Abstract
SF3b155 is an essential spliceosomal protein, highly conserved during evolution. It has been identified as a subunit of splicing factor SF3b, which, together with a second multimeric complex termed SF3a, interacts specifically with the 12S U2 snRNP and converts it into the active 17S form. The protein displays a characteristic intranuclear localization. It is diffusely distributed in the nucleoplasm but highly concentrated in defined intranuclear structures termed “speckles,” a subnuclear compartment enriched in small ribonucleoprotein particles and various splicing factors. The primary sequence of SF3b155 suggests a multidomain structure, different from those of other nuclear speckles components. To identify which part of SF3b155 determines its specific intranuclear localization, we have constructed expression vectors encoding a series of epitope-tagged SF3b155 deletion mutants as well as chimeric combinations of SF3b155 sequences with the soluble cytoplasmic protein pyruvate kinase. Following transfection of cultured mammalian cells, we have identified (i) a functional nuclear localization signal of the monopartite type (KRKRR, amino acids 196–200) and (ii) a molecular segment with multiple threonine-proline repeats (amino acids 208–513), which is essential and sufficient to confer a specific accumulation in nuclear speckles. This latter sequence element, in particular amino acids 208–440, is required for correct subcellular localization of SF3b155 and is also sufficient to target a reporter protein to nuclear speckles. Moreover, this “speckle-targeting sequence” transfers the capacity for interaction with other U2 snRNP components.
A fundamental feature of the mammalian cell nucleus is the presence of morphologically defined structural units often referred to as “nuclear bodies,” including nucleoli, Cajal (coiled) bodies, promyelocytic leukemia bodies, and speckles (1, 2). Most of these nuclear structures have been reported to be involved in the synthesis, processing, and modification of RNA (3) and are thought to be formed in response to gene expression. Moreover, they represent characteristic accumulations of a certain set of nuclear proteins, which, however, rapidly associate and dissociate with these nuclear components, suggesting that they are in continuous flux (4, 5).
Although the amino acid sequences responsible for the nuclear uptake of proteins are well characterized (6), little is still known about the sequence motifs that affect the specific intranuclear distribution of proteins, i.e., accumulation in distinct subnuclear regions. For example, several studies have suggested that a single nucleolar localization signal does not exist (ref. 7 and references therein), and it is widely accepted that targeting of a specific protein to the nucleolus results from direct or indirect interaction with other nucleolar components (8). A similar mechanism can be assumed for targeting a regulatory factor to specific subnuclear sites. The transcription factor AML exhibits a 31-aa element at its C terminus required for the association of the protein to nuclear matrix-associated subnuclear sites where transcription takes place (9, 10), and other specific sequences can direct replication factors to sites at which DNA synthesis occurs (11).
Speckles are intranuclear structures characterized by high concentrations of pre-mRNA splicing factors, which seem to correspond to interchromatin granules originally described in the electron microscope (reviewed in ref. 12). Although the precise function of speckles remains controversial, there is strong evidence that they are sites of splicing component storage and/or assembly (13, 14). The purification and biochemical characterization of speckles has disclosed their extensive complexity with about 150 proteins, although it remains unclear which of these are constitutive components (2, 15).
Recently, we have described the identification and characterization of a highly conserved nuclear 146-kDa protein that shows, in addition to a diffuse nucleoplasmic distribution, a strong enrichment in nuclear speckles. The protein has been identified as a subunit of the U2 snRNP-associated splicing complex SF3b and is now termed SF3b155 and SAP155, respectively (16, 17). Striking features of the primary sequence of SF3b155 are a putative bipartite nuclear localization signal (NLS) and a domain rich in threonine/proline-dipeptide repeats (TP domain). In contrast to a number of non-snRNP splicing factors called SR proteins, SF3b155 contains neither a region enriched in arginine/serine residues (RS domain; reviewed in ref. 18) nor a consensus-type RNA-recognition motif (19), which have been reported to be essential for the proper localization of SR proteins to nuclear speckles (20–22).
In the present study, we have examined the topogenic properties of different protein domains of SF3b155 and have identified molecular segments that mediate its nuclear import and determine its accumulation in nuclear speckles. Our data also indicate that this “speckle-targeting sequence,” i.e., the TP domain of SF3b155, contributes to the association of the protein with the U2 snRNP.
Materials and Methods
Antibodies.
