Abstract
Macrophages display a large variety of surface receptors that are critical for their normal cellular functions in host defense, including finding sites of infection (chemotaxis) and removing foreign particles (phagocytosis). However, inappropriate regulation of these processes can lead to human diseases. Many of these receptors utilize tyrosine phosphorylation cascades to initiate and terminate signals leading to cell migration and clearance of infection. Actin remodeling dominates these processes and many regulators have been identified. This review focuses on how tyrosine kinases and phosphatases regulate actin dynamics leading to macrophage chemotaxis and phagocytosis.
Keywords: phagocytosis, chemotaxis, macrophage, kinase, phosphatase, signal transduction
Introduction
Macrophages function as an essential component of the immune system by migrating in tissue and, in particular, to sites of infection to chase and eliminate pathogens and by antigen presentation to cells of the adaptive immune system. Macrophages use a variety of surface receptors to sense and initiate defensive actions beneficial to the host. However, inappropriate recruitment of macrophages leads to several human diseases, such as atherosclerosis, arthritis and other chronic inflammatory diseases (reviewed in [1]). While phagocytosis is necessary for clearance of foreign particles and cellular debris, phagocytosis is accompanied by the generation of reactive nitrogen intermediates, reactive oxygen intermediates and the production of inflammatory cytokines that can result in tissue damage [2]. In fact, many forms of glomerulonephritis are due to the actions of macrophages attempting to clear deposits of immune complexes in the kidney (reviewed in [3]). Hence, it is essential that signaling pathways leading to macrophage functions be not only coordinated but also tightly regulated. Such delicate balance in signaling may be achieved by modulation of signaling cascades.
A diverse repertoire of receptors expressed by macrophages allows them to respond to a variety of external stimuli (reviewed in [1, 4, 5]). Immune receptors come in a variety of classes and some, such as the well-characterized phagocytic Fcγ receptor (FcγR) and the complement receptor (CR3), are associated with tyrosine kinases. Surface receptors for various chemoattractants require tyrosine kinases for functional responses, such as adhesion and migration. These receptors can be divided into two major classes; one is the receptor tyrosine kinase (RTK) family and the other is G-protein coupled receptor (GPCR) superfamily [Figure 1]. While chemokines act through GPCRs to mediate their biological responses, the signaling that regulates these biological effects is not well understood. However, recent studies indicate that tyrosine kinases, such as Src family kinases (SFKs), also serve as effectors for several GPCRs (reviewed in [5]), demonstrating tyrosine kinases are very important for functional responses mediated by GPCRs as well.
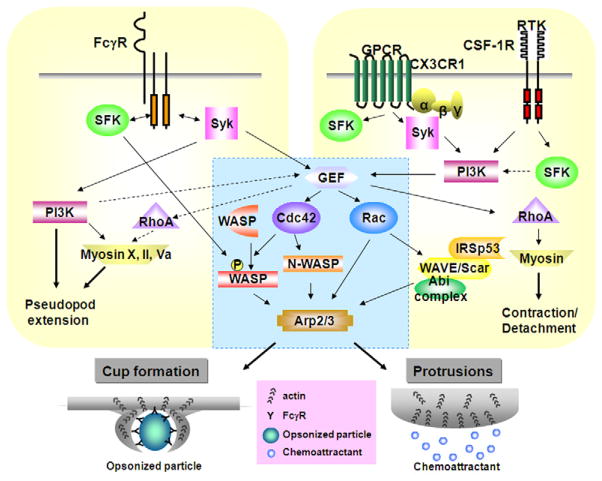
Fig.1.
Signaling in FcγR-mediated phagocytosis and RTK or GPCR-mediated chemotaxis. The Rho GTPases such as Rac and Cdc42 are important in both phagocytosis and chemotaxis. Activated Cdc42 recruits N/WASP, and then WASP is tyrosine phosphorylated by SFK. N/WASP stimulates actin polymerization by the Arp2/3 complex leading to phagocytic cup formation or protrusions. Downstream of Rac, the WAVE/Scar complex contributes to membrane protrusions through binding to IRSp53 and Abi complex, but is not required for phagocytosis. Blue box represents common signaling pathways. See text for details.
A growing body of work implicates various kinases and phosphatases in the regulation of the actin cytoskeleton of macrophages [1, 5]. The actin cytoskeleton plays a central role in the regulation of cell shape, adhesion, migration and phagocytosis, all intimately tied to the execution of macrophage functions in immunity. The precise signaling pathways that connect the activated surface receptors with direct effectors of actin cytoskeleton remodeling have not been completely delineated in macrophages. However, FcγRs, RTKs and GPCRs share common downstream effectors that could be similarly regulated by kinases and phosphatases for coordinated responses leading to the reorganization of actin cytoskeleton. Hence, this review focuses on the involvement of protein kinases and phosphatases in the regulation of receptor mediated signaling events of macrophages in response to phagocytic particles and chemoattractant stimuli leading to actin remodeling required for phagocytosis and migration.
1. Protein kinases and phosphatases in Phagocytic signaling
Macrophages clear foreign particles and cellular debris by phagocytosis which is defined as the mechanism of internalization of particles that are greater than 0.5 μm in diameter [6]. Many receptors are capable of mediating phagocytosis but most studies have focused on FcγR and CR3; much of our understanding of phagocytic signaling is due to studies on the FcγR. Therefore, this review will only focus on FcγR-mediated phagocytosis, which is a beautifully spatially and temporally coordinated series of events initiated by the binding of an opsonised IgG particle with FcγR that leads to actin polymerization and the formation of pseudopods that extend around the particle to form a phagocytic cup. Continued pseudopod extension, requiring exocytic membrane insertion, eventually surrounds the particle (reviewed in [7]). The pseudopod completely surrounds the particle and it becomes sealed to form a phagosome that is then internalized and the contents degraded. For a more complete discussion on phagosome maturation readers are referred to [8].
Three classes of FcγRs have been recognized to date: FcγRI, FcγRII, and FcγRIII and each class consists of several individual receptor isoforms [9]. Most of these FcγR isoforms, including FcγRI, FcγRIIA, and FcγRIIIA, are able to mediate phagocytosis [10]. Interaction of FcγRs with Fc domains of IgG triggers phosphorylation of tyrosine residues of the receptors and plays a key role in the transduction of phagocytic signaling cascades [11]. Immunoreceptor tyrosine-based activation motifs (ITAMs) are conserved sequences of four amino acids repeated twice (YXXI/L-X6–12-YXXI/L) located either in the cytoplasmic domain of FcγRIIA or in γ chains associated with FcγRI and FcγRIIIA. Phosphorylation of these tyrosine residues in ITAMs initiates numerous intracellular events leading to local rearrangement of the actin cytoskeleton and internalization of the particles. Indeed, it has been shown that transfection of non-phagocytic cells with FcγRs enabled these cells to gain the ability to ingest opsonized particles as professional phagocytes do [12]. Moreover, substitution of one of the tyrosine residues in the ITAM inhibited phagocytosis by 50% to 65%, whereas double mutation in this motif eliminated receptor tyrosine phosphorylation and phagocytic activity entirely [12] suggesting the phagocytic abilities of FcγRs depend on ITAMs. These conserved tyrosine residues of FcγRs appear to serve as substrates as well as binding sites for Src homology 2 domain (SH2) of the kinases and initiate downstream signaling cascades leading to phagocytosis (Tables 1 and 2).
Table 1.
Protein tyrosine kinases and phosphatases implicated in the regulation of FcγR-mediated phagocytosis and motility of macrophages.
| Kinases | Phagocytosis | Motility | |
|---|---|---|---|
| SFK | Hck | Hck−/−Fgr−/−Lyn−/− BMM show reduced phagocytosis [30, 32]. | Consititutively active form associated with increased motility in vitro [128] and in vivo [129]. Hck−/− macrophages show reduced motility in vitro and in vivo [126]. |
| Fgr | Negatively regulates phagocytosis in associate with SIRPα-SHP-1 complex [97]. | Hck−/−Fgr−/− show reduced migration ability [127]. | |
| Lyn | Hck−/−Fgr−/−Lyn−/− BMM show reduced phagocytosis [30, 32]. | Hck−/−Fgr−/−Lyn−/− BMM show reduced migration ability [194]. | |
| Src | Phosphorylate FcγRIIA in vitro [25]. | Co-immunoprecipitates with WASP, Pyk2, PTP-PEST and PSTPIP in osteoclasts and expression of constitutively active Src increases their phosphorylation [188]. | |
| Fyn | Phosphorylate FcγRIIA in vitro [26]. | Co-immunoprecipitates with Hck, Fgr, Lyn and Fyn in uPA-R complex involved in adhesion and chemotaxis [195]. | |
| Lck | Phosphorylate Vav [44]. | ? | |
| Syk | Syk−/− BMM fail to ingest IgG-coated particles [31, 32]. γ subunit is not phosphorylated in Syk−/− BMM [31]. |
Required for CX3CL1 chemotaxis but not CSF-1 [193]. | |
| Pyk2/FAK | Pyk2 and FAK are activated during phagocytosis [196, 197]. | Pyk2−/− BMM fail to polarize or ruffle [163]. SDF1-α stimulated Pyk2−/− BMM fail to detach from the underlying substrate [163]. FAK−/− BMM show migration defect toward CSF-1, SDF-1α and MCP-1 [171] |
|
| c-Abl | ? | c-Abl and SFK regulate migration and activation of the small GTPases Cdc42 and Rac [198]. | |
| Csk | Csk abolish phagocytosis signaling in a kinase-dependent manner [15] | Overexpression of CSK show enhance effect of MIF-induced MMP-13 expression [199]. | |
| Phosphatases | |||
| SHP-1 | Overexpression of SHP-1 show reduced phagocytic ability [98]. SHP-1 associates with FcγRIIa and this association appears to suppress total cellular tyrosine phosphorylation [2]. Negatively regulates phagocytosis in association with SIRPα [97]. |
SHP-1 deficiency lead to an abundant and exclusive increase in the infiltration of macrophages into the CNS [200]. | |
| PTP-PEST | ? | Reduced expression of PTP-PEST inhibits osteoclast migration towards osteopontin [188]. | |
| PTP-phi | ? | PTP phi induces cell rounding and ruffle formation and dephosphorylates Paxillin in dorsal ruffles [155]. | |
| CD148 | Double deficient BMM of CD148 and CD45 show reduced phagocytosis [17]. | Anti-CD148 antibody inhibits migration of primary macrophages in response to CSF-1 [201] |
FOOTNOTES : ?, Unknown
Table 2.
