Abstract
Background
Psychiatric illness and anxiety disorders have strong neurodevelopmental components. Environmental insults such as prenatal exposure to stress and genetic differences in stress responses may affect brain development.
Methods
A rat model of random variable prenatal stress was used to study the expression and processing of hippocampal brain-derived neurotrophic factor (BDNF) in the offspring of the stressed rat dams. To account for unknown genetic influences that may play a role in the outcome of this prenatal stress paradigm, three different rat strains with known differences in stress responsivity were studied: Fischer, Sprague-Dawley, and Lewis rats (n=132).
Results
Multiple disparities in mRNA expression levels of BDNF, and transcripts related to its processing and signaling were found in the three strains. Of the numerous splice variants transcribed from the BDNF gene, the transcript containing BDNF exon VI was most aberrant in the prenatally stressed animals. Protein levels of both uncleaved proBDNF and mature BDNF were also altered, as was intra-cellular signaling by phosphorylation of the TrkB receptor and Erk 1/2. Changes were not only dependent on prenatal stress, but were also strain dependent, demonstrating the importance of genetic background.
Conclusions
BDNF signaling provides both positive neurotrophic support for neurons and negative apoptotic effects, both of which may contribute to behavioral or neurochemical outcomes after prenatal exposure to stress. Differential processing of BDNF after prenatal stress in the three rat strains has implications for human subjects where genetic differences may protect or exacerbate the effects of an environmental stressor during fetal development.
Keywords: prenatal stress, schizophrenia, HPA axis, brain-derived neurotrophic factor (BDNF), rat, development
Introduction
Neuropsychiatric disorders, including depression, autism, bipolar disorder and schizophrenia have complex etiologies and are transmitted in a non-Mendelian mode (Risch, 2000;Kennedy et al., 2003). Genetic predisposition alone is not sufficient for the diseases to manifest, implicating environmental factors, such as stress (Bale et al., 2010;Markham and Koenig, 2011). Various animal models have been developed to mimic endophenotypes found in psychiatric disorders (Alleva and Francia, 2009;Dawe et al., 2009;Braff et al., 2007;Freedman et al., 2000) and environmental factors have become a focus of these models (Van Os and Kapur, 2009;Gray and Hannan, 2007;Cantor-Graae, 2007;Koenig et al., 2002). This is particularly true when environmental perturbations occur neurodevelopmentally during the prenatal period.
The neurotrophin gene, BDNF (brain-derived neurotrophic factor), is associated with several neuropsychiatric disorders, and is an important gene of interest in stress research (Correia et al., 2010;Angelucci et al., 2005;Buckley et al., 2007a). For example, schizophrenia patients show decreased BDNF in the brain (Durany et al., 2001;Weickert et al., 2003), reduced BDNF and TrkB positive neurons (Iritani et al., 2003;Weickert et al., 2005), and lower BDNF in serum (Toyooka et al., 2002). Moreover, lower BDNF levels are seen at the onset of psychosis, which may contribute to the manifestation of the disease (Buckley et al., 2007b).
The BDNF gene was originally described as having four noncoding exons (I–IV) and one coding exon (V) (Timmusk et al., 1993), but is now known to contain at least 9 different exons that can generate numerous transcripts through alternative splicing (Aid et al., 2007;Liu et al., 2006). Each BDNF transcript is formed from one of eight 5’ noncoding exons (III and IV are now IV and VI, respectively) and one 3’ protein coding exon (IX, previously V) (Aid et al., 2007). The unique 5’ exons are each transcribed from different promoters, combining with the common 3’ exon to generate the various isoforms. Each transcript is translated into the identical 32 kD precursor protein, proBDNF, which is transported in vesicles bidirectionally to both axons and dendrites. BDNF is primarily released postsynaptically from the dendrites (Matsuda et al., 2009). ProBDNF is cleaved into its mature form, mBDNF, by various proteinases, either intracellularly or extracellularly (Lessmann and Brigadski, 2009).
Unprocessed proBDNF and mBDNF have very different effects on downstream signaling (Lu et al., 2005;Schweigreiter, 2006;Arevalo and Wu, 2006;Nykjaer et al., 2005). In the pro-form, BDNF has a high affinity for the p75 receptor. Binding to p75 has an overall negative effect, inducing cellular responses such as apoptosis, long-term depression, and synaptic retraction. If proBDNF is processed into its mature form, mBDNF activates the TrkB receptor, yielding positive cellular effects. Activated TrkB receptors dimerize, resulting in autophosphorylation and further downstream signaling through PKC, PI3K, AKT, ERK, and CREB. These signals promote survival, synaptic plasticity, and differentiation, and are sufficient for late-phase long-term potentiation (LTP) while inhibiting long-term depression (LTD).
It has been previously shown that prenatal stress in the last week of fetal gestation can produce multiple endophenotypic changes related to schizophrenia in the Sprague-Dawley rat strain (Kinnunen et al., 2003;Koenig et al., 2005;Lee et al., 2007;Taylor et al., 2010). Since little is known about the underlying molecular basis of these phenotypic changes, we evaluated the expression of the BDNF gene in three rat strains to address genetic differences. Fischer rats have a hyperactive HPA-axis, Lewis rats are known to be hypo-responsive to stress, and Sprague-Dawley rats have an intermediate corticosterone response (Dhabhar et al., 1993;Moncek et al., 2001;Kosten and Ambrosio, 2002). Here we show how the BDNF pathway is affected in a rodent model of prenatal stress. The severity of changes in the BDNF pathway responses in the offspring was dependent on the rat strain.
Experimental Procedures
Animals
Timed-pregnant Fischer, Sprague-Dawley, and Lewis rats were purchased from Charles River laboratories, arriving in our facilities on gestational day 2 (G2). Twelve dams were used for each strain (36 timed-pregnant rats in all). All dams were housed individually in static cages, allowed ad libitum access to food and water, and maintained on a 12-h light/dark cycle. In addition, the animals were housed in a private room removed from any other experiment to prevent stress unrelated to the testing. At parturition, dams and their pups were left undisturbed in large static cages until weaning on postnatal day (P) 23–25. At this point, only male offspring were retained, and each was housed with a single littermate until an acute stress and sacrifice on P56. All animal procedures were conducted in accordance with NIH policies and were approved by the University of Colorado Denver and University of Maryland animal care committees.
Prenatal stress procedure
Beginning on G14, all prenatal stress (PS) pregnant dams were exposed to a one week random variable stress paradigm, until G21. Control dams were left in the housing room. All stressors were mild in nature and consisted of both psychological and physiological stress. These stressors consisted of: (1) restraint in a well-ventilated cylindrical plexiglass restrainer (Harvard Bioscience, Boston, MA) for 1 hour, (2) exposure to a cold environment (4°C) for 6 hours, (3) overnight food deprivation for 12 hours, (4) 15 minutes of non-escapable swim in a plexiglass cylinder filled with room temperature water, (5) 24 hour light exposure, and (6) social stress induced by overcrowded housing conditions for 12 hours. Each PS dam received two to three of these stressors per day, which were varied throughout the week to prevent habituation (Kinnunen et al., 2003). After delivery, dams and pups remained undisturbed until weaning.
Postnatal acute stress
On P56, all PS and control male offspring were sacrificed for blood and tissue collection by rapid decapitation without anesthesia. This was done at one of four time points: 1) at baseline without acute stress (baseline), 2) immediately after a 30 min restraint stress (30+0 min), 3) restraint stress + 120 min recovery (30+120 min), or 4) restraint stress plus a 24 hr recovery (30 min+24 hr).
Corticosterone measurement
Trunk blood was collected after decapitation and serum isolated by centrifugation at 3000g before being stored at –80°C. Corticosterone (CORT) was measured using a radioimmunoassay kit (MP Biomedicals, Orangeburg, NY) according to manufacturer’s instructions. The sensitivity of the assay was 3 ng/ml and the intra- and interassay coefficients of variation were less than 10%.
Tissue Preparation
Following decapitation, brains were quickly removed and hippocampi dissected. One hippocampal hemisphere was immediately placed in 2 mls of RNAlater solution (Ambion, Carlsbad, CA) to stabilize RNA, while the other was snap frozen in liquid nitrogen for protein analysis. Tissue was placed in tubes containing homogenization beads, and either TRIzol® lysis buffer (Invitrogen, Carlsbad, CA) for RNA, or radioimmunoprecipitation assay buffer (RIPA) + 3% SDS lysis buffer (Boston Bioproducts, Worcester, MA) with protease inhibitors (Roche diagnostics, Indianapolis, IN) for protein isolation. Homogenization was performed using the Mini Beadbeater-8® (Biospec, Bartlesville, OK).
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
After tissue lysis and RNA precipitation, RNA was purified using an RNeasy® mini kit (Qiagen, Valencia, CA). RNA quality was verified using an Agilent bioanalyzer 2100 (Agilent, Santa Clara, CA). First strand complementary DNA (cDNA) was synthesized by reverse transcription using random hexamers and Superscript III reverse transcriptase (Invitrogen). qRT-PCR was performed on a Bio-rad iCycler IQ® (Bio-rad,) using SYBRgreen supermix (Bio-rad, Hercules, CA). Primers used for the study are shown in Table 1. Each sample was run in triplicate and followed by a heat dissociation step to identify nonspecific products. Triplicates were averaged and then normalized to the housekeeping gene Polr2a (Radonic et al., 2004) using mean normalized expression (MNE) (Simon, 2003) and actual efficiencies of each run.
Table 1.
