vector were obtained from Mayo Clinic RNA Interference Shared Resource. HEK293T cells were cotransfected with pLKO.1-shRhoU or scrambled control with pCMV-ΔR8.91 containing gag, pol, and rev genes and pMD-G(VSV-G) using FuGENE (Roche, Indianapolis, IN). The medium containing the transfection reagent was removed 16 h later and replaced with fresh complete DMEM plus 10% FBS and penicillin/streptomycin. Twenty-four hours later, the culture medium containing lentiviral particles was harvested, filtered, and transferred to a polypropylene storage tube in aliquots at –80°C. PANC1 cells were infected with appropriate amounts of lentiviral particles containing medium. Twenty-four hours later, virus-containing medium was removed and replaced with fresh medium containing 2 μg/ml of puromycin for 10 d. Pooled resistant clones were used as stable PANC1 cell clones after validation of successful RhoU suppression.
Supplementary Material
Acknowledgments
We thank the members of D.D.B.’s laboratory for helpful discussions. We also thank Mark McNiven and Donald Tindall for helpful discussions and providing useful regents. This work was supported by National Cancer Institute SPORE grant in Prostate Cancer CA91956 (J.S.Z.) and SPORE in Pancreatic Cancer CA102701 (D.D.B.). D.D.B. is a Leukemia and Lymphoma Scholar. A.K. is supported by a Mildred Scheel Fellowship of the German Cancer Society.
Abbreviations used:
- AP1
activator protein 1
- CDC42
cell division control protein 42 homologue
- EEA1
early endosome antigen 1
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ERK
extracellular-signal-regulated kinase
- GRB2
growth factor receptor–bound protein 2
- GST
glutathione S-transferase
- JNK
c-Jun N-terminal kinase
- PAK1
p21 activating kinase 1
- PI3K
phosphatidylinositol-3-kinase
- PKC
protein kinase C
- RhoU
Ras homologue gene family, member U
- SH2
SRC Homology 2 domain
- SH3
SRC Homology 3 domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-0969) on April 20, 2011.
REFERENCES
- Alan JK, Berzat AC, Dewar BJ, Graves LM, Cox AD. Regulation of the Rho family small GTPase Wrch-1/RhoU by C-terminal tyrosine phosphorylation requires Src. Mol Cell Biol. 2010;30:4324–4338. doi: 10.1128/MCB.01646-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J. 2004;377:327–337. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzat AC, Buss JE, Chenette EJ, Weinbaum CA, Shutes A, Der CJ, Minden A, Cox AD. Transforming activity of the Rho family GTPase, Wrch-1, a Wnt-regulated Cdc42 homolog, is dependent on a novel carboxyl-terminal palmitoylation motif. J Biol Chem. 2005;280:33055–33065. doi: 10.1074/jbc.M507362200. [DOI] [PubMed] [Google Scholar]
- Brady DC, Alan JK, Madigan JP, Fanning AS, Cox AD. The transforming Rho family GTPase Wrch-1 disrupts epithelial cell tight junctions and epithelial morphogenesis. Mol Cell Biol. 2009;29:1035–1049. doi: 10.1128/MCB.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier H, Pawlak G, Vives V, Blangy A. The Rho GTPase Wrch1 regulates osteoclast precursor adhesion and migration. Int J Biochem Cell Biol. 2009;41:1391–1401. doi: 10.1016/j.biocel.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Brazier H, Stephens S, Ory S, Fort P, Morrison N, Blangy A. Expression profile of RhoGTPases and RhoGEFs during RANKL-stimulated osteoclastogenesis: identification of essential genes in osteoclasts. J Bone Miner Res. 2006;21:1387–1398. doi: 10.1359/jbmr.060613. [DOI] [PubMed] [Google Scholar]
- Chuang YY, Valster A, Coniglio SJ, Backer JM, Symons M. The atypical Rho family GTPase Wrch-1 regulates focal adhesion formation and cell migration. J Cell Sci. 2007;120:1927–1934. doi: 10.1242/jcs.03456. [DOI] [PubMed] [Google Scholar]
- Fritz G, Kaina B. Rho GTPases: promising cellular targets for novel anticancer drugs. Curr Cancer Drug Targets. 2006;6:1–14. [PubMed] [Google Scholar]