The guinea pig antiserum DRSP-M3 directed against a peptide deduced from the human DRS (domain rich in serines) protein has been described (23). The protein is a specific component of nuclear speckles where it colocalizes with the splicing factor SC35 (24). The mAb 9E10 (American Type Culture Collection, CRL 1729) specifically recognizes an epitope in the decapeptide EQKLISEEDL of the human c-myc protein (25). Secondary antibodies used for immunofluorescence microscopy were Texas Red- and Cy2-conjugated goat antibodies against IgG of mouse or guinea pig, respectively (Dianova, Hamburg, Germany).
Construction of Epitope-Tagged Mutant Forms of SF3b155 and Fusion Proteins.
The cDNA constructs pBT myc, pBT mycPK, pBT B2-Xen, and pBT mycB2-Xen have been described (16, 26). To generate the construct pBT mycPK-Δstop, a 1.63-kb subfragment of pBT mycPK encoding the myc-tag and amino acids 17–525 of pyruvate kinase was isolated by HindIII/BstXI digestion and blunt-end ligated into the SmaI site of the Bluescript expression vector (Stratagene). This construct lacking the authentic stop codon contains several restriction sites at the 3′-end and allows the in-frame insertion of isolated cDNA fragments. All N- and C-terminal deletion mutants of SF3b155 (ΔNC, ΔN, ΔC1-C6; cf. Fig. 2) were derived from the wild-type (wt) SF3b155 cDNA clones pBT B2-Xen and pBT mycB2-Xen, respectively, by using appropriate restriction enzymes.
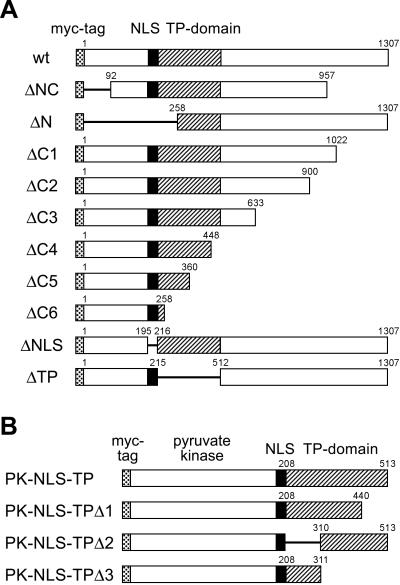
Figure 2.
Mutational analysis of SF3b155. (A) Schematic drawing of the domains of wt protein SF3b155 from X. laevis and various mutated forms (Δ-mutants). The first and the last amino acid residues are indicated by numbers. Hatched areas represent a domain enriched in the dipeptide TP. The myc-tag fused in-frame to the amino terminus of SF3b155 is indicated as a dotted box, whereas a putative NLS is denoted by a black box. (B) Schematic representation of fusion protein constructs containing various parts of the TP domain of SF3b155 fused to pyruvate kinase.
Mutants ΔNLS, ΔTP, and SF3b155-N214N215 were constructed according to the recombinant PCR technique as described (27) by using pBT B2-Xen as template. Mutant ΔNLS was generated by using the “outside” primers out1 (5′-CTGTTAGCGGAAGCTTCGGT-3′) corresponding to bp 35–54 of the B2-Xen cDNA and out2 (5′-CTAGGTGTAGGATCCCAAAT-3′) corresponding to bp 872–853, together with the “inside” primers ins1 (5′-GCACAGCCACCTTCTCTATCGAGCTGGGAT-3′) and ins1′ (complementary to ins1). The latter correspond to the 15 bp each flanking the B2-Xen sequence element encoding the putative NLS (bp 673–732). For the construction of SF3b155-N214N215, primers out1 and out2 were used together with the inside primers ins2 (5′-GGTTCTACTCCAAACCACTATCGAGCTGG-3′) and ins2′ (complementary to ins2) corresponding to bp 715–744 of the B2-Xen cDNA with two base pair exchanges (G729→C, A730→C). ΔTP was generated with out1 and out3 (5′-CTCTGCCAAAGCAGCAAT-3′, bp 2238–2221 of the B2-Xen cDNA) and the primer pair ins3 (5′-TCTACTCCAAAGAAAATGAGGAAGGCTGCC-3′) and ins3′ (complementary to ins3). The latter correspond to the 15 bp each flanking bp 733-1626 of the B2-Xen sequence element, which encodes the TP domain (bp 733-1626). The amplified recombinant PCR fragments were inserted into the B2-Xen cDNA upon digestion with HindIII/BamHI (ΔNLS, SF3b155- N214N215) or HindIII/XcmI fragments (ΔTP) by replacing the corresponding wild-type fragments. For transfection experiments, all constructs were subcloned into the eukaryotic expression vectors pRcCMV and pcDNA3.1(+), respectively (Invitrogen).
