Abstract
Rationale: Neutrophils are usually the first circulating leukocytes to respond during bacterial pneumonia. Their expression of oxidants, proteases, and other mediators present in granules is well documented, but their ability to produce mediators through transcription and translation after migration to an inflammatory site has been appreciated only more recently. Interferon (IFN)-γ is a cytokine with many functions important in host defense and immunity.
Objectives: To examine the expression and function of IFN-γ in bacterial pneumonias.
Methods: IFN-γ mRNA and protein were measured in digests of mouse lungs with 24-hour bacterial pneumonia. Bacterial clearance was studied with IFN-γ–deficient mice.
Measurements and Main Results: Streptococcus pneumoniae and Staphylococcus aureus each induce expression of IFN-γ mRNA and protein by neutrophils by 24 hours. Only neutrophils that have migrated into pneumonic tissue produce IFN-γ. Deficiency of Hck/Fgr/Lyn, Rac2, or gp91phox prevents IFN-γ production. IFN-γ enhances bacterial clearance and is required for formation of neutrophil extracellular traps. In contrast, Pseudomonas aeruginosa and Escherichia coli induce production of IFN-γ mRNA but not protein. During pneumonia induced by E. coli but not S. pneumoniae, neutrophils produce microRNAs that target the 3′ untranslated region of the IFN-γ gene.
Conclusions: S. pneumoniae and S. aureus, but not P. aeruginosa and E. coli, induce emigrated neutrophils to produce IFN-γ within 24 hours. Hck/Fgr/Lyn, Rac2, and NADPH oxidase are required for IFN-γ production. IFN-γ facilitates bacterial clearance at least in part through regulating formation of neutrophil extracellular traps. Differential expression by neutrophils of microRNAs that target the 3′ untranslated region of the IFN-γ gene may contribute to the pathogen-specific regulation of translation.
Keywords: host defense, inflammation, innate immunity, infection
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Neutrophils are widely recognized to produce reactive oxygen species and proteases. However, their ability to regulate the inflammatory process by producing cytokines and other mediators in a highly regulated manner has many unexplored aspects.
What This Study Adds to the Field
In mice, neutrophils produce IFN-γ during pneumonia induced by the bacterial pathogens Streptococcus pneumoniae and Staphylococcus aureus, but not in response to Pseudomonas aeruginosa or Escherichia coli. IFN-γ production uses signaling pathways in neutrophils that include nonreceptor Src tyrosine kinases, Rac2, and NADPH oxidase and appears to be important in regulating the clearance of bacteria.
Interferon-γ (IFN-γ) is a pleiotropic cytokine that exerts its effects through binding to a single receptor, which is expressed on most cell types (1, 2). It is expressed primarily by T lymphocytes (1, 2), although other cell types including neutrophils can produce it (3–10). The effects of IFN-γ include a number of roles in specific immune processes, including modulation of the class I and II antigen–presenting pathways and the development of helper T-cell type 1 (Th1) responses. It plays important roles in controlling the cell cycle, growth, and apoptosis. It also contributes to both innate and specific immunity through activation of microbicidal effector pathways, priming of LPS responses, and leukocyte trafficking.
Bacterial pneumonia is a particularly common and devastating public health problem that is associated with high morbidity, mortality, and cost (11). Many studies from our laboratory and others have focused on the host defense mechanisms that are important in the recognition of pathogens in the lungs and the recruitment of leukocytes to the lung tissue, including the production of chemokines and cytokines such as IL-8, tumor necrosis factor-α, IL-1, and IL-6. The role of IFN-γ, as well as IFN-α and -β, has been documented during viral infections (2, 12). However, its role in acute bacterial infections is less clear, although some of its known functions in facilitating clearance of bacteria and enhancing innate responses predict that it may benefit host defense. The studies presented in this article determine which cell type produces IFN-γ during acute bacterial pneumonia, how its production is regulated, and whether it contributes to clearance of bacteria from the lungs. Initial studies tested the hypotheses that IFN-γ is produced early in the course of acute bacterial pneumonias and that neutrophils are the major source of IFN-γ production at this time. Subsequent studies tested the hypotheses that (1) the leukocyte nonreceptor tyrosine kinases Hck, Fgr, and Lyn, the small GTPase Rac2, and the NADPH oxidase component gp91phox are required for production of IFN-γ and (2) this IFN-γ is important in bacterial clearance from the lungs. Our observations that IFN-γ mRNA is translated to protein in response to only some bacterial species led to the final hypothesis that bacterial species that induce translation of IFN-γ mRNA do not induce microRNA (miR) targeted to the 3′ untranslated region (UTR) of the IFN-γ gene, whereas those that do not cause IFN-γ mRNA translation do induce these miRs.
Two gram-positive and two gram-negative organisms were selected for study: Streptococcus pneumoniae because it is the most common cause of community-acquired pneumonia (13), Staphylococcus aureus because it causes both community-acquired and nosocomial pneumonias, Pseudomonas aeruginosa because it is a common cause of nosocomial pneumonia, and Escherichia coli because it is also a cause of nosocomial pneumonia and pneumonia due to gram-negative sepsis (14) as well as because of the many in vivo studies that have examined this organism or its lipopolysaccharide (LPS). Pulmonary inflammation induced by lipoteichoic acid (LTA) and LPS, important constituents of the cell wall of gram-positive and gram-negative organisms, respectively, was also examined. Some of the results of these studies have been previously reported in the form of abstracts (15–18).
METHODS
Reagents
Reagents used for this study were from the following sources: Dispase II was from Roche Applied Science (Indianapolis, IN); fetal calf serum (FCS), phosphate-buffered saline (PBS), collagenase type 1, TRIzol, the SuperScript III first-strand synthesis system, NuPAGE LDS sample buffer, a colloidal blue staining kit, and Alexa Fluor 546–conjugated goat anti-mouse IgG antibody were from Invitrogen (Carlsbad, CA); DNase I from bovine pancreas, red blood cell lysing buffer, paraformaldehyde, LPS from E. coli O55:B5, LTA from S. aureus, protease inhibitor cocktail, TRI Reagent, and 4′,6-diamidino-2-phenylindole (DAPI) were from Sigma-Aldrich (St. Louis, MO); the RNeasy mini kit was from Qiagen (Valencia, CA). Protein L–agarose beads were from Santa Cruz Biotechnology (Santa Cruz, CA). Cell lysis buffer was from Cell Signaling Technology (Danvers, MA). Reagents for flow cytometry were from the following sources: fluorescein isothiocyanate (FITC)–conjugated rat anti-mouse CD45 antibody (clone 30-F11), FITC-conjugated Armenian hamster anti-mouse CD3ɛ antibody (clone 145-2C11), phycoerythrin (PE)-conjugated rat anti-mouse IFN-γ antibody (clone XMG1.2), PE–cyanine 5 (Cy5)–conjugated rat anti-mouse F4/80 (clone BM8) and Gr-1 (clone RB6–8C5) antibodies, allophycocyanin-conjugated rat anti-mouse CD45 antibody (clone 30-F11), fluorochrome-conjugated isotype control antibodies, brefeldin A, and permeabilization buffer were purchased from eBioscience (San Diego, CA); FITC-conjugated rat anti-mouse Ly-6G antibody (clone 1A8) and rat anti-mouse FcγIII/II receptor antibody (clone 2.4G2) were purchased from BD Pharmingen (San Diego, CA); FITC-conjugated anti-mouse neutrophil antibody (clone 7/4) was purchased from Serotec (Raleigh, NC). Mouse monoclonal anti–pan-histone antibody was purchased from Millipore (Billerica, MA). Mouse monoclonal anti-mouse myeloperoxidase (MPO) antibody (clone 8F4) was purchased from Hycult Biotechnology (Uden, The Netherlands).
Bacteria
Streptococcus pneumoniae (S. pneumoniae; serotype 19, ATCC 49619), Staphylococcus aureus (S. aureus; serotype 3, ATCC 12600), Escherichia coli (E. coli; serotype O55:K59(B5):H-, ATCC 12014), and Pseudomonas aeruginosa (P. aeruginosa; PAO-1, ATCC BAA-47) were purchased from the American Type Culture Collection (Manassas, VA).
Mice
Adult C57BL/6 (wild-type) mice and IFN-γ–deficient mice were purchased from Jackson Laboratory (Bar Harbor, ME). Rac2 null and gp91phox null mice were provided by M. C. Dinauer (Indiana University, Indianapolis, IN). Hck/Fgr/Lyn triply and singly deficient mice were provided by C. A. Lowell (University of California, San Francisco, CA). All deficient mice were backcrossed to the wild-type background. Mice were between 6 and 8 weeks of age in all experiments. All studies were subject to review by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill (Chapel Hill, NC) and conformed to the Guide for the Care and Use of Laboratory Animals by the Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council.
