Abstract
Pax6 plays an important role in embryonic cell (ES) differentiation during embryonic development. Expression of Pax6 undergoes from a low level to high levels following ES cell differentiation to neural stem cells, and then fades away in most of the differentiated cell types. There is a limited knowledge concerning how Pax6 is regulated in ES cell differentiation. We report that Pax6 expression in mouse ES cells was controlled by CCCTC binding factor (CTCF) through a promoter repression mechanism. Pax6 expression was significantly enhanced while CTCF activity was kept in the constant during ES cell differentiation to radial glial cells. Instead, the interaction of CTCF with Pax6 gene was regulated by decreased CTCF occupancy in its binding motifs upstream from Pax6 P0 promoter following the course of ES cell differentiation. Reduced occupancy of CTCF in the binding motif region upstream from the P0 promoter was due to increased DNA methylations in the CpG sites identified in the region. Furthermore, changes in DNA methylation levels in vitro and in vivo effectively altered methylation status of these identified CpG sites, which affected ability of CTCF to interact with the P0 promoter, resulting in increases in Pax6 expression. We conclude that there is an epigenetic mechanism involving regulations of Pax6 gene during ES cell differentiation to neural stem cells, which is through increases or decreases in methylation levels of Pax6 gene to effectively alter the ability of CTCF in control of Pax6 expression, respectively.
Introduction
Regeneration of neural progenitor cells in vitro from embryonic stem (ES) cells is one of the promising methods to meet neurogenesis requirements in neurodegenerative therapy [1], [2], [3]. However, a major problem needs to be solved when inducing ES cells to generate neuronal stem cells is the heterogeneity. Improved methods have been developed to generate a certain type of neuronal stem cells, or called radial glial cells [4], [5], [6]. Radial glial cells have dual functions to produce neurons and to guide migration of the newly formed neurons [7], [8]. Recent studies demonstrate that treating ES cells with retinoid acid (RA) induces ES cell differentiation to become a pure population of the precursor cells that not only express a set of radial glial cell markers, but also have an enhanced expression of Pax6. However, Pax6 rapidly fades away when radial glial cells start terminal differentiation becoming neurons [4], [5]. Due to dynamic changes in Pax6 expression during committed neuronal differentiation of ES cells, Pax6 may be a key factor that controls the specification of radial glial cell differentiation in vitro.
Homeobox transcription factor Pax6 is highly conserved among vertebrate and invertebrate species and is crucial for the development of the eye, pancreatic islet cells and the central nervous system (CNS). Pax6 mutations cause the small eye (Sey) defect in mice and ocular aniridia in humans [9]. During normal CNS development, Pax6 regulates the balance between self-renewal and neurogenesis in neuronal precursors cells in a dose-dependant fashion, which reflects a need for a critical level of Pax6 at the certain stage of neuronal differentiation [10]. However, there is a limited knowledge about regulation of Pax6 in CNS development. Remaining questions are why there is a significant up-regulated Pax6 expression in neural differentiation toward radial glial cells, and what causes down-regulation of Pax6 during the terminal differentiation later. It is important to find answers for these questions in order to understand mechanisms involving regulation of Pax6 and to ultimately control the process of neural differentiation.
In most species, Pax6 transcription is regulated via P0 and P1 promoters [11], [12], [13]. There is a highly conserved transcription control element, termed ectoderm enhancer (EE). EE is located approximately −3.5 kbp upstream from the P0 promoter, which is important for promoting specific expression of Pax6, and to regulate retina- and neuro-specific gene expression [13]. Another regulatory element is located in intron 4 of the gene to direct activities of P0 and P1 promoters in the ciliary body, iris and amacrine cells [13]. The most recent report indicates that Pax6 expression is regulated by CTCF, a zinc finger protein (ZFP) [14], [15], [16]. CTCF is a multivalent eukaryotic transcription factor that interacts with DNA sequences in the region of Pax6 P0 promoter to block the interaction between the EE element and Pax6 P0 promoter [15], [16], [17].
CTCF plays multifunctional roles in epigenetic regulation of DNA imprinting, X chromosome inactivation and transcriptional controls of gene expression [18], [19]. CTCF is initially characterized as a negative and positive regulator because of its capability to bind to DNA motifs in the promoter of various genes, including c-myc, p19ARF, p16INK4a, PIM-1, PLK, BRCA1, TERT and Pax6 [16], [20], [21], [22], [23]. Later, there are new discoveries that demonstrate that CTCF functions as a unique insulator protein to regulate gene expression in both chicken globin and h19/Igf2 loci [24], [25]. It also regulates communications between adjacent regulatory DNA elements in a position-dependent manner, or serves as a barrier to buffer transgenes from position effects caused by spread of the repressive heterochromatin from adjacent sequences [18], [23]. The binding of CTCF to insulator sequences or DNA boundary elements is often sensitive to modification of DNA methylation (CH3) that usually inhibits CTCF binding and eliminates CTCF-dependent actions [21], [26].
Previous study reveals that there is a repressor element located in promoter of Pax6 gene containing five CCCTC motifs within a sequence of 80-bp nuclear acids. All of the five CCCTC motifs in this region are functional for the CTCF action [16]. The repressive effect of CTCF on Pax6 expression is required for growth factor-induced proliferation [27]. In addition, ultraviolet (UV) irradiation inhibits CTCF expression that minimizes CTCF binding to the repressor element of Pax6 [17], [28], [29]. In fact, over-expression of CTCF in transgenic mice suppresses Pax6 gene expression resulting in retardation of embryonic ocular development including the cornea, lens and retina [15], [16]. In the present study, we generated neural stem cells from ES cells and obtained radial glial cells with enhanced Pax6 expression. This in vitro differentiation procedure was applied as a working model to investigate the effect of CTCF on Pax6 expression dynamically, and to further understand the regulatory mechanism underlying Pax6 expression in early stages of ES and radial glial cells. Our data revealed an epigenetic mechanism indicating that increases and decreases in the methylation levels of Pax6 gene effectively alter the ability of CTCF to control Pax6 expression.