Primer sequences
| BDNF coding (f) | GCCCAACGAAGAAAACCA |
| BDNF coding (r) | CCAGCAGAAAGAGCAGAGGA |
| BDNF exon I (f) | GTGTGACCTGAGCAGTGGGCAAAGGA |
| BDNF exon II (f) | GGAAGTGGAAGAAACCGTCTAGAGCA |
| BDNF exon III (f) | CCTTTCTATTTTCCCTCCCCGAGAGT |
| BDNF exon IV (f) | CTCTGCCTAGATCAAATGGAGCTTC |
| BDNF exon V (f) | CTCTGTGTAGTTTCATTGTGTGTTC |
| BDNF exon VI (f) | GCTGGCTGTCGCACGGTCCCCATT |
| BDNF exon VII (f) | CCTGAAAGGGTCTGCGGAACTCCA |
| BDNF exon VIII (f) | GTGTGTGTCTCTGCGCCTCAGTGGA |
| BDNF exon IXa (f) | CCAGAGCTGCTAAAGTGGGAGGAAG |
| BDNF all exons (r) | GAAGTGTACAAGTCCGCGTCCTTA |
| PAI-1 (f) | GTTCAACTACACTGAGTTCACC |
| PAI-1 (r) | GAAGATGTCAGTCATGCCCA |
| plasminogen (f) | TCATCTGCAGGTCATTCCA |
| plasminogen (r) | TCATCATTGTCTGGGTTCCT |
(f) = forward, (r) = reverse
Western blotting
Subsequent to tissue homogenization, protein concentration was assayed using a bicinchoninic acid (BCA) test (Thermo-Fisher Scientific, Rockford, IL). Samples containing 10–20 µgs of total protein were resolved on 4–12% criterion XT bis-tris gels (Bio-rad) under reducing and denaturing conditions, followed by transfer to 45µm nitrocellulose membranes in Tris-glycine transfer buffer containing 20% methanol. Membranes were blocked using 1x TBS, 0.05% Tween-20 (TBS-T) with 5% w/v nonfat dry milk. Primary antibodies were incubated on the membrane overnight at 4°C with agitation in TBS-T and 5% BSA. BDNF antibody AB5555P (Chemicon, Billerica, MA), proBDNF antibody AB5613P (Chemicon), and p75 antibody 07–476 (Upstate-Chemicon, Billerica, MA) were used at a dilution of 1µg/µl. After washing, membranes were incubated with Immun-Star® HRP linked goat anti-rabbit secondary antibody (Bio-rad) in TBS-T before detection with electrochemiluminescence (Bio-rad). Blots were scanned at high resolution and analyzed using Un-scan-it® software (Silk Scientific, Orem, UT). After subtracting local background, band intensity was normalized to beta-tubulin AB6046 (Abcam, Cambridge, MA) as a loading control to assess protein levels.
Phosphorylation Assay
Quantification of phosphorylated TrkB and Erk proteins was determined by Pathscan sandwich ELISA kits (Cell Signaling, Danvers, MA) according to manufacturer’s instructions. Phospho-TrkB ELISA measures phosphorylation at tyrosine 516, while Phospho- Erk1/2 ELISA quantifies threonine 202/tyrosine 204. Samples were added to microplates at a concentration of 0.4 µg/µl. All results were determined spectrophotometrically at 450 nm using a Bio-rad iMark® microplate reader.
Statistical Analysis
A total of 132 male offspring were used in the analyses. Of the 23 independent groups studied (including strain, condition, and time points), each group had an average sample size of 6. Each group measure used only one animal from the same mother. For CORT data and qRT-PCR analyses, all measures are expressed as means of each group ±SEM and compared using a mixed model analysis of variance (ANOVA) with fixed effects of strain, condition, and time group, plus a random effect of the offspring’s dam. All data were calculated using SAS foundation software V9 (SAS, Cary, NC), and differences among experimental groups were considered significant when P<0.05. For western blots, a Student t-test (2 groups) or ANOVA with Tukey-Kramer multiple comparisons test (three or more groups) was used to determine significance at P<0.05. Values are plotted with the control baseline Fischer band normalized to 100%.
Results
Corticosterone response to acute stress differs by strain
Serum CORT was measured on all animals after sacrifice. Baseline levels compared between prenatal stress or rat strain conditions showed no significant differences (Fig. 1). When exposed to a restraint stress for 30 minutes, Lewis control rats and PS animals responded with lower levels of CORT than their Fischer counterparts (P<0.05, t=2.31,df=79). In addition, Lewis controls had significantly higher CORT compared to Lewis PS rats (P<0.05,t=2.39,df=79). No significant differences were seen in the 120 minute or 24 hour groups.
Figure 1.
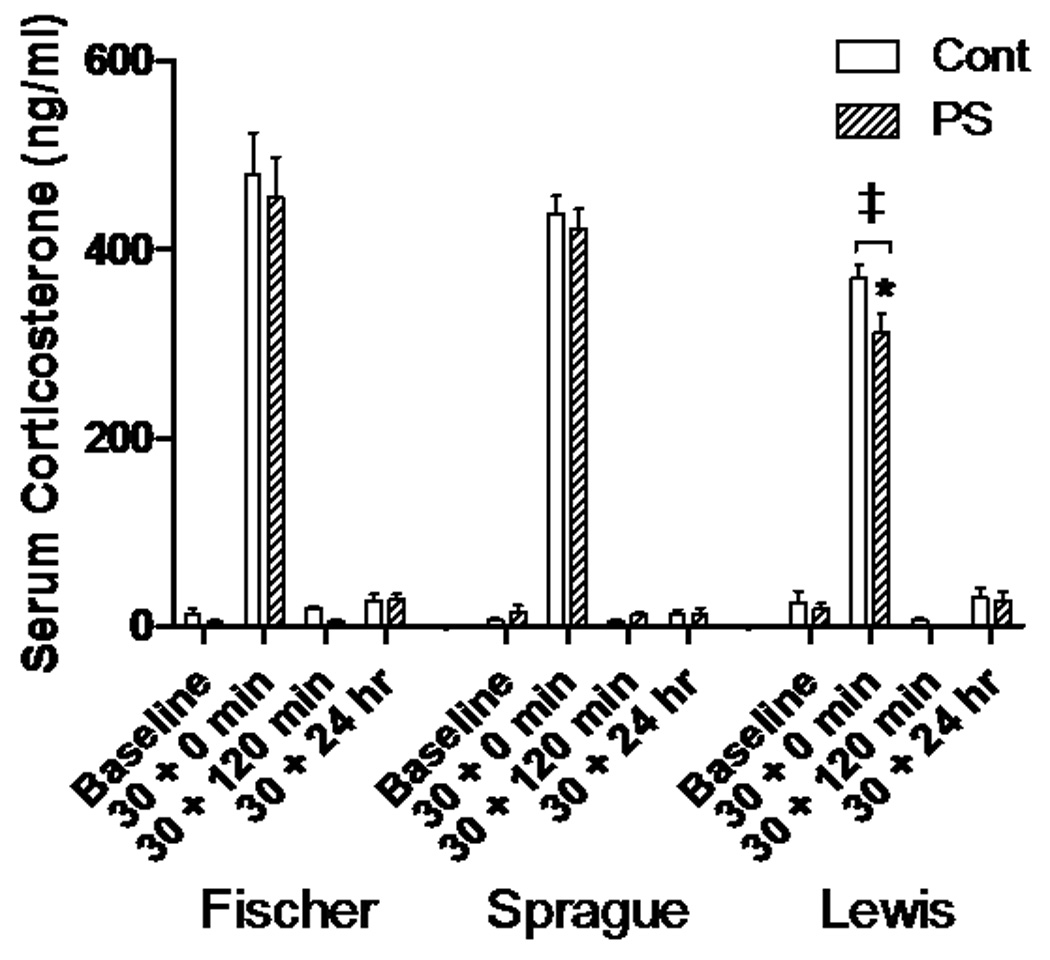
Serum corticosterone responses. After a 30 minute restraint stress, corticosterone levels significantly rise from baseline for each group, but fall back to baseline levels within 120 minutes. Lewis Ctrl and PS animals increase less than the Fischer and Sprague-Dawley groups (‡, p<0.001). Lewis PS concentrations at the 30 + 0 min time point were also significantly less than Lewis Ctrl (*, p<0.001).
Hippocampal BDNF expression is altered in prenatally stressed animals
Sprague-Dawley (P<0.01,t=2.65,df=79) and Lewis PS rats (P<0.01,t=3.43,df=79) had significantly elevated baseline levels of total BDNF mRNA in the hippocampus after PS, compared to their strain controls; total hippocampal BDNF was unchanged by PS at baseline in the Fischer rats (Fig. 2A).
Figure 2.