Induction of Bacterial Pneumonia and LPS- or LTA-induced Lung Inflammation
Bacterial pneumonia and LPS- or LTA-induced lung injury were induced by intratracheal instillation of 50 μl of bacterial suspension, or of LPS (100 μg/mouse) or LTA (100 μg/mouse) solution, into anesthetized mice. After exposure of the trachea through a ventral incision in the neck, a 24-gauge catheter was threaded into the trachea through a tracheostomy and positioned near the bifurcation. The mice were positioned on their left side so that the bacteria instilled entered the left lung. Previous experience in using this method showed that greater than 90% of the stimulus entered the left lung. Concentrations of bacterial suspensions were determined by optical density at 600 nm. Optical density for each bacterial solution was as follows: S. pneumoniae, 1.0; S. aureus, 1.0; E. coli, 0.3; P. aeruginosa, 0.1. These concentrations yield doses that induce similar numbers of neutrophils to migrate into murine lungs. Colony-forming units in bacterial solutions were subsequently determined by serial dilution and colony counting on agar plates. The ranges of colony-forming units were as follows, unless otherwise indicated: S. pneumoniae, 2.8–20 × 106 cfu/mouse; S. aureus, 8.5–13 × 107 cfu/mouse; E. coli, 3.8–18 × 106 cfu/mouse; P. aeruginosa, 4.5–48.3 × 106 cfu/mouse.
Preparation of Single Lung Cells from Lungs
Six or 24 hours after instillation, mice received an overdose of inhaled isoflurane. Their lungs were perfused with PBS via the right ventricles. PBS-perfused lungs were isolated with other mediastinal organs. Dispase II solution was instilled into lungs through the trachea, which was ligated with a silk suture. After incubation for 50 minutes at 37°C, lungs were separated from other mediastinal organs. Left lung samples were minced well and digested in PBS with 0.1% collagenase, 0.01% DNase I, and 5 mM CaCl2 at 37°C for 20 minutes. Cells were suspended in red blood cell lysing buffer to remove red blood cells and washed with PBS. Cells were centrifuged again and suspended in PBS.
Bronchoalveolar Lavage
Bronchoalveolar lavage (BAL) was performed by instilling 1.0 ml of ice-cold PBS into the lungs and then gently aspirating the fluid. BAL was repeated five times. These five fluid samples were pooled and centrifuged. BAL fluid cells were suspended in PBS.
Intracellular Staining for IFN-γ
Isolated lung cells were incubated in medium (10% FCS–RPMI 1640) with brefeldin A (3.0 μg/ml) for 1 hour at 37°C. Cells were centrifuged and resuspended with 2% FCS–PBS. To prevent nonspecific binding of antibody to Fc receptors, cells were incubated with rat anti-mouse CD16/CD32 antibody (mouse Fc Block; BD Biosciences, San Jose, CA). Staining for cell surface antigens was performed with fluorochrome-conjugated antibodies. Cells were washed and fixed with 2% paraformaldehyde. Cells were washed again and suspended with permeabilization buffer. Cells were then incubated with PE-conjugated anti–IFN-γ antibody or isotype control antibody. Cells were washed and resuspended for flow cytometric analysis, using a FACScan or LSR (BD Biosciences). In studies assessing IFN-γ production in neutrophils, the neutrophils were identified with the FITC-conjugated rat anti-mouse Ly-6G antibody (clone 1A8). This antibody is specific for neutrophils and recognizes only Ly6G (19). The anti–Gr-1 antibody (clone RB6-8C5) was used only in experiments where two neutrophil markers were required (intravascular neutrophils and all neutrophils, please see details below). This anti–Gr-1 antibody recognizes both Ly6G and Ly6C, which is expressed at low levels on dendritic cells and some populations of monocytes, as well as on neutrophils (19).
Identification of Proteins Recognized by Anti-Mouse IFN-γ Antibody
Neutrophils were isolated from lung cell suspensions of mice with 24-hour S. pneumoniae pneumonias (purity, >99%), using an anti–Ly-6G MicroBead kit (Miltenyi Biotec, Bergisch Gladbach, Germany). The neutrophils were fixed with 2% paraformaldehyde, washed, and suspended in permeabilization buffer. Cells were then incubated with anti–IFN-γ antibody or isotype control antibody. Cells were washed, and lysed with radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% IGEPAL CA-630 [1% Nonidet P-40] containing 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris-HCl [pH 7.5]), and a protease inhibitor cocktail. IFN-γ–anti–IFN-γ antibody complexes were isolated with protein L–agarose beads that were incubated with the lysate at 4°C overnight. Samples were washed with RIPA buffer five times and incubated with NuPAGE LDS sample buffer to elute. Eluted samples were electrophoresed on a NuPAGE 4–12% gradient bis-Tris sodium dodecyl sulfate–polyacrylamide gel. After electrophoresis, the gel was stained with the colloidal blue staining kit and cut into 11 pieces according to the distribution of stained bands. The gel pieces were digested with trypsin (Promega, Madison, WI) for 18 hours at 37°C. Digests were injected into a liquid chromatography–mass spectrometry system (UltiMate 3000 HPLC Nano; Dionex, Sunnyvale, CA with C18 reverse phase columns and a Fourier transform quadrupole ion trap mass spectrometer [LTQ FT Ultra Hybrid Mass Spectrometer]; Thermo Scientific, Waltham, MA). Data files were then processed with the Mascot search engine (Matrix Science, Boston, MA), using the database NCBInr 20070216 (4,626,804 sequences; 1,596,079,197 residues).
ELISA for Detection of Mouse IFN-γ
Lungs were harvested from mice 24 hours after instillation of S. pneumoniae, E. coli, or PBS, and homogenized with cell lysis buffer supplemented with a protease inhibitor cocktail. Supernatants were used for ELISA, following the manufacturer's instructions (mouse IFN-γ ‘Femto-HS’ high-sensitivity ELISA kit; eBioscience).
Identification of Intravascular and Extravascular Neutrophils in Lungs
Identification of intravascular and extravascular neutrophils in lungs was performed as previously described with modification (20). PE–Cy5–conjugated anti-mouse Gr-1 antibody (10 μg) was injected intravenously and allowed to circulate for 5 minutes. After 5 minutes, mice were killed. The lungs were digested as described previously in the presence of excess unlabeled anti-mouse Gr-1 antibody to prevent possible binding of excess PE–Cy5–conjugated anti–Gr-1 antibody to extravascular neutrophils. Lung cell suspensions were then stained with FITC-conjugated anti-mouse neutrophil antibody (7/4). Intravascular (7/4+Gr-1+) and extravascular (7/4+Gr-1−) neutrophil populations were assessed by flow cytometry.
Bacterial Numbers in Lungs
The number of viable bacteria was quantified by colony-forming assays. Lungs were homogenized in sterile PBS, serially diluted, and plated on Trypticase soy agar with 5% sheep blood (TSA II; BD Diagnostic Systems, Sparks, MD). Bacterial colonies were counted after being incubating overnight at 37°C in 5% CO2. Clearance was calculated as the number of colony-forming units remaining in the lungs at 24 hours divided by the number of colony-forming units instilled.
Immunofluorescence Assays for Neutrophil Extracellular Traps
Mice were given an intrabronchial instillation of S. pneumoniae (1.8–3.1 × 106 cfu/mouse). After 24 hours, bronchoalveolar lavage fluid (BALF) was collected. Cytospin slides of BALF were prepared. Cells were fixed with 4% paraformaldehyde, blocked (4% normal goat serum, 1% bovine serum albumin), and incubated with either the anti-histone antibody or the anti-MPO antibody, which were detected with a secondary antibody conjugated to Alexa Fluor 546. Control slides were stained with isotype-matched controls. For DNA detection, DAPI staining was performed. Neutrophil extracellular traps (NETs) were counted and expressed per 500 neutrophils.
Pulmonary Edema
Plasma extravasation was quantified with radiotracers as described previously (21). Briefly, anesthetized mice received intravenous injections of 125I-labeled human albumin (Mallinckrodt Medical, Hazelwood, MO) 6 hours and 15 minutes before euthanasia and of 51Cr-labeled murine erythrocytes 2 minutes before euthanasia, as markers of plasma and blood content, respectively. Mice were killed by isoflurane overdose 24 hours after the intratracheal instillation of S. pneumoniae. The total volume of plasma equivalents in the lungs was derived from the 125I-labeled albumin activities in the lungs and plasma. The volume of intravascular plasma in the lungs was calculated from the hematocrit (derived from the 125I-labeled albumin activities in the blood and plasma) and the pulmonary blood volume (derived from the 51Cr-labeled red blood cell activity in the lungs and blood). The volume of extravascular plasma equivalents in the lungs was calculated as the total volume of plasma equivalents minus the volume of intravascular plasma. The edema index was calculated as the volume of extravascular plasma equivalents divided by the mouse body weight.
Isolation of Lung Neutrophils
Lung neutrophils were separated from lung cell suspensions, using the anti–Ly-6G MicroBead kit (Miltenyi Biotec) according to the manufacturer's protocol. Purity of isolated neutrophils was evaluated by microscopic and flow cytometric analyses, and was more than 98%.
