Abstract
The viral RNA-dependent RNA polymerase (RdRp) is essential for multiplication of all RNA viruses. The sequence diversity of an RNA virus population contributes to its ability to infect the host. This diversity emanates from errors made by the RdRp during RNA synthesis. The physical basis for RdRp fidelity is unclear but is linked to conformational changes occurring during the nucleotide-addition cycle. To understand RdRp dynamics that might influence RdRp function, we have analyzed all-atom molecular dynamics simulations on the nanosecond timescale of four RdRps from the picornavirus family that exhibit 30–74% sequence identity. Principal component analysis showed that the major motions observed during the simulations derived from conserved structural motifs and regions of known function. The dynamics of residues participating in the same biochemical property, for example, RNA binding, nucleotide binding or catalysis, were correlated even when spatially distant on the RdRp structure. The conserved and correlated dynamics of functional structural elements suggest coevolution of dynamics with structure and function of the RdRp. Crystal structures of all picornavirus RdRps exhibit a template–nascent RNA duplex channel too small to fully accommodate duplex RNA. Simulations revealed opening and closing motions of the RNA and nucleoside triphosphate channels, which might be relevant to nucleoside triphosphate entry, inorganic pyrophosphate exit and translocation. A role for nanosecond timescale dynamics in RdRp fidelity is supported by the altered dynamics of the high-fidelity G64S derivative of PV RdRp relative to wild-type enzyme.
Keywords: MD simulation, polymerase, poliovirus, conformational change, fidelity
Abbreviations used: PV, poliovirus; CVB3, coxsackievirus B3; HRV16, human rhinovirus type 16; FMDV, foot-and-mouth disease virus; RdRp, RNA-dependent RNA polymerase; NTP, nucleoside triphosphate; MD, molecular dynamics; RMSF, root-mean-square fluctuation; PCA, principal component analysis; DCCM, dynamic cross-correlation map; PDB, Protein Data Bank; PC, principal component; WT, wild type; 2D, two-dimensional; Rg, radius of gyration
Graphical Abstract
Research Highlights
► Molecular dynamics simulations of picornavirus RdRps reveal conserved dynamics. ► Conserved motifs and functional regions are major contributors to the observed motions. ► Dynamic regions of RdRp structures are correlated in a manner that could relate to function(s). ► Simulations suggest a role of dynamics in polymerase fidelity. ► Simulation results are in agreement with experimental data from X-ray crystallography and NMR.
Introduction
Positive-strand RNA viruses represent an existing and emerging threat to public health as well as the health of animals. In the past decade alone, the incidence of infections by hepatitis C virus, West Nile virus and dengue virus has increased. Eradication of poliovirus (PV) has been thwarted by the reemergence of virulent strains, most often the result of recombination between a vaccine strain and a circulating enterovirus such as coxsackievirus A.1, 2 Previously unknown viruses such as severe acute respiratory syndrome coronavirus have emerged and spread across the globe, causing morbidity and mortality and highlighting our lack of preparedness to deal with such outbreaks.
Replication of the genomes of positive-strand RNA viruses requires the virus-encoded RNA-dependent RNA polymerase (RdRp). This enzyme uses the RNA genome as a template for polymerization of ribonucleotides to form a negative-strand RNA intermediate, which, in turn, is used as a template for production of progeny genomes. Inhibition of RdRp activity will prevent genome replication and virus multiplication.
The RdRp, like the reverse transcriptase of retroviruses, is a validated target for the development of antiviral therapeutics.3, 4 One class of inhibitors consists of nucleoside analogues that are converted into the triphosphate in the cell and serve as substrates for the RdRp without terminating RNA synthesis.4 Incorporation of this type of nucleotide analogue alters genome sequence, structure and/or function, thus leading to the observed antiviral activity.4, 5 Only one drug, ribavirin, is currently used clinically that can be placed into this class of nucleoside analogues as an antiviral agent targeting the RdRp. Interestingly, ribavirin is the only clinically approved broad-spectrum antiviral nucleoside.4, 6 The clinical efficacy of any inhibitor will be antagonized by the development of resistance. Mechanisms of resistance to ribavirin have been studied most extensively in PV7, 8, 9 and the related foot-and-mouth disease virus (FMDV).10, 11, 12 These studies have revealed that RdRp variants exhibiting altered fidelity of nucleotide incorporation are less sensitive to ribavirin in cell culture.7, 8, 12 However, viruses harboring these ribavirin-resistant alleles of the RdRp are attenuated in animals,13 and these attenuated viruses elicit protective immunity against a lethal challenge with wild-type (WT) virus.14 Together, these studies lead to the suggestion that RdRp fidelity is optimized to provide RNA viruses with population diversity that is apparently required for pathogenesis. Therefore, RdRp fidelity represents an important, untapped target for development of antiviral agents and development of attenuated virus variants that can be used as vaccine strains. Unfortunately, the physical basis for nucleic acid polymerase fidelity is not completely clear.
