Abstract
Rationale: Acute lung injury (ALI) acts as a complex genetic trait, yet its genetic risk factors remain incompletely understood. Large-scale genotyping has not previously been reported for ALI.
Objectives: To identify ALI risk variants after major trauma using a large-scale candidate gene approach.
Methods: We performed a two-stage genetic association study. We derived findings in an African American cohort (n = 222) using a cardiopulmonary disease–centric 50K single nucleotide polymorphism (SNP) array. Genotype and haplotype distributions were compared between subjects with ALI and without ALI, with adjustment for clinical factors. Top performing SNPs (P < 10−4) were tested in a multicenter European American trauma-associated ALI case-control population (n = 600 ALI; n = 2,266 population-based control subjects) for replication. The ALI-associated genomic region was sequenced, analyzed for in silico prediction of function, and plasma was assayed by ELISA and immunoblot.
Measurements and Main Results: Five SNPs demonstrated a significant association with ALI after adjustment for covariates in Stage I. Two SNPs in ANGPT2 (rs1868554 and rs2442598) replicated their significant association with ALI in Stage II. rs1868554 was robust to multiple comparison correction: odds ratio 1.22 (1.06–1.40), P = 0.0047. Resequencing identified predicted novel splice sites in linkage disequilibrium with rs1868554, and immunoblots showed higher proportion of variant angiopoietin-2 (ANG2) isoform associated with rs1868554T (0.81 vs. 0.48; P = 0.038).
Conclusions: An ANGPT2 region is associated with both ALI and variation in plasma angiopoietin-2 isoforms. Characterization of the variant isoform and its genetic regulation may yield important insights about ALI pathogenesis and susceptibility.
Keywords: acute lung injury, acute respiratory distress syndrome, functional genetic polymorphism, genetic association study
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Although an underlying genetic susceptibility to acute lung injury (ALI) is supported by multiple lines of evidence, few candidate genes have shown reproducible associations with ALI, and fewer have been tested in multiple ethnicities.
What This Study Adds to the Field
We performed large-scale candidate gene genotyping in a cohort of critically ill subjects with trauma, and replicated the top associations in a large case-control population. This strategy identified a region of the angiopoietin-2 gene, a gene shown to impact lung permeability in experimental models of lung injury, demonstrating a consistent association with increased risk for trauma-associated ALI in Americans of African and European descent. The intronic ANGPT2 polymorphism was also associated with increased variant angiopoietin-2 isoform in plasma, suggesting that the risk polymorphism tags a splice site enhancer or novel splice site.
Acute lung injury (ALI) and its severe form, the acute respiratory distress syndrome (ARDS), afflict an estimated 190,000 people each year in the United States and carry a mortality of over 35% (1). The two syndromes are characterized by alveolar flooding and profound hypoxemia, with Pao2:FiO2 ratio less than or equal to 200 for ARDS and less than or equal to 300 for ALI (2). The syndromes follow extreme environmental insults, such as sepsis, pneumonia, aspiration, and trauma. However, because only a minority of patients exposed to these predisposing insults manifest ALI, it has been hypothesized that individual genetic variation may contribute to a patient's susceptibility to ALI (3, 4). A number of recent studies have identified genetic variants that may confer differential risk of developing ALI (5–9) or of ALI mortality (7, 10). However, many candidate gene studies have been difficult to replicate, either because of small sample sizes, population stratification, variability of the control population, or heterogeneity of the ALI phenotype (5).
We hypothesized that we could efficiently identify single nucleotide polymorphisms (SNPs) associated with a differential risk for development of ALI by using a large-scale candidate gene-centric platform, a carefully phenotyped critically ill cohort at risk for ALI, and a two-stage genotyping strategy to validate results. To test this hypothesis, we used our most genetically diverse subjects in the discovery phase to narrow the genomic region of association, and investigated potential functional consequences using subjects' plasma. Some of the results of these studies have been previously reported in the form of abstracts (6, 7).
METHODS
Study Populations and Phenotyping
Stage I (Penn) was a single center cohort study of critically ill subjects with trauma admitted to the surgical intensive care unit (8). Details of this institutional review board–approved study have been described previously (9, 10). To be classified as ALI, subjects met all American-European Consensus Conference (AECC) definition criteria within a 24-hour period while mechanically ventilated (2). We derived our results in an African American (AA) cohort adjusted for population stratification (Stage I) and replicated in a large multicenter trauma-associated ALI case-control population of European American (EA) subjects. The discovery population, although smaller than the replication population, had the strength of being a cohort study with patients with ALI and without ALI at risk, and extensive phenotypic detail available for clinical adjustment. Using an African ancestry discovery cohort also allows finer resolution for the genomic region of association, because a higher level of genetic diversity exists among Africans than non-Africans (11, 12).
The replication population (Stage II; Trauma-associated ALI SNP Consortium [TASC]) consisted of EA ALI cases identified from ongoing institutional review board–approved populations with severe trauma at five United States centers applying AECC criteria (10, 13–20). These centers (Harvard University, University of California at San Francisco, University of Pennsylvania, University of Washington, and Vanderbilt University) initiated the TASC (7). Population-based EA control subjects were selected from ongoing cohort studies at the Center for Applied Genomics at the Children's Hospital of Philadelphia (7, 21, 22). Control subjects were largely healthy pediatric subjects.
Genotyping Strategy
For Stage I, we used a 50K SNP genotyping array (HumanCVD BeadChip; Illumina, San Diego, CA) designed to assay SNPs in approximately 2,000 candidate genes affecting cardiopulmonary phenotypes (23). This platform assesses 22 (85%) of the 26 published ALI-associated genes to date (5, 17, 24, 25). The HumanCVD platform was used for discovery given its more complete coverage of the candidate genes included (see Table E1 in the online supplement) and greater coverage of the African variability for these loci compared with genome-wide association (GWA) platforms (23). To confirm associations (26) in Stage II, the Human610-quad BeadChip (Illumina) GWA platform was used on the TASC EA replication population (7) and significant SNPs were confirmed using Taqman (Applied Biosystems, Foster City, CA) genotyping. This platform provides excellent genomic coverage (∼ 90%) of Europeans, and could be relied on to be informative for all polymorphic genomic loci identified in Stage I. DNA was extracted from ethylenediaminetetraacetic acid blood samples. Laboratory personnel were unaware of the ALI status of each sample. Further details and quality control standards are presented in the online supplement.
ANGPT2 Resequencing and In Silico Analysis
DNA from 48 subjects (24 cases and 24 control subjects, divided equally between AA and EA) was selected for sequencing of polymerase chain reaction fragments (27). Focusing on the region associated with ALI, polymerase chain reaction primers were designed using polymerase chain reaction overlap (University of Washington) to generate amplicons 600–800 bp that overlapped by at least 100 bp. Primers were optimized, and then DNA was amplified and sequenced in the forward and reverse direction using a 3730 automated sequencer (Applied Biosystems). Sequencher 4.8 (Gene Codes, Ann Arbor, MI) was used to facilitate secondary peak calls and to compare the sequence data with the NCBI reference sequence. Primer sequences are shown in Table E2. In silico splice site enhancement prediction was performed using the SNP analysis function of Human Splicing Finder 2.4.1 (French Institute of Health and Medical Research, Montpellier, France) (28), a position weight matrix–based package to predict the effect of mutations on consensus splicing signals. The effect of SNPs on consensus splice enhancers was also investigated with alternative position weight matrix splice enhancer matrices (29–32).
