Abstract
Epigenetics is traditionally defined as the study of heritable changes in gene expression caused by mechanisms other than changes in the underlying DNA sequence. There are three main classes of epigenetic marks—DNA methylation, modifications of histone tails, and noncoding RNAs—each of which may be influenced by the environment, diet, diseases, and ageing. Importantly, epigenetic marks have been shown to influence immune cell maturation and are associated with the risk of developing various forms of cancer, including lung cancer. Moreover, there is emerging evidence that these epigenetic marks affect gene expression in the lung and are associated with benign lung diseases, such as asthma, chronic obstructive pulmonary disease, and interstitial lung disease. Technological advances have made it feasible to study epigenetic marks in the lung, and it is anticipated that this knowledge will enhance our understanding of the dynamic biology in the lung and lead to the development of novel diagnostic and therapeutic approaches for our patients with lung disease.
Keywords: epigenetics, DNA methylation, histone modifications, microRNA, noncoding RNA
EPIGENETIC MECHANISMS
The word epigenetics is derived from the Greek word epi, for over or above, and genetics, or the science of heredity. Epigenetics is traditionally defined as the study of heritable changes in gene expression caused by mechanisms other than changes in the underlying DNA sequence (1). However, more recent evidence suggests that the epigenome is dynamic and changes in response to the environment, diet, diseases, and ageing (2, 3). There are three main classes of epigenetic marks: DNA methylation, modifications of histone tails, and more recently discovered noncoding RNAs (Figure 1).
Figure 1.
(a) Epigenetic mechanisms in eukaryotes—DNA methylation, modifications of histone tails, and noncoding RNAs. (b) Effect of epigenetic marks on gene expression. Wide blue arrows indicate cross-talk between DNA methylation and histone modifications.
Methylation of cytosine residues in CpG dinucleotides within the context of CpG islands (stretches of DNA > 200 bp in length with > 50% GC content and observed/expected CpG > 0.6 [4]) is the simplest form of epigenetic regulation in eukaryotes. Hypermethylation of CpG islands in gene promoters generally leads to gene silencing, and hypomethylation leads to active transcription. There are a few exceptions to this general rule, such as methylation of TERT promoter sequence being positively correlated with gene expression and telomerase activity (5). CpG island methylation has long been studied in cancer with findings that hypermethylation of tumor suppressor genes and hypomethylation of oncogenes contribute to the process of carcinogenesis (6, 7). More recent studies have demonstrated that methylation of less CpG-dense regions near CpG islands (“CpG island shores”) controls expression of tissue-specific genes as well as genes relevant to carcinogenesis and lineage-specific cell differentiation (8, 9), suggesting that DNA methylation outside of CpG islands is an important mechanism that controls gene transcription. Moreover, the DNA “methylome” of the H1 human embryonic stem cell line uniquely revealed that nearly one-quarter of all methylation is in non-CpG context (10), suggesting that embryonic stem cells may use different methylation mechanisms to control gene expression. Finally, recent reports are challenging traditional views of DNA methylation being a permanent mark and suggesting that DNA methylation of promoter regions is sometimes transient in nature (11).
Methylation, acetylation, phosphorylation, and ubiquitylation of histone tails (1) occur at specific sites and residues and control gene expression by regulating DNA accessibility to RNA polymerase II and transcription factors. In eukaryotes, DNA is packaged and ordered into nucleosomes, the basic structural unit of chromatin, by wrapping around the octamer consisting of two copies each of histone proteins (H2A, H2B, H3, and H4). The most common amino acids to undergo modifications are basic lysine and arginine residues, with some modifications marking active and some inactive chromatin state. H3K4 trimethylation or trimethylation of lysine 4 on H3 (H3K4me3), for example, is strongly associated with transcriptional activation, whereas H3K27 trimethylation or trimethylation of lysine 27 on H3 (H3K27me3) is frequently associated with gene silencing. Similarly, histone tail acetylation leads to active gene transcription, whereas deacetylation is a repressive mark and leads to gene silencing. Histone acetyltransferases (HATs) are enzymes that acetylate histone tails, whereas histone deacetylases (HDACs) remove acetyl groups from histone tails. Analogous to DNA methylation, dysregulation of these histone modifications has been linked to misregulation of gene expression in cancers (12).