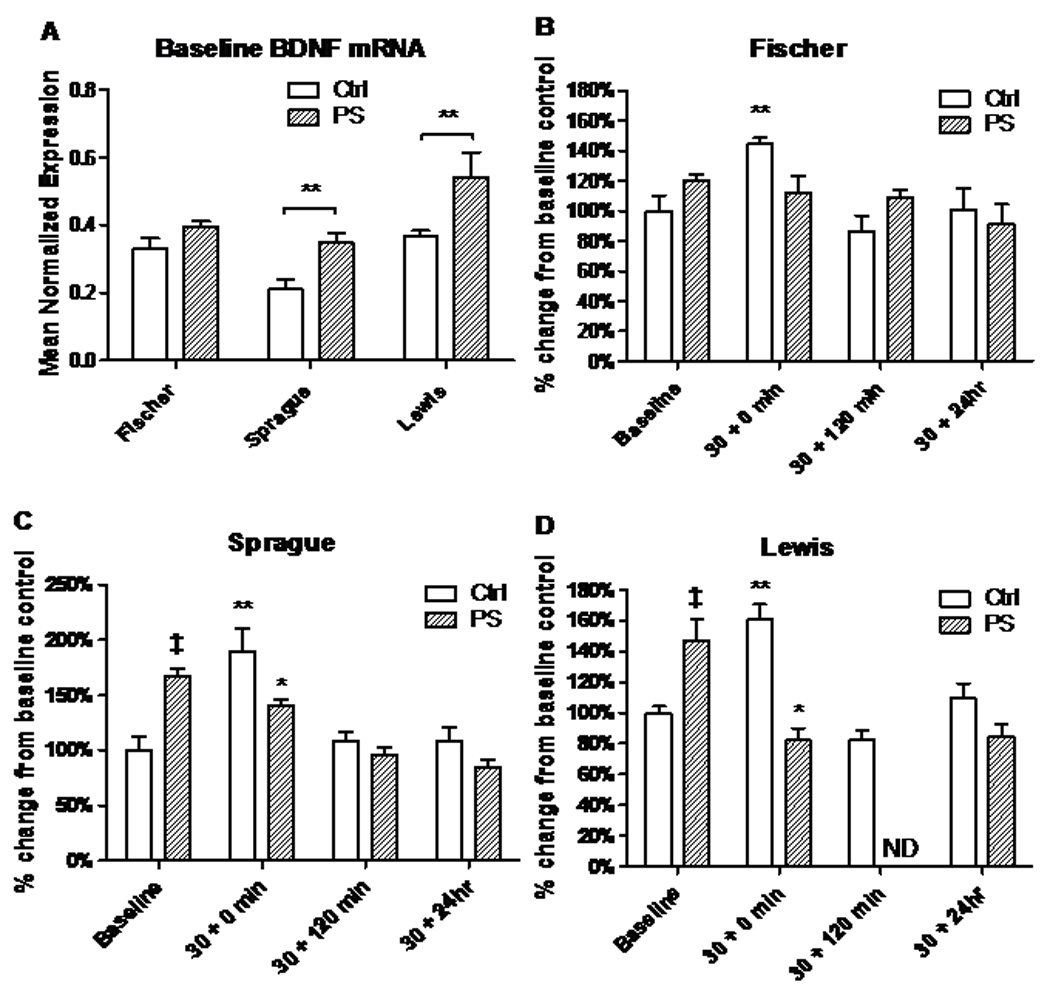
BDNF mRNA response to acute stress is changed by PS in a strain-specific manner in the hippocampus. A. Baseline BDNF transcript levels in three rat strains. In Sprague-Dawley and Lewis rats exposed to PS, BDNF mRNA is up-regulated compared to their non-PS exposed controls. B. Control levels of BDNF mRNA rise significantly in Fischer (**, P<0.001) after a 30 minute restraint stress (30 + 0), but fall back to baseline within a 120 minute recovery (30 + 120). Prenatally stressed (PS) Fischer show no rise after acute stress. C–D. Sprague-Dawley and Lewis control rats also show significant rise after acute stress that falls with recovery. Unlike the Fischer strain, PS significantly increased BDNF transcription in Sprague-Dawley and Lewis rats at baseline (‡, P<0.001). Following 30 minutes of restraint, BDNF mRNA in the hippocampus of the Sprague-Dawley and Lewis control rats increased significantly, while rats of these strains exposed to PS exhibited a significant decline in expression after acute stress when compared to their respective baselines, and lower levels when compared to controls of the same time point (*, P<0.001). ND= Not Determined.
Regardless of strain, total hippocampal BDNF mRNA is up-regulated after a 30 minute restraint stress (30+0 min) in Fischer (P<0.01,t=2.76,df=79), Sprague-Dawley (P<0.01,t=3.39,df=79), and Lewis (P<0.01,t=4.40,df=79) control animals (Fig.2, B–D). BDNF levels do not statistically differ from baseline after 120 minutes of recovery (30+120 min) and remain there 24 hours later (30+24hr). In the control animals, the BDNF mRNA response is similar to the CORT response after an acute stress in that it is up-regulated, but quickly falls back to baseline after 120 minutes of recovery.
In PS rats, a different pattern emerged. In the Fischer strain, hippocampal BDNF expression was unaffected after acute stress, and remains near baseline at all time points (Fig. 2B). In the Sprague-Dawley (P<0.01,t=2.65,df=79) and Lewis strains (P<0.01,t=3.43,df=79), BDNF transcripts were significantly higher at baseline in the PS animals when compared to controls (Fig. 2, C&D). Further, hippocampal mRNA levels in PS animals dropped significantly after the 30 min acute stress instead of increasing, as seen in the controls (Sprague-Dawley: P<0.05,t=2.05,df=79; Lewis: P<0.0001, t=5.47, df=79). In the Sprague-Dawley rats, BDNF continues to drop over a 24 hour period, while Lewis appears to down-regulate immediately, with these transcripts remaining low 24 hours later.
Hippocampal BDNF transcripts containing exon VI increase after stress
To determine which of the exon isoforms of BDNF were responsible for the change in total BDNF expression, qRT-PCR was performed utilizing primers specific for each upstream exon. Of the alternative transcripts analyzed, exons V, VII, and VIII were not significantly expressed in the hippocampus, transcripts containing exon II were poorly expressed, and exons I, III, and IXa were expressed in the hippocampus without significant differences between control and stressed animals. However, transcripts containing exons IV and VI were found to be changed by acute or prenatal stress (Fig. 3). Exon IV transcripts are significantly increased after an acute stress in all three strains (P<0.0001, Fischer t=13.35, Sprague-Dawley t=11.34, Lewis t=7.18, df=79), although no differences were found between the control and PS groups within strains (Fig. 3A). However, for exon VI transcripts, all three strains show a significant rise after an acute stress, but only in the control groups (P<0.0001, Fischer t=20.75, Sprague-Dawley t=5.21, Lewis t=4.84, df=79) (Fig. 3B). Interestingly, the increase is less pronounced in the Sprague-Dawley and Lewis strains than in the Fischer rats. At baseline, PS produces an increase in transcripts containing exon VI in Sprague-Dawley (P<0.0001,t=13.15,df=79), and Lewis rats (P<0.0001,t=10.25,df=79), but not in Fischer. PS also resulted in a change in response to acute stress, where exon VI hippocampal transcripts were less abundant in the PS animals than in the within-strain controls with Fischer (P<0.0001,t=19.210,df=79), and Lewis rats (P<0.0001, t=6.74,df=79).
Figure 3.
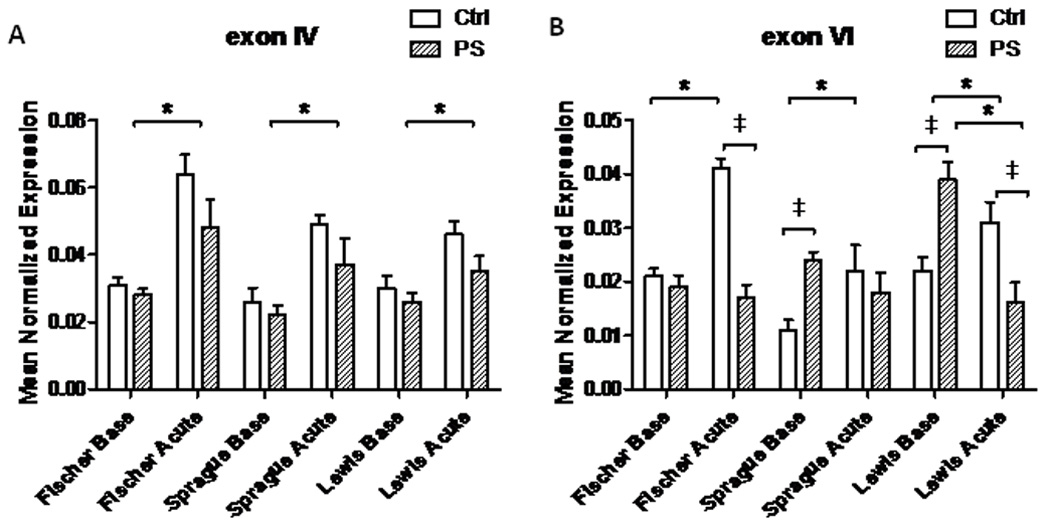
BDNF exon-specific transcript expression at baseline and after an acute stress in the hippocampus. A. Rats of all three strains, regardless of PS exposure, show an increase in exon IV expression after restraint stress (*, P<0.001). B. Exon VI expression is also significantly increased after restraint stress in each strain, but only in controls. Exon VI mRNA is elevated in Sprague-Dawley and Lewis PS animals at baseline vs. their respective controls. The responses of the PS animals to an acute stressor were quite different across the strains. Fischer and Sprague-Dawley PS rats did not increase exon VI containing transcripts after the 30-minute stress, and the PS Lewis strain transcripts were markedly decreased (‡, P<0.001).
