Abstract
Bio-inspired mineralization is an innovative approach to the fabrication of bone biomaterials mimicking the natural bone. Bone mineral hydroxylapatite (HAP) is preferentially oriented with c-axis parallel to collagen fibers in natural bone. However, such orientation control is not easy to achieve in artificial bone biomaterials. To overcome the lack of such orientation control, we fabricated a phage-HAP composite by genetically engineering M13 phage, a non-toxic bio-nanofiber, with two HAP-nucleating peptides derived from one of the non-collagenous proteins, Dentin Matrix Protein-1 (DMP1). The phage is a biological nanofiber that can be mass produced by infecting bacteria and is non-toxic to human beings. The resultant HAP-nucleating phages are able to self-assemble into bundles by forming β-structure between the peptides displayed on their side walls. The β-structure further promotes the oriented nucleation and growth of HAP crystals within the nanofibrous phage bundles with their c-axis preferentially parallel to the bundles. We proposed that the preferred orientation resulted from the stereochemical matching between the negatively charged amino acid residues within the β-structure and the positively charged calcium ions on the (001) plane of HAP crystals. The self-assembly and mineralization driven by the β-structure formation represent a new route for fabricating mineralized fibers that can serve as building blocks in forming bone repair biomaterials and mimic the basic structure of natural bones.
Keywords: phage display, hydroxylapatite, self-assembly, β-sheet, dentin matrix protein-1
Introduction
In recent decades, biomimetism/bioinspiration has been a promising direction for the innovative design of biological materials and systems. The fabrication of novel biomaterials for bone regeneration by mimicking the composition of natural bones is a hot area in the bioengineering field. Natural bone is a hierarchically structured composite material assembled under organism’s fine control. It is mainly composed of organic collagen fibrils, non-collagenous proteins and inorganic carbonated hydroxylapatite (HAP). Collagen molecules self-assemble into triple helical polypeptides and then further assemble into fibrils which facilitate the nucleation and oriented growth of HAP crystals on them. Within these fibrils, non-collagenous proteins play a role in the preferential growth of oriented HAPs with c-axis parallel to the collagen fibrils, 1-5 although other studies also show that the negatively charged noncollagenous proteins may simply inhibit extrafibrillar mineralization and the organization of the minerals inside the fibrils is controlled by collagen molecules.6-8 Mimicking the natural method of HAP biomineralization on collagen fibrils is not only the key to but also a challenge for bone regeneration by biomimicry. To achieve this, several materials such as collagen,9-11 polymers,12-17 and spider silks18, 19 have been employed to demonstrate different levels of control over mineralization and the resultant mineralized structures showed good mechanical support and/or biocompatibility as the artificial bone materials.
M13 phage is a type of virus that specifically infects bacteria. It is a flexible biopolymer fiber with ~7 nm in width and ~1 μm in length which resembles the morphology of collagen fibrils. It has a single stranded genome DNA packed into an outer protein coat composed of 2700 copies of helically aligned major coat proteins (pVIII) on the side walls and 5 copies of various minor coat proteins (including pIII, pVI, pVII, and pIX) at the two distal tips. The N-terminal end of the coat protein is exposed to the external environment and can be genetically engineered to display foreign peptides on the phage surface, termed as phage display technique (Scheme 1). Its main application is to study protein-protein interaction since invented in 1985.20, 21 However, more importantly, M13 phage has also become a good candidate of biomaterials since it would not induce obvious toxicity and immune response in human beings as reported in previous literatures. 22-25 This human friendly property indicates the potential safety of applying phage-inorganic composites into human bodies.
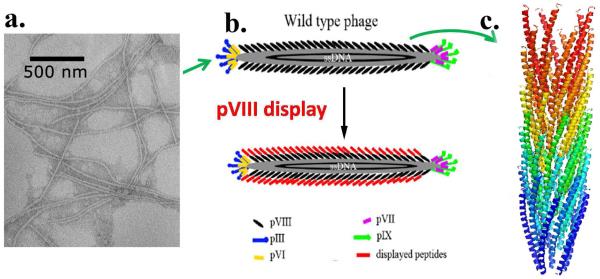
Scheme 1.
Phage display technique. (a) TEM image of wild type M13 phages. (b) Display of a foreign peptide on the side wall of M13 phage by inserting the peptide into the N-terminus of major coat protein (pVIII) of M13 phage. (c) 3D model of pVIII (PDB ID: 1ifd).
Earlier we have found that electrostatic interaction between Ca2+ ions and phage with a negatively charged surface (e.g., M13 phage genetically displaying a negatively charged E8 peptide sequence on the side wall or fd phage with more negatively charged amino acids on the side wall) can drive the assembly of the filamentous phages into bundles.26, 27 In this work, we report our recent findings that when two peptides, which can form β-structure and promote HAP nucleation and growth in their free forms, can still form β-structures once genetically fused to the major coat protein on the side wall of M13 phage, which can further drive the self-assembly of the phage into bundles and favor the nucleation and growth of HAP within the bundles. The driving force of phage self-assembly and mineral nucleation in this work is the formation of β-sheet structure instead of electrostatic interaction which is easily affected by environments, such as pH value and solvents. This work is the first attempt to employ β-sheet structure for achieving phage self-assembly. The β-sheet structure formed in this work can easily form in water, and no additional inducting agent (e.g. Ca2+ ions) is needed. This finding may introduce a novel way for molecular self-assembly and it is an improvement of our previous works.