Results
Induction of radial glial cells from embryonic stem cells
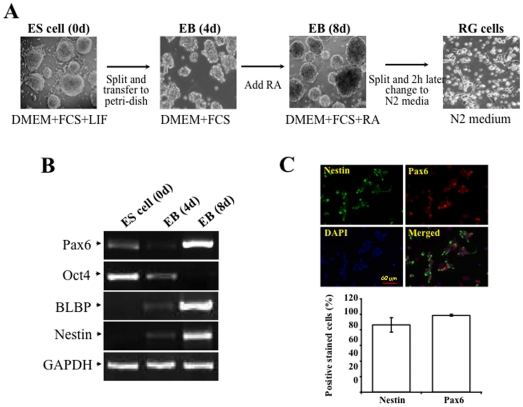
In the present study, a previous established protocol was used to induce ES cell differentiation toward radial glial cells [4], [5]. Cultured mouse ES cells were transferred to petri-dishes and treated with RA. Photos were taken by a Nikon invert microscope to document morphological changes of RA-induced ES cells in different stages of differentiation ( Fig. 1A ). Results of RA-induced ES differentiation followed a pattern that was consistent with previous reports. After formation of embryonic body (EB), RA-induced cells were successfully differentiated to spindle-shaped radial glial cells. Results from RT-PCR revealed that mRNAs of nestin (neural precursor marker) and BLBP (radial glial cell marker) were highly expressed, while expression of Oct4 mRNA (an ES cell marker) was suppressed in 8 days in RA-induced cells ( Fig. 1B ). More importantly, expression of Pax6 mRNA was dramatically increased in the 8th day in RA-induced cells. Nestin- and Pax6-positive cells were identified by immunostaining experiments in RA-induced cells, demonstrating that there was a significantly high conversion of ES cells to the radial glial cells ( Fig. 1C ). These results indicate that Pax6 is regulated during ES cell differentiation toward radial glial cells.
Figure 1. Induction of radial glial cells from embryonic stem cells.
(A) Morphological changes of neural differentiation from ES cells. (B) Altered mRNA expression of ES cell markers (Oct4) and radial gial cells markers (nestin, BLBP and Pax6). (C) Staining for radial gial cells markers. ES cells growing on gelatin-coated plates were dissociated and transferred to nonadherent bacterial dishes to form embryonic body. After 4-day of embryonic body formation, 5 µM retinoid acid was added to the culture media and cells were allowed to grow for another 4 days. Embryonic bodies were dissociated after a total of 8-day induction and plated to poly-D-lysine/laminin-coated dishes in N2 medium. N2 medium is changed after 2 h. Cell samples were collected before and after the 4 days and days induction as shown as ES (0 d), EB (4 d) and EB (8 d) respectively. Photos of RG cells were taken 6 h after passage. Total RNAs were extracted at indicated time points and altered expressions of cell lineage-specific markers were monitored by RT-PCR. Cells were immunostained with antibodies specific to Nestin (green) and Pax6 (red). Cell nuclei were stained by DAPI (Blue). Results were plotted as a percentage of positively stained cells with Nestin and Pax6 antibodies vs numbers of DAPI-positive nuclei.
Regulation of Pax6 expression by CTCF in ES cells
Our previous studies have demonstrated that CTCF is a negative regulator of Pax6 gene in the cornea and retina. To study whether CTCF also involves regulation of Pax6 in mouse ES cells, CTCF-specific siRNA was transfected to ES cells to knock down CTCF mRNA. We found that expression of CTCF at the protein level was decreased about 75% in ES cells transfected with CTCF-specific siRNA at 72 h ( Fig. 2A&2B ). At the same time (72 h), expression of Pax6 protein was significantly increased following knockdown of CTCF ( Fig. 2A&2C ). For control experiments, ES cells were transfected with non-silencing control siRNA following the same procedure and time course as the experimental group. The results demonstrate that altered levels of CTCF by knockdown of CTCF mRNA in ES cells result in up-regulation of Pax6 expression.
Figure 2. Effect of altered CTCF activity on Pax6 expression in ES cells.
(A) Effect of knocking down CTCF mRNA on Pax6 expression. Statistical analysis of suppressed CTCF expression (B) and increased Pax6 expression (C). Data was shown as mean ±S.E. Symbol “*” indicates significant differences (p<0.05, n = 4).