MicroRNAs (miRNAs) are approximately 22-nucleotide–long regulatory RNAs that control gene expression by binding to the 3′ untranslated regions of messenger RNA (mRNA), which leads to either mRNA degradation or inhibition of protein translation (13). miRNAs, which were first identified in 1993 (14), are evolutionarily conserved and are the most extensively studied family of small noncoding RNAs. Mature miRNAs are cleaved from approximately 70-nucleotide hairpin precursor mRNAs (pre-mRNAs) by the enzyme Dicer, whereas the pre-miRNAs are excised from a primary miRNA (pri-miRNA) transcript by the enzyme Drosha (15). Pri-miRNAs are typically transcribed by RNA polymerase II, can be thousands of nucleotides long, and generally contain multiple pre-miRNAs. There are more than 1,000 mature miRNAs in the human genome according to the miRBase (http://www.mirbase.org/), but it is expected that many more miRNAs will be identified in the near future. Alterations of expression of miRNAs contribute to pathogenesis of most malignancies, with miRNAs acting as both oncogenes and tumor suppressor genes (16). Many microRNAs map to chromosome regions that are involved in genomic instability and rearrangements in human cancer.
An emerging paradigm for epigenetic regulation of gene expression is the relationship between DNA methylation and histone modifications. One example of these interactions is binding of DNMT3L, a regulatory factor related in sequence to mammalian de novo methyltransferases DNMT3A and DNMT3B, to the N terminus of histone H3 tail (17, 18). DNMT3L recognizes unmethylated H3 tails at lysine 4 and induces de novo DNA methylation by recruitment or activation of DNMT3A2; these findings establish the N terminus of histone H3 tail with an unmethylated lysine 4 as a chromatin determinant for DNA methylation. Similarly, DNA methyltransferases preferentially target nucleosome-bound DNA (19). The relationship of histones and DNA methylation is bidirectional; in addition to histones playing a role in the establishment of DNA methylation patterns, DNA methylation is important for maintaining patterns of histone modification through cell division (20).
ROLE OF THE ENVIRONMENT IN MODULATING EPIGENETIC MARKS
Unlike an individual's genetic make-up, epigenetic marks can be influenced by exposures, diet, and ageing. Randy Jirtle's seminal experiments showed that maternal diet supplemented with methyl donors (folic acid, vitamin B12, choline, and betaine) shifts coat color distribution of progeny toward the brown pseudoagouti phenotype, and that this shift in coat color resulted from an increase in DNA methylation in a transposon adjacent to the agouti gene (21, 22). These studies also revealed that mice with yellow coat color are obese and are more prone to develop cancer, suggesting for the first time that changes in DNA methylation caused by diet may be linked to disease development.
Other studies have shown that pesticides and fungicides can alter the methylome, resulting in changes in male fertility (23), and that ageing is also associated with changes in DNA methylation and gene expression (2). Decreased DNA methylation (as measured by LINE-1 repeats) was found to be associated with recent exposure to PM2.5 particles among 718 elderly individuals in the Boston area (24), and although this correlated with time-dependent variables, such as day of the week and season, there was no association with air pollution–related health effects.
Several recent studies have examined the relationship between exposure to cigarette smoke and epigenetic marks. Among 384 children, a global reduction in DNA methylation, as measured by the extent of methylation of Alu repeats, and differential methylation of eight specific CpG motifs, was found to be associated with in utero cigarette smoke exposure (25). Normal human airway epithelial cells and immortalized bronchial epithelial cells exposed to cigarette smoke condensate (CSC) identified time- and dose-dependent changes in histone modifications (decrease in H4K16Ac and H4K20Me3, and increase in H3K27Me3) accompanied by decreased DNMT1 and increased DNMT3b expression; these changes are characteristic of lung cancer progression (26).