The β-structure-forming and HAP-nucleating peptides to be displayed on M13 phages are derived from a reported HAP-nucleating domain of dentin matrix protein 1 (DMP-1), one of the non-collagenous proteins that can promote HAP nucleation. 28 DMP-1 can be found in mineralized matrix of bone and dentin.29 It is an acidic protein and processed into a 57kDa C-terminal domain and a 37kDa N-terminal domain. The C-terminus contains both HAP-nucleating domains and collagen-binding domains.28 Two HAP-nucleating sites 386ESQES390 (named pE) and 415QESQSEQDS422 (named pQ) have been identified from the C-terminal fragment of DMP1.28 In the presence of calcium ions, these two HAP-nucleating domains can form an intermolecular β-sheet structure in their free forms and further self-assemble into a microfibril endowing its ability to precisely control the nucleation of HAP crystals.28 By utilizing the phage display technique, these two HAP-nucleating peptides from DMP-1 (pE and pQ) were separately fused into the N terminal of pVIII coat protein of M13 phage so that these two peptides were displayed on the surface of M13 phages. The engineered phage displaying pE peptide (ESQES) was termed as E phage and the one displaying pQ peptide (QESQSEQDS) was termed as Q phage. With the help of displayed HAP-nucleating peptides, E and Q phages are expected to have the ability to nucleate HAP crystals on them. In addition, due to the helically chiral surface of bacteriophage with periodically and precisely aligned HAP-nucleating peptides, the nucleation sites are expected to be orderly aligned on the bacteriophage surface. This fine control is expected to allow us to organize HAP crystals along phage bundles with a preferred orientation.
Our strategy is to create a phage-HAP composite for mimicking the basic structure of natural bone. The methodology includes three main steps: first, two β-structure-forming and HAP-nucleating peptides derived from DMP-1 are genetically displayed onto the surface of M13 phages separately; the intermolecular β-structure formation between the peptides displayed on the neighbouring phages drives the self-assembly of phages into bundles to simulate collagen fibrils in nature bone; finally, HAP crystals are precisely nucleated from HAP supersaturated solution onto the HAP-nucleating β-structures within the formed engineered phage bundles with a bone-like c-axis preferred orientation.
Materials and Methods
Reagents.
Luria broth (LB) medium, tetracycline, chloramphenicol, and polyethylene glycol (PEG) were purchased from Sigma Co. Tris was obtained from Fisher. Kanamycin was obtained from Fluka and sodium chloride was purchased from EMD.
Molecular cloning of M13 bacteriophage PVIII display.
Phage display technique was used to introduce two DMP-1 derived peptides (pE: 386ESQES390 and pQ: 415QESQSEQDS422) into the N-terminus of M13 phages’ major coat protein (PVIII) respectively. Three primers were designed and synthesized by Invitrogen to introduce the desired peptides. pE primer: 5′-ATCCATGGCGGAATCTCAGGAATCTGATCCCG CAAAAGC-3′; pQ primer: 5′-ATCCATGGCGCAGGAATCTCAGTCTGAACAGGACTCTGATCCCGCAAAAGCG-3′; Reverse primers: 5′-GCAAGCTTTTATCAGCTTGCTTTCGAG-3′. Then M13KO7 phage replicative form (RF) DNA was used as template with pE or pQ primer and reverse primer to do PCR amplification. The purified PCR product was digested with appropriate restriction endonucleases (NcoI and HindIII) and fused into linearized phagemid. The recombinant phagemids were confirmed by DNA sequencing (McLab Inc., CA).
The production, amplification, and purification of engineered phage.
The recombinant phage virions (engineered phage) displaying two separate DMP-1 derived peptides were obtained by incubating one tube (5 ml) of recombinant phagemids-transfected Escherichia coli TG-1 strain with helper phage, wild type M13KO7 virions, for one hour at 37°C. LB-agar plates, antibiotics, LB broth, and chloramphenicol antibiotics were used during this process. The 5 ml recombinant phage virions were then transferred to 1L LB medium containing chloramphenicol on a shaking incubator at 37°C. Kanamycin and IPTG were added to make final concentration of 70 μg/ml and 0.1 mM, respectively. After obtaining enough engineered phage, we conducted PEG double precipitation to purify it. The overnight culture was first centrifuged at 3000g for 15 min at 4°C with a Beckman Coulter high-performance centrifuge. The supernatant was collected and recentrifuged at 8200g for 15min at 4°C. The precipitate was suspended by 0.15 volume of 20% PEG/2.5M NaCl solution and stored in a refrigerator overnight. Phage precipitate was collected at 8200g for 1 h at 4°C, dissolved in Tris-Buffered Saline (TBS), and recentrifuged for 10 min to clear. The final phage precipitate was diluted by appropriate volume of double distilled water. The concentrations of both harvested engineered phages were determined via ultraviolet-visible absorption at 269 nm and the suspensions were diluted by ddH2O to have the same value of optical density (OD269nm=1).