Plasma Protein Assessment
Plasma samples from a subset of Stage I subjects (n = 128) were available for analysis. Plasma angiopoietin-2 (ANG2) was measured by sandwich ELISA (R&D Systems, Minneapolis, MN). Six samples with the highest plasma ANG2 concentration for each rs1868554 genotype were normalized to 4 ng/ml concentration based on the ELISA results; immunoprecipitated with anti-human ANG2 antibody (R&D Systems); and then subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis under reducing conditions for immunoblotting using anti-ANG2 primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) (33). Further detail is provided in the online supplement.
Statistical Analysis
A two-stage association study was performed (34). Analyses were ancestry-specific by genetically determined ancestry (Figure E1) as described previously and in the online supplement (35, 36). Haplotypes were inferred using the standard expectation maximization algorithm in Haploview (37, 38). For each SNP or haplotype, ALI incidence was calculated according to genotype and significance of odds ratios (ORs) was determined using the chi-square test in PLINK (35). An additive genetic model was assumed. We used a P value of 10−4 to pass Stage I (39), rather than 10−6 (0.05/50,000 SNPs), given the candidate gene design of the HumanCVD chip and its dense genotyping of covered loci (39). Consensus is lacking for the appropriate significance threshold when using an array containing thousands of hypothesis-driven, densely covered loci. Previous reports using this array have used α thresholds of 1 × 10−5, 5 × 10−5, and 1 × 10−6 (36, 39, 40), at times without a replication population (39). We used a slightly more relaxed Stage I threshold (10−4) to balance the concerns of power adequacy with the potential for false-positives and considered independent replication the most reliable measure of true association (34). We used logistic regression to adjust for potential confounding by clinical factors in Stage I. Age, injury severity score, Acute Physiology and Chronic Health Evaluation III score, blunt mechanism, pulmonary contusion, era of enrollment, and volume of red blood cell transfusion were used as covariates. To investigate the potential effects of misclassification of the ALI phenotype, we performed a sensitivity analysis removing equivocal control subjects, as previously described (41).
In the replication stage, we applied Bonferroni correction for the number of SNPs carried forward for replication, such that α less than 0.0167 was the threshold for Stage II significance. Replication at the SNP level was tested using chi-square statistics assuming an additive model. Imputed genotypes were determined using MACH 1.0 software (University of Michigan, Ann Arbor, MI) (26, 27). Positive associations were subjected to alternate genotyping of all 600 cases (Taqman; Applied Biosystems). For protein investigations, groups were compared by Student t test, Wilcoxon rank-sum, or Kruskal-Wallis testing as appropriate. Please see the online supplement for additional methodologic detail and power calculations.
RESULTS
Stage I: Penn Cohort
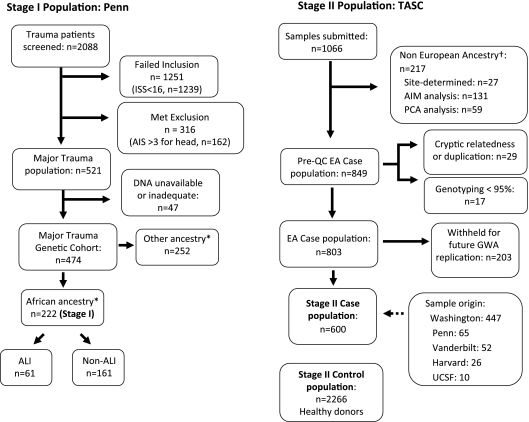
Over 2,000 subjects with trauma were screened for eligibility, of whom 521 met all eligibility criteria and 474 had adequate DNA available. Of the 474 subjects, 222 (47%) subjects were genetically identified as AA. Sixty-one (27%) AA subjects with trauma developed ALI during the first 5 days post-trauma. The study population is described in more detail in Figure 1 and Table 1.
Figure 1.
Subject populations. Stage I consisted of African American subjects with trauma at a single site. *Ancestry confirmed by multidimensional scaling with PLINK (35) using all markers of the HumanCVD BeadChip (36). ISS = injury severity scale; AIS = abbreviated injury severity scale; ALI = acute lung injury. Stage II included European American (EA) trauma-associated ALI cases identified from ongoing ALI cohorts (10, 13–17, 67–69) submitted to Trauma-associated ALI SNP Consortium (TASC). †Non-European subjects were determined by clustering analysis with HapMap samples (AIM analysis) or by a separate analysis by Principle Components Analysis (PCA analysis). TASC sites are abbreviated as follows: Washington (University of Washington); Penn (University of Pennsylvania); Vanderbilt (Vanderbilt University); Harvard (Harvard University/Massachusetts General Hospital); UCSF (University of California, San Francisco).
TABLE 1.
DEMOGRAPHIC AND CLINICAL DATA: STAGE I
| Variable | ALI (n = 61) | Non-ALI (n = 161) | P Value |
|---|---|---|---|
| Age, yr | 37 ± 16 | 32 ± 14 | 0.02 |
| Male, n (%) | 57 (93) | 130 (82) | 0.04 |
| Era of injury 1999–2003, n (%) | 23 (38) | 71 (45) | 0.33 |
| Injury factors | |||
| Blunt trauma, n (%) | 29 (47) | 65 (41) | 0.39 |
| ISS | 25 ± 9 | 22 ± 6 | 0.12 |
| APACHE III* | 64 ± 24 | 56 ± 15 | 0.05 |
| Pulmonary contusion, n (%) | 17 (18) | 28 (18) | 0.10 |
| Treatment factors | |||
| Total pRBC first 24 h, units | 3.95 ± 6.3 | 2.17 ± 3.7 | 0.13 |
| Mechanical ventilation, n (%) | 61 (100) | 124 (77) | <0.001 |
| Outcomes | |||
| Mortality, n (%) | 15 (25) | 8 (5) | <0.001 |
| Hospital length of stay, d | 35.1 ± 49 | 20.9 ± 22 | 0.002 |
Definition of abbreviations: ALI = acute lung injury; APACHE = Acute Physiology and Chronic Health Evaluation; ISS = injury severity score; pRBC = packed red blood cells.
Data are expressed as mean ± SD, unless otherwise specified. Percents are numerators divided by the total number of subjects with ALI or without ALI. The reported P value results from unpaired t test, chi-square, or Wilcoxon rank-sum test as appropriate.
APACHE III score modified to exclude ABG data given collinearity with ALI definition.
SNP associations with ALI.
Genotyping with the HumanCVD BeadChip yielded successful typing of 44,621 polymorphic SNPs (genotyping calls >95%, chi-square test of Hardy-Weinberg equilibrium >10−4 [P value], and minor allele frequency >0.01) in individuals of African ancestry (Table E3). Table 2 displays the polymorphisms in Stage I meeting the threshold P value (≤ 10−4) for association with ALI. The two SNPs demonstrating the strongest association with ALI were in the same gene, ANGPT2: rs2442598 and rs1868554 (P <5 × 10−5). These two ANGPT2 SNPs display marginal linkage disequilibrium (LD) with each other in the HapMap Yoruban population (r2 0.26; D' 0.72) (42). In the Penn cohort, the SNPs displayed modest LD (r2 0.42) and were relatively common, each with minor allele frequency greater than 0.30. Both variants are intronic (42). Of the remaining three variants passing the significance threshold, two (in STAT and GP5) are never observed in the EA population (43). These SNPs, italicized in Table 2, were not amenable to replication in the EA TASC population. Therefore, three SNPs in two genes from Stage I were tested for replication in Stage II.