Hippocampal proBDNF and mature BDNF protein are lower in Sprague-Dawley and Lewis strains than in Fischer rats
Protein concentrations for proBDNF and mature BDNF (mBDNF) were assayed in the hippocampus by western blot. Data are reported as a percent of the control value after normalization to a β-tubulin loading control (Baseline, Figure 4 A, B and C; after an acute stressor, Figure 4 D, E, and F). Each membrane consisted of 6–7 samples per group loaded in duplicate. At baseline, the predominant form of BDNF in the hippocampus of all three strains was the precursor form, proBDNF (Fig. 4A, 32 kD band). In rats exposed to the random variable PS paradigm, both proBDNF and mBDNF were diminished. The differences were particularly marked in the Sprague-Dawley and Lewis PS rats where significant decreases were found in proBDNF levels (Fig. 4B, P<0.0001, Sprague t=9.70, Lewis t=9.62, df=22). The levels of mBDNF were also lower in the PS Sprague-Dawley and Lewis animals, but the differences did not reach significance. When the animals were given an acute 30 minute restraint stress, proBDNF was cleaved to form mBDNF in the hippocampus of controls of all strains (14kd band, Fig. 4D). However, in the PS animals, strain differences in response were found. Similar to the baseline timepoint, PS Sprague-Dawley (P<0.0001,t=5.60,df=22) and Lewis rats (P<0.0001,t=5.71,df=22) show significantly lower levels of proBDNF (Figure 4E). mBDNF was also significantly different in the PS Sprague-Dawley (P<0.0001,t=5.17,df=22) and Lewis rats (P<0.0001,t=7.00,df=22) after an acute stress, with Lewis rats having the lowest levels (Fig. 4F).
Figure 4.
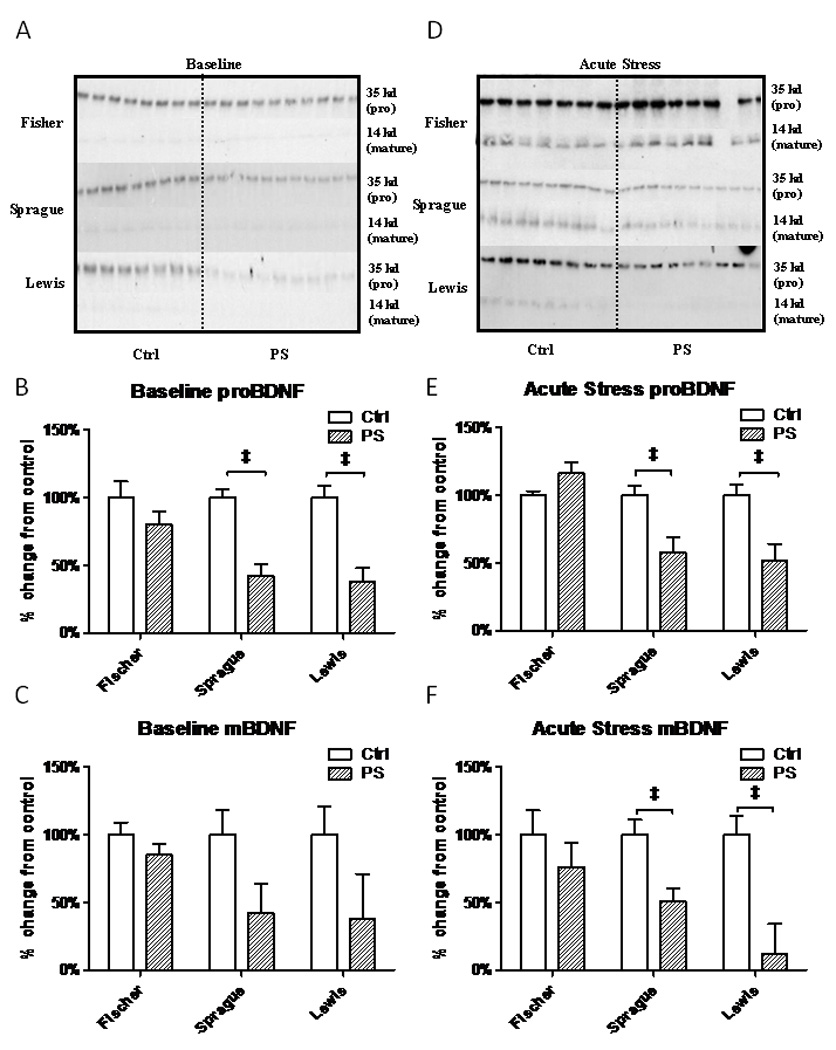
Western blot of proBDNF (32 kD) and mature BDNF (14 kD) proteins in the hippocampus. All values are compared to their strain controls. A. Representative BDNF western blot of all 3 strains at baseline. B. Baseline proBDNF levels (32 kD) are decreased in PS Sprague-Dawley and Lewis rats with respect to their controls. C. Baseline mBDNF levels (14 kD), while lower were not statistically different. D. Representative BDNF western blot in the 3 strains immediately after a 30 minute restraint stress. E. Acute Stress proBDNF levels are decreased in both PS Sprague-Dawley and Lewis rats. F. Acute stress mBDNF protein expression is also decreased in PS Sprague-Dawley and Lewis rats. (‡, P<0.001)
A western blot representing two subjects from each group was done to compare differences between strains on the same gel (Fig. 5A). Relative amounts in each group were compared to the Fischer rat baseline group after normalization to the γ-tubulin loading control. Significant intra-strain differences between control and PS groups were still seen with proBDNF in Sprague-Dawley rats at baseline (P<0.0001,t=7.84,df=22), Lewis rats at baseline (P<0.0001,t=19.50,df=22), and Lewis rats exposed to acute stress (P<0.0001,t=75.92,df=22) (Fig. 5B); and in mBDNF in Sprague-Dawley baseline (P<0.0001,t=10.95,df=22) acutely stressed (P<0.001,t=3.97,df=22) and Lewis baseline (P<0.0001,t=20.77,df=22) and acutely stressed (P<0.0001,t=17.70,df=22) (Fig. 5C). Fischer animals also have less hippocampal proBDNF protein after an acute stress compared to baseline (Fig. 5B). Inter-strain comparisons of proBDNF show Fischer controls and PS animals have more protein at baseline than all other groups, and after acute stress both Fischer control and PS rats have significantly more than Sprague-Dawley and Lewis PS groups (Fig. 5B). In addition, Fischer mBDNF levels for control and PS at baseline and after stress are significantly elevated when compared to Sprague-Dawley and Lewis PS animals (Fig. 5C).
Figure 5.
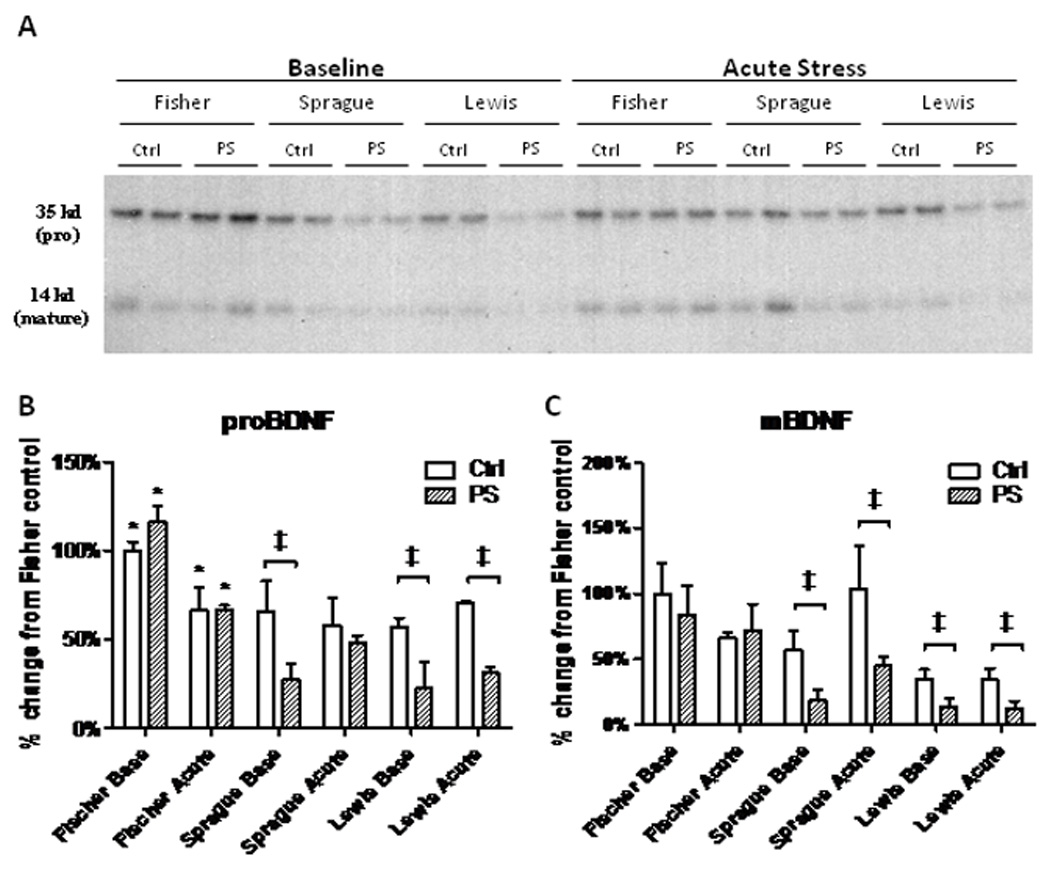
A. Western blot of proBDNF (32kD) and mature BDNF (14 kD) proteins in the hippocampus comparing all strains. B. Quantification of protein levels of proBDNF using the Fischer baseline group as the control. Intra-strain comparisons show significant reductions in Sprague and Lewis baseline groups, and further significant suppression of proBDNF expression in the Lewis rats following restraint stress (‡, P<0.01). More protein is seen in Fischer controls and PS animals at baseline than all other groups, and both Fischer control and PS rats have significantly more than Sprague-Dawley and Lewis PS groups after acute stress (*, P<0.01). C. Protein quantification of mBDNF levels. Significant intra-strain expression decreases are seen again in Sprague-Dawley and Lewis rats at baseline and after acute stress (‡, P<0.01).