The preparation of HAP supersaturated solution.
Supersaturated HAP solution was prepared from HAP powder (Sigma) as described in our previous publications.26, 27 Briefly, a stock solution was prepared first by dissolving HAP powder (Sigma) with 100mM of hydrochloric acid to reach a concentration of 50 mM of calcium. Then the HAP stock solution was diluted by sodium chloride solution to achieve the final concentration of 200 mM sodium chloride and 4 mM of calcium. The pH value was carefully adjusted to 7.01 with 0.05 M potassium hydroxide.
Nucleation of phages in HAP supersaturated solution.
20 μl of 0.2 mg/ml phage suspension was mixed with 1ml supersaturated HAP solution in vials. After 5, 10, 15, and 20 days, 10 ml solution was transferred onto a transmission electron microscopy (TEM) copper grid, rinsed to remove soluble salts and characterized by TEM.
Ultraviolet (UV) Absorption and Circular Dichroism (CD) spectroscopy.
300 μl of engineered phages fused with pE or pQ, were measured using UV absorption and CD spectroscopy. Mixed phages were made by mixing 150 μl E phages and 150 μl Q phages. UV absorption spectra were measured using a spectrophotometer. CD spectra were obtained using a JASCO 750 circular dichrometer. CD spectra were measured using a spectral band width of 1 nm in 0.1 cm cells. Measured CD spectra of the phages were then smoothed and plotted as [θ] in units of deg10−3cm2 dmol−1. Each spectrum was the average of five measurements. The phage concentration used for the analyzed CD measurements is 0.2 mg ml−1. Concentrations of phage solutions were determined from absorbance measurements and published extinction coefficients in unit of mg−1cm2 of 3.84 at 269 nm for M13 phages.30
Transmission Electron Microscopy Characterization.
The TEM images were obtained using a transmission electron microscope (JEOL2000, 200 kV). For pure phage samples, 10 μl of phage solutions was dried on the copper grids and washed by 10 μl ddH2O. Then the samples were negatively stained by 1% uranyl acetate (UA). For nucleation samples, 10 μl mineralized phage solution from each nucleation test was put onto the copper grid and allowed to dry. Then the sample was washed by distilled water twice to remove any soluble salts. Since no staining was done on the nucleation samples, under TEM the filamentous structures could be seen only when the phage and minerals were associated together.
Fourier Transform Infrared Spectroscopy (FT-IR) Characterization.
2 mg of freeze-dried phage particles were mixed with 200 mg KBr and the mixtures were then ground. The pellets were pressed to produce transparent disks about 1 mm thick and 13 mm in diameter. FTIR analysis was conducted on a Nexus FTIR spectrometer from Nicolet.
Results and Discussions
Both pE and pQ peptides were derived from DMP1 and could direct the deposition of HAP crystals by an intermolecular β-sheet structure between them.28 In order to test whether these two peptides displayed on separate phages still function as their free form in DMP1, purified E and Q phages were mixed together and incubated for 1 day (For convenience, we termed mixed E and Q phages as EQ phages.). At the same time, E phages and Q phages were separately incubated for 1 day as control.
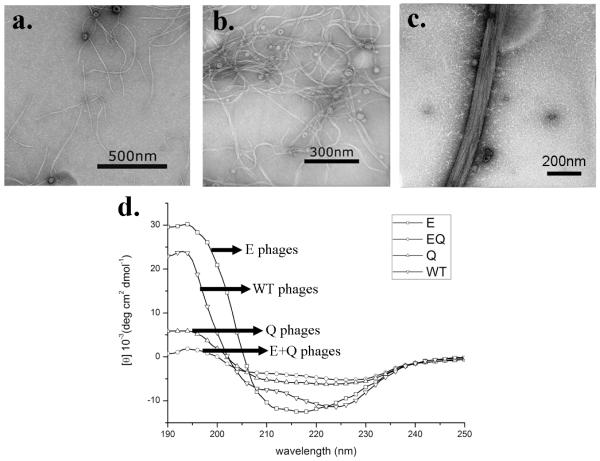
Only individual flexible phage fibers were observed in pure E phage sample (Figure 1a), indicating no interactions occurred between E phages. For Q phages, much to our surprise, they could form small bundles in water by themselves (Figure 1b). The bundles were flexible and randomly distributed, each of which was composed of 3-4 individual Q phages along the width direction. This fact suggests the presence of certain driving force that facilitates the assembly of Q phages into bundles. When mixed together, E and Q phages could self-assemble into larger bundles in which individual phages were arranged parallel in a side-by-side fashion (Figure 1c). These bundles were relatively rigid and longer than Q bundles. About 9-10 phages comprised one bundle of EQ phages along the width direction. These results show that E and Q phages are able to self-assemble into bundles under a stronger driving force compared with the formation of the Q phage bundles.