Effect of RA-induced ES cell differentiation on CTCF activity
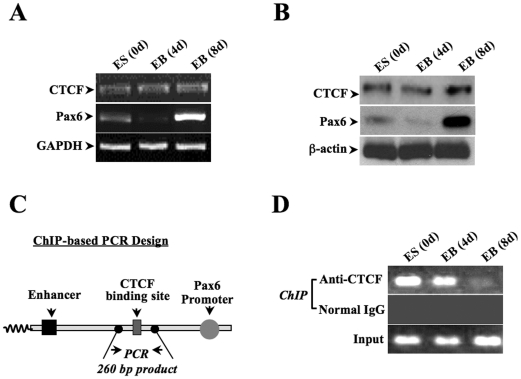
Based on the observation that Pax6 expression was regulated by CTCF in ES cells, we investigated whether CTCF activity was also changed to correlate to the altered Pax6 expression during the time course of ES cell differentiation to radial glial cells. The effect of RA-induced ES differentiation on CTCF activity was investigated by focusing on two respects that include the CTCF expression level and its DNA binding capacity. First, both mRNA and protein expressions of CTCF were monitored following the time course of ES cell differentiation ( Fig. 3A&3B ). We found that Pax6 expression markedly increased on the 8th day of EB formation, and that there were no changes in CTCF expression at either the protein or mRNA level. Second, association of CTCF with the binding motifs in the promoter of Pax6 gene was examined. It has been shown that CTCF binds as a repressor to the region of −1.2 kb upstream from Pax6 P0 promoter to suppress Pax6 transcription [16]. A set of primers was designed accordingly for chromatin immunoprecipitation (ChIP)-based PCR experiments to generate a 260 bp DNA fragment based on consensus DNA sequences upstream from the Pax6 P0 promoter within the region of CTCF binding motifs ( Fig. 3C ). Results of ChIP-based PCR using specific antibodies against CTCF revealed that there were remarkable decreases in CTCF binding on DNA motifs in Pax6 promoter region on the 8th day of EB formation. For controls, non-immunoprecipitated chromatins were used as DNA templates in PCR experiments (labeled as input) ( Fig. 3D ). Taken together, CTCF expression was not affected by ES cell differentiation, instead its binding ability to DNA motifs in Pax6 promoter was diminished, suggesting that increases in Pax6 expression during differentiation to radial glial cells are very likely resulted from disengaging CTCF from its binding motifs and releasing Pax6 promoter from the repressor effect of CTCF.
Figure 3. Correlation of CTCF and Pax6 in ES cell differentiation to radial glial cells.
(A) Expressions of CTCF and Pax6 mRNAs during ES cell differentiation to radial glial cells. Total RNA samples were harvested from undifferentiated ES cells (0 d) and embryonic body (EB) cells after 4 day or 8 day induction. RT-PCR assays were performed to measure the expression of CTCF, Pax6 and GAPDH. (B) Expressions of CTCF and Pax6 proteins during ES cell differentiation to radial glial cells. Western assays were performed to detect expression levels of CTCF, Pax6 and β-actin. (C) Illustration of ChIP-based PCR designs. (D) Detection of specific protein-nucleotide interaction between CTCF and Pax6 gene during ES cell differentiation and EB formation. Chromatin samples were immuno-pulled down by CTCF-specific antibody. ChIP-based PCR was performed to amplify CTCF binding DNA fragment in Pax6 P0 promoter region. All ChIP and pCR experiments were repeated for at least three times.
Methylation-modification within CTCF binding region
It has shown that the interaction of CTCF with DNA sequence can be inhibited if the binding motif is methylated [18]. Next question is whether DNA methylation affects associations of CTCF with Pax6 DNA since CTCF binding capacity to Pax6 DNA decreased during radial glial cell differentiation. First, several potential 5-methylcytosines were located in the 80 bp region of CTCF binding motifs through DNA sequence analyzing ( Fig. 4A ). Genomic DNAs from ES cells were isolated and treated with sodium bisulfite that converts unmethylated cytosine to uracil while leaving 5-methylcytosine unchanged. DNA fragments containing CTCF binding sites were obtained by PCR with primers specific to bisulfite-converted DNA and sequenced for conformation. Normal genomic DNAs without sodium bisulfite treatment were also isolated for control experiments. In the PCR products, only 5-methylcytosines remained as cytosines (C) while un-methylcytosines were converted to thymines (T). Sequencing results revealed that within the tested DNA region, there were mixed C/T pairs occurring in 3 positions (sites 1–3 shown in Fig. 4A ), indicating an uneven methylation of cytosine residues. All of other cytosine residues in CG-rich islands were completely converted to T (data not shown). Sites 1–3 were subjects to further quantitative analysis by performing Ms-SNuPE assays. These sites were differentially mehylated showing that methylations occurred about 15% in site 1, 55% in sties 2 and 3 ( Fig. 4B&4C ). To further verify methylation data, methylation responses of these sites were examined by altering cellular DNA methylation levels. ES cells were treated with 1 µM of 5-azadCyd to suppress methyltransferase resulting in decrease in DNA methylation. In contrast, ES cells were transfected with full-length cDNA encoding methyltranferase Dnmt3a to increase DNA methylation. Results showed: 1) site 1 was about 15% methylated in control cells compared to 12% and 18% methylated in 5-azadCyd treated and Dnmt3a-transfected cells, respectively; and 2) both sites 2 and 3 were approximate 55% methylated in control cells compared to 32% and 80% methylated in 5-azadCyd-treated and Dnmt3a-transfected cells, respectively ( Fig. 4C ). These results suggest that there are at least three cytosine residues that respond to alterations of cellular DNA methylation levels located in the region of CTCF binding motifs.
Figure 4. Identification of methylation sites within CTCF binding motifs.
(A) Putative methylation sites in the region of CTCF binding motifs upstream from Pax6 promoter. (B) Quantitative determination of methylation status at cytosines in CTCF binding sties after inductions of demethylation and methylation. Genomic DNA is treated with sodium bisulfite followed by PCR of the target sequence to generate the template for Ms-SNuPE assays. Methylation statuses were evaluated by the radio of radial incorporation of [32]P-dCTP (representing methylated cytosine) and [32]P-dTTP (representing unmethylated cytosines). DNA methylations were detected by using bisulfite sodium modifications and PCR. Demethylation and methylation were induced by 5-azadCyd (1 µM) and by over-expression of Dnmt3a, respectively. (C) Statistical analysis of methylation percentages in site1–3. Data was shown as mean ±S.E. and represented results from three independent Ms-SNuPE assays. Symbol “*” indicates significant differences comparing with cells without treatment. (p<0.05, n = 3).