Downstream effectors of FcγR-mediated phagocytosis or chemotaxis in macrophage.
| Phagocytosis | Chemotaxis | References | ||
|---|---|---|---|---|
| Serine/Threonine Kinase | PAK | ✓ | ✓ | [202–204] |
| MEK/ERK | ✓ | ✓ | [10, 32, 205–208] | |
| PKC | ✓ | ✓ | [70, 72, 73, 209–211] | |
| Lipid Kinases | PI3K | ✓ | ✓ | [64, 79–81, 83, 88, 94, 133, 212] |
| PI4P-5Kα, γ | ✓ | ? | [62, 63, 213, 214] | |
| PLC | ✓ | ? | [66–69] | |
| PLD | ✓ | ✓ | [215–218] | |
| Lipid Phosphatases | PTEN | ✓ | ✓ | [219, 220] |
| SHIP | ✓ | ✓ | [100, 102, 103, 124, 221] | |
| Ubiquitin Ligase | c-Cbl | ✓ | ✓ | [15, 110, 222–224] |
| GTPases | Cdc42 | ✓ | ✓ | [38–40, 56, 139, 141] |
| Rac | ✓ | ✓ | [38–40, 138, 225] | |
| RhoA | ? | ✓ | [41, 226] | |
| Arf1 | ✓ | ? | [64] | |
| Arf6 | ✓ | ✓ | [60, 61, 64, 227, 228] | |
| Rab11, 5, 7 | ✓ | ? | [229–231] | |
| Dynamin | ✓ | ✓ | [84, 232, 233] | |
| GEFs | Vav | ✓ | ✓ | [44–46, 124, 234, 235] |
| DOCK180 | ✓ | ? | [48] | |
| GAP | p190RhoGAP | ? | ✓ | [164, 236] |
| Actin regulatory proteins | WASP/N-WASP | ✓ | ✓ | [45, 52, 53, 56, 142, 143, 151, 237, 238] |
| WAVE2 | X | ✓ | [59, 140] | |
| Paxillin | ✓ | ✓ | [127, 167, 196, 197, 239, 240] | |
| Cortactin | ? | ✓ | [127, 241] | |
| LIMK | ✓ | ✓ | [211, 242, 243] | |
| Cofilin | ✓ | ✓ | [211, 242, 243] | |
| Arp2/3 | ✓ | ✓ | [55, 244, 245] | |
| Adaptor proteins | LAT | ✓ | ? | [246, 247] |
| Grb2 | ✓ | ✓ | [218, 248–251] | |
| Gab2 | ✓ | ✓ | [249, 252] | |
| Nck | ✓ | ? | [45, 250] | |
| Crk | ✓ | ✓ | [48, 166, 250, 253] | |
| Contractile proteins | Myosin II | ✓ | ✓ | [83, 94, 177, 254, 255] |
| Myosin X, Va | ✓ | ? | [92, 93] | |
| MLCK | ✓ | ✓ | [94, 255] |
FOOTNOTES : ✓, Downstream effector.; ?, Unknown or controversial
Src-family tyrosine kinases (SFKs)
SFKs associate with inactive FcγR and are believed to regulate ITAM tyrosine phosphorylation [13]. SFKs have five conserved domains and the N-terminal domain of SFK is myristoylated, which is responsible for anchoring the kinases to the cell membrane. The remaining four domains consist of SH2 and SH3 domains, the catalytic domain (SH1), and a short C-terminal non-catalytic tail. SFKs are maintained in a quiescent state by phosphorylation of a tyrosine residue in the SFK non-catalytic tail by C-terminal Src kinase (Csk) which inhibits catalytic activity through interaction of the SH2 domain with the SH1 domain [14] and negatively regulates phagocytosis [15]. Upon FcγR cross-linking, the associated SFK becomes activated through dephosphorylation of its inhibitory tyrosine residue by the phosphatases CD45 [16] and CD148. Genetic deletion of either CD45 or CD148 alone has only a mild effect on phagocytosis while deletion of both CD45 and CD148 substantially reduces phagocytosis and suggests functional redundancy exists between CD45 and CD148 [17]. However, SFKs are mainly localized to specialized cholesterol-rich membrane microdomains known as lipid rafts [18] and the majority of CD45 is excluded from these lipid rafts domains [19]. On the other hand, a small fraction of SFKs are found in non-lipid raft fractions [15, 20]. Therefore, it is plausible that CD45 directly regulates the activity of SFKs, yet the possibility of other phosphatases involved in the regulation of activities of SFKs cannot be excluded.
The potential role of SFKs in phosphorylation of ITAM sequences is supported by studies that used a random synthetic peptide library to screen potential Src substrates, demonstrating that YXXL/I, a consensus ITAM sequence, is a preferred substrate for phosphorylation by SFKs [21]. Several studies show that FcγRs are recruited to lipid rafts after FcγR cross-linking [18, 22, 23] and SFKs are enriched in lipid rafts [18]. However, a small fraction of SFKs (e.g., Lyn) are also found in non-lipid raft fractions [15, 20] and SFKs can be constitutively associated with FcγR in both the presence and absence of FcγR clustering [15, 22]. It is possible that the non-raft associated SFKs may be responsible for the initial phosphorylation upon FcγR cross-linking. Following recruitment to lipid rafts, ITAM phosphorylation may then be rapidly amplified by raft associated SFKs.
Several SKFs, such as Src, Fyn, Fgr, Hck, and Lyn have been identified in macrophages [11, 24]. Src and Fyn are able to phosphorylate FcγRIIA in vitro [25, 26] and FcγRI and FcγRII physically and functionally associate with Hck and Lyn in a human THP-1 monocytic cell line [27]. Using the same THP-1 macrophages, Wang et al. also showed that the activation of FcγRI is followed by tyrosine phosphorylation of Hck and Lyn with an increase in these kinases’ activity [28]. Bewarder suggested that Lyn kinase is the most likely candidate for the receptor phosphorylation in vivo [29]. Consistent with the ability of these SFKs to phosphorylate ITAMs and initiate signaling, BMMs derived from hck−/−fgr−/−lyn−/− mice demonstrated a severe reduction in FcγR-mediated tyrosine phosphorylation and phagocytosis [30], indicating that these phosphorylation events are critical for the phagocytic ability of macrophages.
Spleen tyrosine kinase (Syk)
In addition to SFKs, Syk is present in all hematopoietic cells and is important in the regulation of phagocytic signaling. Macrophages and neutrophils derived from the fetal livers of Syk−/− mice failed to ingest IgG-coated particles [31, 32], and a chimeric Syk protein bearing an extracellular fragment of FcγRIII and a cytoplasmic fragment of Syk’s catalytic domain was sufficient to mediate internalization of IgG-opsonized particles when expressed in COS cells [33]. Interestingly, the aforementioned BMMs from hck−/−fgr−/−lyn−/− mice exhibited poor Syk activation upon FcγR clustering, suggesting that the activation of SFKs is a prerequisite for Syk engagement [32]. Syk likely interacts with ITAMs of FcγR by means of its two SH2 domains binding to the phosphorylated tyrosine residues of ITAMs [34]. Once bound to ITAMs, Syk phosphorylates downstream effectors leading to phagocytosis [34]. In support of this model, reduction of endogenous Syk expression did not inhibit phosphorylation of activated FcγRIIA in monocytes [35] but reduced downstream tyrosine phosphorylation and phagocytosis was observed in Syk deficient macrophages [32]. Taken together, these studies suggest that SFKs are responsible for the phosphorylation of FcγRs that then recruits Syk.
However, it is still unclear whether Syk is downstream of SFKs in phagocytic signaling. Even though Syk is not myristoylated and therefore not constitutively bound to the plasma membrane [11], Syk was found to constitutively associate with FcγRIIA in THP-1 cells or with the γ-chain of FcγRI and FcγRIIIA in macrophages [36, 37] and clustering of FcγRs in BMMs derived from Syk−/− mice did not lead to γ subunit phosphorylation [31]. Perhaps Syk itself may phosphorylate the γ subunit of FcγR. BMMs derived from Hck−/−Fgr−/−Lyn−/− mice do not show complete inhibition of phagocytosis when challenged with IgG-coated particles [30, 32]. Moreover, treatment of cells with the tyrosine kinase inhibitor PP1, which inhibits SFKs but not Syk kinases, also did not completely block FcγR-mediated phagocytosis [30]. Taken together, these observations suggest that Syk might be able to phosphorylate the γ subunit of FcγR by directly initiating signaling cascades in the absence of SFKs. However, the possibility of functional redundancy between the different SFKs cannot be excluded. Whether initiated by SFKs or by Syk, tyrosine phosphorylation signaling cascades resulting from FcγR cross-linking lead to actin dependent pseudopod extension.
Downstream effectors of FcγRs signaling and actin assembly
Actin cytoskeletal rearrangements such as actin polymerization, pseudopod extension, and phagosome closure, are required for phagocytosis downstream of the receptor activation. Although the precise signaling events downstream of ITAM phosphorylation are not clear, members of the Rho and Arf families of GTPases, as well as phosphoinositide (PI) regulating proteins such as phospholipase C-γ (PLCγ) have been shown to play critical roles in the regulation of the actin cytoskeleton (Table 2).
Rho and Arf families of GTPases
Both the Rho family GTPases members Rac1 and Cdc42 are required for optimal assembly of actin and phagocytosis [38–40] while the role of Rho in FcγR-mediated phagocytosis is controversial [38, 41]. Also, it is still not clear how Rac1 and Cdc42 are activated following engagement of FcγRs but activation involves guanine nucleotide-exchange factors (GEFs). The GEF Vav is a substrate of Src kinases [42, 43] and phosphorylation of Vav by Lck up-regulates Vav activity towards the GTPase Rac1 [44], and Vav is recruited to sites of the forming phagosome [45, 46]. However, Vav deficient macrophages exhibit no defect in FcγR-mediated phagocytosis [47], which indicates that other GEFs may activate Rac during phagocytosis. In fact, recent evidence indicates that CrkII-DOCK180 complex contributes to Rac activation at the phagocytic cup during internalization of IgG-opsonized particles [48]. DOCK180 associates with the adaptor protein Crk, which mediates recruitment of the GEF to sites of tyrosine phosphorylation [49] and transfection of dominant negative CrkII prevented both the recruitment of DOCK180 and the activation of Rac at the phagocytic cup [48]. Moreover, phagocytosis of IgG-opsonized particles can be inhibited by small interfering RNA reduction of either DOCK180 or CrkII, which is the predominant Crk isoform in macrophages [48]. Since Syk can also phosphorylate Vav [50] and associate with several adaptor molecules such as Crk [51], it is possible that the ITAM recruited Syk may also activate GEFs, dependent or independent of SFKs.
After their activation, both Rac and Cdc42 have been proposed to regulate actin polymerization through the Wiskott-Aldrich syndrome protein/WASP family verprolin-homologous (WASP/WAVE) family of proteins that are nucleation promoting factors [52, 53]. The catalytically active domain of WASP lies in its C-terminus that is conserved among all WASP/WAVE proteins that contains a VCA (verprolinhomology and cofilin-like and acidic region) domain capable of activating the Arp2/3 complex which initiates actin polymerization [54] and is required for phagocytosis [55]. In addition to Cdc42 activation, subsequent tyrosine phosphorylation of WASP is required for phagocytosis [56, 57]. Interestingly, while WAVE/Scar binds to Rac through intermediary proteins such as IRSp53 and the Abi1 complex [58], it does not appear to be required for FcγR-mediated phagocytosis [59]. This suggests that other downstream effectors of Rac are required, such as PAK (see Table 2).
ADP ribosylation factor 6 (ARF6) is another small GTPase that modulates actin assembly during phagocytosis [60]. ARF6 appears to primarily control early events in phagocytosis upstream of PI 3-kinase (PI3K), Cdc42 and Rac1 signaling [61]. While its precise role and signaling still remain unknown, GTP-bound ARF6 can activate lipid-modifying enzymes such as phosphatidylinositol 4-phosphate 5-kinase α (PI4P-5Kα) and PLD [62]. The product of PI4P-5Kα, phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), can facilitate actin polymerization through recruitment of WASP/N-WASP proteins [63]. In addition, ARF1 is also recruited to the phagocytic cup and is required for phagocytosis [64, 65]. However, ARF1 is required to regulate membrane trafficking (discussed in a later section).
Phosphoinositide related proteins
Lipids are important regulators of actin dynamics during phagocytosis. PLCγ is a phosphoinositide-specific phospholipase that cleaves PI(4,5)P2 to generate IP3 and diacylglycerol (DAG). After FcγR cross-linking, PLCγ is activated [66, 67] and inhibition of this enzyme leads to impaired FcγR-mediated actin assembly and phagocytosis in RAW 264.7 macrophage cells [68]. PLCγ is accumulated at the phagocytic cup [68], which was found to be dependent on Syk [69]. Furthermore, activation of PLCγ is prolonged during phagosome formation and dependent on Bruton’s tyrosine kinase (Btk) and Tec, a member of the Tec family of cytoplasmic tyrosine kinases [69]. Syk also activates Bruton’s tyrosine kinase (Btk) and Tec.