ProBDNF processing in the hippocampus is affected by prenatal stress in a strain-dependent manner
In neurons, plasmin is the major proteinase responsible for the conversion of proBDNF to mBDNF. However, plasmin itself must first be activated by conversion of the zymogen, plasminogen, into plasmin by another proteinase, tissue-plasminogen activator (tPA) (Lee et al., 2001). In addition, tPA can be inhibited by various proteins, including the major inhibitor in brain, PAI-1 (plasminogen activator inhibitor 1) (Salles and Strickland, 2002).
These processing enzymes were altered in the hippocampus of Lewis and Sprague-Dawley rats after PS. Plasminogen transcript levels in controls did not change in response to acute stress in any of the three strains (Fig. 6A). However, at both baseline and after a 30 minute stress, we found significantly decreased levels of plasminogen mRNA in the Sprague-Dawley baseline (P<0.0001,t=7.76,df=10), acute (P<0.01,t=4.15,df=10), and Lewis baseline (P<0.0001,t=5.45,df=10) and acute (P<0.0001,t=5.92,df=10) animals when compared to respective controls (Fig. 6A). Consistent with this finding, the tPA inhibitor PAI-1, is significantly over expressed in the PS animals in the Sprague-Dawley (P<0.0001,t=11.69,df=10) and Lewis strains (P<0.0001,t=20.95,df=10) at baseline (Fig. 6B).
Figure 6.
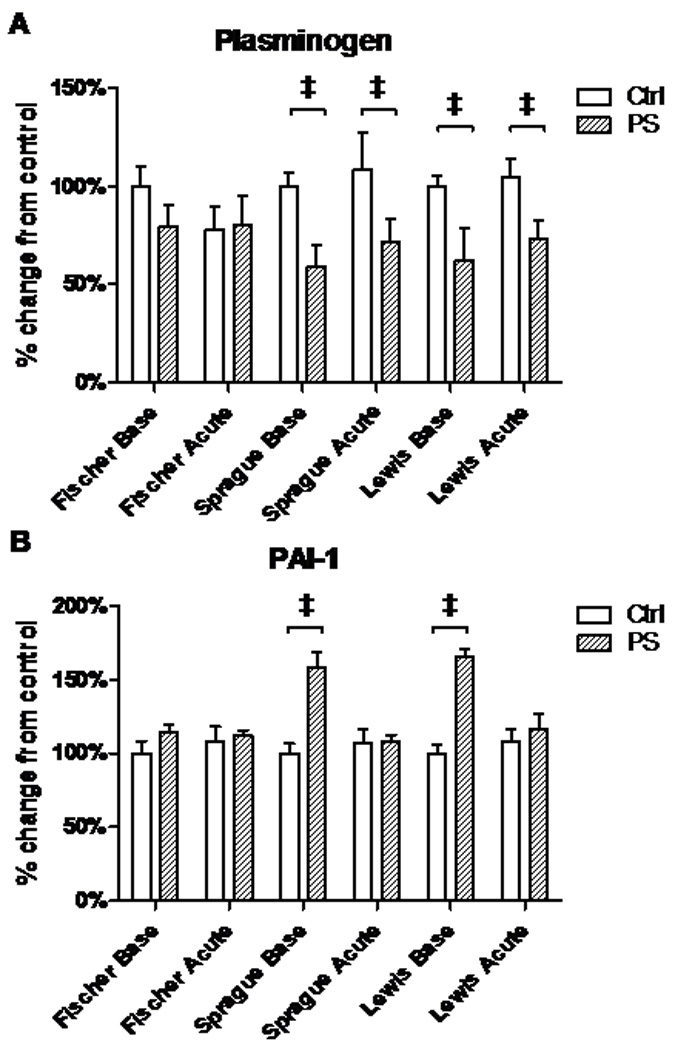
Plasminogen and PAI-1 mRNA expression in the hippocampus. Acute stress does not induce changes in plasminogen or PAI-1 mRNA expression in any of the rat strains under baseline conditions. While no changes were found in Fischer rats for either transcript, PS Sprague-Dawley and Lewis rats have lower levels of plasminogen at baseline and immediately after stress (A), and over-expression of PAI-1 at baseline (B).
Downstream signaling of BDNF through TrkB is decreased in the hippocampus of Sprague-Dawley and Lewis rats
A decrease in mBDNF could result in a decrease in TrkB signaling, which requires TrkB phosphorylation. Total TrkB versus phosphorylated TrkB (pTrkB) at tyrosine 512 was measured by ELISA. In the hippocampus of all control groups at baseline, there was little to no pTrkB and no difference between groups. After the acute restraint stress, phosphorylated TrkB was found in the hippocampus of all groups (Fig. 7A). The PS Lewis rats had significantly less pTrkB compared to controls after acute stress (P<0.0001,t=4.86,df=10). Levels of pTrkB were decreased slightly in Sprague-Dawley rats, but the difference was not significant.
Figure 7.
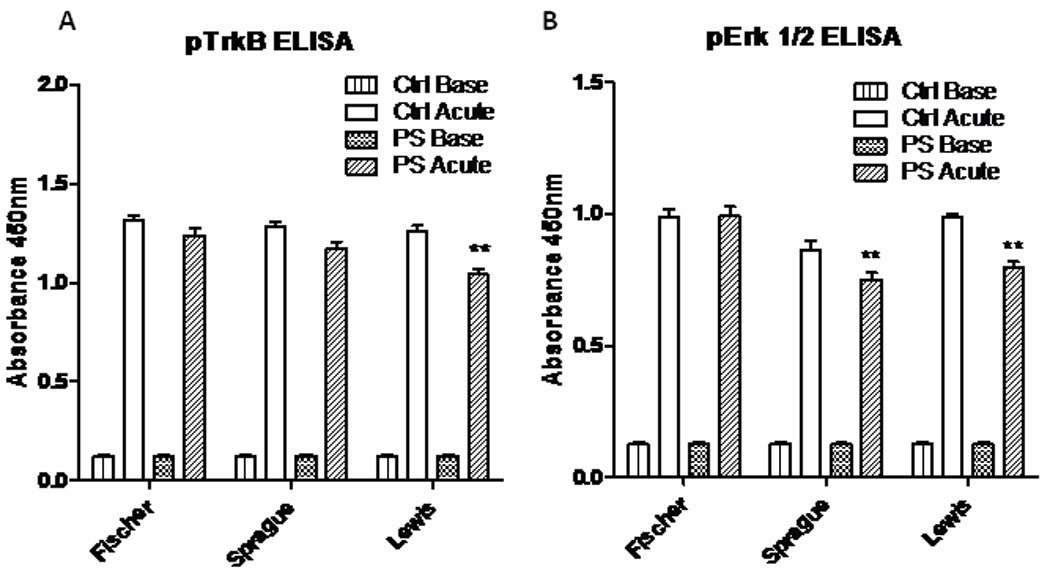
pTrkB and pErk 1/2 phosphorylation at baseline and after a 30-minute restraint stress. A. Baseline phosphorylation of the TrkB receptor was very low but 30 minutes of restraint stress elevated TrkB phosphorylation levels in all animals. However, PS blunted the acute stress elevation in TrkB phosphorylation in Lewis rats. B. Baseline phosphorylation of ERK1/2 was low but 30 minutes of restraint stress elevated ERK phosphorylation levels in all animals. PS exposure attenuated the increase in Erk 1/2 phosphorylation in both Sprague-Dawley and Lewis rats after stress compared to controls. (**, P<0.001)
There are multiple downstream targets of pTrkB. Two of those important for anti-apoptotic events and synaptic plasticity include the MAP/Erk1/2 and PI3-K/Akt cascades. Utilizing ELISA, we found induction of phosphorylated Erk1/2 in the hippocampus after acute stress in controls of all strains (Fig. 7B). However, the Lewis (P<0.0001,t=7.37,df=10) and Sprague-Dawley PS animals (P<0.05,t=2.76,df=10) had significantly less pErk1/2 when compared to their respective strain controls. No differences were seen between control and PS groups for pAkt.
Discussion
The current study demonstrates the sensitivity of the hippocampal BDNF pathway to PS with a strong dependence on genetic background. The male Sprague-Dawley and Lewis strains were most affected, with the Lewis strain being particularly vulnerable to the prenatal stress model. PS alters the normal response of increased mBDNF release after an acute stress. In both Sprague-Dawley and Lewis male rats, PS decreased BDNF mRNA immediately after stress instead of elevating it, decreased the amount of available proBDNF and mature BDNF protein, and changed the activation of TrkB and its downstream signaling cascade. PS in the male Fischer rats did not elicit these changes, suggesting that the Fischer genetic background encodes a developmental environment that is protective during prenatal stress.
BDNF signaling in the hippocampus can follow one of two distinct signaling pathways that have opposite effects on the cell (Fig 8). As proBDNF, it has a high affinity for the p75 receptor, which increases LTD, dendritic atrophy, and cellular apoptosis. For proBDNF to be cleaved into its mature form by plasmin, the zymogen plasminogen must first be activated by tPa. This enzyme can be blocked by numerous serine protease inhibitors such as PAI-1. Once processed, mBDNF can bind to the TrkB receptor either pre- or post-synaptically. In the dendrite, binding induces phosphorylation of Erk 1/2, leading to LTP, synaptic plasticity, cell survival and differentiation.
Figure 8.