Figure 1.
TEM images and circular dichroism spectra of engineered phages. (a) E phages; (b) Q phage bundles; (c) EQ phage bundles. (d) Circular dichroism spectra of wild type, E, Q and EQ phages.
Since pE and pQ are capable of forming intermolecular β-sheet structures in DMP-1,28 we expect that engineered E and Q phage bundle should also form β-sheet structures. In order to demonstrate the secondary structure, CD measurements were performed on engineered phages while wild-type phage served as control. The CD spectrum of wild-type phages confirmed that α-helix existed as the major protein secondary structure typical for intact M13 phage (Figure 1d). Its spectrum had one positive band at 192 nm and two negative bands at 208 nm and 222 nm, corresponding to the well-known π →π*, parallel π→π*, and perpendicular π→π* bands of α-helix, respectively.31 It has been well documented that α-helix is the major structure of pVIII coat protein and each phage has ~2700 copies of pVIII protein, so the presence of α-helix in the spectrum is anticipated. Using the method from Whitmore,32 the α-helical and β-sheet contents of wild type phage were estimated to be 74% and 1%, respectively. The CD spectrum of E phages (Figure 1d) shows a combination of α-helical and β-sheet structures, which were estimated to be 63% and 16%, respectively. Since no obvious interactions between individual E phages could be observed from the TEM, we believed that the increased β-sheet contents of E phages might be due to the hydrogen bonding between displayed pE peptides and neighboring pVIII protein on the same surface of an individual phage. Q phages have a similar CD spectrum demonstrating a combination of α-helix and β-sheet structures, but its β-sheet contents were higher (22%) than those in E phages. In our opinion, the driving force of Q phage bundles is possibly the hydrogen bonding in β-sheet structure formed between displayed pQ peptides on neighboring Q phages. The CD spectrum of mixed E and Q phages (Figure 1d) shows a highest content of β-sheet (29%) and lowest content of α-helix (43%) among all samples. Considering the large bundles formed between E and Q phages (Figure 1c), we conclude that the potential driving force was the hydrogen bonding in β-sheet structure formed between pE and pQ peptides displayed on pE and pQ phages, respectively. In addition, increased β-sheet structures were formed between E and Q phages than between Q phages only. These results are consistent with the earlier finding that pE and pQ from DMP1 can form an intermolecular β-sheet structure.28 Besides CD analysis, FT-IR analysis was also applied to confirm the formation of β-sheet between E and Q phages. It is known that amide I band (1600-1700 cm−1) contains several overlapped bands and each band represents a type of secondary structure.33, 34 The FT-IR spectra characteristic of β-sheet will show a stronger band around 1634 cm−1 and a weaker and broad band near 1680 cm−1 35 while those of α-helix will show a single band around 1656 cm−1.34 Our FT-IR spectra of E, Q and EQ phages in the amide I band range (Figure S1) show that the E phage contains essentially α-helix with a broad absorption band around 1656 cm−1. However, a band with a maxima corresponding to β-sheet shows up for Q phage and further increased for EQ phages, suggesting more β-sheet structures formed between E and Q phages than between Q phages or E phages only. The FT-IR results are consistent with the findings from CD spectra; namely, the β-structure content is increased after E and Q phages are mixed together.