TABLE 2.
SNPS MOST ASSOCIATED WITH ALI IN STAGE I
| Locus (chromosome: base pair) | Case genotypes (MAF) | Noncase genotypes (MAF) | HapMap MAF† |
Odds Ratio (95% CI) | P Value (additive) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Function | HWE p | YRI | ASW | |||||
| rs2442598 | ANGPT2 | 8:6394821 | Intron | 15/21/24 (0.43) | 4/58/97 (0.21) | 0.30 | 0.22 | 0.28 | 2.73 (1.71–4.35) | 2.52E-05 |
| rs1868554 | ANGPT2 | 8:6386747 | Intron | 21/19/20 (0.51) | 8/74/77 (0.28) | 0.37 | 0.40 | — | 2.60 (1.66–4.09) | 3.34E-05 |
| rs6734110 | STAT1 | 2:191838342 | Intron | 0/15/45 (0.13) | 0/7/152 (0.02) | 1 | 0.06 | 0.04 | 7.29 (2.79–19) | 4.92E-05 |
| rs2185479 | GP5 | 3:194119180 | Intron | 0/16/44 (0.13) | 0/9/150 (0.03) | 1 | 0.07 | 0.11 | 6.17 (2.54–15) | 6.01E-05 |
| rs6892794 | PPARGC1 | 5:149124131 | Intron | 23/31/6 (0.64) | 31/70/58 (0.42) | 0.29 | 0.49 | 0.45 | 2.45 (1.57–3.83) | 7.77E-05 |
Definition of abbreviations: ALI = acute lung injury; CI = confidence interval; HWE p = chi-square statistic testing Hardy Weinberg Equilibrium proportions in the trauma cohort; MAF = minor allele frequency; SNP = single nucleotide polymorphism.
The five SNPs associated with ALI at P ≤ 10−4 are displayed by additive model P value. Locus is listed as chromosome:base pair position according to dbSNP build 131. Genotype columns display genotype frequencies stratified by ALI status, categorized as homozygous for the minor allele, heterozygous, or homozygous for the major allele and displayed in that order. Stage I MAF is shown in parentheses. Rows with italic type indicate SNPs with MAF = 0 in the CEU population, Utah residents with Northern and Western European ancestry. Because of their rarity, italicized SNPs were not amenable to replication in the Stage II European American population.
The HapMap MAFs for YRI, a Yoruban population, and ASW, an African American population from the southwestern United States (43), are shown to indicate expected frequency for each SNP. Frequencies for ASW are only available for the ∼ 1.5 million SNPs genotyped as part of HapMap III; rs1868554 was not a typed marker in this phase.
Haplotype associations with ALI.
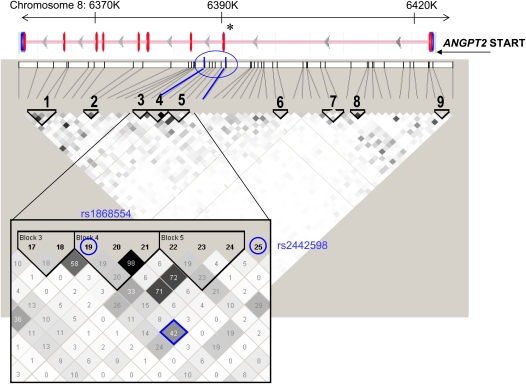
With two SNPs in ANGPT2 demonstrating significant association with ALI, we investigated whether ANGPT2 haplotypes, or combinations of alleles at different loci, also manifested ALI association. We constructed nine haplotype blocks assuming a minimum haplotype frequency greater than 1% (37, 38). Block 4 TCA, in which the first allele is rs1868554T, showed the strongest association with ALI (OR 2.43; P = 3.2 × 10−5; permutation P value 0.0013) (Tables E4 and E5). Considerable linkage was demonstrated between blocks 3, 4, and 5. As shown in the geneview in Figure 2, these LD blocks span the end of the first intron and into the second intron of ANGPT2. A total of four SNPs, with r2 ranging from 0.33 to 0.58 with rs1868554, in ANGPT2 associated with ALI at P less than 0.005 (Figure E2).
Figure 2.
ANGPT2 haplotype structure in Stage I. The linkage disequilibrium (LD) plot of Stage I samples is shown beneath a geneview of ANGPT2. In the geneview, exons are depicted as vertical red lines. The 62 genotyped single nucleotide polymorphisms (SNPs) annotated to ANGPT2 are represented as black vertical lines and arrayed by position along chromosome 8, with tangential lines connecting each SNP to its haplotype block. The LD plot represents blocks of high LD as outlined by triangles and numbered. The LD block coloring reflects pairwise LD (r2) between any two SNPs, with white representing no LD, gray indicating moderate LD (0.70 > r2 > 0.10), and black representing high LD (r2 > 0.70). The r2 value is displayed as a percentage for the magnified region (inset). Blocks 3, 4, and 5 (inset) were each associated with acute lung injury. These blocks span the gene's first and second intron, including the alternatively spliced exon 2 (asterisk). The two SNPs individually identified as acute lung injury–associated (rs1868554, marker 19; rs2442598, marker 25) in Stage I are highlighted in blue, as is the LD between them (blue diamond). SNP rs1868554T defines the TCA haplotype of block 4, whereas rs2442598 is not a haplotype member. The LD plot was generated using Haploview (38).
Clinical variable adjustment.
SNPs passing the Stage I threshold were tested for confounding with known clinical risk factors. Clinical factors previously shown to associate with trauma-associated ALI in this population (9, 44) were tested individually for confounding on the association between genotype and ALI, and then combined in a logistic regression model. The results are shown in Table 3. All five SNPs passing the Stage I threshold (rs1868554T allele, rs2442598A, rs6734110G, rs2185479A, and rs6892794G) remained independently associated with the development of ALI after multivariate adjustment. We performed a sensitivity analysis by excluding all subjects with equivocal classification and comparing only ALI cases with definite non-cases, as we have previously described (41). Despite excluding approximately 40% of equivocal non-cases, the analysis for both ANGPT2 SNPs remained significant (Table E6).
TABLE 3.