Changed DNA methylation in CTCF binding region during differentiation
DNA methylation statuses of 3 cytosine residues within sites 1–3 were monitored during ES cell differentiation to radial glial cells. DNA samples were taken from cells in three stages: un-differentiated ES cells before the induction, on EB 4th day and EB 8th day after the induction. Ms-SNuPE assays were preformed to quantitatively evaluate methylation levels of sites 1–3. Levels of 5-methylcytosines in sites 1–3 were measured by using 32P-dCTP incorporation. There were no significant changes in methylation levels in those measured sites between ES cells and EB at 4th day ( Fig. 5A ). However, methylation levels were significantly increased in all of the three sites on the 8th day of EB formation ( Fig. 5A&5B ). The results demonstrate an increased DNA methylation levels within CTCF binding motifs located in Pax6 promoter region. Increases in DNA methylation in this region following the time course of ES cell differentiation may result in inhibition of CTCF binding to the Pax6 promoter.
Figure 5. Methylation status of CTCF binding region in Pax6 promoter during ES cell differentiation to radial glial cells.
(A) Detecting changes of DNA methylation in CTCF binding sites during ES cell differentiation and EB formation. (B) Statistical analysis of DNA methylation changes in CTCF binding motifs during ES cell differentiation and EB formation. The genomic DNA harvested from ES cells and EB cells at different stages was treated with sodium bisulfite followed by PCR of the target sequence to generate the template for Ms-SNuPE assays. Methylated cytosines at site 1–3 were revealed by [32]P-dCTP incorporation. Symbol “*” indicates the statistical significance determined by Student's t test at p<0.05 (n = 3).
Effect of DNA methylation on CTCF protein-nucleotide interaction
To study the specific effect of DNA methylation on CTCF binding capability to the Pax6 promoter, EMSA/super-shift assays and ChIP-based PCR were performed to evaluate the DNA binding capability of CTCF. First, EMSA and super-shift assays were performed with synthesized and 32P-labelled DNA probes. These probes contained four CTCF binding sites in the 80 bp fragment upstream from the promoter. The probes were un-methylated or methylated in all of the three-cytosine sites. DNA-protein complexes were formed in nuclear extracts when those un-methylated probes were used and the binding between DNA and protein was competed by un-labeled probes, but not by nonspecific control probes ( Fig. 6A ). Furthermore, there was a super-shift band found in cell nuclear extracts incubated with CTCF-specific antibodies, however, there were no super-shift bands in the control sample treated with c-Jun antibodies. In contrast, there were no specific bands found in lanes that were treated with methylated probes, indicating that there was no DNA-protein complex formed ( Figure 6A ). The results demonstrate that DNA methylation negatively affects CTCF-specific binding activity in its motifs upstream from the Pax6 promoter. To further verify the methylation-sensitive interaction between CTCF and the Pax6 promoter, ChIP-based PCR were performed to detect the binding capacity of CTCF with the Pax6 promoter in ES cells in the absence and presence of 5-azadCyd. Consistently, results of ChIP assays demonstrated remarkably enhanced bands in demethylation-induced ES cells when the CTCF specific antibody was used to pull down chromatins ( Fig. 6B ). Results from both EMSA and ChIP-based PCR experiments indicate that there are indeed methylation-sensitive interactions between CTCF and Pax6 gene in ES cells.
Figure 6. Effect of DNA methylation or de-methylation on protein-nucleotide interaction of CTCF and Pax6 gene.
(A) DNA and nuclear protein interactions were detected by EMSA using demethylated and methylated CTCF-specific DNA probes. Anti-CTCF antibody was used to detect DNA-protein complex super-shift. In addition, AP-1 DNA probe and c-Jun antibody were used in control experiments for DNA binding competition and super-shift experiments, respectively. Radioactive signals were visualized by exposure of the EMSA gel to X-ray film. (B) Significant increase in specific protein-nucleotide binding of CTCF to Pax6 gene in demethylated ES cells. Chromatin samples in 1 µM 5-azadCyd-induced ES cells were immuno-pulled down by CTCF-specific antibody. ChIP-based PCR was performed to amplify the CTCF-binding motif DNA fragment in Pax6 promoter region. Data were obtained from three independent ChIP and PCR experiments. Symbol “*” indicates significant differences (p<0.05, n = 3).
Effect of DNA methylation on Pax6 expression
Effects of altered DNA methylation on Pax6 expression in 5-azadCyd-treated and Dmnt3a-transfected cells were monitored by Western analysis. Treatment of 5-azadCyd induced DNA de-methylation down-regulated Pax6 expression ( Fig. 7A ). In contrast, over-expression of Dnmt3a by transfecting cells with full-length cDNA encoding Dnmt3 up-regulated Pax6 expression following a time course ( Fig. 7B ). Statistical analysis showed that Pax6 expression was significantly increased after over-expression of Dnmt3. The result is consistent with the observation that there was an increased DNA methylation in the CTCF binding motifs in the Pax6 promoter region, and that there was an increased Pax6 expression on the 8th day of EB formation during ES cell differentiation to radial glial cells. Taken together, our data support the notion that there are increased DNA methylation in the region of CTCF binding motifs upstream from the Pax6 promoter can exclude CTCF binding in the region during the period of ES cell differentiation to radial glial cells, which abolishes the repressor effect of CTCF on Pax6 promoter activity, resulting in an enhanced Pax6 expression.