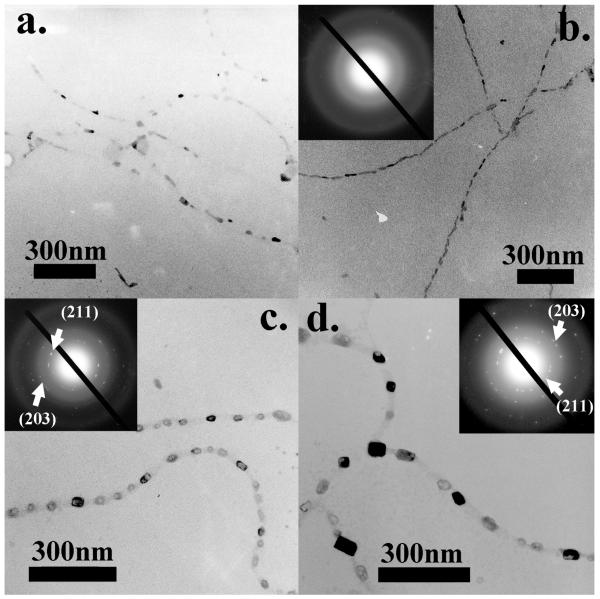
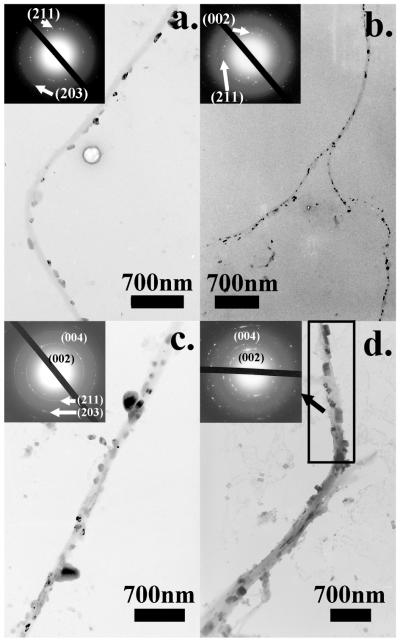
Subsequently, we did a series of in vitro HAP nucleation tests on wild-type, E and Q phages. Specifically, 20 μl of phages were incubated in 1 ml supersaturated HAP (pH=7.01) solution36 at room temperature. Mineral deposited samples were rinsed and characterized under TEM without any staining with 5 days as an interval. Consistent with our earlier findings,26, 27 wild-type phages didn’t show any ability to nucleate HAP minerals. Individual phage particles were observed under TEM only with the help of negative staining (1% PTA) (Figure S2). On E phages, only some amorphous calcium phosphate phase could be observed after incubation for 10 days (Figure 2a). But on day 20, almost all E phage fibers were coated by amorphous calcium phosphate, resulting in a necklace structure (Figure 2b). The amorphous phase of nucleated minerals was confirmed by the electron diffraction (ED) pattern (Figure2b). While on Q phage bundles, small minerals could be found after incubation for 10 days (Figure 2c). The diffraction spots from the (211) and (203) planes of HAP in ED pattern of Q phage samples indicated that the minerals were polycrystalline HAP. At day 20, those HAP crystals had grown into larger crystals but no preferred orientation was observed from the ED pattern (Figure 2d). In addition, Figure 2 c and d suggest that the nucleation sites on Q phage bundles were not distributed uniformly. Similarly, we mixed 10 μl of E phages and 10 μl of Q phages with equal concentrations into 1 ml of HAP supersaturated solution. On day 5 and 10, small HAP crystals appeared on EQ phage bundles. The crystalline structures were verified by the ED patterns. But no obvious preferred orientations of the crystals were found (Figures 3a and b). On day 15 and 20, more and larger HAP crystals appeared along EQ phage bundles (Figures 3c and d). The ED patterns of the bundles show arcs corresponding to (00l) planes of HAP, indicating that HAP crystals on bundles are oriented with their c-axis preferentially parallel to the phage bundles.27 These results show that E and Q phages could co-self-assemble to induce oriented HAP nucleation while individual E phages or Q phages by themselves could not.
Figure 2.
TEM images of nucleation of HAP on E and Q phages after incubation in supersaturated HAP solution for 10 days and 20 days. (a) HAP nucleation on E phages for 10 days. (b) HAP nucleation on E phages for 20 days. (c) HAP nucleation on Q phages for 10 days. (d) HAP nucleation on Q phages for 20 days.
Figure 3.
TEM images of the HAP nucleation on EQ phage bundles after incubation in supersaturated HAP solutions for 5 days (a), 10 days (b), 15 days (c) and 20 days (d).
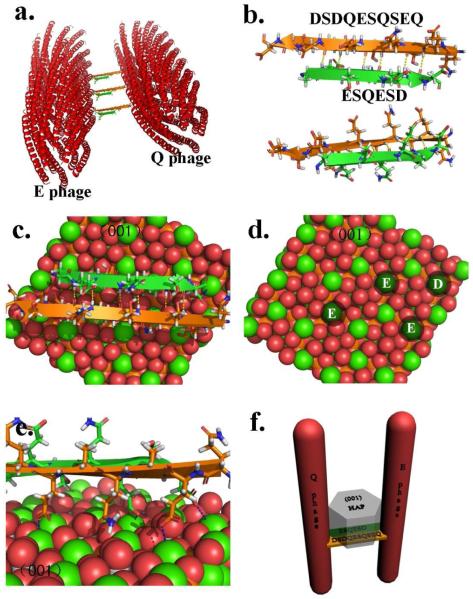
The mechanism of bone biomineralization has been controversial. 5-8 Most recently, a new mechanism was proposed,37 where an amorphous calcium phosphate phase forms “liquid-droplets” that can interact with the non-collagenous proteins and then bind to the preformed collagen matrix. However, we did not find the evidence of converting amorphous calcium phosphate phase into crystalline one in our study. We believe that the orientation control of nucleated HAP crystals on EQ phage bundles was directed by the β-sheets formed between E and Q phages (Figure 4). It is known that pE and pQ could self-assemble into microfibrils by forming intermolecular β-sheets.28 When displayed on phage, pE and pQ peptides are fully exposed and free to interact with other molecules. Therefore, when E and Q phages were mixed, displayed pE and pQ could form β-sheets between neighboring phages which induced the formation of EQ phage bundles (Figure 4a). This was also confirmed by the relatively high content of β-sheet in the CD spectrum of EQ phage bundles (Figure 1d). The possible β-sheet formed was simulated in Figure 4b. Aspartic acid (Asp, D) is the amino acid (originally present in the native pVIII) following the inserted foreign peptides in engineered pVIII proteins of M13 phages. We believe that it is also important for inducing the formation of HAP crystals so it was included in our models. Previous studies found that negatively charged amino acids (D, E or phosphorylated S) had the trend to induce the nucleation of HAP crystals.1, 38 From Figure 4b, we can see that E4, D6 from E1S2Q3E4S5D6 and D1, D3, E5, E9 from D1S2D3Q4E5S6Q7S8E9Q10 are on the same side of β-sheet which could create an attracting center for calcium ions and thus initiate the nucleation of HAP crystal.