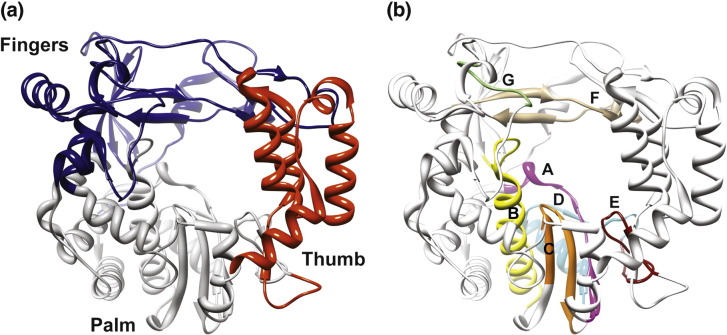
PV and FMDV, distantly related members of the picornavirus family of viruses, are prototypical positive-strand RNA viruses that have emerged as important model systems for relating RdRp structure, function and mechanism to RNA virus biology and pathogenesis.4, 15, 16 The picornaviral RdRp is also known as 3Dpol. The structure of the PV RdRp revealed three subdomains: fingers (including fingertips), thumb and palm (Fig. 1a).17 Interaction between the fingertips and the thumb leads to a completely encircled active site (Fig. 1a). The fingers and thumb subdomains contribute primarily to nascent RNA (primer) and template binding (Table 1 ). The palm subdomain is required for interacting with the primer 3′-end, templating nucleotide, incoming nucleotide substrates and divalent cations, all of which contribute to the nucleotidyl transfer reaction catalyzed by the RdRp (Table 1). The RNA template and nucleoside substrates access the active site through the template and nucleoside triphosphate (NTP) channels, respectively; for the egress of the newly synthesized RNA, the template–nascent RNA duplex channel is used. The recent structure of PV RdRp in complex with RNA substrate clearly showed the template and template–nascent RNA duplex occupying their corresponding channels.20 The conserved functions of the RdRp are mediated by conserved structural motifs (A–G in Fig. 1b) that contain conserved residues (Table 1). Structures are also available for the RdRps from coxsackievirus B3 (CVB3), human rhinovirus type 16 (HRV16) and FMDV.19, 21, 22 The structures of these RdRps are essentially identical with that of the PV RdRp in spite of substantial sequence differences (Fig. S1).
Fig. 1.
Structure of PV RdRp and location of conserved structural motifs. (a) The ribbon diagram shown was created from PDB code 1RA6. The three subdomains are indicated as follows: fingers (blue), palm (gray) and thumb (red). (b) The conserved structural motifs (A–G) necessary for substrate or cofactor binding and catalysis are indicated as follows: A (pink), B (yellow), C (orange), D (cyan), E (dark red), F (tan) and G (light green).
Table 1.