Figure 7. Effect of DNA methylation or de-methylation on Pax6 gene expression.
(A) Effect of decreasing DNA methylation on Pax6 gene expression in ES cells. ES cells were treated with 1 µM 5-azadCyd for 24 and 48 h and subjected to Western analysis to compare the Pax6 expression w/wo 5-azadCyd-mediated de-methylation treatment. (B) Effect of increasing DNA methylation on Pax6 gene expression in ES cells. ES cells were transfected with full-length cDNA encoding Dmnt3a for 24–72 h and protein levels of Pax6, CTCF, Dmnt3a and β-actin were determined by Western analysis. (C) Data was shown as mean ±S.E. obtained from three independent experiments. Symbol “*” indicates significant differences between treated and un-treated groups.
Discussion
Neural progenitor cells derived from ES cells can be a promising candidate for the cell-replacement therapy for neurodegenerative and retinal diseases [3]. To generate a homogenous resource of neural progenitor cells, it is important to identify key factors that can be used not only as an indicator of neural differentiation, but also as an entry point to manipulate the conversion course. Pax6 appears to be a good candidate for this purpose since it is crucial for balancing neural stem cells self-renewal and neurogenesis [10], [30], [31]. There are three types of neural stem cells during CNS development, including neuroepithelial cells, radial glial cells and basal progenitor cells [7], [30]. Pax6 is a key factor in the specification of radial glial cells [5], [32], [33]. Loss function of Pax6 leads to the transition from predominantly radial glial cell-based neurogenesis to basal progenitor cell-based neurogenesis [30]. More importantly, regulation of CNS development by Pax6 is sensitive to Pax6 dosages that is evidenced by the followings: 1) heterozygous human mutation of Pax6 results in aniridia and forebrain abnormalities; 2) homozygous human and mouse mutations of Pax6 lack eyes and have marked microcephaly and absent olfactory bulbs; 3) increased expression of Pax6 in transgenic mice results in microphthalmia and forebrain abnormalities [10]. In addition, during the course of in vitro conversion of ES cells to neural cells via an intermediate radial glial cell stage, Pax6 expression exhibits extensive changes with the peak expression level in RG cells [4], [5]. In the present study, we have monitored Pax6 expression patterns and the control of Pax6 expression by CTCF in an established model in vitro during generating radial glial cells from ES cells. The mechanism involving CTCF occupancy and DNA methylation in CTCF-specific binding location in Pax6 promoter explains how up-regulation of Pax6 occurs in a certain stage of EB formation. All these in vivo and in vitro investigations are consistent with previous study results, indicating that Pax6 expression must be under tight control during neural differentiation.
Differentiation of ES cells is initiated by cell aggregation in non-tissue culture plates, and then by adding retinoid acid on the 4th day of EB formation. Pax6 expression was tremendously enhanced on the 8th day of EB formation accompanied with suppressed Oct4 expression, and with enhanced expressions of BLBP and nestin that are markers for radial glia and neuronal precursor cells ( Fig. 1 ). These results are reminiscent of previous reports and prompted us to investigate what causes the dramatically increase in Pax6 activity in radial glial cells [4], [5]. We consistently observed a slight down-regulated Pax6 expression both at its mRNA and protein levels on the 4th day of RA-induced differentiation. The association between CTCF protein and Pax6 promoter may have not been affected yet on the 4th day of the induction, suggesting that other mechanisms are involved in down-regulating Pax6 expression. One possibility is that Pax6 expression may be regulated by altered microRNA levels at this stage as it has been shown in neuron progenitor cells microRNA-9 indirectly inhibits Pax6 expression [34].
Previous studies found that the epigenetic regulator CTCF negatively controls transcription of the Pax6 gene in ocular tissues [16]. Here, we demonstrate that CTCF also involved in regulation of Pax6 in ES cells by showing that deficiency of CTCF by knocking down CTCF mRNA significantly could enhance Pax6 expression ( Fig. 2 ). Expression levels of CTCF mRNA and protein were not changed in the entire period of monitoring ES cell differentiation to radial glial cells. In stead, capability of CTCF interacting with its binding motif located upstream from Pax6 promoter was remarkably reduced on the 8th day of EB formation, providing a new explanation for increased Pax6 expression at this stage that CTCF releases Pax6 from a negative transcription control. The rest of the study contributed to investigate mechanisms underlying the decreased interaction of CTCF with its binding motif in Pax6 promoter in radial glial cells.
Regulation of CTCF binding with its target DNA sequences is often through a methylation-sensitive mechanism. For example, CTCF epigenetically controls IGF-2/H19 expression by binding to a differentially methylated domain (DMD) determined as imprinted control region (ICR) [26]. Three potential methylation sites (CG-rich islands) are tested inside the region where CTCF binding motifs are found. Site 1 was a low methylated site with approximately 15% methylation, suggesting that site 1 is unlikely to play a major role in methylation-modifed CTCF occupancy in control ES cells. However, methylation levels in sites 2 and 3 are rather higher in control cells and changed from 55% to 32% and 80% after 5-azadCyd-induced demethylation and Dnmt3a transfection-enhanced methylation, respectively ( Fig. 4 ). The results not only demonstrate that there are 3 methylation sites inside of CTCF binding region, but also verify that 5-azadCyd treatment and Dnmt3a transfection can efficiently change methylation status of CTCF occupancy to interact with its binding motifs. Therefore, these results are helpful for us to understand the effect of DNA methylation on CTCF binding capability and control of Pax6 expression. Methylation in all of the three sites has been quantitatively evaluated during ES cell differentiation. On the 8th day of EB formation, hypermethylation occurred in sites 2 and 3 is remarkable, indicating these sites may play major roles in regulating CTCF occupancy and protein-DNA interaction with Pax6 promoter. As reported in RA-induced ES cell differentiation to neuron progenitor cells, there are predominant gains of DNA methylation in promoters of several hundred genes. However, there are a little changes of globle DNA methylation between neuronal progenitors and terminal neurons [35]. This indicates that promoter hypermethylation is less dynamic during a transition to the terminal differentiation. We believed that there might be different mechanism for down-regulating Pax6 expression during terminal differentiation, such as polycomb-mediated repression. Polycomb targets are very dynamic during linage commitment and terminal differentiation [36], [37].