ADJUSTMENT FOR CLINICAL CONFOUNDERS
| rs1868554T (ANGPT2) | rs2442598A (ANGPT2) | rs6734110C (STAT1) | rs2185479T (GP5) | rs6892794G (PPARGC1B) | |
|---|---|---|---|---|---|
| Unadjusted odds ratio (95% CI) | 2.30 (1.49–3.55) | 2.62 (1.65–4.17) | 7.29 (2.79–19) | 6.17 (2.54–15) | 2.30 (1.49–3.56) |
| Multivariate model odds ratio (95% CI) | 2.39 (1.49–3.84) | 2.47 (1.51–4.08) | 7.34 (2.65–20.3) | 5.26 (2.03–13.6) | 2.34 (1.47–3.72) |
The raw and adjusted odds ratios with 95% confidence intervals (CI) are shown assuming an additive model of each single nucleotide polymorphism with the development of acute lung injury (ALI). Each single nucleotide polymorphism from Stage I remained significantly associated with ALI after clinical adjustment (P < 0.001). The multivariable logistic regression model included the potential confounders age; blunt mechanism of trauma (vs. penetrating); presence of pulmonary contusion; injury severity score; modified Acute Physiology and Chronic Health Evaluation III score; volume of packed red blood cell resuscitation during the first 24 hours; and era of enrollment (2005–2007 compared with 1999–2002). The modified Acute Physiology and Chronic Health Evaluation III score omitted Pao2 because hypoxemia was collinear with the diagnosis of ALI.
Stage II: TASC
As of January 2009, a total of 1,066 samples had been submitted by five TASC centers and 803 (73%) passed quality control parameters (Figure 1) (35). Summary characteristics of the case and control populations are presented in Table 4. We genotyped 600 EA ALI cases and 2,266 EA population-based control subjects and filtered the results for the three SNPs passing the significance threshold in Stage I that were not private to African ancestry. Two of the SNPs (rs2442598 in ANGPT2 and rs6892794 in PPARGC1) were directly genotyped on the GWA array, whereas rs1868554 was imputed based on complete LD with a typed marker (45, 46). The overall genomic inflation factor for the imputed dataset was 1.0182.
TABLE 4.
DEMOGRAPHIC AND CLINICAL DATA: STAGE II
| Variable | ALI Cases (n = 600) | Control Subjects (n = 2,266) | P Value |
|---|---|---|---|
| Age, yr | 45 ± 20 | 8 ± 6 | <0.001 |
| Male, n (%) | 413 (70) | 1,287 (57) | <0.001 |
| ISS | 27 ± 10 | NA | |
| Blunt trauma, n (%) missing for 14% | 472 (92) | NA | |
| Site, n (%) | |||
| Harvard | 26 (4) | ||
| Penn | 65 (11) | ||
| UCSF | 10 (2) | ||
| Uwash | 447 (74) | ||
| Vanderbilt | 52 (9) | ||
| CHOP | 2,266 |
Definition of abbreviations: ALI = acute lung injury; CHOP = Children's Hospital of Philadelphia; Harvard = Harvard University Health Systems; ISS = injury severity score; Penn = University of Pennsylvania; UCSF = University of California, San Francisco; UWash = University of Washington; Vanderbilt = Vanderbilt University.
Data are expressed as mean ± SD, unless otherwise specified. Percents are numerators divided by the total number of subjects with ALI or control subjects. The reported P value results from chi-square test (sex) or Wilcoxon rank-sum test (age). Clinical variables pertaining to trauma (ISS, blunt mechanism of injury, site of trauma center) were only available for the case population. Site refers to location from which subject was enrolled.
Replication results are shown in Table 5. Both ANGPT2 SNPs replicated at P less than 0.05 (additive), although only one of these (rs1868554) passed P less than 0.0167, the significance threshold accounting for multiple comparisons. In contrast, the association with PPARGC1B SNP rs6892794 did not replicate. Results for rs1868554 were stratified by TASC site and revealed a similar association and direction of effect, although with small numbers; not all sites were statistically significant (Table E7). To confirm genotyping of positive associations, all 600 cases were typed for rs1868554 by Taqman. Because the correlation (r2) between imputed call and Taqman genotype was 0.998, the imputed result was accepted for the control population. To confirm rs2442598 genotyping, 60 of the 600 samples (10%) underwent SNPlex genotyping with r2 = 1.0.
TABLE 5.
REPLICATION RESULTS OF THE TOP ALI-ASSOCIATED SNPS (STAGE II)
| SNP | Gene | Human 610-quad | Confirmatory Genotyping* | Case MAF (n = 600) | Control MAF (n = 2,027) | Odds Ratio (95% CI) | P Value (additive) |
|---|---|---|---|---|---|---|---|
| rs2442598 | ANGPT2 | Genotyped | SNPlex (10% cases) r2 = 1.0 | 0.31 | 0.27 | 1.16 (1.01–1.33) | 0.038 |
| rs1868554 | ANGPT2 | Imputed† | Taqman (100% cases) r2 = 0.998 | 0.33 | 0.28 | 1.22 (1.06–1.40) | 0.0047‡ |
| rs6892794 | PPARGC1 | Genotyped | N/A | 0.26 | 0.25 | 1.07 (0.77–1.96) | 0.38 |
Definition of abbreviations: ALI = acute lung injury; MAF = minor allele frequency; SNP = single nucleotide polymorphism.
For each SNP passing the Stage I significance threshold (P ≤ 10−4) with MAF greater than or equal to 0.05 in European Americans, the association with ALI is shown. An additive model is assumed.
Confirmatory genotyping of the Human610-quad results were performed on a portion (%) of the Stage II population by SNPlex or Taqman methodology; the subject-level correlation (r2) of genotyping calls is reported.
The ANGPT2 SNP rs1868554 was imputed based on its perfect linkage disequilibrium (r2 1.0; D' 1.0) with the genotyped marker rs734701.
P value less than 0.05/3 = 0.0167.
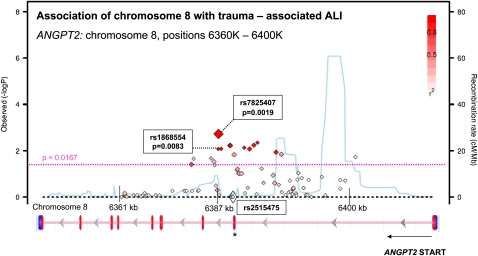
A Manhattan plot of the association for this region of ANGPT2 graphed as chromosomal position versus negative log of the additive model P value is shown in Figure 3 (42). It reveals a peak in association very close to rs1868554 but centered on a SNP 33 bases away, rs7825407, with OR 1.22 (95% confidence interval [CI], 1.06–1.40) and P = 0.0047; the red color indicates the strong degree of LD (r2) between this SNP and rs1868554. The plot also displays (in blue) the HapMap-calculated background recombination rate for this region of chromosome 8, which demonstrates three hotspots of recombination flanking the ALI-associated region.
Figure 3.
Regional association plot of ANGPT2 region with acute lung injury (ALI). Results from the Trauma-associated ALI SNP Consortium (TASC) replication population (Stage II) are plotted as genomic locus versus −log (P value) for the association with trauma-associated ALI. Markers near the y-intercept demonstrate no association with ALI, whereas those above the dashed pink line were significantly associated with ALI (P < 0.017). The P value reflects an additive genetic model for association with ALI. Each locus is also annotated with the background genome recombination rate (blue line), where a spike reflects a population recombination hotspot based on HapMap data (43). Red coloration indicates strong LD (r2 > 0.8) with the most ALI-associated marker (rs7824507). Significant LD can be observed between the single nucleotide polymorphisms crossing the threshold for Stage II significance (dashed pink line). Underneath the regional association plot is a schematic of the ANGPT2 gene with exons represented as vertical red lines. The region of ANGPT2 demonstrating ALI association spans the first and second intron of ANGPT2, the same region as in Stage I. Markers rs1868554 (Stage I) and rs2515475 (previously associated with sepsis-associated ALI [17]) are highlighted (13). There is no LD between these markers, suggesting that their associations with ALI may reflect independent mechanisms. Plot was generated using the single nucleotide polymorphisms Annotation and Proxy Search (SNAP) tool based on phased haplotypes from HapMap release 22 (hg18) in the CEU population, a Utah population of European ancestry (27, 28).