Cigarette smoke exposure has also been shown to have a significant influence on expression of miRNAs (27–29). Comparing current to never smokers, 28 miRNAs were differentially expressed, mostly down-regulated in human bronchial airway epithelium of smokers (27). miR-218 was found to be one of the strongly associated miRNAs with cigarette smoke exposure, and it was further shown that a change in miR-218 expression in primary bronchial epithelial cells and H1299 cell line resulted in a corresponding negatively correlated change in expression of predicted mRNA targets for miR-218. Two recent studies also demonstrated changes in miRNA expression in lungs of mice (29) and rats (28) exposed to cigarette smoke. All three studies showed the predominant effect of smoke exposure is down-regulation of miRNAs, with substantial overlap between mice and rats and some overlap of rodent miRNA expression changes in the lung with those observed in human airway epithelium. However, the mechanisms linking cigarette smoke to any of these epigenetic changes have not been clearly defined, thus raising uncertainty about the cause and effect relationship between cigarette smoke and epigenetic marks.
EPIGENTICS MARKS AND THE IMMUNE SYSTEM
A large body of evidence suggests that epigenetic mechanisms affect the expression of cytokines and binding of transcription factors that control the lineage of Th1, Th2, regulatory T cells (Tregs), and Th17 cells. In the context of Th1/Th2 differentiation, the most extensively studied are the Th1 cytokine IFN-γ, and Th2 cytokines IL-4 and IL-13. It has been shown that the de novo DNA methyltransferase Dnmt3a methylates CpG-53 in the IFN-γ promoter, which inhibits ATF2/c-Jun and CREB transcription factor binding and leads to suppression of IFN-γ transcription in developing Th2 cells in mice (30). Cord blood CD4+ cells enhance the development of Th1 (but not Th2) lineage through progressive demethylation of the IFN-γ promoter (31). Methylation of the IFN-γ promoter was reduced in CD8+ cells from atopic children in the age range in which hyperproduction of IFN-γ occurs, suggesting that DNA methylation at this locus may be a contributing factor in the development of atopy in children.
Importantly, differentiation of human CD4+ cells into the Th2 subtype is accompanied by the appearance of DNase I hypersensitive sites (DHS) and CpG demethylation around these DHS sites within IL-4 and IL-13 promoters (32). Moreover, the IL-4 locus undergoes a complex series of methylation and demethylation steps during Th cell differentiation, and demethylation of the IL-4 locus is strongly associated with efficient expression of the IL-4 transcript by differentiated Th2 cells (33). Similarly, formation of DHS sites and decreased methylation at the proximal IL-13 promoter was observed in human cord blood CD4+ cells (34). Finally, extensive studies of the Th2 cytokine locus control region (LCR) (35) have shown that rad50 hypersensitive site 7 (RHS7) within the Th2 cytokine LCR undergoes rapid demethylation during Th2 differentiation (36).
In addition to DNA methylation, histone modifications are also important in guiding T-cell differentiation. Tbet and GATA-3 transcription factors control lineage-specific histone acetylation of IFN-γ and IL-4 loci during Th1/Th2 differentiation (37). Rapid methylation of H3K9 and H3K27 residues (repressive marks) at the IFN-γ locus are associated with differentiating Th1 cells, whereas demethylation of H3K9 and methylation of H3K27 was associated with Th2 differentiation. In a study of human cord blood CD4+ cells, histone acetylation marks at the proximal IL-13 promoter were selectively observed in Th2 cells (34), suggesting that permissive histone marks together with DNA demethylation lead to expression of IL-13 in Th2 cells. In aggregate, these studies suggest that DNA methylation and histone modifications are highly dynamic and represent important determinants of Th1 and Th2 cell lineages.
Epigenetic mechanisms controlling Treg development are also beginning to be explored. Tregs are a unique T-cell lineage with an important role in immunological tolerance, whose development is primarily regulated by the transcription factor FOXP3. Evidence for the role of DNA methylation and histone modifications in regulation of FOXP3 expression are summarized in two recent reviews (38, 39). Similarly, recent evidence suggests that histone modifications play a role in the development of Th17 cells (40).