RNA Extraction, cDNA Synthesis, and Quantitative Polymerase Chain Reaction
Total RNA was extracted from neutrophils, using the RNeasy mini kit. cDNA was synthesized from total RNA, using the SuperScript III first-strand synthesis system. IFN-γ mRNA was quantified by quantitative real-time polymerase chain reaction (PCR), using a specific primer set (Mm99999071_m1) purchased from Applied Biosystems (Foster City, CA). A primer set specific for 28S rRNA was also used for normalization (sense primer, 5′-TGTGGATGGCGAGAAATACCA-3′; antisense primer, 5′-GCATCAGCCTCCAGTATAGTTGT-3′).
Quantitative Reverse Transcription PCR for MicroRNAs
Quantitative reverse transcription PCR was performed as described previously (22). Briefly, total RNA was isolated with TRI Reagent (Sigma-Aldrich). The integrity of the total RNA was evaluated with a 2100 Bioanalyzer and an RNA 6000 series II Nano kit (Agilent, Santa Clara, CA). The TaqMan microRNA assay system (Applied Biosystems) was used to quantitatively detect the expression of microRNAs, in accordance with the manufacturer's instructions. The primers used in the TaqMan microRNA assay system were specific and recognized the mature form of each microRNA. Real-time quantitative PCR was conducted with the ABI 7500 real-time PCR system (Applied Biosystems). All reactions were done in technical triplicates. The relative expression level of microRNAs was calculated by the 2−ΔΔCt method (23) after normalization to the endogenous small RNA control, small nucleolar RNA (snoRNA) 202.
Statistical Analyses
Data are expressed as means ± SEM. Statistical analyses were performed with Statistica software (StatSoft Inc., Tulsa, OK). Sets containing two groups of data were analyzed by Student t test. Sets containing more than two groups of data were analyzed by one-way analysis of variance with a post hoc test (Scheffé test or Tukey HSD [honestly significantly different] test). P values less than 0.05 were considered significant.
RESULTS
S. pneumoniae and S. aureus Induce Production of IFN-γ in Neutrophils, whereas E. coli, P. aeruginosa, LPS, and LTA Do Not
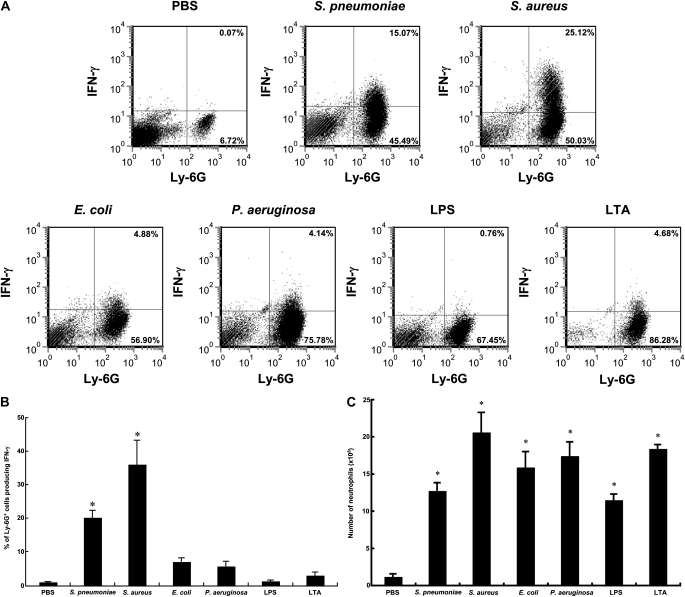
Mice were given pneumonia by instilling into the left lung one bacterial species, LPS, LTA, or PBS. After 24 hours, single-cell lung digests were stained to identify neutrophils using the neutrophil-specific anti-Ly6G antibody (clone 1A8) and IFN-γ using an anti-mouse IFN-γ antibody (clone XMG1.2). S. pneumoniae and S. aureus induced a significant increase in the percentage of neutrophils that contained intracellular IFN-γ, whereas E. coli, P. aeruginosa, LPS from E. coli, and LTA from S. aureus did not (Figures 1A and 1B). Each stimulus induced a significant accumulation of neutrophils, and the number of accumulated neutrophils was similar among inflammatory stimuli (Figure 1C). Furthermore, IFN-γ was present in lung homogenates from pneumonia induced by S. pneumoniae (3.4 ± 0.18 pg/mg protein) but not E. coli (1.1 ± 0.32 pg/mg protein) compared with noninfected controls (1.9 ± 0.05 pg/mg protein), using an ELISA.
Figure 1.
Streptococcus pneumoniae and Staphylococcus aureus induced production of IFN-γ in lung neutrophils by 24 hours, whereas Escherichia coli, Pseudomonas aeruginosa, LPS, and lipoteichoic acid (LTA) did not. (A) Representative flow cytometric analyses to detect intracellular IFN-γ in neutrophils from single lung cells prepared from lung digests of mice given phosphate-buffered saline (PBS) (controls), gram-positive bacteria (S. pneumonia or S. aureus), gram-negative bacteria (E. coli or P. aeruginosa), LPS, or LTA. Twenty-four hours after intraairway instillation of a stimulus, lungs were perfused with PBS via the right ventricle. Dispase II solution was infused into the lungs through the trachea. After 50 minutes at 37°C, the left lungs were minced well and digested in PBS with 0.1% collagenase, 0.01% DNase I, and 5 mM CaCl2 at 37°C for 20 minutes. Red blood cells were lysed, and isolated lung cells were incubated in medium (10% fetal calf serum–RPMI 1640) with brefeldin A (3.0 μg/ml) for 1 hour at 37°C, followed by Fc Block. Neutrophils were identified with fluorescein isothiocyanate–conjugated rat anti-mouse Ly-6G antibody (clone 1A8), an antibody that recognizes only neutrophils (19). Cells were fixed with 2% paraformaldehyde, suspended in permeabilization buffer, and incubated with phycoerythrin-conjugated anti–IFN-γ antibody or isotype control antibody. The percentage of cells in each quadrant is shown. (B) Mean percentage of neutrophils producing IFN-γ. (C) Mean number of neutrophils in pneumonic lungs. The number of mice in each group was as follows: PBS, n = 9; S. pneumoniae, n = 21; S. aureus, n = 6; E. coli, n = 15; P. aeruginosa, n = 6; LPS, n = 9; LTA, n = 6. Shown are means ± SEM of three to seven separate experiments. (*P < 0.0001 vs. the PBS group, using analysis of variance with the Scheffé post hoc test.)
To further ensure the identity of the antigen recognized by the anti–IFN-γ antibody and to document the production of IFN-γ, neutrophils were isolated from S. pneumoniae lung digests (>99% pure) and labeled with this antibody as for flow cytometry. The antibody–antigen complexes were eluted and electrophoresed, and gel segments were subjected to liquid chromatography–mass spectrometry. IFN-γ was identified in the gel segment containing the appropriate molecular mass range (IFN-γ, 18,123 Da) by four peptides: RLFEVLKD, KFEVNNPQVQRQ, RQAFNELIRV, and RVVHQLLPESSLRK (Mascot score, 78; P < 0.05). These four peptides cover 25.8% of the IFN-γ protein and are shown highlighted in italic and bold in the IFN-γ amino acid sequence: MNATHCILALQLFLMAVSGCYCHGTVIESLESLNNYFNSSGIDVEEKSLFLDIWRNWQKDGDMKILQSQIISFYLRLFEVLKDNQAISNNISVIESHLITTFFSNSKAKKDAFMSIAKFEVNNPQVQRQAFNELIRVVHQLLPESSLRKRKRSRC.
The underlined residues were included in two peptides. The only murine peptide/protein that was present in the fragment of polyacrylamide gel that included the IFN-γ molecular mass range was IFN-γ. Furthermore, no peptides mapping to IFN-γ were found in neutrophils of IFN-γ–deficient mice, and neutrophils from mice deficient in the NADPH oxidase component gp91phox, which is required for production of IFN-γ (see below), also did not produce IFN-γ.
To better understand which leukocytes produced IFN-γ during S. pneumoniae pneumonia and the kinetics of IFN-γ expression, single cells from digests of lungs infected with S. pneumoniae were stained with lineage-specific markers and anti–IFN-γ antibody (Table 1). The pan-leukocyte marker CD45 showed that leukocytes were making IFN-γ 24 hours, but not 6 hours, after instillation of S. pneumoniae. At 24 hours, CD3+ T cells, common producers of IFN-γ, did not produce IFN-γ. In fact, no CD45+Ly-6G− cells expressed IFN-γ. Immunostaining of BAL fluid from lungs with S. pneumoniae pneumonia demonstrated that only Ly-6G+ neutrophils, and not F4/80+ macrophages, produced IFN-γ (Table 1). Furthermore, circulating neutrophils in blood from mice instilled with S. pneumoniae did not express IFN-γ at 6 or 24 hours (Table 1), indicating that only neutrophils recruited to the lungs expressed IFN-γ during S. pneumoniae pneumonia. Indeed, studies of 24-hour S. pneumoniae pneumonias, in which intravascular neutrophils were labeled in vivo with intravascular PE–Cy5–conjugated anti-mouse Gr-1 antibody and all pulmonary neutrophils in the lung digest were labeled with FITC-conjugated anti-mouse 7/4, demonstrated that only the extravascular neutrophils (i.e., cells labeled with anti-7/4 but not anti–Gr-1 antibody) produced IFN-γ (33.3 ± 1.3% of extravascular neutrophils vs. 1.9 ± 0.4% of intravascular neutrophils). These observations suggest that neutrophils are the major source of IFN-γ production at 24 hours during S. pneumoniae pneumonia and that only the extravascular neutrophils express IFN-γ. However, the possibililty of an Ly6G− cell type producing IFN-γ cannot be completely excluded.