Sequencing and in silico analysis.
Eight individuals of each race proved difficult to sequence, with more than 50% missing calls at reference SNPs. Thus, our effective sequencing sample size was 16 per race. Sequencing of the region 5 kb upstream and downstream from the ANGPT2 second exon in 32 individuals revealed 87 novel polymorphisms of which 5 were single base deletions. No coding polymorphisms or large copy number variations were identified. Table E8 summarizes the variation found by sequencing. We used Human Splicing Finder to predict splice site variation based on polymorphisms for the region 1,000 bp upstream and downstream of exon 2. Nineteen SNPs in this region were returned for analysis (Table E9), of which 13 were observed in our sequencing. We determined the LD between sequenced polymorphisms and rs1868554 in our individuals using Haploview (38). Results from the in silico analysis are shown in Table E9. Human Splicing Finder predicted the creation or disruption of a splice site for eight SNPs and the splice regulatory machinery matrices predicted many novel or disrupted enhancer elements. Of those with a predicted Human Splicing Finder potential splice site prediction, four SNPs were not observed in our sequencing. Three of the remaining SNPs (rs2515478, rs1031303, and rs17077419) demonstrated significant LD with rs1868554 in individuals of European, African, or both ancestries (Table E9).
Functional assessment.
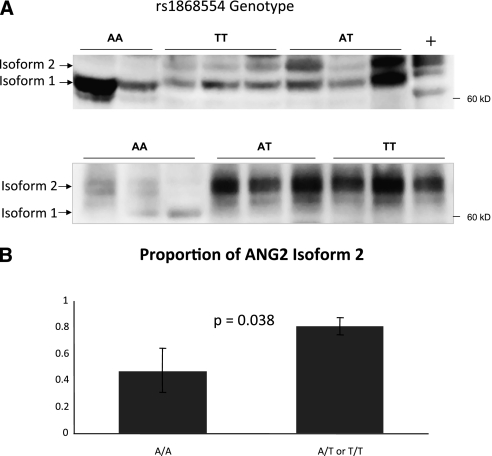
Because ANG2 is a circulating plasma protein known to be elevated in ALI, we measured the ANG2 level of available subject plasma, and confirmed that ANG2 level was significantly higher in ALI cases (P = 0.0041) (Figure E3A). However, total plasma ANG2 did not vary significantly by rs1868554 genotype or by the presence of the rs1868554T allele (Figure E3B). Given the predicted potential alteration of splice sites by our in silico analysis, we performed immunoprecipitation followed by Western blot of ANG2 using subject plasma. Six samples of each genotype were analyzed. A representative blot is shown in Figure 4A, demonstrating an increase in the proportion of a second isoform band (hereafter called “isoform 2”) for carriers of the rs1868554T allele. In some A/A blots, this band was not visualized at all. The proportion of isoform 2 was significantly higher in A/T or T/T individuals (0.81) compared with A/A individuals (0.48, P = 0.038) (Figure 4B). There was no apparent difference in isoforms between A/T and T/T subjects.
Figure 4.
Plasma angiopoietin-2 (ANG2) immunoblotting reveals distinct isoforms. (A) Two representative immunoblots demonstrating markedly different isoform banding patterns in rs1868554 AA subjects relative to AT or TT plasma, with significantly more isoform 2 present in subjects carrying the T allele. A human lung microvascular endothelial cell line, unknown genotype, serves as the positive control (+). All samples were normalized to 4 ng/ml ANG2 before immunoprecipitation. (B) The mean proportion of isoform 2 (± SEM) is shown by rs1868554 genotype, grouping A/T and T/T together given their similar appearance on gels. n = 6 for A/A, A/T, and T/T. The proportion of isoform 2 was significantly higher in the plasma of A/T and T/T subjects (0.48 ± 0.17 vs. 0.81 ± 0.06; P = 0.038).
DISCUSSION
We have identified a genetic variant and region of the ANGPT2 gene demonstrating a consistent association with increased risk of trauma-associated ALI in two separate populations, two ethnicities, and across multiple genotyping platforms. Sequencing of the ALI-associated region revealed no untyped coding variants, and in silico modeling predicted splice site alteration for SNPs in LD with rs1868554. Immunoblotting of ALI subject plasma revealed an alteration in the isoform pattern of ANG2 in carriers of the rs1868554T allele. Thus, we have identified a shift in ANG2 isoform ratio in the plasma of rs1868554T carriers with ALI.
ANG2 protein was first described in 1997 as a naturally occurring antagonist for ANG1, an angiogenic factor essential for normal vascular development (47). In the absence of angiogenic stimuli, ANG2 induces endothelial cell apoptosis and vascular regression, enhances vascular leak, and destabilizes blood vessels (48). In recent years, ANG2 has been implicated in pulmonary vascular leak syndromes including ALI and sepsis in both animal and human studies (49–53). ANG2-rich serum from patients with sepsis disrupts endothelial architecture when applied exogenously (52), and elevated levels of ANG2 have been detected in the blood and bronchoalveolar lavage fluid of patients with ALI (49, 52). Among patients with trauma, plasma ANG2 was among the top performing biomarkers distinguishing patients who did from those who did not develop ALI (20). Other vascular permeability regulating genes, such as MYLK, PBEF1, and VEGFA, have also shown association with ALI, supporting the critical role of endothelial barrier regulation in the pathophysiology of ALI (54–57).
We identified two ANGPT2 SNPs (rs2442598 and rs1868554) strongly associated with the development of ALI in patients with major trauma. The area demonstrating association with ALI was consistent both on haplotype (Stage I) and regional association (Stages I and II) analysis (Figures 2, 3, and E2). This genomic region spans the first to the second intron of ANGPT2 and includes an exon that is variably spliced (58), termed Ang2443 or isoform C (NCBI ref NP_001112360.1). The alternatively spliced isoform, which lacks the second exon and alters the coiled-coil but not the signal sequence or fibrinogen-like domain, is expressed in primary endothelial cell lines at approximately 10% the abundance of isoform A (58). The ratio of ANG2 isoforms in peripheral blood has not previously been published.
To test the in silico prediction that individual SNPs might cause splice variation, we performed ELISA of plasma ANG2 followed by immunoprecipitation and Western blotting. In our samples, plasma ANG2 did not vary predictably by rs1868554 genotype, with a very wide range in values for each genotype. However, we found that carriers of the rs1868554T allele had a shift in the isoform ratio for ANG2. We do not yet know if isoform 1 or 2 described here are ANG2 isoforms C and A, because we did not have subjects' RNA (endothelial or circulating) available to analyze the coding sequence. Nor is it known if the proprietary antibody for the ELISA used to quantitate plasma ANG2 (R&D Systems catalog DANG20) discriminates between isoforms. Isoform C was first described in human endothelial cell lines (58), and we know very little about whether or how circulating ANG2 might differ from intracellular ANG2. We observed a slightly higher migration of ANG2 bands in plasma relative to endothelial cell lysate. This might represent a post-translational modification or some other variation between endothelial and circulating protein. In the future it will be important to analyze the messenger RNA associated with these isoforms and compare them with the reference sequences for ANG2 isoforms A and C, and to characterize circulating versus cellular protein. If isoforms lacking exon 2 enhance vascular permeability relative to the reference protein, the role of ANG2 coiled–coil domain in vascular permeability regulation may warrant reexamination.