Conserved sequence/structure motifs and functionally important regions of PV RdRp
| Subdomain | Conserved structural/functional regions | Sequencea | Function |
|---|---|---|---|
| Fingers | G | 113 STSAGYPY 120 | Template bindingb |
| F | 153 PLVTYVKDELRSKTKVEQGKSRLIEA 178 | NTP and template binding | |
| I | 107 LEALDL 112 | Template binding | |
| II | 184 SVAMRMAFGNLYAAFHK 200 | Template binding | |
| Palm | A | 229 LFAFDYTGYDAS 240 | 2′-OH of NTP and metal B binding |
| B | 293 TSIFNSMINNLIIRTLLLKT 312 | Base of NTP and template binding | |
| C | 323 MIAYGDDVIAS 333 | Primer and metal A binding and catalysis | |
| D | 338 VDASLLAQSGKDYGLTMTPADKSAT 362 | Triphosphate of NTP binding and catalysis | |
| E | 363 FETVTWENVTFLKRFFRA 380 | Nascent RNA 3′-end binding | |
| Thumb | III | 405 KDPRNTQDHVRSLCLL 420 | Nascent RNA duplex binding |
The residues shown in boldface are completely conserved among RdRps; those residues representing signatures of the conserved sequence motifs are shown in boldface and underlined.18
Mechanistic studies of the RdRp from PV have shown that fidelity of nucleotide incorporation is governed by a conformational change step that precedes chemistry and chemistry.23 Substitution of Gly64 of PV RdRp with Ser leads to a high-fidelity variant that is less sensitive to ribavirin.7, 8 Position 64 is remote from the active site but is nevertheless able to selectively alter the equilibrium constant across the conformational change step that is used as a fidelity checkpoint.7 Structural analysis of the G64S derivative of PV RdRp was unable to provide an explanation for the observed difference in fidelity.24 Observations such as this one have led to the conclusion that dynamics may be the missing link to our understanding of RdRp fidelity.25 Moreover, evidence for differences in dynamics between WT PV RdRp and its G64S mutant has recently been detected by using NMR.26
Protein dynamics are now thought to be as fundamental to protein function as protein structure.27 Even more provocative, however, is the notion that conformational sampling by enzymes is not random and, preferentially, corresponds to conformations that will channel the enzyme and its substrate(s) and cofactor(s) through the reaction coordinate with the speed and specificity required for optimal fitness of the organism from which the enzyme derives.28, 29 Although the timescale for most enzyme reactions is in the order of milliseconds, motions on this timescale are now thought to be linked, if not governed, by motions that occur in the picosecond-to-nanosecond range.30
Since the first molecular dynamics (MD) simulation of a protein in 1977,31 this approach has provided invaluable insights into the role of dynamics in protein function.32, 33 Therefore, as a prelude to a more empirical pursuit of the role of dynamics in the function and fidelity of the RdRp, we performed an all-atom MD simulation of PV RdRp. We observed the largest amplitude motions for residues contributing to conserved structural motifs required for nucleic acid and nucleotide binding and catalysis. Correlated motions were also apparent for these dynamic structural motifs. In contrast, conserved structural motifs required for divalent cation binding were the least flexible. Finally, rigid-body movement of the fingers and thumb subdomains relative to the palm subdomain led to expansion and contraction of the RNA and NTP channels, revealing the conformation that likely exists in solution. Given these intriguing findings of potential mechanistic and biological importance, we extended our analysis to RdRps from related viruses, specifically, CVB3, HRV16 and FMDV. Overall, there was congruence in the results obtained for the RdRps from all four viruses. Together, these studies provide compelling evidence for coevolution of dynamics and structure among the picornavirus RdRps and provide support for the hypothesis that RdRp dynamics are required for function. Finally, we used MD simulation of the high-fidelity derivative of PV RdRp (G64S) to show that this single-amino-acid substitution altered the dynamics of elements implicated in binding and conformational rearrangements of the NTP substrate. We conclude that dynamics is an important contributor to RdRp functions, in particular, the fidelity of nucleotide selection.
Results
Validation of MD simulation
We performed a 25-ns all-atom MD simulation of PV RdRp at 300 K using its crystal structure [Protein Data Bank (PDB) code 1RA6; 2 Å resolution17] as the starting coordinates for simulation. The details of the MD simulation are described in Methods. During the simulation, the temperature, energy and density of the system were monitored, and all showed stable MD trajectories with small variations around reasonable average values. The average temperature was 299.97 ± 0.82 K, and the average density was 0.9982 ± 0.0009 g/cm3. The simulated structure maintained its native fold throughout the entire simulation.
In order to determine the useful time frame for the simulation and to exclude portions of the trajectory that may lead to erroneous conclusions, we evaluated the root-mean-square deviation (RMSD) relative to the minimized starting structure as a function of time for both the entire structure and the palm subdomain utilizing the backbone (Cα, C and N) atoms for the calculations (Fig. S2). The palm subdomain is the most conserved subdomain of the structure when polymerases of different classes or from different virus families are compared.34 The PV RdRp simulated system was equilibrated in approximately 2 ns. The RMSD reached plateau values around 2.0 Å for residues of the entire structure and 1.1 Å for residues of the palm (Fig. S2).