In addition, PLCγ-generated DAG is required for protein kinase C (PKC) activation [70]. PKCs are composed of serine/threonine kinases that are activated by DAG, phosphatidylserine, and in the case of some isoenzymes, by Ca2+ [71]. Upon FcγR ligation, PKC is activated in human monocytes [72] and an isozyme of PKC localizes to nascent phagosomes in macrophages [73]. Two PKC substrates, MARCKS (myristoylated alanine-rich C kinase substrate) and the related protein MacMARCKS, associate with phagosomes [73–75]. MARCKS cross-links actin filaments and interacts with the plasma membrane [76]. PKC-dependent phosphorylation displaces MARCKS from the membrane, and its subsequent dephosphorylation is accompanied by its reassociation with the membrane [77]. Therefore PKC and MARCKS may play a role in the regulation of actin structure of the phagosome. In support of these findings, a mutant form of MacMARCKS abolished phagocytosis in J774A.1 macrophage cells [75]. Nevertheless, BMMs derived from MacMARCKS null mice show normal phagocytosis [78] and whether PKC regulates actin assembly during phagocytosis remains to be determined.
Regulation of pseudopod extension and phagosomal closure
Several studies suggest PI3K participates in FcγR-mediated phagocytic signaling in addition to the lipid regulating proteins described above. Cross-linking of FcγRs increases PI3K activity and phagocytosis is inhibited by PI3K inhibitors such as wortmannin and LY294002 [79–81]. Although the block in phagocytosis is correlated with an inhibition of maximal pseudopod extension, this is not due to a decrease in actin polymerization [80]. The mechanism of PI3K action in phagocytosis is likely through exocytosis of vesicles into the growing pseudopod [80, 82]. In addition, phagosomal closure is impaired in macrophages incubated with wortmannin [81, 83]. Interestingly, Dynamin II recruitment to phagocytic cups is controlled by PI3K and is required for phagocytosis in RAW 264.7 cells [84]. Dynamin is a large GTPase involved in membrane trafficking that mediates the fission of the vesicle from the membrane (reviewed in [85, 86]). PI3K mediated increase in PI(3,4,5)P3 levels also leads to the deactivation of Cdc42, which was necessary for completion of phagocytosis [87]. Although these studies suggest that PI3K plays multiple roles in phagocytosis, more studies are required to assess the function of these molecules in phagosome closure.
The PI3K family of proteins can be divided into three main classes based on their in vitro lipid substrate specificity and structure. Class I PI3K can phosphorylate PI, PI(4)P and PI(4,5)P2. The prototypical class IA PI3K is composed of heterodimers consisting of catalytic and regulatory subunits with molecular weights of 110 kDa (p110) and 85 kDa (p85). Mammals have three genes for class IA p110 subunits encoding p110α/β/δ and one gene for the class IB p110 subunit encoding p110γ. The class II PI3K (e.g. PI3K-C2α/β/γ) utilizes predominantly PI and PI(4)P as substrates and the class III PI3K utilizes only PI as a substrate [88]. To date, the specific isoforms of PI3K required for FcγR-mediated phagocytosis have not been identified. Class IA PI3K p110α has been shown to be required for the uptake of IgG-opsonized zymosan in PMA-differentiated THP-1 cells and Raw 264.7 cells [89, 90]. However, in contrast with these results, phagocytosis of IgG-opsonized particles in BMMs is significantly inhibited by injection of anti-p110β antibody and not by anti-p110α antibody [91]. Furthermore, the possibility of functional redundancy of PI3K isoforms cannot be ruled out.
While actin polymerization is the main force that drives phagocytic cup formation, molecular motors that bind to actin filaments also appear to participate in the internalization process. Butanedione monoxime, an uncompetitive inhibitor of myosin II, and perhaps other myosins, prevented later constrictions without inhibiting the initial pseudopod extension [83]. In addition to myosin II, myosin X was also recruited to phagocytic cups in a PI3K-dependent manner [83, 92]. Furthermore, myosin X is needed for membrane-spreading on IgG-opsonized particles [92]. These data suggest myosin X may be the molecular linkage between PI3K for pseudopod extension and particle internalization during phagocytosis. Additionally, myosin IC, myosin V, and myosin IXb are also found at phagosomes [73, 83, 93] and Myosin II is required for phagosome closure [94]. Taken together, these data support roles for multiple myosin isoforms in particle internalization.
Importance of inhibitory signaling during phagocytosis
Phagocytic signaling cascades also need to be inactivated following completion of phagocytosis. In addition, activating signals through ITAMs in macrophages are counterbalanced by inhibitory signals mediated by the inhibitory receptor FcγRIIb to adjust cellular sensitivity to phagocytic signals to prevent tissue damage, which is prevalent in systemic lupus erythematosus (reviewed in [95]). The inhibitory FcγRIIb, contains immunoreceptor tyrosine-based inhibitory motifs (ITIMs) instead of ITAMs in the cytoplasmic tail [96]. ITIMs bind to the SFK member Fgr [97], protein-tyrosine phosphatase SHP-1 [98], or the SH2 domain containing inositol phosphatases (SHIPs) SHIP-1 [99] and SHIP-2 [100]. Under high opsonization conditions FcγR-mediated phagocytosis is suppressed, probably due to the action of the SFK Fgr since phagocytosis is enhanced in Fgr−/− BMMs [97]. This negative regulation may be due to the activation of SHP-1 (also known as SH-PTP1, HCP, and PTP1C), a phosphatase predominantly expressed in hematopoetic cells [97]. SHP-1 associates with the phosphorylated N-terminal ITAM tyrosine residue of FcγRIIa and this association appears to suppress total cellular tyrosine phosphorylation [2]. Consistent with a suppressive function, overexpression of SHP-1 in the J774A.1 macrophages reduces phagocytic efficiency [98]. SHIP, a 145-kDa SH2 domain-containing inositol phosphatase that hydrolyses the 5-phosphate of the inositol ring to generate PI(3,4)P2, is a negative regulator of PI3K [101]. SHIP also becomes associated with ITAMs and is activated following FcγRIIa cross-linking [102]. SHIP is recruited to phagocytic cups, and in agreement with its role as a negative regulator, inhibition of SHIP activity by expression of a dominant-negative construct or genetic deletion results in enhanced phagocytosis [102, 103].
Negative regulation of phagocytosis may also occur indirectly. The Cbl E3 ubiquitin ligase appears to interact with many of the molecules that play critical roles in phagocytosis. The mammalian Cbl family consists of three known genes c-cbl, cbl-b, and cbl-c/cbl-3 [104]. c-Cbl is a substrate of Src family kinases and Syk [105, 106], and c-Cbl and Cbl-b are also negative regulators of Syk kinase activity [107, 108]. The phosphotyrosine binding (PTB) domain of Cbl binds cognate phosphopeptide motifs in tyrosine kinases, such as SFKs and Syk, and suppresses kinase activity [109, 110]. In J774A.1 macrophages, Cbl is also a key substrate for SHP-1 and dephosphorylation of Cbl abrogates Cbl-CrkL interaction [98], which is required for Rac activation. Furthermore, Cbl also associates with PI3K and Vav [111]. Confirming a role for Cbl in negative regulation, macrophages from c-Cbl−/− and Cbl-b−/− mice demonstrate enhanced target binding, FcγR receptor-mediated tyrosine phosphorylation and phagocytosis [110]. Thus, Cbl plays a critical role in the regulation of inhibitory signaling by negatively regulating activating molecules and also enhancing inhibitory signals through its interactions. As an E3 ubiquitin ligase, c-Cbl is capable of inducing ubiquitination of Src when Src is in an open active conformation [112]. Cbl-b also targets phospho-Syk for ubiquitination, resulting in its subsequent degradation [108]. Although macrophages from c-Cbl−/− and Cbl-b−/− mice showed similar expression levels of SFKs and Syk proteins, c-Cbl−/−BMMs showed modest increases in surface expression of FcγRI [110]. Since these results also suggest the possibility of functional redundancy between c-Cbl and Cbl-b, or with other E3 ubiquitin ligases, the possible role of ubiquitylating activity of Cbl in the regulation of tyrosine kinases and FcγR activities remains to be determined.
2. Protein kinases and phosphatases in chemotactic signaling
In order to perform their functions, macrophages need to be recruited to specific sites, including sites of infection, by a process called chemotaxis. Chemotaxis refers to directed cell migration towards chemoattractant gradients. The cell detects a gradient and extends actin-rich protrusions towards the chemoattractant [113]. These protrusions persist and are stabilized through the formation of new adhesions to the underlying substratum in two dimensions or the extracellular matrix in three dimensions. Then the cell body moves forward propelled by actin-myosin mediated contraction. Finally, the tail of the cell detaches from the substratum and retracts [114]. The extension of a leading lamellipodium, or protrusion, during chemotaxis bears many similarities in structure to that of a phagocytic cup and involves many of the same signaling pathways. This portion of the review will focus on common signaling pathways utilized for chemotaxis and phagocytosis.
CSF-1 receptor tyrosine kinase and receptor proximal signaling
Colony stimulating factor-1 (CSF-1, also known as M-CSF) is a potent macrophage chemoattractant [115] whose effects are mediated through the tyrosine kinase receptor CSF-1R. The CSF-1R belongs to the class III receptor tyrosine kinase subfamily, which contains a split cytoplasmic kinase domain. CSF-1 binding induces receptor dimerization, kinase activation, and phosphorylation of seven out of twenty tyrosine residues in the cytoplasmic domain. Phosphorylation of these tyrosine residues on the CSF-1R creates binding sites for Src homology 2 (SH2) or phosphotyrosine binding domain (PTB) signaling proteins [116–118], such as SFKs and PI3K, that are known to mediate chemotaxis as well as various cellular responses (reviewed in [1]).
Similar to phagocytosis, many studies implicate the importance of SFK regulation in migration (reviewed in [5]). In fact, several SFKs bind to the activated CSF-1R [119] and specifically to phosphorylated Y559 [120]. Mutation of Y559 in CSF-1R inhibits maximal receptor activation [121]. BMMs expressing a chimeric receptor with the extracellular domain of erythropoietin and the cytoplasmic domain of CSF-1R (EpoR/CSF-1R) have been used to study the role of 559 in CSF-1R-mediated actin polymerization. Wild-type EpoR/CSF-1R chimera supported osteoclast differentiation and macrophage proliferation to the same extent as endogenous CSF-1R [122]. When Y559 was mutated in Epo/CSF-1R, it failed to promote actin polymerization in response to erythropoietin [123], suggesting that Y559 is a functional residue in CSF-1R signaling. CSF-1 chemotaxis can also be blocked using the SFK inhibitor PP2 [124]. Since the SFKs Hck, Fgr, and Lyn have highly redundant functions [125] it is unclear which particular SFK is required for macrophage chemotaxis. However, BMMs from Hck−/− or Hck−/−Fgr−/− macrophages display reduced migratory capacity in vitro and in vivo [126, 127]. In particular, over expression of wild type or constitutively active Hck increases chemotactic ability in the myelomonocytic cell line U937 [128] and macrophages from transgenic mice expressing constitutively active Hck demonstrate enhanced migratory ability in vitro. In vivo these mice develop lung pathology characterized by monocyte infiltration [129]. These studies suggest an important role for SFKs and Hck in particular in both macrophage migration and phagocytosis.
CSF-1 stimulates PI3K activity through association of the p85 regulatory subunit of PI3K with CSF-1R Y721 directly and indirectly through SFKs [130–132]. PI3K, and in particular the delta isoform, is essential for macrophage motility in response to CSF-1 [88, 133, 134]. Both SFKs and PI3K regulate macrophage chemotaxis through the regulation of downstream effectors.