For the construction of mycPK-NLS-TP, a fragment of pBT B2-Xen encoding amino acids 196–511 of SF3b155 followed by a stop codon was PCR amplified with the primer pair 5′-CCCCCCCCAAGCTTAAAACGCAAACGTAGA-3′ and 5′-CCCCCCTCTAGATTATGGAGGGTGCCATT-3′. This PCR fragment digested with HindIII/XbaI and a BamHI/HindIII fragment of pBT mycPK-Δstop encoding amino acids 17–525 of pyruvate kinase were ligated to pcDNA3.1(+) previously linearized with BamHI/XbaI. Chimeric constructs lacking different parts of the TP domain were obtained by using the Quick Change site-directed mutagenesis kit (Stratagene). Constructs PK-NLS-TPΔ1 and PK-NLS-TPΔ3 were generated by the introduction of stop codons into PK-NLS-TP at positions corresponding to amino acid 441 and amino acid 312 of SF3b155, respectively. To create PK-NLS-TPΔ2, two conservative point mutations (A2625→C and G2628→T, respectively) were introduced into PK-NLS-TP, thereby generating an AgeI restriction site downstream of the NLS sequence. Subsequently, a PCR fragment comprising amino acids 310–513 of SF3b155 was amplified with specific primers, redigested with AgeI/XbaI, and finally subcloned into the modified PK-NLS-TP construct previously cut with AgeI/XbaI.
MycPK-NLSmonopartite was generated by PCR amplification of a BsrGI/XbaI fragment of mycPK-NLS-TP with the primer pair 5′-GAGGCCGCAATGTTCCAT-3′ and 5′-GAGAGTCTAGACTATCTACGTTTGCGTTTTAA-3′. This PCR fragment, which encoded amino acids 402–525 of pyruvate kinase followed by amino acids 196–200 of SF3b155 and a stop codon, was reinserted into mycPK-NLS-TP by replacing the corresponding BsrGI/XbaI fragment.
DNA Transfection and Immunofluorescence Microscopy.
Culture conditions of human breast adenocarcinoma cells, line MCF-7, have been described (28). Transient transfections of MCF-7 cells were carried out by using FuGene 6 (Roche Molecular Biochemicals) as the transfection reagent according to the manufacturer's protocol. Immunofluorescence microscopy analysis was performed 24 h after transfection by using standard conditions for fixation and immunocytochemistry (28). For actinomycin D experiments, the cells were incubated for 4 h in fresh medium containing a 5 μg/ml concentration of the drug before fixation.
In Vitro Synthesis of cDNA-Cloned Proteins.
[35S]Methionine-labeled proteins were produced in vitro by using the TNT-coupled reticulocyte lysate system (Promega). Translation products were analyzed by SDS/PAGE and autoradiography (16).
Sucrose Gradient Density Centrifugation.
Reticulocyte lysate (10 μl) containing [35S]methionine-labeled epitope-tagged wt SF3b155 was fractionated by centrifugation in a 5–30% linear sucrose gradient in PBS, and resulting fractions were analyzed by SDS/PAGE and autoradiography (16).
Immunoprecipitation from Cellular Extracts of Transfected Cells.
Cellular extracts from transiently transfected MCF-7 cells were obtained as described (16). For immunoprecipitation experiments, 2-ml cell culture supernatants of mAb 9E10 were coupled to 100 μl of preswollen protein G-Sepharose beads (Amersham Pharmacia) for 1 h at 4°C. Cell extracts were preincubated with protein G-Sepharose (1 h, 4°C) to avoid nonspecific binding. Subsequently, the precleared extracts were incubated with the antibody-protein G-Sepharose complex by end-over-end rotation for 2 h at 4°C. Finally, the Sepharose beads with the bound immune complexes were washed five times with lysis buffer and twice with PBS before analyzing the RNA content.
RNA Analysis.
Total RNA from MCF-7 cells was obtained according to standard protocols. The RNA from immunoprecipitates was extracted by phenol/chloroform and digested with RNase-free DNase I to remove any contaminating traces of genomic DNA. Reverse transcription (RT)-PCR was performed by using the Superscript One-Step RT-PCR system (Life Technologies, Karlsruhe, Germany) with the U2 snRNA-specific primer pair 5′-ATCGCTTCTCGGCCTTTTGGC-3′ and 5′-TGGTGCACCGTTCCTGGAGG-3′. The PCR products were analyzed by 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
Results
Sedimentation Behavior of SF3b155.