Figure 4.
The proposed three dimensional models of possible HAP nucleation mechanism. (a) 3D model of possible EQ phage bundles. (b) The simulation of intermolecular β-sheet formed by two HAP nucleating peptides displayed on the major coat of phage. This model includes the aspartic acid (D) which is the amino acid right following the inserted foreign peptides in pVIII due to its possible contribution to the β-sheet formation. (c) The possible mechanism of the HAP nucleation and growth along c-axis due to the specific match between the negatively charged amino acid in HAP nucleating peptides forming intermolecular β-sheet and the (001) plane of HAP crystals. (d) Schematic illustration showing that the four calcium ions on HAP (001) plane can perfectly match four negatively charged amino acids. (e) A closer look of electrostatic attraction between matched negatively charged amino acids and calcium ions. (f) The 3D model schematically illustrating the possible mechanism of oriented nucleation and growth of HAP crystals triggered by EQ phage bundles. (All the figures were made in PyMOL)
In our experiments, HAP crystals nucleated on EQ bundles were oriented with their c-axis preferentially parallel to the phage bundles (Figure 3c and d). Previous work has demonstrated that β-sheets formed from pE and pQ could nucleate HAP crystals on them.28 We further proposed a model to illustrate how the β-sheets within phage bundles facilitated the oriented nucleation and growth of HAP crystals. Figure 4c, d, and e demonstrate glutamic acids and aspartic acids from both pE and pQ can attract calcium ions and trigger the nucleation process. Specifically, E4 and D6 from pE as well as E5 and E9 from pQ are able to attract 4 calcium ions and those attracted 4 calcium ions on the β-sheet can perfectly match 4 calcium ions in the (001) plane of HAP crystals (Figure 4c and d). As we know, mostly, HAP crystal tends to grow along its [001] direction. Therefore, the electrostatic interaction and stereochemical match will further encourage the nucleation of HAP crystals on the β-sheet with their c-axis parallel to the phage bundles. The nucleated HAP will grow along its c-axis, that is, perpendicular to the plane where the nucleating β-sheet is lying in between neighboring phages. Since the long axis of phage is also perpendicular with the β-sheet, the c-axis of HAP crystals should be parallel to phage bundles (Figure 4f). This structure greatly mimicked the lowest level of hierarchical structure of natural bone in which HAP crystal are nucleated and grown with their c-axis preferentially parallel to the collagen fibrils.1
Our results supported the previous findings that pE and pQ can form β-sheet and nucleate HAP crystals. 28 Moreover, this work suggests the feasibility in the field of bone regeneration of using genetically modifiable biomaterials (e.g. bacteriophage) displaying pE and pQ and inducing HAP deposits by forming β-sheets. Scientists are always looking for a new method to achieve self-assembly. In this study we demonstrate the feasibility by taking advantage of forming β-sheet intermolecular bridge without any artificial inductive or additive reagents. β-sheet structure is a common type of secondary structures of peptides or protein formed via hydrogen bonding. It is a spontaneous and naturally evolved process. If fused on specific positions of genetically engineerable biomaterials, pE and pQ are able to act like leucine zipper peptides39 to assemble biomaterials into hierarchical structures with a precise control by forming β-sheets. We previously found that Ca2+ ions could induce the bundling of negatively charged phages.26, 27 However, the mineralization of such electrostatically bound bundles is faster than that of β-structure-stabilized bundles although both bundles induce the formation of oriented HAP along the bundles. This fact may reflect the different mechanisms involved in the mineralization in this work and suggest that β-structure driven nucleation may be slower.