TABLE 1.
IFN-γ IS PRODUCED BY NEUTROPHILS IN 24-HOUR S. pneumoniae PNEUMONIA
| Percentage of Cells Producing IFN-γ |
|||||
|---|---|---|---|---|---|
| Site of Cells | Lineage-specific Marker | Lineage | Duration of Pneumonia (h) | PBS (Control) | S. pneumoniae |
| Lung digests | CD45 | All leukocytes | 6 | 0.09 ± 0.02, n = 3 | 0.10 ± 0.05, n = 3 |
| 24 | 0.22 ± 0.07, n = 8 | 14.2 ± 0.95,* n = 14 | |||
| CD3 | T cells | 24 | 0.23 ± 0.06, n = 3 | 0.47 ± 0.10,† n = 12 | |
| CD45+, Ly-6G− | Nonneutrophilic leukocytes | 24 | ND | 0.31 ± 0.04,† n = 3 | |
| CD45+, Ly-6G+ | Neutrophils | 24 | ND | 21.98 ± 3.43, n = 6 | |
| BAL fluid | F4/80 | Macrophages | 24 | ND | 0.08 ± 0.11, n = 3 |
| Ly-6G | Neutrophils | 24 | ND | 16.4 ± 1.5,‡ n = 3 | |
| Circulating blood | Ly-6G | Neutrophils | 6 | 0.02 ± 0.04, n = 3 | 0.04 ± 0.02, n = 3 |
| 24 | 0.20 ± 0.20, n = 3 | 0.32 ± 0.16, n = 3 | |||
Definition of abbreviations: BAL = bronchoalveolar lavage; ND = not determined; PBS = phosphate-buffered saline; S. pneumoniae = Streptococcus pneumoniae.
Flow cytometry was performed to detect intracellular IFN-γ levels in all leukocytes (CD45+ cells), T cells (CD3+ cells), nonneutrophils (CD45+Ly-6G− cells), and neutrophils (CD45+Ly-6G+) in the lungs of S. pneumoniae–infected or PBS-treated mice 6 or 24 hours after instillation. Isotype-matched control antibody was used to assess the level of background staining and to set gates. The percentages of cells in each quadrant and the means of the percentage in the double-positive quadrant were calculated for the number of animals indicated. IFN-γ expression was similarly measured in F4/80- and Ly-6G–positive cells from the BAL fluid and circulating blood (n = 6 in each group). Neutrophils in the lung digests or the BAL fluid from pneumonic lungs at 24 hours expressed IFN-γ. None of the other leukocyte subtypes were found to produce IFN-γ using this approach. Data represent means ± SEM, and the number of mice (n) is provided for each study. The results are from three to six separate experiments.
P < 0.001 versus the PBS group.
P < 0.01 versus the CD45 group.
P < 0.05 versus the F4/80 group.
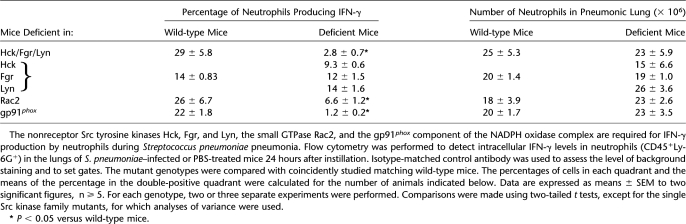
Rac2, Hck/Fgr/Lyn, and gp91phox Are Required for Neutrophils to Produce IFN-γ
Other investigators have suggested that the small GTPase Rac2 may modulate IFN-γ production by lymphocytes, and that the three nonreceptor Src tyrosine kinases Hck, Fgr, and Lyn (Hck/Fgr/Lyn) modulate Rac2 activation in neutrophils (24, 25). A major function of Rac2 is to regulate the assembly of the NADPH oxidase complex (26). The gp91phox component of NADPH oxidase is required for the oxidase to produce superoxide (26). To determine whether Hck/Fgr/Lyn, Rac2, and NADPH oxidase are required for IFN-γ production in neutrophils during S. pneumoniae pneumonia, wild-type mice and mice deficient in the either Hck, Fgr, or Lyn or all three nonreceptor Src tyrosine kinases, the small GTPase Rac2, or the NADPH oxidase component gp91phox were given S. pneumoniae. At 24 hours, the number of neutrophils present in the pneumonic lungs was similar in all mutant genotypes compared with wild-type (Table 2). However, the percentage of lung neutrophils that produced intracellular IFN-γ in response to S. pneumoniae was less in mice missing any one of these molecules compared with wild-type mice (Table 2). Mice singly deficient in either Hck, Fgr, or Lyn had no defect in IFN-γ production. These studies indicate that Hck/Fgr/Lyn, Rac2, and gp91phox are each required for the production of IFN-γ by neutrophils. The molecules in this sequence (Hck/Fgr/Lyn:Rac2:NADPH oxidase) form a well-described intracellular signaling pathway that results in oxidant production, suggesting that oxidative stress is required for IFN-γ production.
TABLE 2.
INTRACELLULAR SIGNALING MOLECULES REQUIRED FOR IFN-γ PRODUCTION BY NEUTROPHILS DURING S. pneumoniae PNEUMONIA
IFN-γ Modulates Bacterial Clearance and Host Defense
To elucidate the function of IFN-γ during host defense against S. pneumoniae, IFN-γ−/− and wild-type mice received varying doses of S. pneumoniae (which induced IFN-γ production) or E. coli (which did not). Significantly more viable S. pneumoniae were present in the lungs of IFN-γ−/− mice compared with wild-type mice 24 hours after instillation of the lowest dose (2.8–5.0 × 106 cfu/mouse) of S. pneumoniae (Table 3). At a mid-level dose of 7.4–11 × 106 cfu or a high dose of 13–66 × 106 cfu, there was no significant difference between wild-type and IFN-γ−/− mice. IFN-γ−/− mice given E. coli, which did not induce intracellular IFN-γ protein expression by neutrophils, had no defect in clearance of either dose (Table 3). These data suggest that IFN-γ is required for host defense against S. pneumoniae, particularly at low doses, but not E. coli.
TABLE 3.
CLEARANCE OF BACTERIA, ACCUMULATION OF NEUTROPHILS IN PNEUMONIC LUNGS, CIRCULATING NEUTROPHIL COUNTS, AND PULMONARY EDEMA IN THE LUNGS OF IFN-γ–DEFICIENT AND WT MICE DURING BACTERIAL PNEUMONIA
| Component of Host Defense | Organism | Dose | Wild-type Mice | IFN-γ–deficient Mice |
|---|---|---|---|---|
| Bacterial clearance (ratio of remaining to inoculated cfu) | S. pneumoniae | LowMid | 1.2 ± 0.3, n = 73.3 ± 1.1, n = 8 | 8.0 ± 0.7,* n = 72.2 ± 0.6, n = 6 |
| High | 20.2 ± 1.8, n = 5 | 20.8 ± 5.1, n = 7 | ||
| E. coli | Low | 0.16 ± 0.05, n = 7 | 0.24 ± 0.08, n = 6 | |
| High | 1.4 ± 0.3, n = 6 | 0.90 ± 0.3, n = 6 | ||
| Neutrophil accumulation in the lungs (× 106 per lung) | S. pneumoniae | LowHigh | 17.6 ± 1.7, n = 630.6 ± 3.4, n = 4 | 30.7 ± 4.5,* n = 635.9 ± 7.8, n = 4 |
| E. coli | Low | 8.4 ± 1.1, n = 6 | 8.2 ± 1.1, n = 6 | |
| High | 17.4 ± 1.3, n = 6 | 28.6 ± 3.2,* n = 8 | ||
| Circulating neutrophil counts (× 106/ml blood, n = 5) | S. pneumoniae | LowHigh | 1.7 ± 0.31.4 ± 0.2 | 3.4 ± 0.5*2.4 ± 0.5 |
| Edema index (n = 5) | S. pneumoniae | Low | 0.61 ± 0.08 | 0.60 ± 0.08 |
| High | 1.42 ± 0.45 | 0.85 ± 0.1 |
Definition of abbreviations: cfu = colony-forming units; E. coli = Escherichia coli; S. pneumoniae = Streptococcus pneumoniae.