The formation of heterotypic complexes between (endogenous) wild-type and (transfected) mutant proteins represents a notorious problem with mutational analyses of subcellular protein trafficking (7). Hence, it was important first to determine whether or not SF3b155 displays a propensity to oligomerize. [35S]Methionine-labeled SFb3155, obtained by in vitro translation in a rabbit reticulocyte lysate, was analyzed on linear sucrose gradients (Fig. 1). The bulk of the protein sedimented in fractions 7 to 9; for comparison, BSA (4.3S) peaked in fraction 5, catalase (11.3S) in fraction 11, and thyroglobulin (16.5S) in fraction 19. No SF3b155 was detectable in later fractions, indicating that the bulk of the protein exists predominantly as a monomer with a sedimentation coefficient of ≈7S. This allows the conclusion that oligomerization of SF3b155 would not complicate the interpretation of the results obtained from our systematic mutational analysis of sequence requirements for nuclear accumulation and, in particular, for enrichment in nuclear speckles.
Figure 1.
Sedimentation behavior of SF3b155. [35S]Methionine-labeled SF3b155 translated in vitro from the cDNA clone pBT B2-Xen was fractionated after sucrose gradient centrifugation, separated by SDS/PAGE, and processed for autoradiography. Fraction numbers are indicated on top of the lanes (the top of the gradient is on the left). Bars indicate the peak positions of the reference proteins BSA (4.3S), catalase (11.3S), and thyroglobulin (16.5S). R, reference proteins 205, 116, 97.4, and 66 kDa (from top to bottom). Unfractionated reticulocyte lysate is shown on the right (IVT).
Identification of a Monopartite NLS in SF3b155.
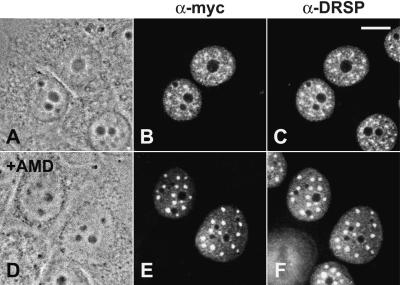
To determine the elements in SF3b155 responsible for the accumulation of the protein in nuclear speckles, we constructed a series of mutants of the Xenopus laevis cDNA (16) suitable for transfection experiments (schematically shown in Fig. 2A). The most characteristic feature of this protein are clusters of the dipeptide TP, which probably represent CDK phosphorylation sites (29), in a domain located between amino acids 208 and 512 (hatched boxes), where they are repeated 29 times. A putative bipartite NLS was noted between amino acids 196–215 [KRKRR (x)13KK; indicated by a black box]. All constructs carried the myc-tag at their N terminus (dotted boxes) to allow the detection of the ectopically expressed proteins upon transient expression in human cultured cells. The results shown in Fig. 3 clearly demonstrate that the myc-tag did not alter the distribution of the wt protein (see also ref. 16). When analyzed by immunofluorescence microscopy, cells expressing wt SF3b155 showed, in addition to a diffuse nucleoplasmic staining, a bright fluorescence of nuclear speckles (Fig. 3 B and E). To facilitate the identification of nuclear speckles, we (i) performed colocalization studies with an antibody against the DRS protein, which is characterized by a domain exceptionally rich in serine residues, and for which a specific enrichment in these structures has been demonstrated (refs. 23, 24; Fig. 3 C and F), and (ii) studied the distribution of the proteins in the presence of the transcription inhibitor actinomycin D (Fig. 3 E and F), known to cause a significant enlargement of nuclear speckles (30).
Figure 3.
Intracellular localization of myc-tagged wt SF3b155 from X. laevis transiently expressed in human MCF-7 cells in the absence (A–C) and presence (D–F) of actinomycin D. Phase contrast micrographs (A and D), immunofluorescence micrographs upon incubation with the anti-myc antibody (B and E), and the antibody directed against the DRS protein (23), respectively (C and F). Scale bar is 10 μm.