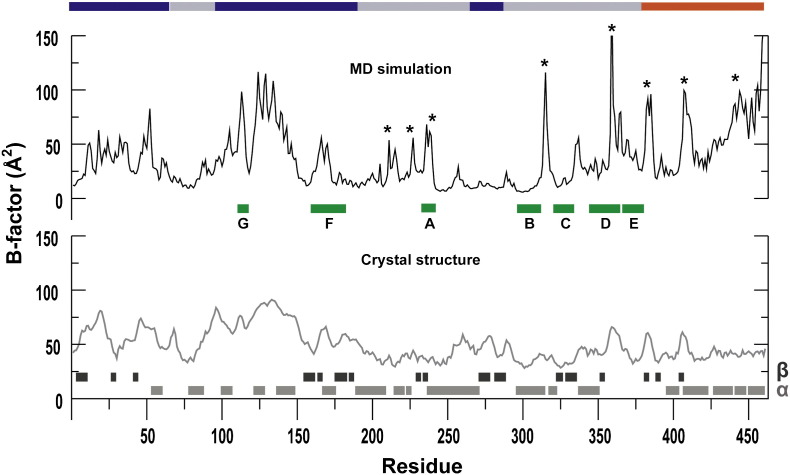
B-factors from MD simulation are consistent with those from the crystal structure but reveal unexpected mobility in some regions of PV RdRp
In order to provide an objective assessment of the output from the simulation, we calculated the B-factors for the backbone atoms and compared these B-factors to those derived from X-ray crystal structure data (Fig. 2 ). B-factors are representative of the amount of disorder and/or the positional atomic fluctuations present in the crystal, providing information on the flexibility of the molecule.
Fig. 2.
B-factor analysis of the PV RdRp validates MD simulation and reveals dynamic regions not seen in the crystal structure. Plots of the B-factors averaged over the protein backbone atoms calculated from simulation (top, black line) and the experimental B-factors extracted from the crystal structure (bottom, gray line) are shown. The calculated B-factors are in good agreement with those obtained from the crystal structure. However, differences were observed at some regions of the structure (marked with asterisks). Most of these differences can be attributed to crystal packing. The secondary structural elements are marked as bars at the bottom of the plots. The conserved motifs A–G are indicated by the green bars. The regions of the sequence corresponding to the different subdomains are labeled and marked with the colored bar at the top: fingers (blue), palm (gray) and thumb (red).
Overall, the pattern of the calculated B-factors was very similar to that of the experimental B-factors. For the most flexible regions, the magnitude of the calculated B-factors was, on average, greater than observed for the experimental B-factors. The simulation revealed higher B-factors in the palm subdomain (214–217, 226–227, 235–239 of motif A, 314–317 and 358–361 of motif D) and thumb subdomain (382–386, 406–412 and 438–461) than observed for B-factors derived from the crystal structure data (Fig. 2). The observed differences might be due to restricted motion in the crystal; in some cases, the crystal lattice provides a plausible explanation for the observed differences. Residues 314–317 of the palm and residues of the thumb are clearly restricted by the crystal lattice. Residues 214–217, 235–239, 226–227 and 358–362 are solvent accessible. In addition, these regions could indirectly be influenced by the restricted motion of the thumb. Region 214–217 is physically linked with the thumb and appeared positively correlated with regions 226–227 and 235–239 of motif A, which, in turn, is correlated with residues 358–361 of motif D.
The fingers subdomain was the most flexible, and the palm subdomain was the least flexible (Fig. 2). Consistent with the flexibility of the fingers subdomain is the high content of solvent-accessible loops. Conserved structural motifs involved in primer, template and/or nucleotide binding (motifs A, D, E, F and G) were all quite flexible, even those located in the palm subdomain (Fig. 2). In contrast, conserved structural motifs whose primary role is to serve as ligands to the divalent cations required for nucleotidyl transfer (portion of motif A harboring Asp233; motif C) were much less flexible. Also, motif B, which plays an indirect role in selection of rNTP over dNTP, was less flexible.