We did not observe a difference in isoform ratio between heterozygous and homozygous carriers of rs1868554T, suggesting that there are additional factors beyond genotype regulating the splicing of this protein. Although we used an additive model based on its superior power relative to a dominant model, rs1868554 was associated with ALI in each stage assuming a dominant model for the T allele, although the association was less pronounced (OR, 1.88 and 95% CI, 1.01–3.49 in Stage I; OR, 1.26 and 95% CI, 1.05–1.51 in Stage II).
We focused our bioinformatic investigation on splice site regulation given the proximity of our association signal to the variably spliced exon and because transcriptional regulation seemed less likely given the distance (35 kb) of our signal from the transcription start site. SNP analysis of variants in LD with rs1868554 predicted the creation of a novel splice site for rs2515478, a variant with strong LD to rs1868554 in EA and weak LD in AA subjects, and for rs1301303, a variant showing complete LD (r2 = 1.0) with rs1868554 in EA subjects. It may be that one of these SNPs, or one or more of the SNPs we did not uncover during sequencing, is the functional polymorphism. Alternatively, there may yet be a more obvious splice disruption signal within exon 2 that our sequencing could not adequately capture. We found no coding variation in this exon, despite HapMap data reporting one synonymous SNP with relatively high frequency (rs6559167).
Ours represents the second report of ANGPT2 genetic variants associated with the development of ALI. Previously, Su and coworkers (17) reported that two ANGPT2 tagging SNPs and their haplotype block were significantly associated with the development of ARDS in a primarily septic population (haplotype OR, 1.42; 95% CI, 1.09–1.85). Our study may be complimentary in that our main finding, rs1868554, resides in the same haplotype block with rs2515475, the strongest SNP reported by Su and coworkers (17). However, despite residing in the same block, the pairwise LD between rs1868554 and rs2515475 is marginal in the African (r2 0.40) and absent in the European (r2 0.08) ancestral population (42). Stage II did not replicate an association between ALI and rs2515475 (OR, 1.0; P = 0.97), although there was a strong association between ALI and SNPs in close proximity to rs2515475. Possible explanations for this discrepancy include the presence of recombination hotspots within the ANGPT2 gene close to our region of association, which could account for neighboring loci failing to display tight LD and failing to demonstrate a consistent association with the ALI phenotype, or that the observed ALI associations for rs1868554 and rs2515475 are completely independent.
The OR for the ANGPT2 variants in Stage I were further from the null than in Stage II, which may reflect an example of the “winner's curse” phenomenon (59), or it may reflect differences caused by the use of healthy control subjects in Stage II, or different clinical or demographic factors between the populations. Most Stage I subjects (53%) experienced penetrating trauma, whereas Stage II cases were more likely to have experienced blunt trauma (92%). It is also possible that despite a similar minor allele frequency (∼ 30%) in both African and European ancestral populations (30), rs1868554 may be more closely linked to the functional variant in subjects of African ethnicity. Our sequencing data revealed expected differences between EA and AA subjects but did not highlight a novel variant more associated with either ancestry.
The major strengths of our approach include the discovery cohort study design, multiethnic investigation with adjustment for population stratification, large replication population, and the association with an alteration in plasma protein (34). The use of a replication stage and functional correlation minimized the risk of false-positive associations. We demonstrated a consistent association with ALI in a large distinct population recruited from five centers across the United States. Although there was some variability by site, the association of rs1868554 showed a consistent direction of effect across all centers (Table E7).
This study has several limitations. The Stage I sample size is relatively small, limiting our power to detect all but the strongest effects (relative risk ≥ 1.8) in the discovery phase. Negative findings from Stage I should be interpreted with caution because this study was not designed to evaluate more modest effect sizes or rarer variants. False-negative gene associations in Stage I may have occurred because of lack of Stage I power. In addition, this study by design could only assess genetic risk factors shared by both African and EAs. Because no appropriate replication population was available for polymorphisms restricted to individuals of African ancestry, we cannot determine whether the Stage I associations observed in STAT1 or GP5 represent false-positives or true associations. Although the rarity of these SNPs may limit their clinical significance, there is evidence suggesting that STAT1 (60, 61) in particular may play a role in lung injury pathogenesis. Furthermore, scientists involved in the design of the HumanCVD chip have suggested that in the AA population, a P value of 1.9 × 10−6 might be considered “chip-wide significance,” analogous to P less than 5 × 10−8 in genome-wide studies (62). Our ANGPT2 Stage I results would not have met this threshold. However, independent replication of the association, coupled with an association with altered plasma isoform ratios, provide good evidence for its legitimacy as a risk variant.
Although using a trauma-specific cohort diminishes heterogeneity caused by different precipitating factors of ALI, the generalizability of our findings to other at-risk populations may be limited. Our sequencing did not definitively identify the causal variant leading to this splice variation, but suggests that splice enhancer variation or novel splice site creation is possible for SNPs in this region. Given the observed recombination hotspots close to the ALI-associated region of ANGPT2, future studies may seek to perform high-density genotyping of this region in subjects with sepsis- or pneumonia-associated ALI.
Our phenotype was based on the AECC definition of ALI, and this definition may be problematic (63, 64). We performed a sensitivity analysis in our most densely phenotyped population (Stage I) to test the extent to which phenotypic misspecification might influence our findings, and found the associations between ANGPT2 variants and ALI were robust to a more stringent control definition, despite lower sample size.
Subjects comprising the control group for Stage II were not critically ill patients with trauma, and they were predominantly children. Despite the theoretical risk for selection bias when using population-based control subjects, accumulating genome-wide data support the use of population-based control subjects provided that the phenotype of interest is rare in the general population (65). We used the Stage II population for replication only, choosing to limit our focus to SNPs already manifesting association with ALI in a critically ill cohort to minimize the risk of significant confounding between gene variants and severe trauma. The use of pediatric control subjects might introduce a survival bias when studying an outcome in adults, although previous genome-wide studies have not identified significant bias associated with birth cohort populations (66), and the control group used in this study has performed well in other genome-wide analyses of adult traits (22). Furthermore, if significant misclassification were to occur because of control subjects never being exposed to an ALI-precipitating event, one might expect a weakening of power to refute the null hypothesis. This should be considered in our inability to replicate the PPARGC1B SNP rs6892794.