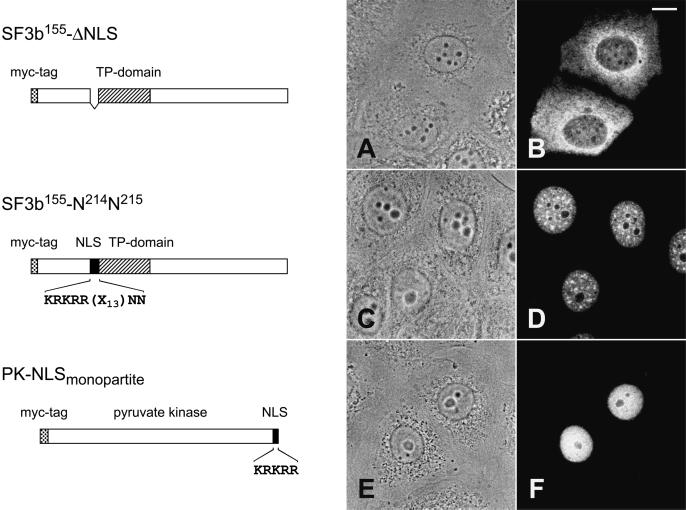
We first concentrated on the identification of the NLS of SF3b155 (Fig. 4). Analyses of the various deletion mutants, in particular of mutant ΔN lacking the N-terminal amino acids 1–257, indicated that a putative NLS is restricted within this region (data not shown). A putative bipartite NLS had been already noted by inspection of the primary sequence; indeed, the corresponding deletion mutant ΔNLS was no longer able to enter the nucleus but remained almost exclusively in the cytoplasm (Fig. 4 A and B). To distinguish the presence of a mono- versus a bipartite NLS (31), mutant SF3b155-N214N215 was generated, in which the second basic cluster was neutralized by changing the lysine residues to asparagine. The resulting mutant protein was still found in the nucleus, indicating that the SF3b155 protein contains a monopartite NLS (Fig. 4 C and D). Moreover, fusion of this sequence element (KRKRR; amino acids 196–200) to pyruvate kinase led to a chimeric protein, which was found exclusively in the nucleus. It should be noted, however, that this fusion protein was diffusely distributed in the nucleoplasm and did not show any enrichment in nuclear speckles (Fig. 4 E and F).
Figure 4.
Identification of a monopartite NLS present in SF3b155. (Left) A schematic description of the analyzed constructs is shown. In SF3b155-ΔNLS, the putative bipartite NLS (KRKRR(x)13KK; amino acid residues 196–216) has been deleted, whereas in mutant SF3b155-N214N215, the second basic cluster was modified by replacing the lysine residues with asparagine. In the fusion protein construct PK-NLS, amino acid residues 196–200 (first basic cluster) of protein SF3b155 were fused in-frame to the cytoplasmic protein pyruvate kinase. (Right) The intracellular distribution of the corresponding proteins expressed in MCF-7 cells upon transient transfection was detected with the anti-myc antibody. Phase contrast (A, C, and E) and corresponding immunofluorescence micrographs (B, D, and F) are shown. Scale bar is 10 μm.
Identification of a Sequence Element Necessary and Sufficient for Localization to Speckles.
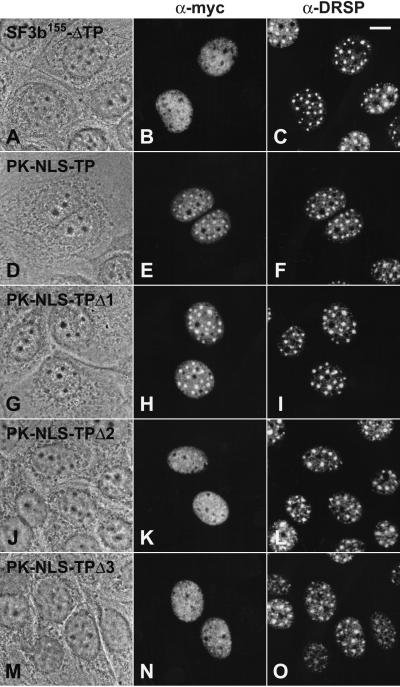
Having identified the NLS of SF3b155, we examined possible subnuclear targeting functions of various molecular regions present in the protein. Analyses of deletion mutants lacking different amino acid numbers at the C terminus (ΔC1–C6) or the N terminus (ΔNC, ΔN) indicated that the TP domain might be required for targeting the protein to nuclear speckles. All mutants containing the entire TP domain in addition to the functional NLS showed a prominent enrichment in speckles, indicating that the C-terminal region is not required for the proper intranuclear targeting of the protein. By contrast, mutated proteins lacking parts of this sequence element showed a reduced capacity to accumulate in nuclear speckles (mutant ΔC5; data not shown). Particularly, mutant ΔC6 lacking amino acids 258-1307 was dispersed throughout the nucleoplasm, whereas a significant accumulation in speckles was not detectable (data not shown). These results suggested that the presence of the TP domain might be essential for the specific subnuclear localization of SF3b155. This was confirmed by an internal deletion mutant ΔTP, lacking amino acids 216–511, which was still able to enter the nucleus by its NLS but did not accumulate in speckles (Fig. 5 A–C). To further analyze the speckle-targeting capacity of the TP domain, we generated a fusion protein with pyruvate kinase. Remarkably, cells expressing this chimeric protein containing the entire TP domain (amino acids 208–513; PK-NLS-TP) showed a strong fluorescence of nuclear speckles when probed with the myc antibody (Fig. 5 D–F), indicating that indeed this domain does play an important role for the characteristic subnuclear distribution of SF3b155. Because this domain is a rather large sequence element, we generated further fusion proteins lacking different parts of the TP domain to map the targeting sequence more precisely (see also Fig. 2B). Analysis of the chimeric protein PK-NLS-TPΔ1, which contains amino acids 208–440 of SF3b155, revealed that the presence of this sequence element is sufficient for a significant enrichment in nuclear speckles (Fig. 5 G–I). In contrast, other truncations of this protein domain (constructs PK-NLS-TPΔ2 and PK-NLS-TPΔ3, respectively) led to fusion proteins that localized throughout the nucleoplasm and did not further accumulate in nuclear speckles in significant amounts (Fig. 5 J–O).