More evidence shows that DNA hypermethylation occurring within this region is parallel to decrease in CTCF occupancy and to increase in Pax6 expression, suggesting that methylation of Pax6 gene plays a functional role in regulating Pax6 transcription that associates with ES cell differentiation. To establish the connection between releases of CTCF from its binding sites and hypermethylation in the same region, it requires more evidence to show whether mthylation/de-methylation regulates CTCF binding capability. To address this question, we performed EMSA and ChIP-based PCR to demonstrate the role that DNA methylation plays in the region of Pax6 promoter to modulate the interaction between CTCF and its binding motifs ( Fig. 6 ). It believes in general that methylation of CpG islands is a dynamic epigenetic marker that undergoes extensive changes during ES cells differentiation, which contributes to turn-on and turn-off of particular genes that are closely related to ES cell pluripotency and self-renewal [38], [39]. Thus, DNA methylation has become a target for developing pharmacological agents to manipulate directional differentiation of ES cells [40]. In the present study, hypermethylation in CTCF binding motifs in the Pax6 promoter region occurring during ES cell differentiation functionally regulates Pax6 activity associated with altering CTCF binding capacity. In summary, the present work provides a novel mechanism underlying the enhanced Pax6 expression during induction of neural progenitor cells from ES cells. The methylation-sensitive regulation of Pax6 during neural differentiation provides theoretical evidences for future development of techniques to better control neural progenitor cell generation from ES cells.
Materials and Methods
ES cell culture and treatments
Mouse J1 ES cells (purchased from ATCC) were cultured in Knockout™ D-MEM supplemented with 10% Knockout™ serum, 2 mM L-glutamine, 100 µM non-essential amino acid, 100 U/ml penicillin/100 µg/ml streptomycin and 1,000 U/ml ESGRO® in 0.1% gelatin coated dishes in a humidified incubator at 37°C with 5% CO2. All reagents for cell culture except for ESGRO® (Chemicon International Inc, CA) were purchased from Invitrogen™ Life Technologies, (Grand Island, NY). For ES differentiation experiments, ES cells were grown in non-adherent dishes in suspension to form embryoid bodies (EB). On the forth day of the EB formation, retinoic acid (5 µM) was added for additional 4-day period. EB cells were dissociated after the 8th day, and cultured in poly-D-lysine/laminin-coated dished for 2 h in a complete DMEM medium supplemented with N2 supplement and human basic fibroblast growth factor (10 ng/ml). (Invitrogen, CA). Neuronal precursor cells were induced by further culturing EB cells in the neuronal precursors medium (Invitrogen, CA). The stock solution of 5-Aza-2′-deoxycytidine (5-AzadCyd, Sigma, St. Louis, MO) was made at a concentration of 1 mM. The final concentration of 5-AzadCyd used in treating ES cells was at 1 µM.
Chromatin-immunoprecipitation (ChIP) and PCR
ES and EB cells were cross-linked in 1% formaldehyde for 10 min before the reaction was quenched by glycine at a final concentration of 125 mM for 5 min at room temperature. Cells were then homogenized in 1 ml cell lysis buffer to release nuclei. Isolated nuclei were re-suspended in 1 ml nuclear lysis buffer and genomic DNAs were broken down into 1∼2 kb fragments by sonication. Chromatin samples were immuno-precipitated by using CTCF-specific antibody (Millipore, Billerica, MA). The antibody-protein-DNA complexes were pulled down by protein A/G agarose pre-absorbed with salmon sperm DNA. Samples were then treated with proteinase K at 55°C overnight. Purified ChIP-DNA samples were analyzed by PCR using a pair of primers targeted to CTCF binding sites in the Pax6 P0 promoter region (sense: 5′- TGTCGGGGGAGGAGCAAGAACC′ and antisense: 5′- CCTGGAGGGGCGGGAGACT-3′).
Gene transfections and RNA interference
The plasmid pCMV-SPORT6 containing the cDNA fragment encoding full-length mouse cytosine-5-methyltransferase 3a (Dnmt3a) was purchased from ATCC® (MGC-5662). The pcDNA4 plasmid containing the cDNA fragment encoding full-length mouse CTCF was constructed in our lab previously [27]. Plasmids (20 µg per sample) were transfected into ES cells (1×106 suspended in 600 µl of PBS solution) through electroporation at a setting of 250 V, 500 µF, and 75 Ω for 10 ms. Transfected cells were placed in full-growth medium and transferred to two 0.1% gelatin coated 60 mm culture dishes. Vectors without inserts were also transfected into ES cells for control experiments. For RNA interference experiments, specific primers that were complementary to CTCF mRNA (21 nucleotides plus other 8 nucleotides complementary to T7 promoter) were designed in our lab [27]. Briefly, a pair of primers was used with a sense strand sequence of “aaggaaugucuucuuuacacc”, and an antisense strand sequence of “aagguguaaagaagacauucc”. In addition, a nonsilencing control siRNA double-strand siRNA (sense: “aacauucgguagauuccucgc” and antisense: “aagcgaggaaucuaccgaaug”) was also synthesized and applied by using the same method. Sequence homologies of siRNA primers were examined by using a NIH Blast program. SiRNA was synthesized by in vitro transcription using a Silencer® siRNA Construction Kit (Ambion, Inc., TX). Double-strand RNA was synthesized by in vitro transcription using T7 RNA polymerase followed by RNase digestions to obtain siRNA. SiRNAs that were pre-mixed with Lipofectamine were used to transfect ES cells. None-specific siRNAs were synthesized using the same kit and transfected into ES cells for control experiments.