Downstream effectors of CSF-1R signaling mediating actin assembly
Unlike FcγR signaling, PI3K plays an important role in actin polymerization downstream of CSF-1R signaling. PI3K may regulate macrophage migration through activation of the GEFs leading to activation of Rho family GTPases required for cell migration [135–137]. Following CSF-1 exposure, Vav is phosphorylated and recruited to the plasma membrane in a PI3K dependent manner. Confirming Vav’s role, a dominant negative mutant of Vav blocks macrophage chemotaxis toward CSF-1 [124]. Cdc42 and Rac are also required for CSF-1-elicited protrusions [40, 138] and chemotaxis [139]. Rac and WAVE form a complex of proteins that mediate CSF-1-induced actin polymerization and macrophage motility [59, 140] while Cdc42 appears to mediate CSF-1-induced directional motility [139] and possibly function upstream of Rac [141]. Cdc42 appears to be the major activator of WASP downstream of CSF-1R activation [142] and WASP−/− macrophages are defective in chemotaxis toward CSF-1 [143]. In addition, WASP is downstream of signaling events by GPCRs and is required for chemotaxis to formylmethionylleucylphenylalanine (fMLP) [143–145], macrophage chemoattractant protein-1 (MCP-1) [145], and macrophage inflammatory protein-1α (MIP-1α) [145] indicating that WASP may be a common regulator of chemotaxis.
WASP activity can be regulated by multiple mechanisms in addition to Cdc42 and tyrosine phosphorylation of WASP and introduction of a phosphomimicking mutation has been shown to stimulate actin polymerization in vitro [146, 147]. However, a recent study clearly demonstrated SFKs are not required for the initial activation of WASP in response to CSF-1 [142], although a more subtle role of WASP phosphorylation for WASP activation cannot be ruled out [148, 149]. Hck has been proposed to phosphorylate and activate WASP in macrophages [146] and the regulation of WASP tyrosine phosphorylation is required for CSF-1 and CX3CL1-mediated chemotaxis in macrophages [150, 151]. These studies suggest that similar to phagocytosis, tyrosine phosphorylation of WASP after activation by Cdc42 is required for chemotaxis [151].
Overall, signaling by the CSF-1R leads to activation of actin polymerization that results in a protrusion towards the source of chemoattractant (Figure 1). Once extended, the directional protrusion needs to be stabilized by attachment to the substratum.
Kinases implicated in the regulation of adhesion
Cells need a certain level of adhesiveness to generate traction and move forward; however, too much adhesiveness prevents cell migration [152, 153]. Since integrins mediate cell–cell and cell–matrix interactions, they play a very important role in cell adhesion and migration [154]. Extracellular matrix (ECM) and integrin interactions lead to tyrosine phosphorylation of FAK, Pyk2 and cytoskeletal molecules such as paxilliin and cortactin [154]. These molecules are found in integrin-dependent focal adhesions and also associate with tyrosine kinase growth factor receptors [154]. Therefore, interactions between adhesion- and growth factor–initiated signal transduction appears to allow the convergence between cell spreading and migration. Macrophages utilize different adhesion structures to attach to the underlying substratum rather than the large focal adhesions observed in other cell types. Monocyte-derived cells employ small phospho-paxillin-rich point complexes, focal complexes, and podosomes to adhere and migrate [155, 156]. Focal complexes are 0.5- to 1-μm dot-like contacts localized along the lamellipodia that often mature into focal adhesions attached to stress fibers. Focal adhesions are mostly composed of β1 and β3 integrins [157]. In contrast, podosomes are defined by a diameter of ~0.5-μm with an actin-rich core, where proteins involved in actin nucleation such as WASP, Arp2/3, and cortactin are found. Integrins show an isotype-specific localization in podosomes. β1 integrins localize preferentially to the actin-rich core, whereas β2 and β3 integrins are found in the outer ring structure of podosomes [156]. In addition, assembly and disassembly of podosomes are much more dynamic than those of focal adhesions [158]. Although podosomes share similar components of focal adhesions, such as Pyk2, FAK and SFKs, their distinct properties suggest these structures have specific regulation and functions in macrophage biology (reviewed in [159]).
Focal Adhesion Kinase (FAK) family
The FAK family of non-receptor tyrosine kinases, consisting of the ubiquitously expressed FAK and the neuronal and hematopoietic expressed proline-rich tyrosine kinase 2 (Pyk2), play roles in adhesion and migration of multiple cell types. In particular, FAK influences cell attachment and movement [160] as well as establishment of a proper leading edge and maintenance of the polarity of moving cells [161, 162]. Pyk2−/− macrophages display morphological alterations with impaired motility [163]. Pyk2 interacts with Fyn in murine BMM [164] and associates with Src and Cbl in osteoclasts [165, 166]. Pyk2 associates with the activated CSF-1R in osteoclasts [166] and functions in combination with SFKs to phosphorylate its binding partner paxillin during macrophage spreading and adhesion [155, 167]. FAK is reported to associate with PI3K after CSF-1 treatment in THP-1 macrophages [168] and SFKs associate with FAK and paxillin through SH2 and SH3 domains [169, 170]. These reports indicate that both FAK and Pyk2 are activated downstream of CSF-1R signaling.
Consistent with a role in macrophage chemotaxis, BMMs from FAK−/− mice showed altered adhesion dynamics, elevated protrusive activity, and a marked inability to form stable lamellipodia in response to CSF-1 which lead to impaired chemotaxis [171]. Interestingly, these FAK−/− BMMs exhibited high basal levels of Rac1-GTP compared to WT cells, which may explain their elevated protrusive activity and implicates FAK in the persistence of protrusion and not in the formation of the protrusions themselves. Furthermore, FAK−/− BMMs showed migration defects towards chemokines such as SDF-1α and MCP-1, which are ligands for specific GPCRs. Therefore, these results suggest that the requirement of FAK in chemotaxis is not limited to CSF-1/RTK signaling. Also, SDF1-α stimulated Pyk2−/− macrophages extend multiple protrusions in several directions and show minimal net migration [163], clearly demonstrating the crucial role of FAK kinases in adherence and directional migration. Interestingly, even though FAK−/− BMMs have a very similar migratory defect compared to Pyk2−/− BMMs, the combined loss of FAK and Pyk2 did not result in greater impairment in their migratory ability than the loss of either molecule alone [171], suggesting that FAK and Pyk2 likely play redundant roles within the same signaling pathways in the regulation of macrophage adhesion and migration.
Kinases and phosphatases implicated in cell body contraction and detachment
In order for the cell to move forward the cell body must move into the extended protrusion, which is often regulated by Rho GTPase activity. Rho is required for CSF-1-mediated chemotaxis [172] by regulating myosin phosphorylation through effector molecules such as Rho-kinase (ROCK), which phosphorylates the myosin light chain [173]. Activated Rho also interacts with the myosin binding subunit of myosin phosphatase to reduce phosphatase activity [174, 175], suggesting both processes contribute to increased myosin light chain phosphorylation. RhoA activity and ROCK negatively regulate integrin adhesions and regulate tail traction during migration of monocytes through endothelial monolayers [137]. While monocytes have sufficient adhesion to drive forward movement in the absence of RhoA activity [152, 153, 176], Rho-kinase inhibitors suppressed cell migration of THP-1 macrophages and human monocytes [137]. Rho-kinase inhibitors also blocked murine peritoneal macrophage migration in response to MCP-1 [177]. Similarly, the myosin II-specific inhibitor blebbistatin [178] inhibited macrophage migration with specificity similar to that of Rho-kinase inhibitors [177].
Protein-tyrosine phosphatases in chemotaxis
The balance of cellular phosphorylation is controlled by the coordinated actions of kinases and protein tyrosine phosphatases (PTPs). Several phosphatases are activated downstream of CSF-1R signaling, such as SHP-1 and PTP-phi (PTPφ), but little is known about the role of these and other phosphatases in CSF-1 chemotaxis. However, several of these phosphatases are known to play negative roles in cell migration and therefore are included in this review. For example, SHP-1 becomes phosphorylated downstream of CSF-1R signaling [179] and it also negatively regulates macrophage chemotaxis to SDF-1 [180]. PTPφ is an isoform that is selectively expressed in macrophages and has increased expression following removal of CSF-1 [181]. Expression of PTPφ in murine BAC1.2F5 macrophages reduces cell spreading and adhesion [155]. Interestingly, even though PTPφ associates with paxillin and Pyk2 in vitro, in vivo PTPφ appears to only reduce paxillin tyrosine phosphorylation levels and not Pyk2. Whether PTPs participate in the dephosphorylation of other kinases and their substrates that regulate signal transduction requires further investigation.
Proline-, glutamic acid-, serine- and threonine-rich (PEST) family of protein tyrosine phosphatases (PTP-PESTs)
PTP-PESTs are implicated in the regulation of migration (reviewed in [182]). PTP-PEST has been shown to interact with WASP via a proline, serine, threonine phosphatase interacting protein (PSTPIP) that functions as a scaffold protein between PTP-PEST and WASP. This interaction inhibits TCR-mediated actin polymerization in T cells [183] possibly through PTP-PEST dephosphorylation of WASP [184]. PTP-PEST also appears to play a key role in dephosphorylation of paxillin by regulating the tyrosine phosphorylation state of PSTPIP [185]. As mentioned earlier, PTP-PEST has been reported to associate with proteins involved in the organization of the cytoskeleton required for cell migration, such as WASP, paxillin [186], and Pyk2 [187]. Small interfering RNA directed against PTP-PEST inhibits osteoclast migration towards osteopontin, which also acts as macrophage chemotactic factor [188]. Cycles of actin polymerization and depolymerization require turnover of protein tyrosine phosphorylation to allow protrusion of the leading edge of the cell. Therefore, PTP-PEST may regulate cell migration through actin reorganization by dephosphorylation of cytoskeleton-associated proteins.
SHIP
SHIP is regulated by SH2 domain recruitment to phosphorylated tyrosine residues of activated receptors (reviewed in [189]). SHIP becomes phosphorylated in a SFK dependent manner [190]. Although the role of SHIP in chemotaxis is still unclear, a study using SHIP−/− BMMs clearly shows that SHIP negatively regulates CSF-1 induced migration of macrophages [124]. Since SHIP is an important regulator of intracellular levels of PI(3,4,5)P3 [190], this result suggests SHIP may regulate macrophage chemotaxis by hydrolysis of PI(3,4,5)P3.
Differences in the requirements of kinase signaling molecules by GPCRs
Chemokines are an important class of molecules that mediate chemotaxis of monocytes and macrophages. Nonetheless, the signaling pathways leading to efficient chemotaxis for these various chemokines are not well characterized in macrophages. However, an increase in tyrosine kinase activity has been commonly observed in response to the stimulation of GPCRs in different cells. As described above, tyrosine kinases such as SFKs, FAK and Pyk2 regulate similar functions in response to the stimulation of GPCRs and CSF-1. Also, there is evidence that phosphatases, which antagonize tyrosine kinase signaling, appear to play overlapping and complementary roles in both RTK and GPCR signaling pathways. For example, SHP-1 negatively regulates macrophage chemotaxis towards the chemokine SDF-1 [180] while it also becomes activated downstream of CSF-1R signaling [179].
However, not all chemotactic receptors have the same signaling requirements. Interestingly, the Syk appears to play non-redundant roles in RTK and GPCR signaling in macrophages. Syk appears to regulate macrophage migration in response to fMLP and MCP-1 in an integrin dependent manner in human THP-1 macrophages [191] and Syk is critical for leukocyte adhesion during inflammation in vivo [192]. Surprisingly, using siRNA approaches it appears that Syk is not required for chemotaxis to CSF-1 but is required for actin cytoskeleton remodelling and migration of macrophages in response to the chemokine CX3CL1 [193]. In fact, Syk was required for the activation of PI3K-dependent Cdc42/WASP and Rac/WAVE2 pathways in CX3CL1-induced migration [151]. Taken together, these data suggest the possibility that Syk acts as a specific regulator of GPCR and not RTK signal transduction in macrophage migration.