Conclusions
In summary, we have used M13 bacteriophage with pE and pQ derived from DMP1 protein genetically displayed on the surface to form self-assembled bundles, which can induce the formation of HAP crystals with c-axis preferentially oriented along the bundles. Displayed pE and pQ formed a β-sheet structure, inducing the self-assembly of E and Q phages into EQ phage bundles. This formation of β-sheet can be applied as a general method to induce precise assemblies between engineerable biomaterials. We also found HAP crystals could be nucleated on EQ bundles with their c-axis preferentially parallel to the phage bundles. This structure greatly mimicked the lowest level of hierarchical structure of natural bone in which HAP crystal are nucleated and grown with their c-axis preferentially parallel to the collagen fibrils. The oriented nucleation and growth arose from the fact that glutamic acids and aspartic acids from pE and pQ attracted calcium ions with a pattern matching the organization of calcium ions in the (001) plane of HAP crystals. Such specific match further induced the nucleation of HAP crystals on the β-sheet and directed the nucleated HAP crystal to grow along their c axis. Our work demonstrated a new route for fabricating bone repair materials through biomimicking and bioinspiration.
Supplementary Material
ACKNOWLEDGMENT:
This publication was made possible by Grant Number CBET-0854414 from National Science Foundation and Grant Numbers R01HL092526-01A2 and R03AR056848-01 from National Institutes of Health. We would also like to thank the financial support from National Science Foundation (DMR-0847758 and CBET-0854465), National Institutes of Health (R21EB009909-01A1), and Oklahoma Center for the Advancement of Science and Technology (HR06-161S). AG would like to thank the financial support from the National Institutes of Health by grant DE11657. We greatly appreciate Dr. Karla Rodgers in University of Oklahoma Health Center for kindly help in Circular Dichroism measurement.
Footnotes
SUPPORTING INFORMATION: FT-IR spectra of E, Q and EQ phages as well as mineralization of wild-type M13 phages can be found in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mann S. Molecular Recognition in Biomineralization. Nature. 1988;332(6160):119–124. [Google Scholar]
- 2.Traub W, Arad T, Weiner S. 3-Dimensional Ordered Distribution of Crystals in Turkey Tendon Collagen-Fibers. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(24):9822–9826. doi: 10.1073/pnas.86.24.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman A, Ahn DJ, Lio A, Salmeron M, Reichert A, Charych D. Total Alignment of Calcite at Acidic Polydiacetylene Films - Cooperativity at the Organic-Inorganic Interface. Science. 1995;269(5223):515–518. doi: 10.1126/science.269.5223.515. [DOI] [PubMed] [Google Scholar]
- 4.Roach H. Why does bone matrix contain non-collagenous proteins? The possible roles of osteocalcin, osteonectin, osteopontin and bone sialoprotein in bone mineralisation and resorption. Cell Biol. Int. 1994;18:617–628. doi: 10.1006/cbir.1994.1088. [DOI] [PubMed] [Google Scholar]
- 5.Boskey AL. Matrix proteins and mineralization: an overview. Connect. Tissue Res. 1996;35:357–363. doi: 10.3109/03008209609029212. [DOI] [PubMed] [Google Scholar]
- 6.Price P, Toroian D, Lim J. Mineralization by inhibitor exclusion: the calcification of collagen with fetuin. J. Biol. Chem. 2009;284:17092–17101. doi: 10.1074/jbc.M109.007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nudelman F, Pieterse K, George A, Bomans PHH, Friedrich H, Brylka LJ, Hilbers PAJ, With G. d., Sommerdijk NAJM. The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. Nature Mater. 2010;9:1004–1009. doi: 10.1038/nmat2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silver FH, Landis WJ. Deposition of apatite in mineralizing vertebrate extracellular matrices: a model of possible nucleation sites on type I collagen. Connect. Tissue Res. 2011;52:1–13. doi: 10.3109/03008207.2010.551567. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande AS, Beniash E. Bioinspired synthesis of mineralized collagen fibrils. Crystal Growth & Design. 2008;8(8):3084–3090. doi: 10.1021/cg800252f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Liao SS, Cui FZ. Hierarchical self-assembly of nano-fibrils in mineralized collagen. Chemistry of Materials. 2003;15(16):3221–3226. [Google Scholar]
- 11.Wang Y, Yang C, Chen X, Zhao N. Adv. Eng. Mater. 2006;8:97–100. [Google Scholar]