Although miRNAs were discovered relatively recently, there is already a growing body of evidence for the role of miRNAs in the development and function of the immune system (41, 42). Early studies of the role miRNAs in the immune system used conditional inactivation of Dicer in T or B cells in mice to demonstrate that miRNAs are critical for lymphocyte development and differentiation. Deletion of Dicer in immature (double-negative 3 or DN3 stage) T cells led to a dramatic decrease in more mature CD4+CD8+, CD4+, and CD8+ subsets (43). A number of differentially expressed miRNAs have been identified in response to innate and adaptive immune stimuli with many commonalities in miRNA expression (miR-21, -103, -155, and -204) (41). miRNA-155 is the most often identified differentially regulated noncoding RNA in studies involving the immune system (41, 42); a very recent study revealed that miR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte–associated antigen 4 (44). Finally, there is clear evidence that miRNAs are involved in the development and function of Treg (43) and Th17 (45) cell lineages.
In addition to being crucial to the development of the immune system, epigenetic marks play an important role in cellular and tissue development in general and specifically in the development of the lung, as reviewed in References 46–48.
EPIGENETICS AND LUNG DISEASE
Given the evidence for a strong influence of environmental exposures on epigenetic marks and the role of epigenetic regulation in T-cell differentiation, it is becoming clear that epigenetic changes may be one of the factors to explain the increasing prevalence of asthma, especially in developed countries (49). Epidemiological studies are beginning to point to the role of maternal diet on the development of atopy (50) and asthma (51, 52) in children. Our group hypothesized that hypermethylation of the genome during gestation skews toward a Th2 phenotype and enhances the risk of allergic airway disease. To test this hypothesis, we conducted a study in which pregnant female mice were fed either a low- or high-methylation diet, and progeny were sensitized and challenged with ovalbumin to directly study the role of maternal diet on the development of allergic airway disease (53). We observed an increase in airway inflammation (eosinophil recruitment and concentrations of IL-4 and IL-13), serum IgE, and airway hyperresponsiveness (AHR) in pups of mothers who were fed a high-methylation diet compared with those of mothers on a low-methylation diet. Furthermore, we demonstrated hypermethylation of 82 gene-associated CpG islands throughout the genome, including extensive hypermethylation of the promoter region of Runx3, a gene known to regulate allergic airway disease in mice. Importantly, a direct link between epigenetic control of the Th2 cytokine locus and development of allergic airway diseases was further demonstrated in mice with deficiency in the Th2 LCR in CD4 cells that have general loss of H3 acetylation, H3K4 methylation, and DNA demethylation in the Th2 cytokine locus; these mice also exhibit marked reduction in airway inflammation and AHR when sensitized and challenged with ovalbumin (54). Two subsequent independent epidemiological studies have demonstrated that in utero exposure to folate (a methylating agent) is associated with the risk of developing asthma in humans (51, 52). In utero exposure to cigarette smoke is also associated with the development of asthma in children (55, 56), and given the recent evidence for modulation of epigenetic marks by cigarette smoke, it is possible that changes in DNA methylation or specific histone modifications mediate the effect of prenatal exposure to cigarette smoke on the development of allergic airway disease; no publications to date have demonstrated this link.
Acetylation of histones may also play a role in asthma. Increased acetylation of H4 has been demonstrated in individuals with asthma and is associated with an increase in expression of several inflammatory genes in the lung (57). It has also been shown that increased acetylation of histones is associated with decreased HDAC activity and/or increased HAT activity, which may be responsible for enhanced expression of inflammatory genes. In addition, glucocorticoids appear to suppress inflammation by altering acetylation of histones that regulate inflammatory and antiinflammatory genes; these studies are described in detail in a recent review (58) and suggest that targeting histone acetylation (and possibly other epigenetic marks) may lead to novel antiinflammatory therapies, especially in corticosteroid-resistant cases of asthma.