Conclusions
Macrophages are versatile cells that play many roles in immune function. Both phagocytosis and migration require robust and coordinated rearrangements of the actin cytoskeleton. Since the activation of signaling molecules results in dynamic changes of the cytoskeleton, many of these molecules are in inhibitory states when the cell is quiescent and they need precise spatial and temporal coordination in order to complete localized actin polymerization leading to the extension of a protrusion whether it is directed around an attached particle or towards a chemotactic gradient. The delicate fine-tuning of these signaling pathways appears to be maintained by kinases and phosphatases through activation and the modulation of activities of the signaling molecules. Not surprisingly, many similarities exist in the properties of the kinases and phosphatases in the regulation of the actin cytoskeleton in either phagocytosis or migration (Table 1 and Figure 1). However, kinases may also have unique roles in specific signaling pathways, such as Syk. Challenges remain ahead to determine the precise roles and interaction of these proteins in order to better understand the regulation of macrophage immune functions.
Acknowledgments
This work was funded by the NIH (GM071828 to DC). We would like to thank Michael Cammer for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Haein Park, Email: haein.park@einstein.yu.edu.
Dan Ishihara, Email: dan.ishihara@med.einstein.yu.edu.
References
- 1.Pixley FJ, Stanley ER. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Ganesan LP, Fang H, Marsh CB, Tridandapani S. J Biol Chem. 2003;278:35710–35717. doi: 10.1074/jbc.M305078200. [DOI] [PubMed] [Google Scholar]
- 3.Tarzi RM, Cook HT. Nephron Exp Nephrol. 2003;95:e7–12. doi: 10.1159/000073018. [DOI] [PubMed] [Google Scholar]
- 4.Korade-Mirnics Z, Corey SJ. J Leukoc Biol. 2000;68:603–613. [PubMed] [Google Scholar]
- 5.Baruzzi A, Caveggion E, Berton G. Cell Mol Life Sci. 2008;65:2175–2190. doi: 10.1007/s00018-008-8005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aderem A, Underhill DM. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 7.Swanson JA, Hoppe AD. J Leukoc Biol. 2004;76:1093–1103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- 8.Vieira OV, Botelho RJ, Grinstein S. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravetch JV, Bolland S. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Garcia E, Rosales C. J Leukoc Biol. 2002;72:1092–1108. [PubMed] [Google Scholar]
- 11.Strzelecka A, Kwiatkowska K, Sobota A. FEBS Lett. 1997;400:11–14. doi: 10.1016/s0014-5793(96)01359-2. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell MA, Huang MM, Chien P, Indik ZK, Pan XQ, Schreiber AD. Blood. 1994;84:1753–1759. [PubMed] [Google Scholar]
- 13.Isakov N. J Leukoc Biol. 1997;61:6–16. doi: 10.1002/jlb.61.1.6. [DOI] [PubMed] [Google Scholar]
- 14.Cooper JA, Howell B. Cell. 1993;73:1051–1054. doi: 10.1016/0092-8674(93)90634-3. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Kono H, Hirose N, Okada M, Yamamoto T, Yamamoto K, Honda Z. J Immunol. 2000;165:473–482. doi: 10.4049/jimmunol.165.1.473. [DOI] [PubMed] [Google Scholar]
- 16.Adamczewski M, Numerof RP, Koretzky GA, Kinet JP. J Immunol. 1995;154:3047–3055. [PubMed] [Google Scholar]
- 17.Zhu JW, Brdicka T, Katsumoto TR, Lin J, Weiss A. Immunity. 2008;28:183–196. doi: 10.1016/j.immuni.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field KA, Holowka D, Baird B. J Biol Chem. 1997;272:4276–4280. doi: 10.1074/jbc.272.7.4276. [DOI] [PubMed] [Google Scholar]
- 19.Mustelin T, Tasken K. Biochem J. 2003;371:15–27. doi: 10.1042/BJ20021637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang ML, Shen L, Wade WF. J Immunol. 1999;163:5391–5398. [PubMed] [Google Scholar]
- 21.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 22.Katsumata O, Hara-Yokoyama M, Sautes-Fridman C, Nagatsuka Y, Katada T, Hirabayashi Y, Shimizu K, Fujita-Yoshigaki J, Sugiya H, Furuyama S. J Immunol. 2001;167:5814–5823. doi: 10.4049/jimmunol.167.10.5814. [DOI] [PubMed] [Google Scholar]
- 23.Kwiatkowska K, Frey J, Sobota A. J Cell Sci. 2003;116:537–550. doi: 10.1242/jcs.00254. [DOI] [PubMed] [Google Scholar]
- 24.Bolen JB. Cell Growth Differ. 1991;2:409–414. [PubMed] [Google Scholar]
- 25.Huang MM, Indik Z, Brass LF, Hoxie JA, Schreiber AD, Brugge JS. J Biol Chem. 1992;267:5467–5473. [PubMed] [Google Scholar]
- 26.Hunter S, Huang MM, Indik ZK, Schreiber AD. Exp Hematol. 1993;21:1492–1497. [PubMed] [Google Scholar]
- 27.Ghazizadeh S, Bolen JB, Fleit HB. J Biol Chem. 1994;269:8878–8884. [PubMed] [Google Scholar]
- 28.Wang AV, Scholl PR, Geha RS. J Exp Med. 1994;180:1165–1170. doi: 10.1084/jem.180.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bewarder N, Weinrich V, Budde P, Hartmann D, Flaswinkel H, Reth M, Frey J. Mol Cell Biol. 1996;16:4735–4743. doi: 10.1128/mcb.16.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzer-Attas CJ, Lowry M, Crowley MT, Finn AJ, Meng F, DeFranco AL, Lowell CA. J Exp Med. 2000;191:669–682. doi: 10.1084/jem.191.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiefer F, Brumell J, Al-Alawi N, Latour S, Cheng A, Veillette A, Grinstein S, Pawson T. Mol Cell Biol. 1998;18:4209–4220. doi: 10.1128/mcb.18.7.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowley MT, Costello PS, Fitzer-Attas CJ, Turner M, Meng F, Lowell C, Tybulewicz VL, DeFranco AL. J Exp Med. 1997;186:1027–1039. doi: 10.1084/jem.186.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenberg S, Chang P, Wang DC, Xavier R, Seed B. Proc Natl Acad Sci U S A. 1996;93:1103–1107. doi: 10.1073/pnas.93.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson SA, Pleiman CM, Pao L, Schneringer J, Hippen K, Cambier JC. J Immunol. 1995;155:4596–4603. [PubMed] [Google Scholar]
- 35.Matsuda M, Park JG, Wang DC, Hunter S, Chien P, Schreiber AD. Mol Biol Cell. 1996;7:1095–1106. doi: 10.1091/mbc.7.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghazizadeh S, Bolen JB, Fleit HB. Biochem J. 1995;305(Pt 2):669–674. doi: 10.1042/bj3050669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darby C, Geahlen RL, Schreiber AD. J Immunol. 1994;152:5429–5437. [PubMed] [Google Scholar]
- 38.Caron E, Hall A. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 39.Massol P, Montcourrier P, Guillemot JC, Chavrier P. Embo J. 1998;17:6219–6229. doi: 10.1093/emboj/17.21.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hackam DJ, Rotstein OD, Schreiber A, Zhang W, Grinstein S. J Exp Med. 1997;186:955–966. doi: 10.1084/jem.186.6.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller RD, Krishna UM, Falck JR, White MA, Broek D. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 43.Gulbins E, Coggeshall KM, Baier G, Katzav S, Burn P, Altman A. Science. 1993;260:822–825. doi: 10.1126/science.8484124. [DOI] [PubMed] [Google Scholar]
- 44.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 45.Coppolino MG, Krause M, Hagendorff P, Monner DA, Trimble W, Grinstein S, Wehland J, Sechi AS. J Cell Sci. 2001;114:4307–4318. doi: 10.1242/jcs.114.23.4307. [DOI] [PubMed] [Google Scholar]
- 46.Patel JC, Hall A, Caron E. Mol Biol Cell. 2002;13:1215–1226. doi: 10.1091/mbc.02-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall AB, Gakidis MA, Glogauer M, Wilsbacher JL, Gao S, Swat W, Brugge JS. Immunity. 2006;24:305–316. doi: 10.1016/j.immuni.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Lee WL, Cosio G, Ireton K, Grinstein S. J Biol Chem. 2007;282:11135–11143. doi: 10.1074/jbc.M700823200. [DOI] [PubMed] [Google Scholar]
- 49.Buday L. Biochim Biophys Acta. 1999;1422:187–204. doi: 10.1016/s0304-4157(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 50.Bertagnolo V, Brugnoli F, Marchisio M, Celeghini C, Carini C, Capitani S. Cell Signal. 2004;16:423–433. doi: 10.1016/j.cellsig.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 51.Gotoh A, Takahira H, Geahlen RL, Broxmeyer HE. Cell Growth Differ. 1997;8:721–729. [PubMed] [Google Scholar]
- 52.Lorenzi R, Brickell PM, Katz DR, Kinnon C, Thrasher AJ. Blood. 2000;95:2943–2946. [PubMed] [Google Scholar]
- 53.McGee K, Zettl M, Way M, Fallman M. FEBS Lett. 2001;509:59–65. doi: 10.1016/s0014-5793(01)03139-8. [DOI] [PubMed] [Google Scholar]
- 54.Takenawa T, Suetsugu S. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 55.May RC, Caron E, Hall A, Machesky LM. Nat Cell Biol. 2000;2:246–248. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- 56.Park H, Cox D. Mol Biol Cell. 2009;20:4500–4508. doi: 10.1091/mbc.E09-03-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuboi S, Meerloo J. J Biol Chem. 2007;282:34194–34203. doi: 10.1074/jbc.M705999200. [DOI] [PubMed] [Google Scholar]
- 58.Miki H, Suetsugu S, Takenawa T. Embo J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abou-Kheir W, Isaac B, Yamaguchi H, Cox D. J Cell Sci. 2008;121:379–390. doi: 10.1242/jcs.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q, Cox D, Tseng CC, Donaldson JG, Greenberg S. J Biol Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- 61.Niedergang F, Colucci-Guyon E, Dubois T, Raposo G, Chavrier P. J Cell Biol. 2003;161:1143–1150. doi: 10.1083/jcb.200210069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 63.Miki H, Miura K, Takenawa T. Embo J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 64.Beemiller P, Hoppe AD, Swanson JA. PLoS Biol. 2006;4:e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braun V, Deschamps C, Raposo G, Benaroch P, Benmerah A, Chavrier P, Niedergang F. Mol Biol Cell. 2007;18:4921–4931. doi: 10.1091/mbc.E07-04-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao F, Shin HS, Rhee SG. Proc Natl Acad Sci U S A. 1992;89:3659–3663. doi: 10.1073/pnas.89.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Z, Lin CT, Unkeless JC. J Immunol. 1994;152:3017–3023. [PubMed] [Google Scholar]