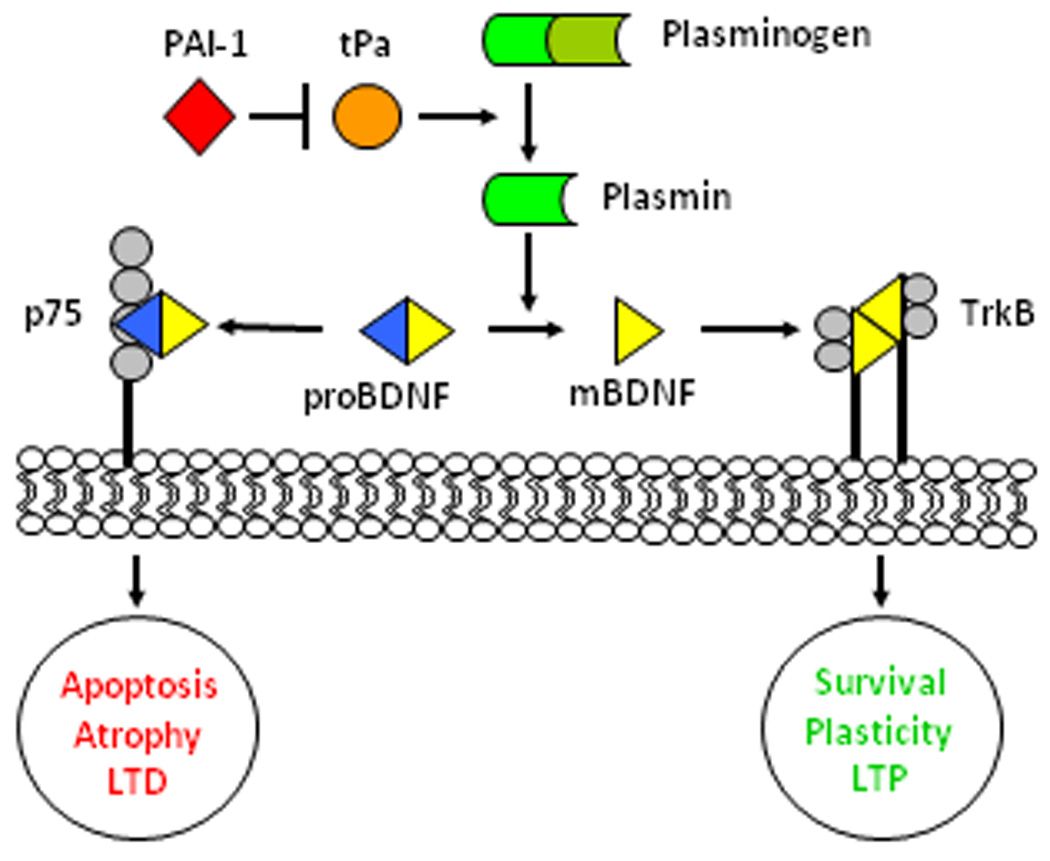
BDNF signaling. proBDNF released from dendrites has a high affinity for the p75 receptor if not first cleaved by plasmin to its mature form (mBDNF). Plasmin is processed from plasminogen by tissue plasminogen activator (tPa). This can be inhibited by serine protease inhibitors, such as PAI-1. mBDNF has a higher affinity for TrkB receptors. TrkB and p75 signaling result in opposing functions in the cell, with mBDNF promoting survival, synaptic plasticity, and increased LTP; while proBDNF and p75 signal apoptosis, atrophy, and increased LTD.
Stress is known to change neuronal architecture during development and cause atrophy in the brain (Kawamura et al., 2006;Vyas et al., 2002). It can compromise normal hippocampal connectivity and reduce hippocampal size, diminishing cognitive function (Lemaire et al., 2000;Szuran et al., 2000;Nishio et al., 2001). BDNF is active within the first two weeks of the rodent embryonic period and peaks 10–14 days into the postnatal period, with highest levels in the hippocampus (Nikolaou et al., 2006;KatohSemba et al., 1997). mBDNF promotes cell survival through TrkB binding and downstream pathways involving Erk1–2. Our data suggest that prenatal stress alters the conventional patterns of these ongoing functions.
The BDNF gene is clearly regulated by stress and HPA axis activation (Tsai, 2006;Mackin, 2005;Malka et al., 2005;Yulug et al., 2009). It is widely known that chronic stress or chronically elevated glucocorticoids can decrease hippocampal BDNF mRNA expression (Chao et al., 1998;Schaaf et al., 1998;Fuchikami et al., 2009), and that acute stress temporarily increases BDNF transcription (Molteni et al., 2008;Marmigere et al., 2003). Our findings suggest male Sprague-Dawley and Lewis PS animals have a hippocampal BDNF response that more closely mimics what is seen in chronic stress, where instead of a brief increase in BDNF mRNA, as seen in the control rats after acute stress, BDNF declines immediately. If PS does indeed alter the animal’s response to acute stress as if it were a chronic one, then this might contribute to the memory deficits seen in models of PS (Lordi et al., 1997;Yaka et al., 2007;Wu et al., 2007;Taylor et al., 2010). In addition, BDNF can induce and is sufficient for long-term potentiation (LTP) (Pang et al., 2004;Pastalkova et al., 2006;Kang et al., 1997), while it’s uncleaved pro-form enhances long-term depression (LTD) (Yang et al., 2009;Woo et al., 2005;Rosch et al., 2005). BDNF can also induce either synaptic enhancement or synaptic depression (Yang et al., 2009;Zagrebelsky et al., 2005;Ghosh et al., 1994;Soule et al., 2006), just as low levels of glucocorticoids can increase synaptic plasticity (Komatsuzaki et al., 2005), or high levels can prevent it (Kumamaru et al., 2008). Interestingly, a single acute stress can facilitate LTP and memory (Vouimba et al., 2004;Barnes and Thomas, 2008) while chronic stress enhances LTD (Diamond et al., 2005). Different BDNF isoforms are thought to be responsible for tissue and stimulus specific functions. Transcripts are transported within the cell, directed in part by the 5’ exons. BDNF transcripts can be targeted to various cellular locations including the soma and proximal or distal dendrites. Transcripts containing exon VI are targeted to the distal dendritic compartments, whereas those with exons I and IV remain within or proximal to the soma (Chiaruttini et al., 2009). Of all of the BDNF isoforms in the hippocampus, exon VI transcription was the most highly changed by exposure to the repeated variable PS paradigm. Although exon IV was also changed by acute stress in the adult rats, no significant differences were found due to PS, though both are known to be influenced by epigenetic mechanisms (Lubin et al., 2008;Murgatroyd et al., 2009). CORT can selectively regulate the transcription of BDNF exon VI (Hansson et al., 2006). This transcript is transported directly to the dendrites, where it is locally transcribed, packaged into vesicles, and released in response to stress, neuronal activity, or other stimuli (Chiaruttini et al., 2009).The BDNF exon VI isoform can also be influenced by MEK and α-CaMKII (Takeuchi et al., 2002), downstream signals of TrkB that may be involved in feedback and regulation of the gene. PS increased transcription of the exon VI isoform in male Sprague-Dawley and Lewis rats at baseline, suggesting that dendritic localization might be altered. In Fischer rats, PS did not produce this change. PS also altered the response of exon VI transcription after acute stress in the Sprague-Dawley and Lewis animals, where a decrease in mRNA levels was seen after the 30 minute stressor. The Sprague-Dawley and Lewis animals have smaller pools of proBDNF protein available at baseline and may, therefore, be more susceptible to both prenatal and acute stress.
The use of multiple strains in this study illustrates the importance of genetic background in the response to prenatal environmental manipulations. Male Fischer rats, which are high responders to stress, show only mild differences in the BDNF pathway due to PS. Male Sprague-Dawley and Lewis rats, which have a more blunted response to stress, demonstrate considerable alterations in hippocampal BDNF mRNA, protein, processing, and signaling. Although the underlying basis for this protection in Fischer rats is unknown, the early developmental period may be the source of differences in the offspring. If this is the case, the protection could be from the mother’s genetically determined response to stress or the fetal response to stress signals from the mother.
Of the major neuropsychiatric disorders, gene X environment interactions are most clearly implicated in schizophrenia (Cannon et al., 2003;Leboyer et al., 2008;Van Os et al., 2008). Thus, in the prenatal stress neurodevelopmental model employed in this study, the strain differences have direct implications for human disease, where genetic heterogeneity between individuals can complicate the study of a disorder. This was also revealed in studies in genetically altered mice exposed to prenatal stress (Oliver and Davies, 2009;Abazyan et al., 2010). Changes in BDNF and its downstream signaling pathways may be part of the complex gene and environment interactions in both animal models of schizophrenia and in patients suffering from this disease and other neuropsychiatric disorders.