- Giehl K, Skripczynski B, Mansard A, Menke A, Gierschik P. Growth factor-dependent activation of the Ras-Raf-MEK-MAPK pathway in the human pancreatic carcinoma cell line PANC-1 carrying activated K-ras: implications for cell proliferation and cell migration. Oncogene. 2000;19:2930–2942. doi: 10.1038/sj.onc.1203612. [DOI] [PubMed] [Google Scholar]
- Gomez TS, et al. HS1 functions as an essential actin-regulatory adaptor protein at the immune synapse. Immunity. 2006;24:741–752. doi: 10.1016/j.immuni.2006.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann MJ, Lubking CM, Luchini DN, Billadeau DD. Asef2 functions as a Cdc42 exchange factor and is stimulated by the release of an autoinhibitory module from a concealed C-terminal activation element. Mol Cell Biol. 2007;27:380–393. doi: 10.1128/MCB.01608-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases in transformation and metastasis. Adv Cancer Res. 2002;84:57–80. doi: 10.1016/s0065-230x(02)84003-9. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jiang X, Huang F, Marusyk A, Sorkin A. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol Biol Cell. 2003;14:858–870. doi: 10.1091/mbc.E02-08-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- Nolz JC, Nacusi LP, Segovis CM, Medeiros RB, Mitchell JS, Shimizu Y, Billadeau DD. The WAVE2 complex regulates T cell receptor signaling to integrins via Abl- and CrkL-C3G-mediated activation of Rap1. J Cell Biol. 2008;182:1231–1244. doi: 10.1083/jcb.200801121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S, Brazier H, Blangy A. Identification of a bipartite focal adhesion localization signal in RhoU/Wrch-1, a Rho family GTPase that regulates cell adhesion and migration. Biol Cell. 2007;99:701–716. doi: 10.1042/BC20070058. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian GRB2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- Ruusala A, Aspenström P. The atypical Rho GTPase Wrch1 collaborates with the nonreceptor tyrosine kinases Pyk2 and Src in regulating cytoskeletal dynamics. Mol Cell Biol. 2008;28:1802–1814. doi: 10.1128/MCB.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saras J, Wollberg P, Aspenström P. Wrch1 is a GTPase-deficient Cdc42-like protein with unusual binding characteristics and cellular effects. Exp Cell Res. 2004;299:356–369. doi: 10.1016/j.yexcr.2004.05.029. [DOI] [PubMed] [Google Scholar]
- Schiavone D, Dewilde S, Vallania F, Turkson J, Di Cunto F, Poli V. The RhoU/Wrch1 Rho GTPase gene is a common transcriptional target of both the gp130/STAT3 and Wnt-1 pathways. Biochem J. 2009;421:283–292. doi: 10.1042/BJ20090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutes A, Berzat AC, Cox AD, Der CJ. Atypical mechanism of regulation of the Wrch-1 Rho family small GTPase. Curr Biol. 2004;14:2052–2056. doi: 10.1016/j.cub.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Singh G, Singh SK, König A, Reutlinger K, Nye MD, Adhikary T, Eilers M, Gress TM, Fernandez-Zapico ME, Ellenrieder V. Sequential activation of NFAT and c-Myc transcription factors mediates the TGF-beta switch from a suppressor to a promoter of cancer cell proliferation. J Biol Chem. 2010;285:27241–27250. doi: 10.1074/jbc.M110.100438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Stähle M, Veit C, Bachfischer U, Schierling K, Skripczynski B, Hall A, Gierschik P, Giehl K. J Cell Sci. 2003;116(18):3835–3846. doi: 10.1242/jcs.00679. [DOI] [PubMed] [Google Scholar]
- Tao W, Pennica D, Xu L, Kalejta RF, Levine AJ. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Zaal K, Hailey D, Presley J, Lippincot-Schwartz J, Samelson LE. Role of Grb2 in EGF-stimulated EGFR internalization. J Cell Sci. 2002;115:1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Gong A, Cheville JC, Smith DI, Young CY. AGR2, an androgen-inducible secretory protein overexpressed in prostate cancer. Genes Chromosomes Cancer. 2005;43:249–259. doi: 10.1002/gcc.20188. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Gong A, Young CY. ZNF185, an actin-cytoskeleton-associated growth inhibitory LIM protein in prostate cancer. Oncogene. 2007;26:111–122. doi: 10.1038/sj.onc.1209769. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.