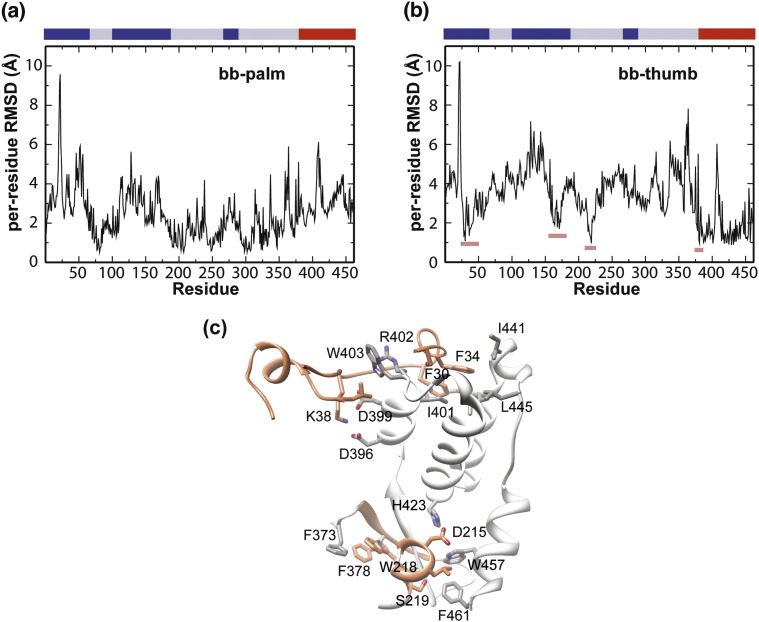
Per-residue RMSD analysis suggests motion of the fingers and thumb subdomains relative to the palm subdomain
To determine the extent of structural changes in PV RdRp during simulation relative to the starting structure, we performed a per-residue RMSD analysis using the last 10 ns of the trajectory (Fig. 3a and b). In this analysis, the average positional deviations of each residue were calculated relative to the minimized starting structure after superposition using either palm or thumb residue as a reference. The per-residue RMSD values provide some indication for the relative movement of subdomains.
Fig. 3.
Per-residue RMSD analysis of PV RdRp suggests substantial conformational changes. The averaged per-residue RMSD was calculated using frames extracted from the last 10 ns of the trajectory at 10-ps intervals. Prior to calculation, the snapshots were superimposed onto the minimized starting structure using the backbone atoms of (a) the palm or (b) the thumb. High per-residue RMSD values for regions outside the subdomain employed for the superimposition indicate substantial conformational changes in the PV RdRp structure during simulation. In (b), segments of low per-residue RMSD values in regions other than the thumb subdomain are highlighted with short colored bars The low per-residue RMSD values suggest that residues of the highlighted regions move concertedly with residues of the thumb subdomain. (c) The residues highlighted in (b) are shown on the PV RdRp structure. These residues are involved in direct interactions with the thumb.
We performed this analysis twice. In the first trial, the palm residues (69–95, 191–268 and 287–380) were used for superposition (Fig. 3a). In the second trial, the thumb residues (381–461) were employed for superposition (Fig. 3b). We observed 4- to 10-Å changes for residues in the fingers relative to the palm or thumb and the thumb relative to the palm (maxima in Fig. 3a and b). Residues 18–26 of the template channel in the fingers exhibited the most substantial conformational changes. Interestingly, this substantial change in residues 18–26 was observed relative to both the palm and the thumb (Fig. 3a and b). These high per-residue RMSD values suggest substantial structural changes in the relative positions of subdomains.