Western analysis
ES cells were harvested by centrifugation at 2,000 g for 5 min. Cell pellets were resuspended in lysis buffer containing (in mM): 20 Tris-HCl-pH 7.5, 137 NaCl, 1.5 MgCl2, 2 EDTA, 10 sodium pyrophosphate, 25 β-glycerophosphate, 10% glycerol, 1% Triton X-100, 0.1% aprotinin, 0.1% leupeptin, 1 sodium orthovanadate, 1 phenylmethylsulfonyl fluoride. Equal amounts of denatured lysates (15 µg) were fractionated by SDS-PAGE and fractionated proteins in SDS-PAGE were electro-transferred onto a PVDF membrane. The membranes were incubated with primary antibodies in Tris-buffered saline containing 0.1% Tween 20 (TBST) containing 5% nonfat milk overnight at 4°C. All protein extracts were displayed in Coomassic blue-stained gels to ensure the equal amount of proteins being loaded. Antibodies against CTCF (sc-5916) and Pax6 (sc-11357) were purchased from Santa Cruz Biotechnology. Antibody against Dnmt3a was purchased from Calbiochem (Cat. 317282). Mouse monoclonal antibody against β-actin (Sigma, St. Louis, MO) was diluted at a ratio of 1∶10,000. Specific signals were detected by using Western Blotting Luminol reagents (Santa Cruz Biotechnology, CA).
Electrophoretic mobility shift assay (EMSA)
Two sets of double-stranded oligonucleotides (80 bp) with the specific sequence of Pax6 promoter region contained all CTCF binding sites. Three of the binding sites were modified with/without replacements of 5-methylated cytosines (Operon Biotechnologies, Huntsville, AL). The probes were labeled with γ-32P-deoxy-ATP (∼3,000 mCi/mmol) using a T4 polynucleotide kinase labeling kit (Promega, WI) and purified with QIAquick nucleotide removal kit (Qiagen, CA). Nuclear protein extracts (10 µg) were incubated in the binding buffer containing 10 mM Tris-HCl (pH 7.5), 0.05 mg/ml poly (dI-dC), 0.5 mM EDTA, 50 mM NaCl, 1 mM MgCl2, 4% glycerol, and 0.5 mM DTT on ice for 15 min. Each binding reaction (20 µl) was initiated by adding 68.8 fmol probes and incubated on ice for 30 min. Gels were vacuum-dried and exposed to X-ray film for autoradiography. Competition experiments were performed by pre-incubating nuclear protein extracts with 1 pM of unlabeled probe on ice for 30 min and by adding different isotope-labeled probes. For super-shift assays, DNA-protein complexes were incubated for an additional 30 min using antibodies specific to CTCF and to negative control proteins.
DNA preparation and bisulfite modifications
Cells were digested in lysis buffer (100 mM Tris-HCl [pH 8.5], 5 mM EDTA, 0.2% sodium dodecyl sulfate (SDS), 200 mM NaCl, 100 µg of proteinase K/ml) at 55°C followed by phenol-chloroform extraction. Total genomic DNAs were obtained by ethanol precipitated and cleaved by EcoR I digestion. The DNA fragments were modified by a bisulfite treatment following a method described previously [41], [42]. In brief, denatured DNA was mixed with freshly prepared 30 µl of 10 mM hydryquinone (Sigma, MO) and 520 µl of 3 M Sodium bisulfite (Sigma, MO) at pH 5.0. Samples were immerged in the mineral oil and incubated at 55°C for 6 h. Modified DNA was purified using the Wizard DNA Purification Resin kit (Promega, WI), and eluted into 50 µl of dd-H2O. Modification was completed by NaOH treatment for 10 min at 37°C and by ethanol precipitation. DNA was re-suspended in 20 µl of dd-H2O and used immediately or stored at −20°C.
PCR amplification and DNA sequencing
Bisulfate-treated DNA (2 µl) was used as a template in strand-specific PCR amplification for the region of interest. Primer pairs were purchased from Operon Biotechnologies (Huntsville, AL). The PCR fragments were amplified under standard conditions with a total 35 cycles at different annealing temperatures followed by a final extension at 72°C for 10 min. Normal genomic DNA without demethylation treatment was used as a control. PCR product was purified and confirmed by DNA sequencing.
Quantitative methylation analysis with Ms-SNuPE
Ms-SNuPE reactions were performed following the method described by Gonzalgo, et al. [43]. Briefly, purified PCR product (10 ng) was used in each reaction containing: 1× PCR buffer, 1 µM of each Ms-SNuPE primer, 1 µCi of 32P-labeled dCTP or dTTP and 1 U of Taq polymerase. Specific primers for Ms-SNuPE were: (site 1) 5′-TTTTTTTTTTTATTGTTTTTATT-3′, (site 2) 5′-TTTTATTT(C)GGTTAAGGAG TG-3′, and (site 3) 5′-TTTGGTTAA GGAGTGC(T)AGG. All samples were treated at 95°C for 1 min, 50–54°C for 2 min, 72°C for 1 min. Reaction products were loaded onto 15% denaturing polyacrylamide gels with 7 M urea. Results were visualized by exposure of X-ray films.