The role of miRNAs in asthma and atopy is also emerging. Although no detectable differences in expression of miRNAs from airway biopsies were observed between subjects with mild asthma and normal subjects (59, 60), only subjects with mild asthma were included in this study, and the number of miRNAs examined was limited. However, this study demonstrated cell-type specific expression of miRNAs in cells isolated from airways and lung tissue, suggesting a possible role for miRNAs in asthma (59, 60). In a recent study in mice, house dust mite sensitization and challenge resulted in up-regulation of miR-16, -21, and -126 in the lung (60). Moreover, selective blockade in miR-126 resulted in diminished Th2 response, inflammation, and AHR; these effects were shown to be mediated by activation of the MyD88 innate immune signaling pathway. The results of this study suggest that focusing on specific exposures relevant to the asthma phenotype may be an effective way to identify the role of miRNAs in allergic airway disease.
Epigenetic mechanisms are likely to be involved in the control of gene expression in chronic lung diseases, such as idiopathic pulmonary fibrosis (IPF) and chronic obstructive pulmonary disease, especially given the association of these diseases with cigarette smoking and the relationship between cigarette smoke and changes in DNA methylation, histone modifications, and miRNAs. Although very few studies have been published to date, we expect this to become a major area of research in the near future. Two published studies point to the role of miRNAs in IPF; let-7d (61, 62) and miR-21 (61, 62) appear to be important epigenetic regulators of fibroproliferation. Moreover, 625 CpG islands were recently reported to be differentially methylated between IPF and control lungs, further supporting a role for epigenomic changes in IPF (63). Several targeted studies have shown that epigenetic modulation regulates expression of genes involved in pathogenesis of IPF. Defective histone acetylation is responsible for the repression of expression of two antifibrotic genes, cyclooxygenase-2 (COX2) (64) and chemokine IP-10 (65). Similarly, Thy-1 (CD90) is an important regulator of cell–cell and cell–matrix interactions that is expressed on normal lung fibroblasts, but its expression is absent in myofibroblasts within fibroblastic foci in IPF. Thy-1 down-regulation in rat lung fibroblasts is controlled by both promoter DNA hypermethylation (66) and histone modifications (67). In chronic obstructive pulmonary disease, airway biopsies demonstrate an increase in histone acetylation in promoters of inflammatory genes that increases with disease severity (68). Importantly, extensive evidence exists for the role of epigenetic regulation in lung cancer and is summarized in these recent reviews: DNA methylation (69), histone methylation (12), and miRNAs (16, 41).
EPIGENOMIC PROFILING
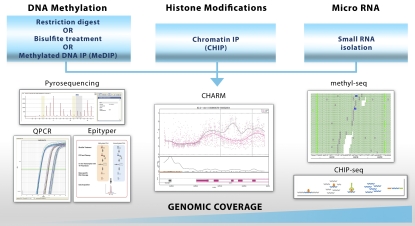
Epigenetic marks can be studied using focused and genome-wide approaches (Figure 2). Pyrosequencing and Epityper assays on the Sequenom MassARRAY platform are commonly used techniques for interrogation of a small number of CpG sites, whereas quantitative PCR methods are typically used for focused studies of histone modifications and miRNAs. Microarrays have been the method of choice for profiling epigenetic marks on a genomic scale, with several platforms and protocols available for DNA methylation: bisulfate conversion of methylated cytosines, methylated DNA immunoprecipitation (MeDIP) using an antimethylcytosine antibody, and digestion of DNA with restriction enzymes specific for methylated or unmethylated cytosines (70). Array platforms have also been used to examine histone modification by chromatin immunoprecipitation followed by hybridization on microarrays (CHIP-chip) (70) and for miRNAs (71). One recently developed microarray-based method that has allowed for major advancements in the area of DNA methylation profiling is the Comprehensive Analysis of Relative DNA Methylation (CHARM) platform (72). CHARM arrays examine not only CpG islands but also other areas of the genome using 2.1 million probes that cover 4.6 million CpG sites in the human genome.
Figure 2.