In addition to observing regions of substantial increase in per-residue RMSD using superposition of thumb subdomain, some regions outside of the thumb showed low (< 2.0 Å) per-residue RMSD values (minima marked by the colored bars in Fig. 3b). These regions include residues 28–42 and 162–173 of the fingers and residues 214–219 and 376–380 of the palm. We interpret this observation to mean that these regions are moving in a concerted manner with the thumb subdomain. When mapped onto the structure, the correlated nature of the movement is understood (Fig. 3c). These regions are all physically linked. The fingertips interact with the top of the thumb. Ser28 forms a hydrogen bond with the backbone of Arg402. Ala29 interacts with Ile441. Phe30 and Phe34 are anchored to the thumb via hydrophobic interactions with Ile401, the aliphatic portion of Lys435, Ile436 and Leu445. Lys38 forms a salt bridge with Glu396 and Glu399. Residues 162–173 are the “ring finger.”17 Leu162 participates in a hydrophobic interaction with the side chain of Trp403. The residues of the ring finger also interact with residues of the fingertips. Palm subdomain residues 214–219 interact with the carboxy-terminal helix of the thumb. Asp215 forms a hydrogen bond with His423. Leu216 is engaged in hydrophobic interactions with Trp457 and Phe461. Ser219 forms a hydrogen bond with the backbone carbonyl of Ile388. Palm subdomain residues 376–380 form an antiparallel β-sheet with thumb residues 388–392. Finally, the aliphatic portion of Arg373 and Phe378 participate in hydrophobic interactions with Trp218. All of these interactions were maintained during the entire time frame of the simulation.
Cluster analysis35 of the PV RdRp trajectory yielded four clusters (C1–C4), shown in Fig. S3, that represent different conformations of PV RdRp sampled over the course of simulation. The regions that showed high per-residue RMSD values (Fig. 3) were the same regions that showed the most structural variability in the cluster analysis (Fig. S3). The largest structural variations between the different clusters were mainly in region 375–391, the helices of the thumb, regions 16–70 and 103–118 and motif F of the fingers and motif A of the palm. Cluster analysis provides additional support for movement of fingers and thumb relative to the palm described above. While cluster analysis represents a rigorous approach to define distinct conformations during the MD simulation, the collective motions observed here are also evident by merely evaluating snapshots from the beginning (t o), middle (t 1) and end (t 2) of the simulation (Fig. S4).
Taken together, this analysis suggests that large intersubdomain conformational changes occur on the nanosecond timescale.
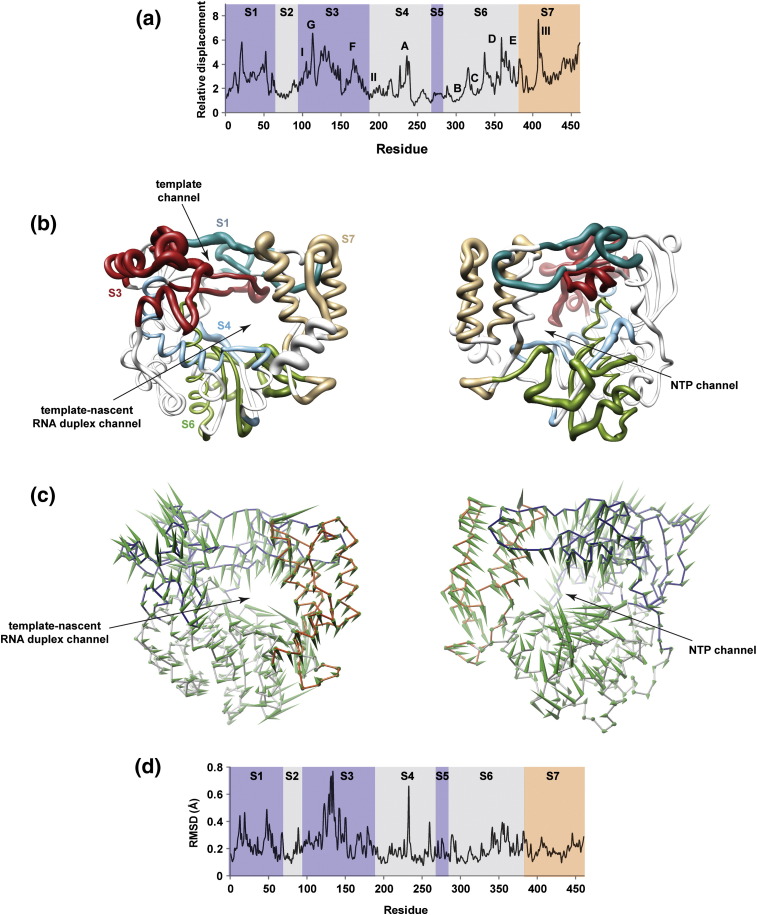
Principal component analysis reveals conserved sequence/structural motifs as contributors to the major motions observed for PV RdRp
Both the B-factor and per-residue RMSD analysis showed PV RdRp to be flexible. To yield information on components of the PV RdRp structure that are major contributors to the motions observed during the simulation, we performed a principal component analysis (PCA)36, 37 using the trajectory of the entire production run. This analysis considers motions as the variance of atomic fluctuations around the average structure. Here, we only considered the variance (mean-square displacements) of Cα atoms. The number of principal components (PCs or modes) is equal to 3 × Cα atoms (461 atoms in the case of PV RdRp). Every mode is associated with an eigenvalue, which corresponds to the magnitude of the fluctuation along that particular mode and contains different contributions from the analyzed atoms. The first mode represents the largest atomic fluctuation in the sampled conformations. The second mode represents the next largest atomic fluctuation orthogonal to the axis of the previous mode and so on. A majority (∼ 70%) of the total variance sampled during the simulation could be accounted for by the top six PCs. Therefore, we restricted our focus to these PCs. We calculated the displacement of each Cα atom in each of the top six modes, which was then summed to produce the total displacement for the individual residues represented by Cα atoms. In order to render the PCA in a manner that would facilitate comparisons with other simulated systems, we normalized the per-residue sum of the displacements of the top six modes to the average of the least flexible 5% of residues in the enzyme.