- 68.Botelho RJ, Teruel M, Dierckman R, Anderson R, Wells A, York JD, Meyer T, Grinstein S. J Cell Biol. 2000;151:1353–1368. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jongstra-Bilen J, Puig Cano A, Hasija M, Xiao H, Smith CI, Cybulsky MI. J Immunol. 2008;181:288–298. doi: 10.4049/jimmunol.181.1.288. [DOI] [PubMed] [Google Scholar]
- 70.Larsen EC, Ueyama T, Brannock PM, Shirai Y, Saito N, Larsson C, Loegering D, Weber PB, Lennartz MR. J Cell Biol. 2002;159:939–944. doi: 10.1083/jcb.200205140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mochly-Rosen D, Gordon AS. Faseb J. 1998;12:35–42. [PubMed] [Google Scholar]
- 72.Zheleznyak A, Brown EJ. J Biol Chem. 1992;267:12042–12048. [PubMed] [Google Scholar]
- 73.Allen LH, Aderem A. J Exp Med. 1995;182:829–840. doi: 10.1084/jem.182.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li J, Aderem A. Cell. 1992;70:791–801. doi: 10.1016/0092-8674(92)90312-z. [DOI] [PubMed] [Google Scholar]
- 75.Zhu Z, Bao Z, Li J. J Biol Chem. 1995;270:17652–17655. doi: 10.1074/jbc.270.30.17652. [DOI] [PubMed] [Google Scholar]
- 76.Aderem A. Cell. 1992;71:713–716. doi: 10.1016/0092-8674(92)90546-o. [DOI] [PubMed] [Google Scholar]
- 77.Wang JK, Walaas SI, Sihra TS, Aderem A, Greengard P. Proc Natl Acad Sci U S A. 1989;86:2253–2256. doi: 10.1073/pnas.86.7.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Underhill DM, Chen J, Allen LA, Aderem A. J Biol Chem. 1998;273:33619–33623. doi: 10.1074/jbc.273.50.33619. [DOI] [PubMed] [Google Scholar]
- 79.Ninomiya N, Hazeki K, Fukui Y, Seya T, Okada T, Hazeki O, Ui M. J Biol Chem. 1994;269:22732–22737. [PubMed] [Google Scholar]
- 80.Cox D, Tseng CC, Bjekic G, Greenberg S. J Biol Chem. 1999;274:1240–1247. doi: 10.1074/jbc.274.3.1240. [DOI] [PubMed] [Google Scholar]
- 81.Araki N, Johnson MT, Swanson JA. J Cell Biol. 1996;135:1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bajno L, Peng XR, Schreiber AD, Moore HP, Trimble WS, Grinstein S. J Cell Biol. 2000;149:697–706. doi: 10.1083/jcb.149.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swanson JA, Johnson MT, Beningo K, Post P, Mooseker M, Araki N. J Cell Sci. 1999;112(Pt 3):307–316. doi: 10.1242/jcs.112.3.307. [DOI] [PubMed] [Google Scholar]
- 84.Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. J Exp Med. 1999;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doherty GJ, McMahon HT. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 86.Durieux AC, Prudhon B, Guicheney P, Bitoun M. J Mol Med. 88:339–350. doi: 10.1007/s00109-009-0587-4. [DOI] [PubMed] [Google Scholar]
- 87.Beemiller P, Zhang Y, Mohan S, Levinsohn E, Gaeta I, Hoppe AD, Swanson JA. Mol Biol Cell. 21:470–480. doi: 10.1091/mbc.E08-05-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vanhaesebroeck B, Jones GE, Allen WE, Zicha D, Hooshmand-Rad R, Sawyer C, Wells C, Waterfield MD, Ridley AJ. Nat Cell Biol. 1999;1:69–71. doi: 10.1038/9045. [DOI] [PubMed] [Google Scholar]
- 89.Lee JS, Nauseef WM, Moeenrezakhanlou A, Sly LM, Noubir S, Leidal KG, Schlomann JM, Krystal G, Reiner NE. J Leukoc Biol. 2007;81:1548–1561. doi: 10.1189/jlb.0906564. [DOI] [PubMed] [Google Scholar]
- 90.Tamura N, Hazeki K, Okazaki N, Kametani Y, Murakami H, Takaba Y, Ishikawa Y, Nigorikawa K, Hazeki O. Biochem J. 2009;423:99–108. doi: 10.1042/BJ20090687. [DOI] [PubMed] [Google Scholar]
- 91.Leverrier Y, Okkenhaug K, Sawyer C, Bilancio A, Vanhaesebroeck B, Ridley AJ. J Biol Chem. 2003;278:38437–38442. doi: 10.1074/jbc.M306649200. [DOI] [PubMed] [Google Scholar]
- 92.Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, Greenberg S. Nat Cell Biol. 2002;4:469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- 93.Al-Haddad A, Shonn MA, Redlich B, Blocker A, Burkhardt JK, Yu H, Hammer JA, 3rd, Weiss DG, Steffen W, Griffiths G, Kuznetsov SA. Mol Biol Cell. 2001;12:2742–2755. doi: 10.1091/mbc.12.9.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Araki N, Hatae T, Furukawa A, Swanson JA. J Cell Sci. 2003;116:247–257. doi: 10.1242/jcs.00235. [DOI] [PubMed] [Google Scholar]
- 95.Kavai M, Szegedi G. Autoimmun Rev. 2007;6:497–502. doi: 10.1016/j.autrev.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 96.Clynes R, Maizes JS, Guinamard R, Ono M, Takai T, Ravetch JV. J Exp Med. 1999;189:179–185. doi: 10.1084/jem.189.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gresham HD, Dale BM, Potter JW, Chang PW, Vines CM, Lowell CA, Lagenaur CF, Willman CL. J Exp Med. 2000;191:515–528. doi: 10.1084/jem.191.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kant AM, De P, Peng X, Yi T, Rawlings DJ, Kim JS, Durden DL. Blood. 2002;100:1852–1859. [PubMed] [Google Scholar]
- 99.D’Ambrosio D, Fong DC, Cambier JC. Immunol Lett. 1996;54:77–82. doi: 10.1016/s0165-2478(96)02653-3. [DOI] [PubMed] [Google Scholar]
- 100.Pengal RA, Ganesan LP, Fang H, Marsh CB, Anderson CL, Tridandapani S. J Biol Chem. 2003;278:22657–22663. doi: 10.1074/jbc.M302907200. [DOI] [PubMed] [Google Scholar]
- 101.Phee H, Jacob A, Coggeshall KM. J Biol Chem. 2000;275:19090–19097. doi: 10.1074/jbc.M001093200. [DOI] [PubMed] [Google Scholar]
- 102.Nakamura K, Malykhin A, Coggeshall KM. Blood. 2002;100:3374–3382. doi: 10.1182/blood-2002-03-0787. [DOI] [PubMed] [Google Scholar]
- 103.Cox D, Dale BM, Kashiwada M, Helgason CD, Greenberg S. J Exp Med. 2001;193:61–71. doi: 10.1084/jem.193.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Swaminathan G, Tsygankov AY. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 105.Feshchenko EA, Langdon WY, Tsygankov AY. J Biol Chem. 1998;273:8323–8331. doi: 10.1074/jbc.273.14.8323. [DOI] [PubMed] [Google Scholar]
- 106.Deckert M, Elly C, Altman A, Liu YC. J Biol Chem. 1998;273:8867–8874. doi: 10.1074/jbc.273.15.8867. [DOI] [PubMed] [Google Scholar]
- 107.Ota Y, Samelson LE. Science. 1997;276:418–420. doi: 10.1126/science.276.5311.418. [DOI] [PubMed] [Google Scholar]
- 108.Sohn HW, Gu H, Pierce SK. J Exp Med. 2003;197:1511–1524. doi: 10.1084/jem.20021686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lupher ML, Jr, Rao N, Lill NL, Andoniou CE, Miyake S, Clark EA, Druker B, Band H. J Biol Chem. 1998;273:35273–35281. doi: 10.1074/jbc.273.52.35273. [DOI] [PubMed] [Google Scholar]
- 110.Dale BM, Traum D, Erdjument-Bromage H, Tempst P, Greenberg S. J Immunol. 2009;182:5654–5662. doi: 10.4049/jimmunol.0803942. [DOI] [PubMed] [Google Scholar]
- 111.Thien CB, Langdon WY. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 112.Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, Zhang H, Baron R. J Biol Chem. 2001;276:35185–35193. doi: 10.1074/jbc.M102219200. [DOI] [PubMed] [Google Scholar]
- 113.Fukata M, Nakagawa M, Kaibuchi K. Curr Opin Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 114.Ridley AJ. J Cell Sci. 2001;114:2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 115.Sasaki Y, Ohsawa K, Kanazawa H, Kohsaka S, Imai Y. Biochem Biophys Res Commun. 2001;286:292–297. doi: 10.1006/bbrc.2001.5388. [DOI] [PubMed] [Google Scholar]
- 116.Sengupta A, Liu WK, Yeung YG, Yeung DC, Frackelton AR, Jr, Stanley ER. Proc Natl Acad Sci U S A. 1988;85:8062–8066. doi: 10.1073/pnas.85.21.8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Joos H, Trouliaris S, Helftenbein G, Niemann H, Tamura T. J Biol Chem. 1996;271:24476–24481. doi: 10.1074/jbc.271.40.24476. [DOI] [PubMed] [Google Scholar]
- 118.Wilhelmsen K, Burkhalter S, van der Geer P. Oncogene. 2002;21:1079–1089. doi: 10.1038/sj.onc.1205166. [DOI] [PubMed] [Google Scholar]
- 119.Courtneidge SA, Dhand R, Pilat D, Twamley GM, Waterfield MD, Roussel MF. EMBO J. 1993;12:943–950. doi: 10.1002/j.1460-2075.1993.tb05735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rohde CM, Schrum J, Lee AW. J Biol Chem. 2004;279:43448–43461. doi: 10.1074/jbc.M314170200. [DOI] [PubMed] [Google Scholar]
- 121.Xiong Y, Song D, Cai Y, Yu W, Yeung YG, Stanley ER. J Biol Chem. doi: 10.1074/jbc.M110.166702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feng X, Takeshita S, Namba N, Wei S, Teitelbaum SL, Ross FP. Endocrinology. 2002;143:4868–4874. doi: 10.1210/en.2002-220467. [DOI] [PubMed] [Google Scholar]
- 123.Faccio R, Takeshita S, Colaianni G, Chappel J, Zallone A, Teitelbaum SL, Ross FP. J Biol Chem. 2007;282:18991–18999. doi: 10.1074/jbc.M610937200. [DOI] [PubMed] [Google Scholar]
- 124.Vedham V, Phee H, Coggeshall KM. Mol Cell Biol. 2005;25:4211–4220. doi: 10.1128/MCB.25.10.4211-4220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lowell CA, Soriano P, Varmus HE. Genes Dev. 1994;8:387–398. doi: 10.1101/gad.8.4.387. [DOI] [PubMed] [Google Scholar]
- 126.Cougoule C, Le Cabec V, Poincloux R, Al Saati T, Mege JL, Tabouret G, Lowell CA, Laviolette-Malirat N, Maridonneau-Parini I. Blood. 115:1444–1452. doi: 10.1182/blood-2009-04-218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Suen PW, Ilic D, Caveggion E, Berton G, Damsky CH, Lowell CA. J Cell Sci. 1999;112(Pt 22):4067–4078. doi: 10.1242/jcs.112.22.4067. [DOI] [PubMed] [Google Scholar]