Technologies for the assessment of epigenetic marks. Samples are prepared and the extent of epigenetic modification assessed using pyrosequencing for DNA methylation or quantitative PCR for histone modifications and miRNAs (single locus), epityper assays (1-40 loci, DNA methylation only), microarrays (a thousand to a few million loci; CHARM interogates 4.6M individual CpGs) or next-generation sequencing (entire genome). Epityper image used with permission from Sequenom (www.sequenom.com) and CHIP-seq image with permission from Life Technologies (www.lifetechnologies.com). Methyl-seq image reprinted from Bormann Chung PLoS One 2010;22:e9320 under open-access license.
However, the most substantial advance to assess epigenetic marks on the genome scale has been the introduction of next-generation sequencing technologies, reviewed in (73). Short-read sequencers manufactured by Life Technologies (SOLiD) and Illumina (Genome Analyzer and HiSeq) are capable of generating 100 to 300 GB mappable sequences per run. Several protocols for methylome sequencing (74), CHIP-seq (70), and miRNA-seq (75) are available, and publications using these new technologies are beginning to appear. Although there are no publications using these epigenetic tools to study lung disease, a few publications that are of relevance to respiratory diseases are worth mentioning. CHIP-Seq technology was recently used to map the distribution of histone methylation marks along with STAT4 and STAT6 binding, and combined with gene expression profiles to determine the entire spectrum of STAT4- and STAT6-occupied genes and to link STAT binding to corresponding gene transcription and epigenetic regulation in polarized Th1 and Th2 cells (76). Globally, STAT4 was shown to have a more dominant role in promoting active epigenetic marks, whereas STAT6 had a more prominent role in antagonizing repressive marks. In another recent study, small RNAs from 31 normal and malignant human B cell samples were sequenced using miRNA-seq to identify expression of 333 known and 286 candidate novel miRNAs in B cells (77). This same study also revealed a novel miRNA cluster containing six novel miRNAs that regulate transforming growth factor-β signaling.
CHALLENGES IN UNDERSTANDING THE LUNG EPIGENOME
There are several challenges that we will be facing in understanding the role of epigenetic regulation of gene expression in lung diseases. First, epigenetic marks are cell-type specific, yet much of the research in this area will be done on the whole lung tissue because isolation of enough material for specific cell types is often not feasible in human subjects. One approach to address this concern may be to identify epigenetic marks in the whole lung and then attribute them to specific types using immunohistochemistry or confocal microscopy with antibodies to specific epigenetic marks. The disadvantage of this approach is that many important epigenetic marks may be missed in the initial screen because the change in the whole lung may be below detection limits of assays used. This same issue arises in gene expression data, and a recent publication suggested a method to decompose whole tissue expression into cell-specific components (78); this approach may prove useful in identifying cell-specific epigenetic marks in complex tissues. Second, peripheral blood may be the only accessible sample, such as in studies involving children with asthma. To understand the relevance of these distant markers of lung disease, paired lung-blood samples should be analyzed to identify epigenetic marks that carry over from the lung to peripheral blood. If such epigenetic marks are identified, development of peripheral blood biomarkers based on epigenetic marks would represent a major advance in the evaluation of lung disease. A third challenge is the dynamic nature of epigenetic marks that are context dependent. Thus, the epigenome needs to be considered in the context of other diseases, exposures, diet, and age. Finally, a major challenge with epigenomics is to experimentally and analytically integrate the epigenetic mechanisms that affect transcription and translation. Evidence for cross-talk between DNA methylation and histone modifications has been rapidly accumulating (17–20). miRNAs most likely act independently of the other two epigenetic mechanisms; however, expression of a gene may be controlled by methylation of CpG motifs in the promoter region, changes in histone marks that coincide with DNA methylation, and miRNAs that bind to the 3′ untranslated regions. Each of the three epigenetic mechanisms is independently complex, but when combined, the complexity of these interactions presents unique experimental and analytic challenges. Sophisticated imaging approaches, such as those that image HDAC activity in vivo (79), will have to be developed to allow us to understand the lung epigenome in health and disease. Better treatments for lung disease based on epigenetic marks can only be developed once we have overcome these challenges. The NIH Roadmap Epigenomics Program (http://nihroadmap.nih.gov/epigenomics/) is a major initiative that is providing funding opportunities for better understanding of the role of epigenetics in human health and disease. Using epigenetics to understand the dynamic biology in the lung and applying this knowledge to the development of novel diagnostic and therapeutic approaches represent promising opportunities for our patients with lung disease.