Figure 5.
Identification of a sequence element required for accumulation in speckles. Mutated variant of SF3b155 lacking the entire TP domain as well as fusion proteins consisting of pyruvate kinase fused to the NLS/TP domain of SF3b155 of various lengths (compare with Fig. 2B) were analyzed upon transfection into MCF-7 cells. Phase contrast micrographs (A, D, G, J, and M), immunofluorescence micrographs by using the anti-myc antibody (B, E, H, K, and N), and antibodies against the DRS protein (C, F, I, L, and O), respectively, are shown. Transfected cells were analyzed upon actinomycin D treatment. Scale bar is 10 μm.
The TP Domain of SF3b155 Also Mediates Association with the U2 snRNP.
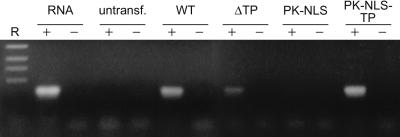
To investigate a possible correlation between the accumulation of SF3b155 in nuclear speckles and its integration into the U2 snRNP, various forms of the transiently expressed protein were immunoprecipitated from transfected cells with the myc antibody. Coprecipitated RNA was subsequently analyzed by RT-PCR using U2-specific primers, and amplified cDNAs were analyzed by agarose gel electrophoresis (Fig. 6). The primers amplified a DNA band of the expected size (186 nt) from total cellular RNA, and the myc antibody coprecipitated the U2 snRNA from cells transfected with wt SF3b155, in accordance with our previous data (16). By contrast, a significantly reduced amount was amplified from cells expressing the mutant protein lacking the TP domain (ΔTP), i.e., the protein showing a strikingly reduced accumulation in speckles (Fig. 5 D–F). Interestingly, fusion of the TP domain to pyruvate kinase (PK-NLS-TP) led to the incorporation of the fusion protein into the U2 snRNP as revealed by the specific coprecipitation of the U2 snRNA (Fig. 6).
Figure 6.
Coimmunoprecipitation of the U2 snRNA. Various forms of SF3b155 were immunoprecipitated in the presence (+) or absence (−) of anti-myc antibody from transfected cells. Total RNAs were extracted, and the U2 snRNA was amplified by RT-PCR with U2-specific primers. In control experiments, immunoprecipitations were done with untransfected cells, and the RT-PCR was performed in the presence or absence of total cellular RNA. The resulting RT-PCR products were separated on agarose gels and detected by ethidium bromide. DNA-size markers of 517, 396, 356, and 247 bp, respectively, are shown in R.
Discussion
As the RS domain is the most prominent feature shared by proteins that so far have been localized to intranuclear structures called speckles, it has been hypothesized that it is this sequence element that directs the specific subnuclear topogenesis (32). In the present study, we have examined the intranuclear distribution of various mutants of the U2 snRNP-associated protein SF3b155, which is not a member of the SR protein family, but nevertheless accumulates in nuclear speckles (16).
We have found that the topogenesis of this protein is determined by two separate sequence elements. Whereas the nuclear uptake of SF3b155 is mediated by an NLS of the monopartite type, its accumulation in speckles depends on a sequence motif located in an extended non-RS-type domain that is highly enriched in the dipeptide threonine/proline, hence designated TP domain (amino acids 208–513). Analyses of a number of chimeric proteins revealed that the presence of amino acids 208–440 is sufficient for targeting a reporter protein to nuclear speckles. Moreover, our immunoprecipitation experiments indicate that the TP domain mediates the association of SF3b155 with the U2 snRNP complex. The coprecipitation of U2 snRNA with antibodies reacting with a hybrid protein combining pyruvate kinase with the TP domain in similar levels than the wt SF3b155 protein confirms the presence of the fusion protein in the U2 snRNP complex. Vice versa, the corresponding deletion mutant of SF3b155 (ΔTP) shows a remarkable reduced binding affinity to this RNP complex, as judged from the much lower amount of U2 snRNA found in the coprecipitate. On the other hand, our observation that at least some U2 snRNA is still bound to this deletion protein suggests that either some additional sequences might be involved in the SF3b155 binding or that the removal of the TP domain generally weakens the interaction of SF3b155 with the U2 snRNA.