Bacterial clearance at 24 hours was decreased in IFN-γ–deficient mice given low-dose S. pneumoniae (2.8–5.0 × 106 cfu/mouse). No defect in clearance was observed after mid-dose S. pneumoniae (7.4–11 × 106 cfu/mouse) or high dose S. pneumoniae (13–66 × 106 cfu/mouse) or either dose of E. coli (low dose, 1.9–2.8 × 106 cfu/mouse; high dose, 9.5–24 × 106 cfu/mouse). Neutrophil recruitment was enhanced in IFN-γ–deficient mice during S. pneumoniae pneumonia induced by low-dose (1.0–6.0 × 106 cfu/mouse) but not high-dose (34–48 × 106 cfu/mouse) S. pneumoniae at 24 hours, paralleling the defect in clearance of S. pneumoniae. No defect in neutrophil accumulation was observed with low-dose E. coli (1.2–2.6 × 106 cfu/mouse), also similar to the lack of defect in clearance of this organism, but enhanced accumulation was found in IFN-γ–deficient mice with high-dose (19–29 × 106 cfu/mouse) E. coli pneumonia. Circulating neutrophil counts and pulmonary edema during pneumonia induced with either low-dose (1.8–2.0 × 106 cfu/mouse) or high-dose (17–20 × 106 cfu/mouse) S. pneumoniae were similar in IFN-γ–deficient and wild-type mice. Data represent means ± SEM, and the number of mice is indicated. For each study, two separate experiments were performed. Statistical significance was determined by analyses of variance and the Tukey HSD (honestly significantly different) post hoc test.
P < 0.05 compared with wild-type mice.
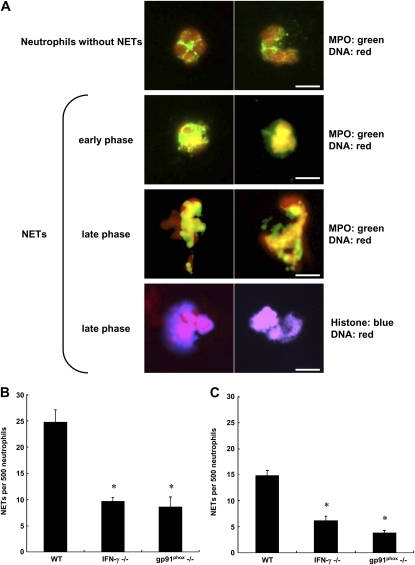
There are several mechanisms through which IFN-γ may be facilitating bacterial clearance. Neutrophil extracellular traps (NETs) were described by Brinkmann and colleagues in 2004 (27). This structure is extruded by neutrophils, most likely as a form of cell death, and contains nuclear material and granular contents such as myeloperoxidase and elastase but not actin. NETs trap microorganisms and have bactericidal capabilities, at least in part due to the histones (27). Their production requires NADPH oxidase (28), and IFN-γ has been suggested to modulate NET formation (29). Our studies tested the hypothesis that IFN-γ produced early in the course of S. pneumoniae pneumonia by neutrophils facilitated NET formation and hence the clearance of bacteria. NETs were found by immunostaining and colocalization of either DNA plus MPO or DNA plus histones (Figure 2A). Significantly fewer NETs were present in the BAL fluid of IFN-γ−/− mice than in that of wild-type mice. This defect in NET formation observed in IFN-γ mice was similar in degree to the defect in mice missing the gp91phox component of NADPH oxidase (which do not produce IFN-γ in response to S. pneumoniae), whether evaluated by quantifying the number of NETs by colocalization of DNA plus MPO (Figure 2B) or DNA plus histones (Figure 2C). Thus, IFN-γ is modulating bacterial clearance at least in part through its effects on NET formation.
Figure 2.
IFN-γ is required for host defense against Streptococcus pneumoniae. (A) Representative immunomicrographs of neutrophils and neutrophil extracellular traps (NETs) after immunofluorescence and 4′,6-diamidino-2-phenylindole staining to detect histones or myeloperoxidase (MPO) and DNA in neutrophils from bronchoalveolar lavage fluid 24 hours after instillation of low-dose S. pneumoniae (1.4–4.1 × 106 cfu/mouse). Top row: Naïve neutrophils showed DNA (red) and MPO (green) located in nuclei and cytoplasm, respectively. The usual lobular structure of nuclei was present. MPO stained in a granular cytoplasmic pattern. Second row: In pneumonic lungs, neutrophils in the early phase of NET formation showed colocalization (yellow) of nuclear DNA and MPO. Cells were losing their nuclear lobules, and MPO was present in the nucleus. Third and fourth rows: In the late phase of NET formation, neutrophils released NETs. Immunomicroscopy showed colocalization of nuclear DNA and MPO (yellow) and of nuclear DNA and histone (pink) in released NETs. Scale bars: 10 μm. (B and C) Quantification of NETs identified by colocalization of (B) DNA and MPO or (C) DNA and histones in wild-type, IFN-γ−/−, and gp91phox−/− mice. IFN-γ was required for the production of NETs, when identified by either DNA plus MPO or DNA plus histone staining. The defect in NET formation in IFN-γ−/− mice was similar to that in mice missing gp91phox, a subunit of NADPH oxidase known to be required for NET formation. Shown are means ± SEM, n = 6 mice of each genotype studied in two separate experiments. (*P < 0.05 vs. wild-type mice, using analysis of variance with the Tukey HSD [honestly significantly different] post hoc test.)
The number of neutrophils recruited to the lungs 24 hours after instillation of lower dose S. pneumoniae in IFN-γ−/− mice was higher than in wild-type mice (Table 3), demonstrating that the impairment of S. pneumoniae clearance in IFN-γ−/− mice is not due to the attenuated infiltration of neutrophils. The number of circulating neutrophils in the blood of IFN-γ−/− mice was also greater than in wild-type mice (Table 3). Lung injury measured as the accumulation of edema (extravascular 125I-labeled albumin accumulation) in the last 6 hours of S. pneumoniae pneumonia was similar between genotypes (Table 3), suggesting that the increased number of neutrophils did not enhance lung injury. The number of neutrophils recruited to high-dose E. coli pneumonias was also increased despite no defect in clearance. Because these studies raised the possibility that IFN-γ expression occurs in response to higher (19–29 × 106 cfu/mouse) doses of E. coli, additional studies were performed using 25 × 106 cfu/mouse. However, only 5.4 ± 1.0% of neutrophils expressed IFN-γ in these pneumonias, which is similar to the value of 5.7 ± 0.8% of IFN-γ+ neutrophils in the lungs of mice given lower doses in Figure 1B (3.8–18 × 106 cfu/mouse), and to mice given PBS (Figure 1B). The mechanism for this increase in neutrophil accumulation is thus not yet clear.
IFN-γ mRNA and Protein Are Differentially Expressed
High-quality RNA was extracted from neutrophils isolated from lung digests (>98% purity), and IFN-γ mRNA was measured by real-time quantitative PCR. Although IFN-γ protein was expressed only by neutrophils from S. pneumoniae and S. aureus pneumonias, IFN-γ mRNA was significantly induced 7- to 40-fold in lung neutrophils by all inflammatory stimuli at 24 hours, including S. pneumoniae, S. aureus, E. coli, P. aeruginosa, LPS, and LTA (Table 4). These studies suggest that each stimulus induces transcription of IFN-γ by neutrophils, but that translation occurs only in S. pneumoniae and S. aureus pneumonia. Furthermore, neutrophils from gp91phox-deficient mice given S. pneumoniae pneumonia did not express IFN-γ mRNA (9.6 ± 0.68-fold vs. 0.72 ± 0.088-fold change in IFN-γ mRNA compared with PBS-treated control in wild-type and gp91phox-deficient mice, respectively), demonstrating that NADPH oxidase is important in the transcription of IFN-γ.
TABLE 4.
IFN-γ mRNA IN LUNG NEUTROPHILS AS DETECTED BY QUANTITATIVE PCR
| Organism or Bacterial Component | Relative Amount of IFN-γ mRNA* |
|---|---|
| PBS | 1.0 ± 0.0, n = 18 |
| S. pneumoniae | 15.9 ± 0.9,† n = 24 |
| S. aureus | 37.7 ± 2.8,‡ n = 6 |
| E. coli | 39.0 ± 5.7,‡ n = 21 |
| P. aeruginosa | 26.2 ± 7.1,‡ n = 12 |
| LPS | 7.2 ± 1.2,† n = 15 |
| LTA | 19.2 ± 0.7,† n = 6 |
Definition of abbreviations: E. coli = Escherichia coli; LTA = lipoteichoic acid; P. aeruginosa = Pseudomonas aeruginosa; PBS = phosphate-buffered saline; S. aureus = Staphylococcus aureus; S. pneumoniae = Streptococcus pneumoniae.
IFN-γ mRNA or 28S rRNA (internal control) in each sample of lung neutrophils was detected by quantitative PCR. Data are expressed as the fold increase in IFN-γ mRNA expression compared with PBS-treated control. Data represent means ± SEM of IFN-γ mRNA levels in lung neutrophils. Statistical significance was determined by analysis of variance with the Scheffé post hoc test. The number of mice (n) is provided. For each stimulus, three to eight separate experiments were performed.
PBS = 1.
p < 0.05 versus PBS group.
p < 0.001 versus PBS group.