Supported by the National Heart, Lung and Blood Institute grants RO1-HL101251 and RC2-HL101715 (D.A.S.) and the National Institute of Allergy and Infectious Diseases grant N01-AI90052.
Author Disclosure: I.V.Y. is employed by National Jewish Health and formerly by the National Institutes of Health. D.S. received expert witness fees from Wallace and Graham, Brayton and Purcell, Weitz and Luxemberg, and Waters and Kraus. D.S. is employed by National Jewish Health and formerly by the National Institutes of Health.
References
- 1.Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009.
- 2.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005;102:10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA 2008;299:2877–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987;196:261–282. [DOI] [PubMed] [Google Scholar]
- 5.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer 2002;101:335–341. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007;447:433–440. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143–153. [DOI] [PubMed] [Google Scholar]
- 8.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 2009;41:1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 2010;467:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature 2008;452:112–115. [DOI] [PubMed] [Google Scholar]
- 12.Chi P, Allis CD, Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 2010;10:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 2008;9:831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 15.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 2007;8:93–103. [DOI] [PubMed] [Google Scholar]
- 16.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL. The n-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci USA 2009;106:22187–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007;448:714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010;466:388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295–304. [DOI] [PubMed] [Google Scholar]
- 21.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003;23:5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 2009;179:572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med 2009;180:462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, Davis S, Zhang Y, Hussain M, Xi S, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010;29:3650–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Bowers J, Vaziri C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA 2009;106:2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J 2009;23:806–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J 2009;23:3243–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J 2006;25:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White GP, Hollams EM, Yerkovich ST, Bosco A, Holt BJ, Bassami MR, Kusel M, Sly PD, Holt PG. CpG methylation patterns in the IFNgamma promoter in naive T cells: variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol 2006;17:557–564. [DOI] [PubMed] [Google Scholar]
- 32.Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol 2002;169:1893–1903. [DOI] [PubMed] [Google Scholar]
- 33.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 2002;16:649–660. [DOI] [PubMed] [Google Scholar]
- 34.Webster RB, Rodriguez Y, Klimecki WT, Vercelli D. The human IL-13 locus in neonatal CD4+ T cells is refractory to the acquisition of a repressive chromatin architecture. J Biol Chem 2007;282:700–709. [DOI] [PubMed] [Google Scholar]
- 35.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 2006;24:369–379. [DOI] [PubMed] [Google Scholar]
- 36.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci USA 2007;104:17052–17057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol 2002;169:647–650. [DOI] [PubMed] [Google Scholar]
- 38.Huehn J, Polansky JK, Hamann A. Epigenetic control of foxp3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol 2009;9:83–89. [DOI] [PubMed] [Google Scholar]
- 39.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of foxp3 expression. Blood 2009;114:3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 2010;32:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB. Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med 2009;179:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell 2009;136:26–36. [DOI] [PubMed] [Google Scholar]
- 43.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, et al. A role for Dicer in immune regulation. J Exp Med 2006;203:2519–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol 2010;126:581–589.e1–20. [DOI] [PubMed] [Google Scholar]
- 45.Wei B, Pei G. MicroRNAs: critical regulators in Th17 cells and players in diseases. Cell Mol Immunol 2010;7:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meehan RR. DNA methylation in animal development. Semin Cell Dev Biol 2003;14:53–65. [DOI] [PubMed] [Google Scholar]
- 47.Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev 2004;14:692–696. [DOI] [PubMed] [Google Scholar]