- 12.Daculsi G, Pilet P, Cottrel M, Guicheux G. Role of fibronectin during biological apatite crystal nucleation: Ultrastructural characterization. Journal of Biomedical Materials Research. 1999;47(2):228–233. doi: 10.1002/(sici)1097-4636(199911)47:2<228::aid-jbm13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 13.Leonor IB, Baran ET, Kawashita M, Reis RL, Kokubo T, Nakamura T. Growth of a bonelike apatite on chitosan microparticles after a calcium silicate treatment. Acta Biomaterialia. 2008;4(5):1349–1359. doi: 10.1016/j.actbio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Murphy WL, Kohn DH, Mooney DJ. Growth of continuous bonelike mineral within porous poly(lactide-co-glycolide) scaffolds in vitro. Journal of Biomedical Materials Research. 2000;50(1):50–58. doi: 10.1002/(sici)1097-4636(200004)50:1<50::aid-jbm8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Filmon R, Grizon F, Basle MF, Chappard D. Effects of negatively charged groups (carboxymethyl) on the calcification of poly(2-hydroxyethyl methacrylate) Biomaterials. 2002;23(14):3053–3059. doi: 10.1016/s0142-9612(02)00069-8. [DOI] [PubMed] [Google Scholar]
- 16.Boduch-Lee KA, Chapman T, Petricca SE, Marra KG, Kumta P. Design and synthesis of hydroxyapatite composites containing an mPEG-dendritic poly(L-lysine) star polycaprolactone. Macromolecules. 2004;37(24):8959–8966. [Google Scholar]
- 17.Verma D, Katti K, Katti D. Bioactivity in in situ hydroxyapatite-polycaprolactone composites. Journal of Biomedical Materials Research Part A. 2006;78A(4):772–780. doi: 10.1002/jbm.a.30774. [DOI] [PubMed] [Google Scholar]
- 18.Kino R, Ikoma T, Monkawa A, Yunoki S, Munekata M, Tanaka J, Asakura T. Deposition of bone-like apatite on modified silk fibroin films from simulated body fluid. Journal of Applied Polymer Science. 2006;99(5):2822–2830. [Google Scholar]
- 19.Cao B, Mao C. Oriented nucleation of hydroxylapatite crystals on spider dragline silks. Langmuir. 2007;23(21):10701–10705. doi: 10.1021/la7014435. [DOI] [PubMed] [Google Scholar]
- 20.Smith GP. Filamentous Fusion Phage - Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 21.Smith GP, Petrenko VA. Phage display. Chemical Reviews. 1997;97(2):391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 22.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 23.Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 24.Frenkel D, Solomon B. Filamentous phage as vector-mediated antibody delivery to the brain. Proc. Natl. Acad. Sci. USA. 2002;99:5675–5679. doi: 10.1073/pnas.072027199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla GS, Krag DN. Selection of tumor-targeting agents on freshly excised human breast tumors using a phage display library. Oncology Reports. 2005;13(4):757–764. [PubMed] [Google Scholar]
- 26.He T, Abbineni G, Cao B, Mao CB. Nanofibrous bio-inorganic hybrid structures formed through self-assembly and oriented mineralization of genetically engineered phage nanofibers. Small. 2010;6:2230–2235. doi: 10.1002/smll.201001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang FK, Cao BR, Mao CB. Bacteriophage Bundles with Prealigned Ca2+ Initiate the Oriented Nucleation and Growth of Hydroxylapatite. Chemistry of Materials. 2010;22(12):3630–3636. doi: 10.1021/cm902727s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He G, Dahl T, Veis A, George A. Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein, 1. Nature Materials. 2003;2(8):552–558. doi: 10.1038/nmat945. [DOI] [PubMed] [Google Scholar]
- 29.George A, Sabsay B, Simonian PAL, Veis A. Characterization of a Novel Dentin Matrix Acidic Phosphoprotein - Implications for Induction of Biomineralization. Journal of Biological Chemistry. 1993;268(17):12624–12630. [PubMed] [Google Scholar]
- 30.Berkowitz SA, Day LA. Mass, Length, Composition and Structure of Filamentous Bacterial Virus Fd. Journal of Molecular Biology. 1976;102(3):531–547. doi: 10.1016/0022-2836(76)90332-6. [DOI] [PubMed] [Google Scholar]
- 31.Clack BA, Gray DM. A CD determination of the alpha-helix contents of the coat proteins of four filamentous bacteriophages: fd, IKe, Pf1, and Pf3. Biopolymers. 1989;28(11):1861–1873. doi: 10.1002/bip.360281103. [DOI] [PubMed] [Google Scholar]
- 32.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89(5):392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 33.Byler DM, Susi H. Examination of the Secondary Structure of Proteins by Deconvolved FTIR Spectra. Biopolymers. 1986;25(3):469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 34.Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochimica et Biophysica Sinica. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 35.Sarver RW, Krueger WC. Protein secondary structure from Fourier transofrm infrared spectroscopy: a data base analysis. Anal. Biochem. 1991;194:89–100. doi: 10.1016/0003-2697(91)90155-m. [DOI] [PubMed] [Google Scholar]
- 36.Silverstone LM, Wefel JS, Zimmerman BF, Clarkson BH, Featherstone MJ. Remineralization of Natural and Artificial Lesions in Human Dental Enamel Invitro - Effect of Calcium-Concentration of the Calcifying Fluid. Caries Research. 1981;15(2):138. doi: 10.1159/000260512. &. [DOI] [PubMed] [Google Scholar]
- 37.Cölfen H. Biomineralization: a crystal-clear view. Nature Mater. 2010;9:960–961. doi: 10.1038/nmat2911. [DOI] [PubMed] [Google Scholar]
- 38.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294(5547):1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 39.Landschulz WH, Johnson PF, Mcknight SL. The Leucine Zipper - a Hypothetical Structure Common to a New Class of DNA-Binding Proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.