- 128.Chiaradonna F, Fontana L, Iavarone C, Carriero MV, Scholz G, Barone MV, Stoppelli MP. EMBO J. 1999;18:3013–3023. doi: 10.1093/emboj/18.11.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ernst M, Inglese M, Scholz GM, Harder KW, Clay FJ, Bozinovski S, Waring P, Darwiche R, Kay T, Sly P, Collins R, Turner D, Hibbs ML, Anderson GP, Dunn AR. J Exp Med. 2002;196:589–604. doi: 10.1084/jem.20020873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reedijk M, Liu X, van der Geer P, Letwin K, Waterfield MD, Hunter T, Pawson T. Embo J. 1992;11:1365–1372. doi: 10.1002/j.1460-2075.1992.tb05181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee AW, States DJ. Mol Cell Biol. 2000;20:6779–6798. doi: 10.1128/mcb.20.18.6779-6798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yeung YG, Stanley ER. Mol Cell Proteomics. 2003;2:1143–1155. doi: 10.1074/mcp.R300009-MCP200. [DOI] [PubMed] [Google Scholar]
- 133.Jones GE, Prigmore E, Calvez R, Hogan C, Dunn GA, Hirsch E, Wymann MP, Ridley AJ. Exp Cell Res. 2003;290:120–131. doi: 10.1016/s0014-4827(03)00318-5. [DOI] [PubMed] [Google Scholar]
- 134.Papakonstanti EA, Zwaenepoel O, Bilancio A, Burns E, Nock GE, Houseman B, Shokat K, Ridley AJ, Vanhaesebroeck B. J Cell Sci. 2008;121:4124–4133. doi: 10.1242/jcs.032763. [DOI] [PubMed] [Google Scholar]
- 135.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 136.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 137.Worthylake RA, Burridge K. Curr Opin Cell Biol. 2001;13:569–577. doi: 10.1016/s0955-0674(00)00253-2. [DOI] [PubMed] [Google Scholar]
- 138.Allen WE, Jones GE, Pollard JW, Ridley AJ. J Cell Sci. 1997;110(Pt 6):707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- 139.Allen WE, Zicha D, Ridley AJ, Jones GE. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kheir WA, Gevrey JC, Yamaguchi H, Isaac B, Cox D. J Cell Sci. 2005;118:5369–5379. doi: 10.1242/jcs.02638. [DOI] [PubMed] [Google Scholar]
- 141.Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. J Cell Sci. 2004;117:1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- 142.Cammer M, Gevrey JC, Lorenz M, Dovas A, Condeelis J, Cox D. J Biol Chem. 2009;284:23302–23311. doi: 10.1074/jbc.M109.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zicha D, Allen WE, Brickell PM, Kinnon C, Dunn GA, Jones GE, Thrasher AJ. Br J Haematol. 1998;101:659–665. doi: 10.1046/j.1365-2141.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 144.Wittmann S, Frohlich D, Daniels S. Br J Pharmacol. 2002;135:1375–1382. doi: 10.1038/sj.bjp.0704592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Badolato R, Sozzani S, Malacarne F, Bresciani S, Fiorini M, Borsatti A, Albertini A, Mantovani A, Ugazio AG, Notarangelo LD. J Immunol. 1998;161:1026–1033. [PubMed] [Google Scholar]
- 146.Cory GO, Garg R, Cramer R, Ridley AJ. J Biol Chem. 2002;277:45115–45121. doi: 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- 147.Suetsugu S, Hattori M, Miki H, Tezuka T, Yamamoto T, Mikoshiba K, Takenawa T. Dev Cell. 2002;3:645–658. doi: 10.1016/s1534-5807(02)00324-6. [DOI] [PubMed] [Google Scholar]
- 148.Torres E, Rosen MK. Mol Cell. 2003;11:1215–1227. doi: 10.1016/s1097-2765(03)00139-4. [DOI] [PubMed] [Google Scholar]
- 149.Torres E, Rosen MK. J Biol Chem. 2006;281:3513–3520. doi: 10.1074/jbc.M509416200. [DOI] [PubMed] [Google Scholar]
- 150.Dovas A, Gevrey JC, Grossi A, Park H, Abou-Kheir W, Cox D. J Cell Sci. 2009;122:3873–3882. doi: 10.1242/jcs.051755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Park H, Cox D. J Biol Chem. 2011 doi: 10.1074/jbc.M110.185181. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huttenlocher A, Ginsberg MH, Horwitz AF. J Cell Biol. 1996;134:1551–1562. doi: 10.1083/jcb.134.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 154.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 155.Pixley FJ, Lee PS, Condeelis JS, Stanley ER. Mol Cell Biol. 2001;21:1795–1809. doi: 10.1128/MCB.21.5.1795-1809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Linder S, Aepfelbacher M. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 157.Zamir E, Geiger B. J Cell Sci. 2001;114:3577–3579. doi: 10.1242/jcs.114.20.3577. [DOI] [PubMed] [Google Scholar]
- 158.Block MR, Badowski C, Millon-Fremillon A, Bouvard D, Bouin AP, Faurobert E, Gerber-Scokaert D, Planus E, Albiges-Rizo C. Eur J Cell Biol. 2008;87:491–506. doi: 10.1016/j.ejcb.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 159.Dovas A, Cox D. Cell Signal. 2011 doi: 10.1016/j.cellsig.2011.02.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mitra SK, Hanson DA, Schlaepfer DD. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 161.Tilghman RW, Slack-Davis JK, Sergina N, Martin KH, Iwanicki M, Hershey ED, Beggs HE, Reichardt LF, Parsons JT. J Cell Sci. 2005;118:2613–2623. doi: 10.1242/jcs.02380. [DOI] [PubMed] [Google Scholar]
- 162.Owen JD, Ruest PJ, Fry DW, Hanks SK. Mol Cell Biol. 1999;19:4806–4818. doi: 10.1128/mcb.19.7.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Okigaki M, Davis C, Falasca M, Harroch S, Felsenfeld DP, Sheetz MP, Schlessinger J. Proc Natl Acad Sci U S A. 2003;100:10740–10745. doi: 10.1073/pnas.1834348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Continolo S, Baruzzi A, Majeed M, Caveggion E, Fumagalli L, Lowell CA, Berton G. Exp Cell Res. 2005;302:253–269. doi: 10.1016/j.yexcr.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 165.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne WC, Baron R. J Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Elsegood CL, Zhuo Y, Wesolowski GA, Hamilton JA, Rodan GA, Duong le T. Int J Biochem Cell Biol. 2006;38:1518–1529. doi: 10.1016/j.biocel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 167.Williams MA, Newland AC, Kelsey SM. Leuk Res. 2000;24:317–330. doi: 10.1016/s0145-2126(99)00189-7. [DOI] [PubMed] [Google Scholar]
- 168.Hatch WC, Ganju RK, Hiregowdara D, Avraham S, Groopman JE. Blood. 1998;91:3967–3973. [PubMed] [Google Scholar]
- 169.Cobb BS, Schaller MD, Leu TH, Parsons JT. Mol Cell Biol. 1994;14:147–155. doi: 10.1128/mcb.14.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brown MC, Perrotta JA, Turner CE. J Cell Biol. 1996;135:1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Owen KA, Pixley FJ, Thomas KS, Vicente-Manzanares M, Ray BJ, Horwitz AF, Parsons JT, Beggs HE, Stanley ER, Bouton AH. J Cell Biol. 2007;179:1275–1287. doi: 10.1083/jcb.200708093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jones GE, Allen WE, Ridley AJ. Cell Adhes Commun. 1998;6:237–245. doi: 10.3109/15419069809004479. [DOI] [PubMed] [Google Scholar]
- 173.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 174.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 175.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 176.Rottner K, Hall A, Small JV. Curr Biol. 1999;9:640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 177.Nakayama M, Amano M, Katsumi A, Kaneko T, Kawabata S, Takefuji M, Kaibuchi K. Genes Cells. 2005;10:107–117. doi: 10.1111/j.1365-2443.2005.00823.x. [DOI] [PubMed] [Google Scholar]
- 178.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 179.Yeung YG, Wang Y, Einstein DB, Lee PS, Stanley ER. J Biol Chem. 1998;273:17128–17137. doi: 10.1074/jbc.273.27.17128. [DOI] [PubMed] [Google Scholar]
- 180.Kim CH, Qu CK, Hangoc G, Cooper S, Anzai N, Feng GS, Broxmeyer HE. J Exp Med. 1999;190:681–690. doi: 10.1084/jem.190.5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pixley FJ, Lee PS, Dominguez MG, Einstein DB, Stanley ER. J Biol Chem. 1995;270:27339–27347. doi: 10.1074/jbc.270.45.27339. [DOI] [PubMed] [Google Scholar]
- 182.Veillette A, Rhee I, Souza CM, Davidson D. Immunol Rev. 2009;228:312–324. doi: 10.1111/j.1600-065X.2008.00747.x. [DOI] [PubMed] [Google Scholar]
- 183.Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA. J Exp Med. 2004;199:99–112. doi: 10.1084/jem.20030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cote JF, Chung PL, Theberge JF, Halle M, Spencer S, Lasky LA, Tremblay ML. J Biol Chem. 2002;277:2973–2986. doi: 10.1074/jbc.M106428200. [DOI] [PubMed] [Google Scholar]
- 185.Angers-Loustau A, Cote JF, Charest A, Dowbenko D, Spencer S, Lasky LA, Tremblay ML. J Cell Biol. 1999;144:1019–1031. doi: 10.1083/jcb.144.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shen Y, Lyons P, Cooley M, Davidson D, Veillette A, Salgia R, Griffin JD, Schaller MD. J Biol Chem. 2000;275:1405–1413. doi: 10.1074/jbc.275.2.1405. [DOI] [PubMed] [Google Scholar]
- 187.Gupta R, Luan S. Plant Physiol. 2003;132:1149–1152. doi: 10.1104/pp.103.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chellaiah MA, Kuppuswamy D, Lasky L, Linder S. J Biol Chem. 2007;282:10104–10116. doi: 10.1074/jbc.M608957200. [DOI] [PubMed] [Google Scholar]
- 189.Coggeshall KM. Curr Dir Autoimmun. 2002;5:1–29. doi: 10.1159/000060554. [DOI] [PubMed] [Google Scholar]
- 190.Baran CP, Tridandapani S, Helgason CD, Humphries RK, Krystal G, Marsh CB. J Biol Chem. 2003;278:38628–38636. doi: 10.1074/jbc.M305021200. [DOI] [PubMed] [Google Scholar]
- 191.Schymeinsky J, Then C, Walzog B. J Cell Physiol. 2005;204:614–622. doi: 10.1002/jcp.20323. [DOI] [PubMed] [Google Scholar]
- 192.Frommhold D, Mannigel I, Schymeinsky J, Mocsai A, Poeschl J, Walzog B, Sperandio M. BMC Immunol. 2007;8:31. doi: 10.1186/1471-2172-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gevrey JC, Isaac BM, Cox D. J Immunol. 2005;175:3737–3745. doi: 10.4049/jimmunol.175.6.3737. [DOI] [PubMed] [Google Scholar]
- 194.Meng F, Lowell CA. EMBO J. 1998;17:4391–4403. doi: 10.1093/emboj/17.15.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bohuslav J, Horejsi V, Hansmann C, Stockl J, Weidle UH, Majdic O, Bartke I, Knapp W, Stockinger H. J Exp Med. 1995;181:1381–1390. doi: 10.1084/jem.181.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kedzierska K, Vardaxis NJ, Jaworowski A, Crowe SM. J Leukoc Biol. 2001;70:322–328. [PubMed] [Google Scholar]