By design, our associations were limited to those genes and SNPs assayed in Stage I by the HumanCVD BeadChip. This platform, designed for cardiovascular, metabolic, and pulmonary conditions, provided coverage for 85% of previous ALI-associated genes (Table E1), suggesting it is a reasonable candidate gene platform for the ALI phenotype. Because of the chip's design, there may be important genetic variation, such as copy number variation or structural variation, which we did not detect. Our sequencing makes copy number variation unlikely as a cause for the ANGPT2 association, but other candidate genes may be significantly influenced by structural variation. An alternative strategy is to use the whole genome analysis in the first stage, which could potentially highlight novel loci or other candidates of interest. The rationale for our chosen genotyping strategy was to have dense genotyping in AAs to use the ethnically diverse Penn trauma cohort to its fullest extent, and to have an adequately powered replication population.
ALI remains a significant source of morbidity and mortality in patients experiencing major trauma. The development of ALI in critically ill patients with trauma is associated with an almost threefold increased risk of mortality compared with those who do not develop ALI (9). A molecular model of ALI susceptibility may aid in the development of specific, targeted therapy for high-risk individuals. Further characterization of ANGPT2 genetic variation and expression, and further mechanistic investigation into the effects of ANG2 isoform variation, may lead to novel therapeutic paradigms in trauma-associated ALI.
Supplementary Material
Supported by National Institutes of Health grants HL081619, HL079063, HL090833, GM085689, HL081332, HL060710, HL090021, HL102254, and GM066946.
Conception and design: N.M., J.C., M.L., B.F., P.L., S.A., R.A., M.B., C.C., M.C., J.F.P., D.C., G.O.K., L.W., A.M., M.W., and H.H. Analysis and interpretation: N.M., J.C., M.L., R.F., J.B., R.G., S.B., M.R., E.A., E.A.V., M.B., and H.H. Drafting the manuscript for important intellectual content: N.M., J.C., M.L., R.F., R.A., M.B., C.C., D.C., G.O.K., L.W., M.W., and H.H.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201005-0701OC on January 21, 2011
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med 2005;353:1685–1693. [DOI] [PubMed] [Google Scholar]
- 2.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818–824. [DOI] [PubMed] [Google Scholar]
- 3.Gong MN. Genetic epidemiology of acute respiratory distress syndrome: implications for future prevention and treatment [abstract]. Clin Chest Med 2006;27:705–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes KC. Genetic determinants and ethnic disparities in sepsis-associated acute lung injury. Proc Am Thorac Soc 2005;2:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao L, Barnes KC. Recent advances in genetic predisposition to clinical acute lung injury. Am J Physiol Lung Cell Mol Physiol 2009;296:L713–L725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer NJ, Li M, Shah CV, Gallop R, Localio AR, Bellamy S, Fuchs B, Lanken PN, Christie JD. Large scale genotyping in an African American trauma population identifies angiopoietin-2 variants associated with ALI. Am J Respir Crit Care Med 2009;179:A3879. [Google Scholar]
- 7.Christie JD, Wurfel MM, O'Keefe GE, Christiani D, Calfee CS, Meyer NJ, Bradfield J, Kim C, Li M, Feng R, et al. Genome wide association (GWA) identifies functional susceptibility loci for acute lung injury [abstract]. Am J Respir Crit Care Med 2010;181:A1025. [Google Scholar]
- 8.Civil ID, Schwab CW. The abbreviated injury scale, 1985 revision: a condensed chart for clinical use. J Trauma 1988;28:87–90. [DOI] [PubMed] [Google Scholar]
- 9.Shah CV, Localio AR, Lanken PN, Kahn JM, Bellamy S, Gallop R, Finkel B, Gracias VH, Fuchs BD, Christie JD. The impact of development of acute lung injury on hospital mortality in critically ill trauma patients. Crit Care Med 2008;36:2309–2315. [DOI] [PubMed] [Google Scholar]
- 10.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JG. Variation in the mylk gene is associated with development of acute lung injury after major trauma. Crit Care Med 2008;36:2794–2800. [DOI] [PubMed] [Google Scholar]
- 11.Campbell MC, Tishkoff SA. The evolution of human genetic and phenotypic variation in Africa. Curr Biol 2010;20:R166–R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McElroy JP, Cree BAC, Caillier SJ, Gregersen PK, Herbert J, Khan OA, Freudenberg J, Lee A, Bridges SL, Hauser SL, et al. Refining the association of MHC with multiple sclerosis in African Americans. Hum Mol Genet 2010;19:3080–3088;36:2794-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, et al. Functional polymorphisms in the transcription factor nrf2 in humans increase the risk of acute lung injury. FASEB J 2007;21:2237–2246. [DOI] [PubMed] [Google Scholar]
- 14.Reddy AJ, Christie JD, Aplenc R, Fuchs B, Lanken PN, Kleeberger SR. Association of human nad(p)h:Quinone oxidoreductase 1 (nqo1) polymorphism with development of acute lung injury. J Cell Mol Med 2009;13:1784–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajwa EK, Yu CJ, Gong MN, Thompson BT, Christiani DC. Pbef gene polymorphisms influence the risk of developing ARDS. Proc Am Thorac Soc 2006;3:A272. [Google Scholar]
- 16.Sheu CC, Zhai R, Su L, Tejera P, Gong MN, Thompson BT, Chen F, Christiani DC. Sex-specific association of epidermal growth factor gene polymorphisms with acute respiratory distress syndrome. Eur Respir J 2009;33:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su L, Zhai R, Sheu CC, Gallagher DC, Gong MN, Tejera P, Thompson BT, Christiani DC. Genetic variants in the angiopoietin-2 gene are associated with increased risk of ARDS. Intensive Care Med 2009;35:1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalhub S, Junker CE, Imahara SD, Mindrinos MN, Dissanaike S, O'Keefe GE. Variation in the tlr4 gene influences the risk of organ failure and shock posttrauma: a cohort study. J Trauma Inj Infect Crit Care 2009;66:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen MJ, Brohi K, Calfee CS, Rahn P, Chesebro BB, Christiaans SC, Carles M, Howard M, Pittet JF. Early release of high mobility group box nuclear protein 1 after severe trauma in humans: role of injury severity and tissue hypoperfusion. Crit Care 2009;13:R174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fremont RD, Koyama T, Calfee CS, Wu W, Dossett LA, Bossert FR, Mitchell D, Wickersham N, Bernard GR, Matthay MA, et al. Acute lung injury in patients with traumatic injuries: utility of a panel of biomarkers for diagnosis and pathogenesis. J Trauma 2010;68:1121–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, et al. A genome-wide association study identifies kiaa0350 as a type 1 diabetes gene. Nature 2007;448:591–594. [DOI] [PubMed] [Google Scholar]
- 22.Van Deerlin VM, Sleiman PMA, Martinez-Lage M, Chen-Plotkin A, Wang L-S, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with tdp-43 inclusions. Nat Genet 2010;42:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE 2008;3:e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores C, Pino-Yanes Mdel M, Villar J. A quality assessment of genetic association studies supporting susceptibility and outcome in acute lung injury. Crit Care 2008;12:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glavan BJ, Holden TD, Goss CH, Black RA, Neff MJ, Nathens AB, Martin TR, Wurfel MM, ARDSnet Investigators. Genetic variation in the fas gene and associations with acute lung injury. Am J Respir Crit Care Med 2011;83:356–363. [DOI] [PMC free article] [PubMed]