These studies led to the hypothesis that differential expression of microRNAs induced by E. coli but not S. pneumoniae results in inhibition of IFN-γ translation. MicroRNAs are 18- to 25-nucleotide noncoding RNAs that bind to the noncoding 3′ UTR and either suppress translation or result in RNA degradation (30, 31). Using the two databases, miRBase (32, 33) and microRNA.org (34, 35), microRNAs were identified that (1) recognized the noncoding region of the murine IFN-γ gene and (2) were expressed by neutrophils (36). Searches revealed 17 microRNAs that fit these two criteria (Table 5), and the expression of these 17 microRNAs was quantified by real-time PCR in neutrophils from lungs exposed to S. pneumoniae, E. coli, and PBS. In addition to these 17 microRNAs, miR-9 and miR-122 also recognize the noncoding region of IFN-γ, but they are not produced by neutrophils. miR-9 was included because it is highly expressed in other cell types, though not in CD15+ granulocytes (36). miR-122 was included because it is down-regulated by IFN-β, which is expressed in S. pneumoniae but not E. coli pneumonia (our unpublished data). The data show that RNA isolated from lung neutrophils was high in quality, as assessed with the 2100 bioanalyzer (Agilent). As expected, miR-9 and miR-122 were not expressed in neutrophils from lungs exposed to PBS, S. pneumoniae, or E. coli. S. pneumoniae induced production of two microRNAs (miR-29b and miR-29c), compared with PBS, whereas E. coli induced production of these two microRNAs and eight others (asterisk, relative amount; Table 6). Most importantly, seven microRNAs were expressed to a greater degree in E. coli than S. pneumoniae pneumonia, as measured by the fold change from levels found in control PBS lungs (dagger, Table 6; miR-15b, miR-26a, miR-26b, miR-29a, miR-30b, miR-93, and miR-106b). The increase in expression induced by E. coli compared with S. pneumoniae ranged from 56 to 228% (Table 6). None of the measured microRNAs were expressed only in S. pneumoniae but not E. coli. Thus, these observations show that during pneumonia, differential expression of multiple microRNAs that target the noncoding region of IFN-γ occurs. We postulate that this differential expression provides a mechanism to explain the failure of neutrophils to translate IFN-γ mRNA to protein in E. coli pneumonia.
TABLE 5.
microRNAs THAT TARGET THE 3′ UNTRANSLATED REGION OF IFN-γ mRNA AND ARE EXPRESSED IN LUNG NEUTROPHILS*
| miRNA | Binding Site of miRNA on 3′ UTR of IFN-γ mRNA | miRNA Gene Location | P Value Comparing S. pneumoniae and E. coli |
|---|---|---|---|
| miR-15a | 376 | 14: 62250864–62250947 [–] | 0.046 |
| miR-15b | 375 | 3: 68813694–68813757 [+] | 0.040 |
| miR-26a | 36 | 9: 118940914–118941003 [+] | 0.021 |
| miR-26b | 37 | 1: 74440884–74440968 [+] | 0.019 |
| miR-29a | 503 | 6: 31012660–31012747 [–] | 0.008 |
| miR-30b | 393 | 15: 68168977–68169072 [–] | 0.008 |
| miR-106b | 410 | 5: 138606965–138607046 [–] | 0.031 |
| miR-424 | 376 | X: 50407432–50407526 [–] | 0.043 |
| miR-16 | 376 | 14: 62250717–62250809 [–]3: 68813824–68813918 [+] | P ≥ 0.05 |
| miR-17 | 408 | 14: 115442893–115442976 [+] | P ≥ 0.05 |
| miR-27a | 247, 531 | 8: 86732571–86732657 [+] | P ≥ 0.05 |
| miR-27b | 247, 531 | 13: 63402020–63402092 [+] | P ≥ 0.05 |
| miR-29b | 506 | 6: 31013023–31013093 [–] | P ≥ 0.05 |
| miR-29c | 503 | 1: 196863741–196863828 [+] | P ≥ 0.05 |
| miR-93 | 412 | 5: 138606751–138606838 [–] | P ≥ 0.05 |
| miR-99b | 271 | 17: 17967152–17967221 [+] | P ≥ 0.05 |
| miR-186 | 226 | 3: 157207243–157207313 [+] | P ≥ 0.05 |
Definition of abbreviations: E. coli = Escherichia coli; miRNA = microRNA; S. pneumoniae = Streptococcus pneumoniae; UTR = untranslated region.
P values were calculated by comparing the ΔCt values by analysis of variance and Scheffé post hoc test. Data represent means ± SEM of microRNA levels in lung neutrophils from nine mice studied in three separate experiments.
TABLE 6.
EXPRESSION OF microRNAs IN LUNG NEUTROPHILS DURING PNEUMONIA
| ΔCt |
Relative Amount of miRNA |
Percentage Difference in E. coli Compared with S. pneumoniae | |||||
|---|---|---|---|---|---|---|---|
| miRNA | PBS | S. pneumoniae | E. coli | PBS | S. pneumoniae | E. coli | |
| miR-15a | 4.59 ± 0.04 | 4.48 ± 0.11 | 4.19 ± 0.06*† | 1.00 ± 0.03 | 1.10 ± 0.09 | 1.32 ± 0.06* | 22 |
| miR-15b | 2.09 ± 0.03 | 2.14 ± 0.13 | 1.03 ± 0.49† | 1.00 ± 0.02 | 1.00 ± 0.08 | 3.28 ± 1.05*† | 228 |
| miR-16 | −1.06 ± 0.02 | −1.05 ± 0.16 | −1.42 ± 0.12 | 1.00 ± 0.02 | 1.05 ± 0.13 | 1.32 ± 0.11 | 27 |
| miR-17 | 3.26 ± 0.02 | 3.56 ± 0.40 | 3.11 ± 0.31 | 1.00 ± 0.01 | 1.14 ± 0.34 | 1.30 ± 0.21 | 16 |
| miR-26a | 1.65 ± 0.03 | 1.90 ± 0.20 | 1.12 ± 0.24† | 1.00 ± 0.02 | 0.92 ± 0.14 | 1.60 ± 0.22*† | 68 |
| miR-26b | 3.80 ± 0.04 | 3.74 ± 0.16 | 3.15 ± 0.17*† | 1.00 ± 0.02 | 1.10 ± 0.13 | 1.66 ± 0.21*† | 56 |
| miR-27a | 4.05 ± 0.03 | 4.15 ± 0.14 | 3.83 ± 0.27 | 1.00 ± 0.02 | 0.96 ± 0.08 | 1.34 ± 0.25 | 38 |
| miR-27b | 5.13 ± 0.03 | 5.51 ± 0.32 | 5.06 ± 0.26 | 1.00 ± 0.02 | 0.92 ± 0.20 | 1.17 ± 0.17 | 25 |
| miR-29a | 3.42 ± 0.03 | 3.08 ± 0.19 | 2.46 ± 0.10*† | 1.00 ± 0.02 | 1.36 ± 0.20 | 1.98 ± 0.14*† | 64 |
| miR-29b | 6.83 ± 0.05 | 5.92 ± 0.05* | 5.99 ± 0.23* | 1.01 ± 0.04 | 1.90 ± 0.07* | 1.99 ± 0.34* | 9 |
| miR-29c | 7.81 ± 0.04 | 7.25 ± 0.14* | 6.97 ± 0.08* | 1.00 ± 0.03 | 1.53 ± 0.17* | 1.82 ± 0.10* | 29 |
| miR-30b | 1.50 ± 0.02 | 1.60 ± 0.09 | 0.95 ± 0.22*† | 1.00 ± 0.01 | 0.94 ± 0.06 | 1.59 ± 0.22*† | 65 |
| miR-93 | 1.57 ± 0.02 | 2.08 ± 0.26 | 1.27 ± 0.35 | 1.00 ± 0.02 | 0.80 ± 0.15 | 1.50 ± 0.28† | 70 |
| miR-99b | 6.60 ± 0.04 | 7.29 ± 0.53 | 6.55 ± 0.29 | 1.00 ± 0.03 | 1.04 ± 0.35 | 1.22 ± 0.25 | 18 |
| miR-106b | 4.22 ± 0.02 | 4.18 ± 0.15 | 3.60 ± 0.20*† | 1.00 ± 0.02 | 1.08 ± 0.12 | 1.65 ± 0.20*† | 57 |
| miR-186 | 8.82 ± 0.07 | 9.41 ± 0.23 | 8.75 ± 0.23 | 1.01 ± 0.05 | 0.74 ± 0.12 | 1.15 ± 0.15 | 41 |
| miR-424 | 5.18 ± 0.02 | 5.19 ± 0.14 | 4.81 ± 0.11† | 1.00 ± 0.01 | 1.03 ± 0.11 | 1.32 ± 0.10* | 29 |
Definition of abbreviations: E. coli = Escherichia coli; miRNA = microRNA; PBS = phosphate-buffered saline; S. pneumoniae = Streptococcus pneumoniae.
Data represent means ± SEM of microRNA levels in lung neutrophils from nine mice in each group (PBS, S. pneumoniae, E. coli). The ΔCt value was calculated by normalizing for the expression of snoRNA 202 in each sample. Statistical significance was determined by analysis of variance and Scheffé post hoc test for ΔCt values and the relative amounts of miRNA.
P < 0.05 versus the PBS group.
P < 0.05 versus the Streptococcus pneumoniae group.