Although SR proteins with a single RNA-recognition motif require an RS domain for proper localization in speckles (SC35, SRp20), SR proteins with two RNA-recognition motifs, such as protein SF2/ASF, do not. In the latter, weak speckle targeting signals are present in all three structural elements, which function additively (22). However, the RS domain proposed to function as a speckle-localization signal was always found to overlap with a nucleoplasmin-type, bipartite NLS. In contrast, we find that in SF3b155 the nuclear localization depends on the presence of a monopartite NLS and can be uncoupled from its localization to speckles. Overall, this kind of speckle-targeting sequence, the TP domain, clearly differs from the RS domain described before. We assume that the mechanism of nuclear speckle localization is most likely a complex process, by analogy to the pathways described for localization in other subnuclear components, e.g., Cajal bodies and nucleoli (7, 33), which involve complex signals rather than simple sequence motifs. Several studies of this kind suggested a mechanism for subnuclear localization based on protein–protein interactions mediated by protein-specific domains, e.g., RS domain or, as described here, the TP domain.
Nevertheless, the TP domain and RS domains might have one common feature. Reversible phosphorylation of SR proteins by specific kinases at sites located in the RS domain is likely to serve as a regulatory mechanism of splicing in vitro (34, 35) as well as in intranuclear distribution (36). As noted before, the TP dipeptides found in the speckle-targeting sequence of SF3b155 are putative phosphorylation sites (16). In this regard, it is interesting that SF3b155 associates with cyclin E-cdk2 in vivo (37) and can be phosphorylated concomitant with the splicing reaction (17). However, a mutated form of PK-NLS-TPΔ1 carrying point mutations in five threonine residues (T212PKK, T238PGR, T270PGR, T357PGK, T429PAR), which perfectly match the consensus CDK phosphorylation site (S/TxK/R; ref. 29), showed the same intracellular distribution as the unmutated chimeric protein, i.e., a typical speckled pattern. Moreover, exposure of HeLa cells to low concentrations of the specific Ser/Thr protein phosphatase inhibitor okadaic acid did not cause a significant change in the intranuclear distribution of the endogenous SF3b155 protein as previously reported for coilin and various snRNPs (ref. 38; unpublished results). Clearly, the possible influence of phosphorylation/dephosphorylation events on the intranuclear topogenesis of SF3b155 requires further investigations.
Among the proteins closely interacting with SF3b155 are additional components of the SF3b complex, notably proteins SF3b145 (39) and SF3b130 (40). Both proteins are not members of the SR protein family, and it remains to be shown whether they contribute to the intranuclear targeting of SF3b155. Another SF3b155-interacting protein has been identified as splicing factor U2AF, a well known SR protein, which consists of two subunits referred as U2AF65 and U2AF35. A distinct subdomain of SF3b155 extending from amino acid 267 to 369, i.e., partially overlapping with the TP domain required for speckle targeting, has been reported to be necessary and sufficient for the interaction with U2AF (41). On the other hand, the RS domain of U2AF65 is not required for the enrichment of the protein in nuclear speckles (42), and we have not detected U2AF65 by Western blot analysis in our immunoprecipitates with SF3b155-specific antibodies. Instead, U2AF65 remained exclusively in the supernatant fractions (our unpublished results). In conclusion, so far there is no indication that SF3b155 is directed to speckles via its interaction with other proteins carrying an RS domain.
Speckles can be regarded as regions of splicing factor storage and spliceosome preassembly, whereas splicing occurs at the sites of transcription (2, 3). One might speculate that the TP domain found in SF3b155 serves as a multifunctional protein–protein interaction domain that is crucial for the subnuclear localization of the protein as well as for its integration into the U2 snRNP. In all cases, this domain seems to mediate the assembly of multicomponent complexes. Meanwhile, it is widely accepted that targeting sequences required for the accumulation of a given protein in specific intranuclear structures, i.e., the nucleolus, Cajal bodies, and speckles, may represent interaction domains with other nuclear subdomain-specific molecules. Therefore, the TP domain is an interesting target for future studies to investigate interactions of SF3b155 with other components of the splicing apparatus.