- 197.Pan XQ, Darby C, Indik ZK, Schreiber AD. Clin Immunol. 1999;90:55–64. doi: 10.1006/clim.1998.4644. [DOI] [PubMed] [Google Scholar]
- 198.Baruzzi A, Iacobucci I, Soverini S, Lowell CA, Martinelli G, Berton G. FEBS Lett. 584:15–21. doi: 10.1016/j.febslet.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Onodera S, Nishihira J, Iwabuchi K, Koyama Y, Yoshida K, Tanaka S, Minami A. J Biol Chem. 2002;277:7865–7874. doi: 10.1074/jbc.M106020200. [DOI] [PubMed] [Google Scholar]
- 200.Christophi GP, Hudson CA, Panos M, Gruber RC, Massa PT. J Virol. 2009;83:522–539. doi: 10.1128/JVI.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Dave RK, Hume DA, Elsegood C, Kellie S. Exp Cell Res. 2009;315:1734–1744. doi: 10.1016/j.yexcr.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 202.Jankowski A, Zhu P, Marshall JG. Anal Biochem. 2008;380:235–248. doi: 10.1016/j.ab.2008.05.047. [DOI] [PubMed] [Google Scholar]
- 203.Diakonova M, Bokoch G, Swanson JA. Mol Biol Cell. 2002;13:402–411. doi: 10.1091/mbc.01-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Weiss-Haljiti C, Pasquali C, Ji H, Gillieron C, Chabert C, Curchod ML, Hirsch E, Ridley AJ, Hooft van Huijsduijnen R, Camps M, Rommel C. J Biol Chem. 2004;279:43273–43284. doi: 10.1074/jbc.M402924200. [DOI] [PubMed] [Google Scholar]
- 205.Hayashi A, Ohnishi H, Okazawa H, Nakazawa S, Ikeda H, Motegi S, Aoki N, Kimura S, Mikuni M, Matozaki T. J Biol Chem. 2004;279:29450–29460. doi: 10.1074/jbc.M400950200. [DOI] [PubMed] [Google Scholar]
- 206.Sanchez-Mejorada G, Rosales C. J Biol Chem. 1998;273:27610–27619. doi: 10.1074/jbc.273.42.27610. [DOI] [PubMed] [Google Scholar]
- 207.Selvaraj SK, Giri RK, Perelman N, Johnson C, Malik P, Kalra VK. Blood. 2003;102:1515–1524. doi: 10.1182/blood-2002-11-3423. [DOI] [PubMed] [Google Scholar]
- 208.Fine JS, Byrnes HD, Zavodny PJ, Hipkin RW. Inflammation. 2001;25:61–67. doi: 10.1023/a:1007152903135. [DOI] [PubMed] [Google Scholar]
- 209.Melendez AJ, Harnett MM, Allen JM. Immunology. 1999;96:457–464. doi: 10.1046/j.1365-2567.1999.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Breton A, Descoteaux A. Biochem Biophys Res Commun. 2000;276:472–476. doi: 10.1006/bbrc.2000.3511. [DOI] [PubMed] [Google Scholar]
- 211.Guo H, Ma Y, Zhang B, Sun B, Niu R, Ying G, Zhang N. J Leukoc Biol. 2009;85:911–918. doi: 10.1189/jlb.0708429. [DOI] [PubMed] [Google Scholar]
- 212.Marshall JG, Booth JW, Stambolic V, Mak T, Balla T, Schreiber AD, Meyer T, Grinstein S. J Cell Biol. 2001;153:1369–1380. doi: 10.1083/jcb.153.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mao YS, Yamaga M, Zhu X, Wei Y, Sun HQ, Wang J, Yun M, Wang Y, Di Paolo G, Bennett M, Mellman I, Abrams CS, De Camilli P, Lu CY, Yin HL. J Cell Biol. 2009;184:281–296. doi: 10.1083/jcb.200806121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Szymanska E, Sobota A, Czurylo E, Kwiatkowska K. Eur J Immunol. 2008;38:260–272. doi: 10.1002/eji.200737170. [DOI] [PubMed] [Google Scholar]
- 215.Kusner DJ, Hall CF, Jackson S. J Immunol. 1999;162:2266–2274. [PubMed] [Google Scholar]
- 216.Bodman-Smith KB, Melendez AJ, Campbell I, Harrison PT, Allen JM, Raynes JG. Immunology. 2002;107:252–260. doi: 10.1046/j.1365-2567.2002.01481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Corrotte M, Nyguyen AP, Harlay ML, Vitale N, Bader MF, Grant NJ. J Immunol. 185:2942–2950. doi: 10.4049/jimmunol.0903138. [DOI] [PubMed] [Google Scholar]
- 218.Knapek K, Frondorf K, Post J, Short S, Cox D, Gomez-Cambronero J. Mol Cell Biol. 30:4492–4506. doi: 10.1128/MCB.00229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cao X, Wei G, Fang H, Guo J, Weinstein M, Marsh CB, Ostrowski MC, Tridandapani S. J Immunol. 2004;172:4851–4857. doi: 10.4049/jimmunol.172.8.4851. [DOI] [PubMed] [Google Scholar]
- 220.Sugatani T, Alvarez U, Hruska KA. J Biol Chem. 2003;278:5001–5008. doi: 10.1074/jbc.M209299200. [DOI] [PubMed] [Google Scholar]
- 221.Kamen LA, Levinsohn J, Cadwallader A, Tridandapani S, Swanson JA. J Immunol. 2008;180:7497–7505. doi: 10.4049/jimmunol.180.11.7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Huang ZY, Barreda DR, Worth RG, Indik ZK, Kim MK, Chien P, Schreiber AD. J Leukoc Biol. 2006;80:1553–1562. doi: 10.1189/jlb.0106019. [DOI] [PubMed] [Google Scholar]
- 223.Caveggion E, Continolo S, Pixley FJ, Stanley ER, Bowtell DD, Lowell CA, Berton G. J Cell Physiol. 2003;195:276–289. doi: 10.1002/jcp.10236. [DOI] [PubMed] [Google Scholar]
- 224.Lesniewski LA, Hosch SE, Neels JG, de Luca C, Pashmforoush M, Lumeng CN, Chiang SH, Scadeng M, Saltiel AR, Olefsky JM. Nat Med. 2007;13:455–462. doi: 10.1038/nm1550. [DOI] [PubMed] [Google Scholar]
- 225.Ueyama T, Eto M, Kami K, Tatsuno T, Kobayashi T, Shirai Y, Lennartz MR, Takeya R, Sumimoto H, Saito N. J Immunol. 2005;175:2381–2390. doi: 10.4049/jimmunol.175.4.2381. [DOI] [PubMed] [Google Scholar]
- 226.Pixley FJ, Xiong Y, Yu RY, Sahai EA, Stanley ER, Ye BH. J Cell Sci. 2005;118:1873–1883. doi: 10.1242/jcs.02314. [DOI] [PubMed] [Google Scholar]
- 227.Uchida H, Kondo A, Yoshimura Y, Mazaki Y, Sabe H. J Exp Med. 2001;193:955–966. doi: 10.1084/jem.193.8.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nishiya N, Kiosses WB, Han J, Ginsberg MH. Nat Cell Biol. 2005;7:343–352. doi: 10.1038/ncb1234. [DOI] [PubMed] [Google Scholar]
- 229.Cox D, Lee DJ, Dale BM, Calafat J, Greenberg S. Proc Natl Acad Sci U S A. 2000;97:680–685. doi: 10.1073/pnas.97.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Duclos S, Diez R, Garin J, Papadopoulou B, Descoteaux A, Stenmark H, Desjardins M. J Cell Sci 113 Pt. 2000;19:3531–3541. doi: 10.1242/jcs.113.19.3531. [DOI] [PubMed] [Google Scholar]
- 231.Huynh KK, Gershenzon E, Grinstein S. J Biol Chem. 2008;283:35745–35755. doi: 10.1074/jbc.M806232200. [DOI] [PubMed] [Google Scholar]
- 232.McNiven MA, Baldassarre M, Buccione R. Front Biosci. 2004;9:1944–1953. doi: 10.2741/1348. [DOI] [PubMed] [Google Scholar]
- 233.Tsuboi S, Takada H, Hara T, Mochizuki N, Funyu T, Saitoh H, Terayama Y, Yamaya K, Ohyama C, Nonoyama S, Ochs HD. J Biol Chem. 2009;284:8548–8556. doi: 10.1074/jbc.M805638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sakai H, Chen Y, Itokawa T, Yu KP, Zhu ML, Insogna K. Bone. 2006;39:1290–1301. doi: 10.1016/j.bone.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 235.Whetton AD, Lu Y, Pierce A, Carney L, Spooncer E. Blood. 2003;102:2798–2802. doi: 10.1182/blood-2002-12-3635. [DOI] [PubMed] [Google Scholar]
- 236.Zeng H, Yoshida T, Kurosaki T, Yamamura H, Oshima A, Kitamura D, Watanabe T, Morikawa M. J Biochem. 1995;118:1166–1174. doi: 10.1093/oxfordjournals.jbchem.a125003. [DOI] [PubMed] [Google Scholar]
- 237.Ridley AJ. FEBS Lett. 2001;498:168–171. doi: 10.1016/s0014-5793(01)02481-4. [DOI] [PubMed] [Google Scholar]
- 238.Isaac BM, Ishihara D, Nusblat LM, Gevrey JC, Dovas A, Condeelis J, Cox D. Exp Cell Res. 316:3406–3416. doi: 10.1016/j.yexcr.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Greenberg S, Chang P, Silverstein SC. J Biol Chem. 1994;269:3897–3902. [PubMed] [Google Scholar]
- 240.Achuthan A, Elsegood C, Masendycz P, Hamilton JA, Scholz GM. Cell Signal. 2006;18:2252–2261. doi: 10.1016/j.cellsig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 241.Boyle SN, Michaud GA, Schweitzer B, Predki PF, Koleske AJ. Curr Biol. 2007;17:445–451. doi: 10.1016/j.cub.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 242.Matsui S, Matsumoto S, Adachi R, Kusui K, Hirayama A, Watanabe H, Ohashi K, Mizuno K, Yamaguchi T, Kasahara T, Suzuki K. J Biol Chem. 2002;277:544–549. doi: 10.1074/jbc.M110153200. [DOI] [PubMed] [Google Scholar]
- 243.Zhang B, Ma Y, Guo H, Sun B, Niu R, Ying G, Zhang N. Eur J Immunol. 2009;39:894–901. doi: 10.1002/eji.200838809. [DOI] [PubMed] [Google Scholar]
- 244.Jones GE. J Leukoc Biol. 2000;68:593–602. [PubMed] [Google Scholar]
- 245.Linder S, Higgs H, Hufner K, Schwarz K, Pannicke U, Aepfelbacher M. J Immunol. 2000;165:221–225. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- 246.Syed SN, Konrad S, Wiege K, Nieswandt B, Nimmerjahn F, Schmidt RE, Gessner JE. Eur J Immunol. 2009;39:3343–3356. doi: 10.1002/eji.200939884. [DOI] [PubMed] [Google Scholar]
- 247.Tridandapani S, Lyden TW, Smith JL, Carter JE, Coggeshall KM, Anderson CL. J Biol Chem. 2000;275:20480–20487. doi: 10.1074/jbc.M909462199. [DOI] [PubMed] [Google Scholar]
- 248.Khandani A, Eng E, Jongstra-Bilen J, Schreiber AD, Douda D, Samavarchi-Tehrani P, Harrison RE. J Leukoc Biol. 2007;82:417–428. doi: 10.1189/jlb.0706469. [DOI] [PubMed] [Google Scholar]
- 249.Gu H, Botelho RJ, Yu M, Grinstein S, Neel BG. J Cell Biol. 2003;161:1151–1161. doi: 10.1083/jcb.200212158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Izadi KD, Erdreich-Epstein A, Liu Y, Durden DL. Exp Cell Res. 1998;245:330–342. doi: 10.1006/excr.1998.4259. [DOI] [PubMed] [Google Scholar]
- 251.Rucci N, DiGiacinto C, Orru L, Millimaggi D, Baron R, Teti A. J Cell Sci. 2005;118:3263–3275. doi: 10.1242/jcs.02436. [DOI] [PubMed] [Google Scholar]
- 252.Yu WM, Hawley TS, Hawley RG, Qu CK. Blood. 2002;99:2351–2359. doi: 10.1182/blood.v99.7.2351. [DOI] [PubMed] [Google Scholar]
- 253.Erdreich-Epstein A, Liu M, Kant AM, Izadi KD, Nolta JA, Durden DL. J Leukoc Biol. 1999;65:523–534. doi: 10.1002/jlb.65.4.523. [DOI] [PubMed] [Google Scholar]
- 254.Olazabal IM, Caron E, May RC, Schilling K, Knecht DA, Machesky LM. Curr Biol. 2002;12:1413–1418. doi: 10.1016/s0960-9822(02)01069-2. [DOI] [PubMed] [Google Scholar]
- 255.Wilson AK, Gorgas G, Claypool WD, de Lanerolle P. J Cell Biol. 1991;114:277–283. doi: 10.1083/jcb.114.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]