Statistical Analysis
For Western and Chip analysis, signals in the films and agarose gels were scanned digitally and optical densities (OD) were quantified by using the Image Calculator software. The relative OD in Western analysis was calculated by normalizing the signals from target proteins against intensities of loading controls. In Chip assays, the relative density was calculated by normalizing PCR signals from immunopricipitated target chromatins against signals from the whole cell chromatin DNA. Data were plotted as Mean±SE. Significant differences between the control and treated groups were determined by One-way ANOVA and Student's t test at P<0.05.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from National Institutes of Health (NIH) (EY015282) to L.L. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fukuda H, Takahashi J. Embryonic stem cells as a cell source for treating Parkinson's disease. Expert Opin Biol Ther. 2005;5:1273–1280. doi: 10.1517/14712598.5.10.1273. [DOI] [PubMed] [Google Scholar]
- 2.Mitjavila-Garcia MT, Simonin C, Peschanski M. Embryonic stem cells: meeting the needs for cell therapy. Adv Drug Deliv Rev. 2005;57:1935–1943. doi: 10.1016/j.addr.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Sugaya K, Alvarez A, Marutle A, Kwak YD, Choumkina E. Stem cell strategies for Alzheimer's disease therapy. Panminerva Med. 2006;48:87–96. [PubMed] [Google Scholar]
- 4.Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 5.Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 6.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 7.Gotz M, Barde YA. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Malatesta P, Appolloni I, Calzolari F. Radial glia and neural stem cells. Cell Tissue Res. 2008;331:165–178. doi: 10.1007/s00441-007-0481-8. [DOI] [PubMed] [Google Scholar]
- 9.Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 10.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plaza S, Dozier C, Saule S. Quail Pax-6 (Pax-QNR) encodes a transcription factor able to bind and trans-activate its own promoter. Cell Growth Differ. 1993;4:1041–1050. [PubMed] [Google Scholar]
- 12.Plaza S, Dozier C, Turque N, Saule S. Quail Pax-6 (Pax-QNR) mRNAs are expressed from two promoters used differentially during retina development and neuronal differentiation. Mol Cell Biol. 1995;15:3344–3353. doi: 10.1128/mcb.15.6.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, et al. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- 14.Canto-Soler MV, Huang H, Romero MS, Adler R. Transcription factors CTCF and Pax6 are segregated to different cell types during retinal cell differentiation. Dev Dyn. 2008;237:758–767. doi: 10.1002/dvdy.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Lu Z, Lu L. Pax6 regulation in retinal cells by CCCTC binding factor. Invest Ophthalmol Vis Sci. 2006;47:5218–5226. doi: 10.1167/iovs.06-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Lu Z, Lu L. Regulation of eye development by transcription control of CCCTC binding factor (CTCF). J Biol Chem. 2004;279:27575–27583. doi: 10.1074/jbc.M313942200. [DOI] [PubMed] [Google Scholar]
- 17.Wu D, Li T, Lu Z, Dai W, Xu M, et al. Effect of CTCF-binding motif on regulation of PAX6 transcription. Invest Ophthalmol Vis Sci. 2006;47:2422–2429. doi: 10.1167/iovs.05-0536. [DOI] [PubMed] [Google Scholar]
- 18.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 21.Renaud S, Loukinov D, Abdullaev Z, Guilleret I, Bosman FT, et al. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–1256. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 25.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 26.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Lu L. Epidermal growth factor-induced proliferation requires down-regulation of Pax6 in corneal epithelial cells. J Biol Chem. 2005;280:12988–12995. doi: 10.1074/jbc.M412458200. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Lu L. Functional role of CCCTC binding factor (CTCF) in stress-induced apoptosis. Exp Cell Res. 2007;313:3057–3065. doi: 10.1016/j.yexcr.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu L, Wang L, Li T, Wang J. NF-kappaB subtypes regulate CCCTC binding factor affecting corneal epithelial cell fate. J Biol Chem. 285:9373–9382. doi: 10.1074/jbc.M109.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 33.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 34.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahhoseini M, Taei A, Mehrjardi NZ, Salekdeh GH, Baharvand H. Epigenetic analysis of human embryonic carcinoma cells during retinoic acid-induced neural differentiation. Biochem Cell Biol. 2010;88:527–538. doi: 10.1139/o09-181. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15:2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- 39.Yeo S, Jeong S, Kim J, Han JS, Han YM, et al. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2007;359:536–542. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 40.Alexanian AR. Epigenetic modifiers promote efficient generation of neural-like cells from bone marrow-derived mesenchymal cells grown in neural environment. J Cell Biochem. 2007;100:362–371. doi: 10.1002/jcb.21029. [DOI] [PubMed] [Google Scholar]
- 41.Warnecke PM, Mann JR, Frommer M, Clark SJ. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics. 1998;51:182–190. doi: 10.1006/geno.1998.5371. [DOI] [PubMed] [Google Scholar]
- 42.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalgo ML, Jones PA. Quantitative methylation analysis using methylation-sensitive single- nucleotide primer extension (Ms-SNuPE). Methods. 2002;27:128–133. doi: 10.1016/s1046-2023(02)00064-6. [DOI] [PubMed] [Google Scholar]