- 48.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007;447:425–432. [DOI] [PubMed] [Google Scholar]
- 49.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med 2006;355:2226–2235. [DOI] [PubMed] [Google Scholar]
- 50.Shaheen SO, Northstone K, Newson RB, Emmett PM, Sherriff A, Henderson AJ. Dietary patterns in pregnancy and respiratory and atopic outcomes in childhood. Thorax 2009;64:411–417. [DOI] [PubMed] [Google Scholar]
- 51.Whitrow MJ, Moore VM, Rumbold AR, Davies MJ. Effect of supplemental folic acid in pregnancy on childhood asthma: a prospective birth cohort study. Am J Epidemiol 2009;170:1486–1493. [DOI] [PubMed] [Google Scholar]
- 52.Haberg SE, London SJ, Stigum H, Nafstad P, Nystad W. Folic acid supplements in pregnancy and early childhood respiratory health. Arch Dis Child 2009;94:180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, Bailey N, Potts EN, Whitehead G, Brass DM, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest 2008;118:3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Koh BH, Hwang SS, Kim JY, Lee W, Kang MJ, Lee CG, Park JW, Flavell RA, Lee GR. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc Natl Acad Sci USA 2010;107:10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henderson AJ, Newson RB, Rose-Zerilli M, Ring SM, Holloway JW, Shaheen SO. Maternal Nrf2 and gluthathione-S-transferase polymorphisms do not modify associations of prenatal tobacco smoke exposure with asthma and lung function in school-aged children. Thorax 2010;65:897–902. [DOI] [PubMed] [Google Scholar]
- 56.Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc Am Thorac Soc 2009;6:660–662. [DOI] [PubMed] [Google Scholar]
- 57.Ito K, Lim S, Caramori G, Cosio B, Chung KF, Adcock IM, Barnes PJ. A molecular mechanism of action of theophylline: induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci USA 2002;99:8921–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barnes PJ. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc 2009;6:693–696. [DOI] [PubMed] [Google Scholar]
- 59.Williams AE, Larner-Svensson H, Perry MM, Campbell GA, Herrick SE, Adcock IM, Erjefalt JS, Chung KF, Lindsay MA. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS ONE 2009;4:e5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of Th2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA 2009;106:18704–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010;182:220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. MiR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2010;207:1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabinovich E, Yakhini Z, Benos P, Pandit K, Milosevic J, Kapetanak M, Richards T, Chensny L, Kaminski N. Human CpG islands arrays reveal changes in global methylation patterns in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010;181:A2017. [Google Scholar]
- 64.Coward WR, Watts K, Feghali-Bostwick CA, Knox A, Pang L. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol Cell Biol 2009;29:4325–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coward WR, Watts K, Feghali-Bostwick CA, Jenkins G, Pang L. Repression of IP-10 by interactions between histone deacetylation and hypermethylation in idiopathic pulmonary fibrosis. Mol Cell Biol 2010;30:2874–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 2008;39:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol (In press). [DOI] [PMC free article] [PubMed]
- 68.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med 2005;352:1967–1976. [DOI] [PubMed] [Google Scholar]
- 69.Heller G, Zielinski CC, Zochbauer-Muller S. Lung cancer: from single-gene methylation to methylome profiling. Cancer Metastasis Rev 2010;29:95–107. [DOI] [PubMed] [Google Scholar]
- 70.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet 2008;9:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu CG, Spizzo R, Calin GA, Croce CM. Expression profiling of microRNA using oligo DNA arrays. Methods 2008;44:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM). Curr Protoc Hum Genet 2010;Chapter 20:Unit 20 21 21–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet 2010;11:31–46. [DOI] [PubMed] [Google Scholar]
- 74.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 2010;11:191–203. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 2010;32:840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jima DD, Zhang J, Jacobs C, Richards KL, Dunphy CH, Choi WW, Au WY, Srivastava G, Czader MB, Rizzieri DA, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood 2010;116:e118–e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nat Methods 2010;7:287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sankaranarayanapillai M, Tong WP, Yuan Q, Bankson JA, Dafni H, Bornmann WG, Soghomonyan S, Pal A, Ramirez MS, Webb D, et al. Monitoring histone deacetylase inhibition in vivo: noninvasive magnetic resonance spectroscopy method. Mol Imaging 2008;7:92–100. [PubMed] [Google Scholar]