Acknowledgments
We thank Astrid Hofmann for expert technical assistance, Andreas Hunziker for competent sequencing work, Eva Ouis for arranging the typescript, and Maria Carmo-Fonseca for providing mAb MC3 directed against U2AF65. We also acknowledge Werner W. Franke for critical reading of the manuscript. This study was supported by Deutsche Forschungsgemeinschaft Grant Schm 862/3–2 (to M.S.S.-Z.).
Abbreviations
- TP domain
domain rich in threonine/proline-dipeptide repeats
- NLS
nuclear localization signal
- RS domain
region enriched in arginine/serine residues
- DRS
domain rich in serine
- wt
wild type
- RT
reverse transcription
- PK
pyruvate kinase
References
- 1.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- 3.Lewis J D, Tollervey D. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 4.Kruhlak M J, Lever M A, Fischle E, Verdin D P, Bazett-Jones D P, Hendzel M J. J Cell Biol. 2000;150:441–451. doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phair R D, Misteli T. Nature (London) 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 6.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 7.Zirwes R F, Kouzmenko A P, Peters J-M, Franke W W, Schmidt-Zachmann M S. Mol Biol Cell. 1997;8:231–248. doi: 10.1091/mbc.8.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmo-Fonseca M, Mendes-Soares L, Campos I. Nat Cell Biol. 2000;2:107–112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 9.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, et al. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng C, McNeil S, Pockwinse S, Nicherson J, Shopland L, Lawrence J B, Penman S, Hiebert S, Lian J, Wijnen A J, et al. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard H, Rahn H P, Cardoso M C. J Cell Biochem. 1998;30–31,Suppl.:243–249. [PubMed] [Google Scholar]
- 12.Spector D L. Annu Rev Cell Biol. 1993;9:2265–2315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 13.Puvion E, Puvion-Dutilleul F. Exp Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- 14.Spector D L. Exp Cell Res. 1996;229:189–197. doi: 10.1006/excr.1996.0358. [DOI] [PubMed] [Google Scholar]
- 15.Mintz P J, Patterson S D, Neuwald A F, Spahr C S, Spector D L. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt-Zachmann M S, Knecht S, Krämer A. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Chua K, Seghezzi W, Lees E, Gorzani O, Reed R. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krämer A. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 19.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Bingham P M. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 21.Hedley M L, Amrein H, Maniatis T. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cáceres J F, Misteli T, Screaton G R, Spector D L, Krainer A R. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandner J M, Reidenbach S, Franke W W. Differentiation. 1997;62:119–127. doi: 10.1046/j.1432-0436.1997.6230119.x. [DOI] [PubMed] [Google Scholar]
- 24.Brandner J M, Reidenbach S, Kuhn C, Franke W W. Eur J Cell Biol. 1998;75:295–308. doi: 10.1016/S0171-9335(98)80063-0. [DOI] [PubMed] [Google Scholar]
- 25.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt-Zachmann M S, Nigg E A. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- 27.Higuchi R. In: PCR Protocols. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 177–183. [Google Scholar]
- 28.Zirwes R F, Eilbracht J, Kneissel S, Schmidt-Zachmann M S. Mol Biol Cell. 2000;11:1153–1167. doi: 10.1091/mbc.11.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nigg E A. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 30.Lamond A I, Carmo-Foseca M. Mol Biol Rep. 1993;18:127–133. doi: 10.1007/BF00986767. [DOI] [PubMed] [Google Scholar]
- 31.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 32.Fu X-D. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 33.Bohmann K, Ferreira J A, Lamond A I. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mermoud J E, Cohen P T, Lamond A I. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tazi J, Kornstadt U, Rossi F, Jeanteur P, Cathala G, Brunel C, Lührmann R. Nature (London) 1993;363:283–286. doi: 10.1038/363283a0. [DOI] [PubMed] [Google Scholar]
- 36.Yeakley J M, Tronchère H, Olesen J, Dyck J A, Wang H-Y, Fu X-D. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seghezzi W, Chua K, Shannahan F, Gozani O, Reed R, Lees E. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyon C E, Bohmann K, Sleeman J, Lamond A I. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- 39.Haynes Pauling M, McPheeters D S, Ares M., Jr Mol Cell Biol. 2000;20:2176–2185. doi: 10.1128/mcb.20.6.2176-2185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das B K, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Mol Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gozani O, Potashkin J, Reed R. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gama-Carvalho M, Krauss R D, Chiang L, Valcárcel J, Green M R, Carmo-Fonseca M. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]