DISCUSSION
These studies demonstrate the surprising and unexpected finding that neutrophils are producing IFN-γ early in the inflammatory response during pneumonia induced by some bacteria, but not others. Although the possibility that another cell type also contributes to IFN-γ production by this 24-hour time point cannot be completely excluded, the only CD45+ leukocyte producing IFN-γ expresses the neutrophil-specific surface marker, Ly6G (19). S. pneumoniae and S. aureus induced the production of IFN-γ, whereas P. aeruginosa and E. coli did not. Neither LPS nor LTA, two lipid substances within the wall of gram-negative and gram-positive organisms, respectively, induced IFN-γ production by neutrophils, suggesting that other components of bacteria were regulating IFN-γ production either directly or through host defense molecules. IFN-γ was produced only by neutrophils that had emigrated into the tissue at 24 hours and was not present at 6 hours postinfection. Neither circulating neutrophils nor neutrophils within the pulmonary microvasculature that had not yet migrated expressed IFN-γ. Whether the process of migration is critical for IFN-γ production or whether the time needed to migrate is required for the production of this cytokine remains a question.
IFN-γ production was documented by three different approaches. Flow cytometry was used to document intracellular IFN-γ and the cell type that was producing it. To confirm that the anti–IFN-γ antibody was specifically identifying intracellular IFN-γ, the antigen recognized by the antibody was determined by mass spectroscopy and confirmed to be murine IFN-γ. The presence of IFN-γ in the tissue was documented by sensitive ELISAs. Last, IFN-γ mRNA was produced by neutrophils. These data clearly document that neutrophils were producing IFN-γ and that this cytokine was expressed early in the inflammatory response to these two gram-positive organisms.
Our studies are not the first to demonstrate that IFN-γ is produced by neutrophils. In 1998, Yeaman and colleagues demonstrated that neutrophils in the stroma of the uterus stain for IFN-γ at all stages of the cycle, as do circulating neutrophils from the blood treated ex vivo with IL-12 (3). Kirby and colleagues demonstrated that splenic neutrophils and macrophages produced IFN-γ in response to Salmonella infection (4, 37, 38). During early graft loss of pancreatic islet transplants, NK T cells stimulate production of IFN-γ by neutrophils (10). Early in renal ischemia–reperfusion injury, NK T cells also induced expression of IFN-γ in neutrophils within the kidney (9). Furthermore, Ellis and Beaman demonstrated that neutrophils within the lungs produced IFN-γ in response to Nocardia asteroides (7). Most recently, Nance and colleagues demonstrated that neutrophils generate IFN-γ mRNA and protein in response to intrapulmonary Saccharopolyspora rectivirgula, an infection that mimics hypersensitivity pneumonitis (39). Our studies are the first to identify neutrophils as a source of IFN-γ in acute bacterial pneumonias, to demonstrate that neutrophils transcribe mRNA, and to address both the function of IFN-γ and the mechanisms through which it is regulated.
These studies also examined the pathways required to produce this IFN-γ. The nonreceptor Src tyrosine kinases Fgr/Hck/Lyn, the small GTPase Rac2, and the NADPH oxidase component gp91phox were also each required for the production of IFN-γ. Other studies have demonstrated that outside/in intracellular signaling initiated by ligation of CD18 can result in the activation of these Src kinases (40, 41). In turn, these Src kinases activate Rac2 (25), which is required for activity of the assembled NADPH oxidase complex, resulting in the production of oxidants (26). Whether this CD18:Fgr/Hck/Lyn:Rac2:NADPH oxidase pathway is the signaling pathway critical in the production of IFN-γ, or whether each of these molecules is critical in other pathways resulting in IFN-γ production, remains to be determined. However, our data clearly show that Rac2, Fgr/Hck/Lyn, and NADPH oxidase are each required for IFN-γ production by neutrophils. Taken together, these data suggest that production of oxidants and the result oxidative stress may regulate production of IFN-γ mRNA.
Curiously, all four organisms, as well as LPS and LTA, result in the production of IFN-γ mRNA, but only S. pneumoniae and S. aureus resulted in the expression of protein from the message. These data suggest that the translation of IFN-γ mRNA to protein is the critical step in the mechanism underlying this differential expression. Our studies examined the hypothesis that differential expression of microRNAs targeted to the noncoding region of IFN-γ may be the mechanism through which translation is differentially regulated, by quantifying the expression of 17 microRNAs identified through database searches as having sequences that potentially recognize the noncoding region of the IFN-γ mRNA and are produced by neutrophils. Our results show that eight microRNAs are induced in E. coli but not S. pneumoniae pneumonias (Tables 5 and 6), and the expression of these microRNAs correlates with the lack of translation of IFN-γ mRNA. None of these 17 microRNAs was differentially induced only in S. pneumoniae pneumonia, in which both IFN-γ mRNA and protein are produced. Although these data do not prove that these microRNAs are the mechanism for the differential expression of IFN-γ protein from mRNA induced by S. pneumoniae and S. aureus compared with E. coli, P. aeruginosa, LPS, and LTA, they do provide evidence that this may in fact be the mechanism. This inhibition of translation may be due to the cumulative effect of the 56–228% increases in these differentially expressed microRNAs. Furthermore, five of the eight differentially expressed microRNAs target sites within region 375–412 of the IFN-γ gene (Table 5), which may also enhance the effect of each. Moreover, the ability of E. coli to induce the production of these microRNAs targeted to IFN-γ may represent a novel strategy by which this pathogen can evade host defenses, which could also be shared by other pathogens toward IFN-γ or other host genes.
How the genes for these microRNAs are regulated is an important question. The gene miR-106b is regulated by the E2F1 transcription factor (42). The expression of miR-15a and miR-26a is modulated by c-Myc (43, 44). However, to our knowledge, these are the only studies examining the regulation of these differentially regulated miR genes, and determining whether a common regulator might be increased in E. coli but not S. pneumoniae pneumonia is not yet possible until more information about the transcription factors modulating these genes is available.
Most importantly, our studies demonstrate that IFN-γ production modulates host defense against pneumonia. Bacterial clearance of S. pneumoniae (2.8–5.0 × 106 cfu/mouse) from the lungs is eightfold less in IFN-γ–deficient compared with wild-type mice. This poor clearance is accompanied by an increase in neutrophils, which is most likely the result of increased numbers of organisms. In contrast, during E. coli pneumonia, in which IFN-γ is not produced, there is no difference in bacterial clearance between these two genotypes. Studies to understand how IFN-γ is altering bacterial clearance demonstrate that the production of NETs was highly dependent on the production of IFN-γ. NETs are an important bactericidal mechanism in neutrophils, as important or more important than phagocytosis in sepsis and likely in later stages of the neutrophil life span (27, 28). Studies have demonstrated that neutrophils from neonates have no defect in IL-8 production but do have a defect in NET formation compared with neutrophils from adults, and this defect correlated with a deficit in extracellular bacterial killing (45). In addition, the production of NETs was much greater in response to S. aureus than E. coli in vitro (46), which, when considered in light of our observation that S. aureus but not E. coli induces production of IFN-γ, further suggests that IFN-γ is required for robust generation of NETs. Thus, our studies suggest that IFN-γ is regulating the formation of NETs, a highly bactericidal effector mechanism by neutrophils, and that this NET formation is responsible for the poor clearance of S. pneumoniae in IFN-γ–deficient animals. Other functions for IFN-γ also seem likely, for example, in regulating the balance between Th1 and Th2 responses and the production of granulomas (7, 37–39, 47).
In summary, these studies demonstrate that neutrophils produce IFN-γ in response to S. pneumoniae and S. aureus pneumonias in mice and that this IFN-γ production is important in host defense and the clearance of organisms from the lungs. These studies add to a growing list of critically important functions of neutrophils not only in releasing preformed granular contents and in generating reactive oxygen species but also in synthesizing regulatory molecules that modulate the inflammatory and innate immune responses (48, 49). IFN-γ production is highly regulated, and these studies begin to identify the signaling molecules that are important in the production of IFN-γ by these cells. The clinical relevance of the production of IFN-γ by neutrophils at this early time in the evolution of bacterial pneumonia is demonstrated by the defect in host defense that results when IFN-γ cannot be produced. These studies also suggest that host defense may be enhanced by the administration of exogenous IFN-γ in susceptible patient populations. In fact, IFN-γ is already used in patients with chronic granulomatous disease, in which NADPH oxidase is genetically dysfunctional. Thus, the induction of IFN-γ and its function have important therapeutic implications.
Acknowledgments
The authors are grateful to Kathleen Lundberg and Mark Chance, Ph.D., at the Case Western Reserve University Center for Proteomics and Bioinformatics for the studies identifying IFN-γ as the antigen for the antibody. The authors are very grateful to Nancy A. Rebert, Sarah Dawson, and Jessica Martin for their technical expertise, and Lisa Brown for help with the manuscript.
Supported by NIH grants HL048160, HL052466, and HL077370 (C.M.D.) and by NIH grant HL045635 (M.C.D.). M.Y. was supported by Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (2006). J.C.G. was supported by T32 007415.
Originally Published in Press as DOI: 10.1164/rccm.201004-0592OC on December 17, 2010
Author Disclosure: M.Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.C.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.E.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.A.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.C.D. was a consultant for InterMune and she receives royalties from Springer. D.P.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.M.D. received grant support from the NIH.