- 26.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, et al. Replicating genotype-phenotype associations. Nature 2007;447:655–660. [DOI] [PubMed] [Google Scholar]
- 27.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet 2001;27:234–236. [DOI] [PubMed] [Google Scholar]
- 28.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science 2002;297:1007–1013. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 2004;18:1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Li WH, Krainer AR, Zhang MQ. RNA landscape of evolution for optimal exon and intron discrimination. Proc Natl Acad Sci USA 2008;105:5797–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. Esefinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003;31:3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atochina-Vasserman EN, Gow AJ, Abramova H, Guo C-J, Tomer Y, Preston AM, Beck JM, Beers MF. Immune reconstitution during pneumocystis lung infection: disruption of surfactant component expression and function by s-nitrosylation. J Immunol 2009;182:2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satagopan JM, Elston RC. Optimal two-stage genotyping in population-based association studies. Genet Epidemiol 2003;25:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. Plink: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cappola TP, Li M, He J, Ky B, Gilmore J, Qu L, Keating B, Reilly M, Kim CE, Glessner J, et al. Common variants in hspb7 and frmd4b associated with advanced heart failure. Circ Cardiovasc Genet 2010;3:147–154. [DOI] [PMC free article] [PubMed]
- 37.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of ID and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 39.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, et al. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD beadchip. Am J Hum Genet 2009;85:628–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, et al. Genetic variants associated with lp(a) lipoprotein level and coronary disease. N Engl J Med 2009;361:2518–2528. [DOI] [PubMed] [Google Scholar]
- 41.Shah CV, Lanken PN, Localio AR, Gallop R, Bellamy S, Ma SF, Flores C, Kahn JM, Finkel B, Fuchs BD, et al. An alternative method of acute lung injury classification for use in observational studies. Chest 2010;138:1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. Snap: a web-based tool for identification and annotation of proxy SNPS using hapmap. Bioinformatics 2008;24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPS. Nature 2007;449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Netzer G, Shah CV, Iwashyna TJ, Lanken PN, Finkel B, Fuchs B, Guo W, Christie JD. Association of RBC transfusion with mortality in patients with acute lung injury. Chest 2007;132:1116–1123. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Willer C, Sanna S. Abecasis Ga. Genotype imputation. Annu Rev Genomics Hum Genet 2009;10:387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biernacka JM, Tang R, Li J, McDonnell SK, Rabe KG, Sinnwell JP, Rider DN, de Andrade M, Goode EL, Fridley BL. Assessment of genotype imputation methods. BMC Proc 2009;3(Suppl. 7):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, Compton D, McClain J, Aldrich TH, Papadopoulos N, et al. Angiopoietin-2, a natural antagonist for tie2 that disrupts in vivo angiogenesis. Science 1997;277:55–60. [DOI] [PubMed] [Google Scholar]
- 48.Bhandari V, Elias JA. The role of angiopoietin 2 in hyperoxia-induced acute lung injury. Cell Cycle 2007;6:1049–1052. [DOI] [PubMed] [Google Scholar]
- 49.Gallagher DC, Parikh SM, Balonov K, Miller A, Gautam S, Talmor D, Sukhatme VP. Circulating angiopoietin 2 correlates with mortality in a surgical population with acute lung injury/adult respiratory distress syndrome. Shock 2008;29:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giuliano JS Jr, Lahni PM, Harmon K, Wong HR, Doughty LA, Carcillo JA, Zingarelli B, Sukhatme VP, Parikh SM, Wheeler DS. Admission angiopoietin levels in children with septic shock. Shock 2007;28:650–654. [PMC free article] [PubMed] [Google Scholar]
- 51.Parikh SM, Mammoto T, Schultz A, Yuan HT, Christiani D, Karumanchi SA, Sukhatme VP. Excess circulating angiopoietin-2 may contribute to pulmonary vascular leak in sepsis in humans. PLoS Med 2006;3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhandari V, Choo-Wing R, Lee CG, Zhu Z, Nedrelow JH, Chupp GL, Zhang X, Matthay MA, Ware LB, Homer RJ, et al. Hyperoxia causes angiopoietin 2-mediated acute lung injury and necrotic cell death. Nat Med 2006;12:1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orfanos SE, Kotanidou A, Glynos C, Athanasiou C, Tsigkos S, Dimopoulou I, Sotiropoulou C, Zakynthinos S, Armaganidis A, Papapetropoulos A, et al. Angiopoietin-2 is increased in severe sepsis: correlation with inflammatory mediators. Crit Care Med 2007;35:199–206. [DOI] [PubMed] [Google Scholar]
- 54.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JG. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med 2008;36:2794–2800. [DOI] [PubMed] [Google Scholar]
- 55.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol 2006;34:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A+, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, et al. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 2005;171:361–370. [DOI] [PubMed] [Google Scholar]
- 57.Medford AR, Keen LJ, Bidwell JL, Millar AB. Vascular endothelial growth factor gene polymorphism and acute respiratory distress syndrome. Thorax 2005;60:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim I, Kim J-H, Ryu YS, Jung SH, Nah JJ, Koh GY. Characterization and expression of a novel alternatively spliced human angiopoietin-2. J Biol Chem 2000;275:18550–18556. [DOI] [PubMed] [Google Scholar]
- 59.Kraft P. Curses–winner's and otherwise–in genetic epidemiology. Epidemiology 2008;19:649–651; discussion 657–648. [DOI] [PubMed] [Google Scholar]
- 60.Battle TE, Lynch RA, Frank DA. Signal transducer and activator of transcription 1 activation in endothelial cells is a negative regulator of angiogenesis. Cancer Res 2006;66:3649–3657. [DOI] [PubMed] [Google Scholar]
- 61.Wincewicz A, Sulkowska M, Rutkowski R, Sulkowski S, Musiatowicz B, Hirnle T, Famulski W, Koda M, Sokol G, Szarejko P. Stat1 and stat3 as intracellular regulators of vascular remodeling. Eur J Intern Med 2007;18:267–271. [DOI] [PubMed] [Google Scholar]
- 62.Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG, Keating BJ, Yang Q, Chen MH, Lapchyk N, et al. Candidate gene association resource (care): design, methods, and proof of concept. Circ Cardiovasc Genet 2010;3:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esteban A, Fernandez-Segoviano P, Frutos-Vivar F, Aramburu JA, Najera L, Ferguson ND, Alia I, Gordo F, Rios F. Comparison of clinical criteria for the acute respiratory distress syndrome with autopsy findings. Ann Intern Med 2004;141:440–445. [DOI] [PubMed] [Google Scholar]
- 64.de Hemptinne Q, Remmelink M, Brimioulle S, Salmon I, Vincent J-L. ARDS. Chest 2009;135:944–949. [DOI] [PubMed] [Google Scholar]
- 65.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–369. [DOI] [PubMed] [Google Scholar]
- 66.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med 2008;178:710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calfee CS, Eisner MD, Parsons PE, Thompson BT, Conner ER Jr, Matthay MA, Ware LB. Soluble intercellular adhesion molecule-1 and clinical outcomes in patients with acute lung injury. Intensive Care Med 2009;35:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ware LB, Matthay MA, Parsons PE, Thompson BT, Januzzi JL, Eisner MD. Pathogenetic and prognostic significance of altered coagulation and fibrinolysis in acute lung injury/acute respiratory distress syndrome. Crit Care Med 2007;35:1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.