Acknowledgements
This research was funded by NIH Grants: DA009457, MH081177, MH073826, MH082999, and the Veterans Affairs Medical Research Service.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abazyan B, Nomura J, Kannan G, Ishizuka K, Tamashiro KL, Nucifora F, Pogorelov V, Ladenheim B, Yang CX, Krasnova IN, Cadet JL, Pardo C, Mori S, Kamiya A, Vogel MW, Sawa A, Ross CA, Pletnikov MV. Prenatal Interaction of Mutant DISC1 and Immune Activation Produces Adult Psychopathology. Biological Psychiatry. 2010;68:1172–1181. doi: 10.1016/j.biopsych.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. Journal of Neuroscience Research. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleva E, Francia N. Psychiatric vulnerability: Suggestions from animal models and role of neurotrophins. Neuroscience and Biobehavioral Reviews. 2009;33:525–536. doi: 10.1016/j.neubiorev.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- Arevalo JC, Wu SH. Neurotrophin signaling: many exciting surprises! Cellular and Molecular Life Sciences. 2006;63:1523–1537. doi: 10.1007/s00018-006-6010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, Nemeroff CB, Reyes TM, Simerly RB, Susser ES, Nestler EJ. Early Life Programming and Neurodevelopmental Disorders. Biological Psychiatry. 2010;68:314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P, Thomas KL. Proteolysis of proBDNF is a key regulator in the formation of memory. PLoS ONE. 2008;3:e3248. doi: 10.1371/journal.pone.0003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: An overview of the use of endophenotypes in order to understand a complex disorder. Schizophren Bull. 2007;33:21–32. doi: 10.1093/schbul/sbl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley PF, Mahadik S, Pillai A, Terry A. Neurotrophins and schizophrenia. Schizophrenia Research. 2007a;94:1–11. doi: 10.1016/j.schres.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Buckley PF, Pillai A, Evans D, Stirewalt E, Mahadik S. Brain derived neurotropic factor in first-episode psychosis. Schizophrenia Research. 2007b;91:1–5. doi: 10.1016/j.schres.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, van Erp TG, Bearden CE, Loewy R, Thompson P, Toga AW, Huttunen MO, Keshavan MS, Seidman LJ, Tsuang MT. Early and late neurodevelopmental influences in the prodrome to schizophrenia: contributions of genes, environment, and their interactions. Schizophr Bull. 2003;29:653–669. doi: 10.1093/oxfordjournals.schbul.a007037. [DOI] [PubMed] [Google Scholar]
- Cantor-Graae E. The contribution of social factors to the development of schizophrenia: A review of recent findings. Canadian Journal of Psychiatry-Revue Canadienne de Psychiatrie. 2007;52:277–286. doi: 10.1177/070674370705200502. [DOI] [PubMed] [Google Scholar]
- Chao HM, Sakai RR, Ma LY, McEwen BS. Adrenal steroid regulation of neurotrophic factor expression in the rat hippocampus. Endocrinol. 1998;39:3112–3118. doi: 10.1210/endo.139.7.6114. [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia CT, Coutinho AM, Sequeira AF, Sousa IG, Venda LLO, Almeida JP, Abreu RL, Lobo C, Miguel TS, Conroy J, Cochrane L, Gallagher L, Gill M, Ennis S, Oliveira GG, Vicente AM. Increased BDNF levels and NTRK2 gene association suggest a disruption of BDNF/TrkB signaling in autism. Genes Brain and Behavior. 2010;9:841–848. doi: 10.1111/j.1601-183X.2010.00627.x. [DOI] [PubMed] [Google Scholar]
- Dawe GS, Hwang EHJ, Tan CH. Pathophysiology and Animal Models of Schizophrenia. Ann Acad Med Singapore. 2009;38:425–430. [PubMed] [Google Scholar]
- Dhabhar FS, McEwen BS, Spencer RL. Stress-Response, Adrenal-Steroid Receptor Levels and Corticosteroid-Binding Globulin Levels - A Comparison Between Sprague-Dawley, Fischer-344 and Lewis Rats. Brain Res. 1993;616:89–98. doi: 10.1016/0006-8993(93)90196-t. [DOI] [PubMed] [Google Scholar]
- Diamond DM, Park CR, Campbell AM, Woodson JC. Competitive interactions between endogenous LTD and LTP in the hippocampus underlie the storage of emotional memories and stress-induced amnesia. Hippocampus. 2005;15:1006–1025. doi: 10.1002/hipo.20107. [DOI] [PubMed] [Google Scholar]
- Durany N, Michel T, Zochling R, Boissl KW, Cruz-Sanchez FF, Riederer P, Thome J. Brain-derived neurotrophic factor and neurotrophin 3 in schizophrenic psychoses. Schizophrenia Research. 2001;52:79–86. doi: 10.1016/s0920-9964(00)00084-0. [DOI] [PubMed] [Google Scholar]
- Freedman R, Adams CE, Adler LE, Bickford PC, Gault J, Harris JG, Nagamoto HT, Olincy A, Ross RG, Stevens KE, Waldo M, Leonard S. Inhibitory neurophysiological deficit as a phenotype for genetic investigation of schizophrenia. Am J Med Genet. 2000;97:58–64. doi: 10.1002/(sici)1096-8628(200021)97:1<58::aid-ajmg8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Kurata A, Yamamoto S, Yamawaki S. Single immobilization stress differentially alters the expression profile of transcripts of the brain-derived neurotrophic factor (BDNF) gene and histone acetylation at its promoters in the rat hippocampus. Int J Neuropsychopharmacol. 2009;12:73–82. doi: 10.1017/S1461145708008997. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME. Requirement for BDNF in activity-dependent survival of cortical neurons. Science. 1994;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- Gray L, Hannan AJ. Dissecting cause and effect in the pathogenesis of psychiatric disorders: Genes, environment and behaviour. Current Molecular Medicine. 2007;7:470–478. doi: 10.2174/156652407781387064. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Sommer WH, Metsis M, Stromberg I, Agnati LF, Fuxe K. Corticosterone actions on the hippocampal brain-derived neurotrophic factor expression are mediated by exon IV promoter. Journal of Neuroendocrinology. 2006;18:104–114. doi: 10.1111/j.1365-2826.2005.01390.x. [DOI] [PubMed] [Google Scholar]
- Iritani S, Niizato K, Nawa H, Ikeda K, Emson PC. Immunohistochemical study of brain-derived neurotrophic factor and its receptor, TrkB, in the hippocampal formation of schizophrenic brains. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:801–807. doi: 10.1016/S0278-5846(03)00112-X. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Welcher AA, Shelton D, Schuman EM. Neurotrophins and time: Different roles for TrkB signaling in hippocampal long-term potentiation. Neuron. 1997;19:653–664. doi: 10.1016/s0896-6273(00)80378-5. [DOI] [PubMed] [Google Scholar]
- KatohSemba R, Takeuchi IK, Semba R, Kato K. Distribution of brain-derived neurotrophic factor in rats and its changes with development in the brain. Journal Of Neurochemistry. 1997;69:34–42. doi: 10.1046/j.1471-4159.1997.69010034.x. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Chen J, Takahashi T, Ichitani Y, Nakahara D. Prenatal stress suppresses cell proliferation in the early developing brain. Neuroreport. 2006;17:1515–1518. doi: 10.1097/01.wnr.0000236849.53682.6d. [DOI] [PubMed] [Google Scholar]
- Kennedy JL, Farrer LA, Andreasen NC, Mayeux R, St George-Hyslop P. The genetics of adult-onset neuropsychiatric disease: Complexities and conundra? Science. 2003;302:822–826. doi: 10.1126/science.1092132. [DOI] [PubMed] [Google Scholar]
- Kinnunen AK, Koenig JI, Bilbe G. Repeated variable prenatal stress alters pre- and postsynaptic gene expression in the rat frontal pole. J Neurochem. 2003;86:736–748. doi: 10.1046/j.1471-4159.2003.01873.x. [DOI] [PubMed] [Google Scholar]
- Koenig JI, Elmer GI, Shepard PD, Lee PR, Mayo C, Joy B, Hercher E, Brady DL. Prenatal exposure to a repeated variable stress paradigm elicits behavioral and neuroendocrinological changes in the adult offspring: potential relevance to schizophrenia. Behavior Brain Res. 2005;156:251–261. doi: 10.1016/j.bbr.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Koenig JI, Kirkpatrick B, Lee P. Glucocorticoid hormones and early brain development in schizophrenia. Neuropsychopharmacol. 2002;27:309–318. doi: 10.1016/S0893-133X(01)00396-7. [DOI] [PubMed] [Google Scholar]
- Komatsuzaki Y, Murakami G, Tsurugizawa T, Mukai H, Tanabe N, Mitsuhashi K, Kawata M, Kimoto T, Ooishi Y, Kawato S. Rapid spinogenesis of pyramidal neurons induced by activation of glucocorticoid receptors in adult male rat hippocampus. Biochemical and Biophysical Research Communications. 2005;335:1002–1007. doi: 10.1016/j.bbrc.2005.07.173. [DOI] [PubMed] [Google Scholar]
- Kosten TA, Ambrosio E. HPA axis function and drug addictive behaviors: insights from studies with Lewis and Fischer 344 inbred rats. Psychoneuroendocrinol. 2002;27:35–69. doi: 10.1016/s0306-4530(01)00035-x. [DOI] [PubMed] [Google Scholar]
- Kumamaru E, Numakawa T, Adachi N, Yagasaki Y, Izumi A, Niyaz M, Kudo M, Kunugi H. Glucocorticoid prevents brain-derived neurotrophic factor-mediated maturation of synaptic function in developing hippocampal neurons through reduction in the activity of mitogen-activated protein kinase. Molecular Endocrinology. 2008;22:546–558. doi: 10.1210/me.2007-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboyer M, Meyer-Lindenberg A, Stetanis N, Rutten BPF, Arango C, Jones P, Kapur S, Lewis S, Murray R, Owen MJ. Schizophrenia aetiology: Do gene-environment interactions hold the key? Schizophrenia Research. 2008;102:21–26. doi: 10.1016/j.schres.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Lee PR, Brady DL, Shapiro RA, Dorsa DM, Koenig JI. Prenatal stress generates deficits in rat social behavior: Reversal by oxytocin. Brain Res. 2007;1156:152–167. doi: 10.1016/j.brainres.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Kermani P, Teng K, Hempstead B. Regulation of Cell Survival by Secreted Proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- Lemaire V, Koehl M, Le MM, Abrous DN. Prenatal stress produces learning deficits associated with an inhibition of neurogenesis in the hippocampus. Proc Natl Acad Sci U S A. 2000;97:11032–11037. doi: 10.1073/pnas.97.20.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessmann V, Brigadski T. Mechanisms, locations, and kinetics of synaptic BDNF secretion: An update. Neurosci Res. 2009;65:11–22. doi: 10.1016/j.neures.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Research. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lordi B, Protais P, Mellier D, Caston J. Acute Stress in Pregnant Rats: Effects on Growth Rate, Learning, and Memory Capabilities of the Offspring. Physiology & Behavior. 1997;62:1087–1092. doi: 10.1016/s0031-9384(97)00261-8. [DOI] [PubMed] [Google Scholar]
- Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nature Reviews Neuroscience. 2005;6:603–614. doi: 10.1038/nrn1726. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic Regulation of bdnf Gene Transcription in the Consolidation of Fear Memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin P. Brain-derived neurotrophic factor (BDNF), stress and affective disorders. Journal of Psychopharmacology. 2005;19:A60. [Google Scholar]
- Malka Y, Kozlovsky N, Kaplan Z, Matar MA, Cohen H. Endogenous and exogenous BDNF mediate resilience to stress exposure: The efficacy of training on synaptic plasticity, cognition and posttraumatic stress responses. Reviews in the Neurosciences. 2005;16:S43–S44. [Google Scholar]
- Markham JA, Koenig JI. Prenatal stress: Role in psychotic and depressive diseases. Psychopharmacology. 2011;214:89–106. doi: 10.1007/s00213-010-2035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigere F, Givalois L, Rage F, Arancibia S, Tapia-Arancibia L. Rapid induction of BDNF expression in the hippocampus during immobilization stress challenge in adult rats. Hippocampus. 2003;13:646–655. doi: 10.1002/hipo.10109. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Lu H, Fukata Y, Noritake J, Gao HF, Mukherjee S, Nemoto T, Fukata M, Poo MM. Differential Activity-Dependent Secretion of Brain-Derived Neurotrophic Factor from Axon and Dendrite. J Neurosci. 2009;29:14185–14198. doi: 10.1523/JNEUROSCI.1863-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni R, Calabrese F, Cattaneo A, Mancini M, Gennarelli M, Racagni G, Riva MA. Acute Stress Responsiveness of the Neurotrophin BDNF in the Rat Hippocampus is Modulated by Chronic Treatment with the Antidepressant Duloxetine. Neuropsychopharmacology. 2008;34:1523–1532. doi: 10.1038/npp.2008.208. [DOI] [PubMed] [Google Scholar]
- Moncek F, Kvetnansky R, Jezova D. Differential responses to stress stimuli of Lewis and Fischer rats at the pituitary and adrenocortical level. Endocr Regul. 2001 [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OFX, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nikolaou KE, Malamitsi-Puchner A, Boutsikou T, Economou E, Boutsikou M, Puchner KP, Baka S, Hassiakos D. The Varying Patterns of Neurotrophin Changes in the Perinatal Period. Ann NY Acad Sci. 2006;1092:426–433. doi: 10.1196/annals.1365.041. [DOI] [PubMed] [Google Scholar]
- Nishio H, Kasuga S, Ushijima M, Harada Y. Prenatal stress and postnatal development of neonatal rats--sex-dependent effects on emotional behavior and learning ability of neonatal rats. Int J Dev Neurosci. 2001;19:37–45. doi: 10.1016/s0736-5748(00)00070-8. [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE, Petersen CM. p75(NTR)-live or let die. Current Opinion in Neurobiology. 2005;15:49–57. doi: 10.1016/j.conb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Oliver PL, Davies KE. Interaction between environmental and genetic factors modulates schizophrenic endophenotypes in the Snap-25 mouse mutant blind-drunk. Human Molecular Genetics. 2009;18:4576–4589. doi: 10.1093/hmg/ddp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P, Teng H, Zaitsev E, Woo N, Sakata K, Zhen S, Teng K, Yung W, Hempstead B, Lu B. Cleavage of proBDNF by tPA/Plasmin Is Essential for Long-Term Hippocampal Plasticity. Science. 2004;306:487–491. doi: 10.1126/science.1100135. [DOI] [PubMed] [Google Scholar]
- Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science. 2006;313:1141–1144. doi: 10.1126/science.1128657. [DOI] [PubMed] [Google Scholar]
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Rosch H, Schweigreiter R, Bonhoeffer T, Barde YA, Korte M. The neurotrophin receptor p75(NTR) modulates long-term depression and regulates the expression of AMPA receptor subunits in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7362–7367. doi: 10.1073/pnas.0502460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles F, Strickland S. Localization and regulation of the tissue plasminogen activator-plasmin system in the hippocampus. J Neurosci. 2002;22:2125–2134. doi: 10.1523/JNEUROSCI.22-06-02125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf MJM, de Jong J, de Kloet ER, Vreugdenhil E. Downregulation of BDNF mRNA and protein in the rat hippocampus by corticosterone. Brain Res. 1998;813:112–120. doi: 10.1016/s0006-8993(98)01010-5. [DOI] [PubMed] [Google Scholar]
- Schweigreiter R. The dual nature of neurotrophins. Bioessays. 2006;28:583–594. doi: 10.1002/bies.20419. [DOI] [PubMed] [Google Scholar]
- Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- Soule J, Messaoudi E, Bramham CR. Brain-derived neurotrophic factor and control of synaptic consolidation in the adult brain. Biochem Soc Trans. 2006;34:600–604. doi: 10.1042/BST0340600. [DOI] [PubMed] [Google Scholar]
- Szuran TF, Pliska V, Pokorny J, Welzl H. Prenatal stress in rats: effects on plasma corticosterone, hippocampal glucocorticoid receptors, and maze performance. Physiol Behav. 2000;71:353–362. doi: 10.1016/s0031-9384(00)00351-6. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Miyamoto E, Fukunaga K. Analysis on the promoter region of exon IV brain-derived neurotrophic factor in NG108-15 cells. Journal Of Neurochemistry. 2002;83:67–79. doi: 10.1046/j.1471-4159.2002.01096.x. [DOI] [PubMed] [Google Scholar]
- Taylor AR, Markham JA, Taylor SB, Brady-Bell D, Koenig JI. Characterization of the cognitive impairments induced by prenatal exposure to stress in the rat. Frontiers Behav Neurosci. 2010;4:173. doi: 10.3389/fnbeh.2010.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Toyooka K, Asama K, Watanabe Y, Muratake T, Takahashi M, Someya T, Nawa H. Decreased levels of brain-derived neurotrophic factor in serum of chronic schizophrenic patients. Psychiatry Research. 2002;110:249–257. doi: 10.1016/s0165-1781(02)00127-0. [DOI] [PubMed] [Google Scholar]
- Tsai SJ. The possible role of tissue-type plasminogen activator and the plasminogen system in the pathogenesis of major depression. Medical Hypotheses. 2006;66:319–322. doi: 10.1016/j.mehy.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- Van Os J, Rutten BPF, Poulton R. Gene-Environment Interactions in Schizophrenia: Review of Epidemiological Findings and Future Directions. Schizophrenia Bulletin. 2008;34:1066–1082. doi: 10.1093/schbul/sbn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouimba RM, Yaniv D, Diamond D, Richter-Levin G. Effects of inescapable stress on LTP in the amygdala versus the dentate gyrus of freely behaving rats. Eur J Neurosci. 2004;19:1887–1894. doi: 10.1111/j.1460-9568.2004.03294.x. [DOI] [PubMed] [Google Scholar]
- Vyas A, Mitra R, Shankaranarayana Rao BS, Chattarji S. Chronic stress induces contrasting patterns of dendritic remodeling in hippocampal and amygdaloid neurons. J Neurosci. 2002;22:6810–6818. doi: 10.1523/JNEUROSCI.22-15-06810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Ligons DL, Romanczyk T, Ungaro G, Hyde TM, Herman MM, Weinberger DR, Kleinman JE. Reductions in neurotrophin receptor mRNAs in the prefrontal cortex of patients with schizophrenia. Molecular Psychiatry. 2005;10:637–650. doi: 10.1038/sj.mp.4001678. [DOI] [PubMed] [Google Scholar]
- Woo N, Teng H, Siao C, Chiaruttini C, Pang P, Milner T, Hempstead B, Lu B. Activation of p75(NTR) by proBDNF facilitates hippocampal long-term depression. Nature Neuroscience. 2005;8:1069–1077. doi: 10.1038/nn1510. [DOI] [PubMed] [Google Scholar]
- Wu J, Song TB, Li YJ, He KS, Ge L, Wang LR. Prenatal restraint stress impairs learning and memory and hippocampal PKCbeta1 expression and translocation in offspring rats. Brain Research. 2007;1141:205–213. doi: 10.1016/j.brainres.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Yaka R, Salomon S, Matzner H, Weinstock M. Effect of varied gestational stress on acquisition of spatial memory, hippocampal LTP and synaptic proteins in juvenile male rats. Behavioural Brain Research. 2007;179:126–132. doi: 10.1016/j.bbr.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Yang F, Je HS, Ji YY, Nagappan G, Hempstead B, Lu B. Pro-BDNF-induced synaptic depression and retraction at developing neuromuscular synapses. Journal of Cell Biology. 2009;185:727–741. doi: 10.1083/jcb.200811147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yulug B, Ozan E, Gonul AS, Kilic E. Brain-derived neurotrophic factor, stress and depression: A minireview. Brain Research Bulletin. 2009;78:267–269. doi: 10.1016/j.brainresbull.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Zagrebelsky M, Holz A, Dechant G, Barde Y, Bonhoeffer T, Korte M. The p75 Neurotrophin Receptor Negatively Modulates Dendrite Complexity and Spine Density in Hippocampal Neurons. J Neurosci. 2005;25:9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