References
- 1.Farrar MA, Schreiber RD. The molecular cell biology of interferon-γ and its receptor. Annue Rev Immunol 1993;11:571–611. [DOI] [PubMed] [Google Scholar]
- 2.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol 2004;75:163–189. [DOI] [PubMed] [Google Scholar]
- 3.Yeaman GR, Collins JE, Currie JK, Guyre PM, Wira CR, Fanger MW. IFN-γ is produced by polymorphonuclear neutrophils in human uterine endometrium and by cultured peripheral blood polymorphonuclear neutrophils. J Immunol 1998;160:5145–5153. [PubMed] [Google Scholar]
- 4.Kirby AC, Yrlid U, Wick MJ. The innate immune response differs in primary and secondary salmonella infection. J Immunol 2002;169:4450–4459. [DOI] [PubMed] [Google Scholar]
- 5.Hwang SJ, Kim S, Park WS, Chung DH. IL-4–secreting NKT cells prevent hypersensitivity pneumonitis by suppressing IFN-γ–producing neutrophils. J Immunol 2006;177:5258–5268. [DOI] [PubMed] [Google Scholar]
- 6.Ethuin F, Gerard B, Benna JE, Boutten A, Gougereot-Pocidalo MA, Jacob L, Chollet-Martin S. Human neutrophils produce interferon γ upon stimulation by interleukin-12. Lab Invest 2004;84:1363–1371. [DOI] [PubMed] [Google Scholar]
- 7.Ellis TN, Beaman BL. Murine polymorphonuclear neutrophils produce interferon-γ in response to pulmonary infection with Nocardia asteroides. J Leukoc Biol 2002;72:373–381. [PubMed] [Google Scholar]
- 8.Bogdan C, Schleicher U. Production of interferon-γ by myeloid cells—fact or fancy? Trends Immunol 2006;27:282–290. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Huang L, Sung SS, Lobo PI, Brown MG, Gregg RK, Engelhard VH, Okusa MD. NKT cell activation mediates neutrophil IFN-γ production and renal ischemia–reperfusion injury. J Immunol 2007;178:5899–5911. [DOI] [PubMed] [Google Scholar]
- 10.Yasunami Y, Kojo S, Kitamura H, Toyofuku A, Satoh M, Nakano M, Nabeyama K, Nakamura Y, Matsuoka N, Ikeda S, et al. Vα14 NK T cell–triggered IFN-γ production by Gr-1+CD11b+ cells mediates early graft loss of syngeneic transplanted islets. J Exp Med 2005;202:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med 2008;358:716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katze MG, He Y, Gale M Jr. Viruses and interferon: a fight for supremacy. Nat Rev Immunol 2002;2:675–687. [DOI] [PubMed] [Google Scholar]
- 13.Torres A, Rello J. Update in community-acquired and nosocomial pneumonia. Am J Respir Crit Care Med 2009;181:782–787. [DOI] [PubMed] [Google Scholar]
- 14.American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 2005;171:388–416. [DOI] [PubMed] [Google Scholar]
- 15.Gomez JC, Yamada M, Rebert NA, Doerschuk CM. IFN-γ production by neutrophils during S. pneumoniae pneumonia requires NADPH oxidase [abstract]. FASEB J 2008;22:47.10.17666454 [Google Scholar]
- 16.Yamada M, Doerschuk CM. Production of interferon-γ (IFN-γ) by neutrophils during bacterial pneumonia [abstract]. Am J Respir Crit Care Med 2007;175:A772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada M, Doerschuk CM. IFN-γ is required for the host defense against S. pneumoniae, but not E. coli during bacterial pneumonia [abstract]. Am J Respir Crit Care Med 2008;177:A36. [Google Scholar]
- 18.Yamada M, Doerschuk CM. Interferon-γ (IFN-γ) contributes to the formation of neutrophil extracellular traps (NETs) for host defense against S. pneumoniae during pneumonia [abstract]. Am J Respir Crit Care Med 2009;179:A5748. [Google Scholar]
- 19.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol 2008;83:64–70. [DOI] [PubMed] [Google Scholar]
- 20.Reutershan J, Basit A, Galkina EV, Ley K. Sequential recruitment of neutrophils into lung and bronchoalveolar lavage fluid in LPS-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol 2005;289:L807–L815. [DOI] [PubMed] [Google Scholar]
- 21.Mizgerd JP, Lupa MM, Hjoberg J, Vallone JC, Warren HB, Butler JP, Silverman ES. Roles for early response cytokines during Escherichia coli pneumonia revealed by mice with combined deficiencies of all signaling receptors for TNF and IL-1. Am J Physiol Lung Cell Mol Physiol 2004;286:L1302–L1310. [DOI] [PubMed] [Google Scholar]
- 22.O'Hara AJ, Vahrson W, Dittmer DP. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood 2008;111:2347–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 24.Li B, Yu H, Zheng W, Voll R, Na S, Roberts AW, Williams DA, Davis RJ, Ghosh S, Flavell RA. Role of the guanosine triphosphatase Rac2 in T helper 1 cell differentiation. Science 2000;288:2219–2222. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Yamauchi A, Marchal CC, Molitoris JK, Quilliam LA, Dinauer MC. Chemoattractant-stimulated Rac activation in wild-type and Rac2-deficient murine neutrophils: preferential activation of Rac2 and Rac2 gene dosage effect on neutrophil functions. J Immunol 2002;169:5043–5051. [DOI] [PubMed] [Google Scholar]
- 26.Hordijk PL. Regulation of NADPH oxidases: the role of Rac proteins. Circ Res 2006;98:453–462. [DOI] [PubMed] [Google Scholar]
- 27.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science 2004;303:1532–1535. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 2007;176:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinelli S, Urosevic M, Daryadel A, Oberholzer PA, Baumann C, Fey MF, Dummer R, Simon HU, Yousefi S. Induction of genes mediating interferon-dependent extracellular trap formation during neutrophil differentiation. J Biol Chem 2004;279:44123–44132. [DOI] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 31.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 2006;25:6163–6169. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S. MiRBase: the microRNA sequence database. Methods Mol Biol 2006;342:129–138. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. MiRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.Org resource: targets and expression. Nucleic Acids Res 2008;36:D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol 2004;2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007;129:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirby AC, Sundquist M, Wick MJ. In vivo compartmentalization of functionally distinct, rapidly responsive antigen-specific T-cell populations in DNA-immunized or Salmonella enterica serovar typhimurium–infected mice. Infect Immun 2004;72:6390–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirby AC, Yrlid U, Svensson M, Wick MJ. Differential involvement of dendritic cell subsets during acute salmonella infection. J Immunol 2001;166:6802–6811. [DOI] [PubMed] [Google Scholar]
- 39.Nance S, Cross R, Yi AK, Fitzpatrick EA. IFN-γ production by innate immune cells is sufficient for development of hypersensitivity pneumonitis. Eur J Immunol 2005;35:1928–1938. [DOI] [PubMed] [Google Scholar]
- 40.Giagulli C, Ottoboni L, Caveggion E, Rossi B, Lowell C, Constantin G, Laudanna C, Berton G. The Src family kinases Hck and Fgr are dispensable for inside-out, chemoattractant-induced signaling regulating β2 integrin affinity and valency in neutrophils, but are required for β2 integrin–mediated outside-in signaling involved in sustained adhesion. J Immunol 2006;177:604–611. [DOI] [PubMed] [Google Scholar]
- 41.Berton G, Mocsai A, Lowell CA. Src and Syk kinases: key regulators of phagocytic cell activation. Trends Immunol 2005;26:208–214. [DOI] [PubMed] [Google Scholar]
- 42.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2f1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008;13:272–286. [DOI] [PubMed] [Google Scholar]
- 43.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 2008;112:4202–4212. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Li JY. MicroRNA gene expression in malignant lymphoproliferative disorders. Chin Med J (Engl) 2007;120:996–999. [PubMed] [Google Scholar]
- 45.Yost CC, Cody MJ, Harris ES, Thornton NL, McInturff AM, Martinez ML, Chandler NB, Rodesch CK, Albertine KH, Petti CA, et al. Impaired neutrophil extracellular trap (NET) formation: a novel innate immune deficiency of human neonates. Blood 2009;113:6419–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Ziffle JA, Lowell CA. Neutrophil-specific deletion of Syk kinase results in reduced host defense to bacterial infection. Blood 2009;114:4871–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tateda K, Moore TA, Deng JC, Newstead MW, Zeng X, Matsukawa A, Swanson MS, Yamaguchi K, Standiford TJ. Early recruitment of neutrophils determines subsequent T1/T2 host responses in a murine model of Legionella pneumophila pneumonia. J Immunol 2001;166:3355–3361. [DOI] [PubMed] [Google Scholar]
- 48.Dale DC, Boxer L, Liles WC. The phagocytes: neutrophils and monocytes. Blood 2008;112:935–945. [DOI] [PubMed] [Google Scholar]
- 49.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol 2006;6:173–182. [DOI] [PubMed] [Google Scholar]