The result of the PCA of PV RdRp simulation is shown in Fig. 4a. In Table 2 , the information on the flexibility of PV RdRp provided by the PCA is presented in the context of our understanding of functions of the segments (S1–S7) of the enzyme and residues within these segments that have been implicated in these functions. The relative displacements of Cα atoms are plotted as a function of residue number. Regions in the structure distributed across the fingers, palm and thumb subdomains all showed contributions to the major motions observed during simulation. In order to add a structural perspective to the information on the dynamics provided by the PCA, we rendered the structure as a tube, the radius of which is equivalent to the magnitude of the relative displacement (Fig. 4b).
Fig. 4.
Functional motifs of PV RdRp largely contribute to the major motions observed during the simulation. (a) The relative displacements of Cα atoms observed in the sum of the first six PCs (modes) are plotted for each residue. Peaks correspond to regions contributing to the major motions observed during the MD simulations. The structure has been divided into seven segments (S1–S7). The fingers subdomain is shaded purple, the palm subdomain is shaded light gray and the thumb subdomain is shaded orange. The conserved motifs A-G are indicated. Other regions of PV RdRp known to have functional significance have been labeled I–III. (b) PV RdRp structure rendered as a tube to indicate flexibility and shown in two views: (left) looking through the template–nascent RNA duplex channel and (right) looking through the NTP channel. The radius of the tube indicates the magnitude of the relative displacement shown in (a). Each segment has a unique color. Only segments S1, S3, S4, S6 and S7 are shown. Interestingly, highly dynamic regions are located around the nascent RNA duplex and NTP channels. (c) The motion content of PC1, the largest mode of PCA, is visualized in two views that show the PV RdRp from the front (left) and the back (right). The starting structure of PV RdRp is depicted as wires with subdomains colored differently: blue (fingers), gray (palm) and red (thumb). The green “porcupine needles” indicate the direction of displacements of motions based on PC1; the size of the needle is proportional to the displacement. From the directions of displacement vectors represented by the needles, expansion and contraction of the RNA and NTP channels can be indicated; interestingly, when the template–nascent RNA duplex channel is open, the NTP channel is closed and vice versa. (d) Structural variation among multiple crystal structures of PV RdRp is represented by RMSD for each residue in PV RdRp, calculated from superimposed crystallographic structures (PDB codes 1RA6, 1RA7, 2IM0, 2IM1, 2IM2, 2IM3, 2ILY and 2ILZ). The structures were superimposed using Cα atoms of motifs B and C; superimposition was performed using Chimera. Residues that demonstrate large structural variations, indicated by high RMSD values, are implied to be flexible. Structural variations among different crystal structures qualitatively showed a pattern similar to that displayed in (a) for residues in segments S1–S5. The smaller amount of variability of residues in segment S6 could be a result of using residues within this segment for superimposition. Differences between experimental data and simulation observed for residues in segment S7 are attributable to the crystal packing effect where mobility of these residues is masked inside crystals; such an effect is absent from simulation.
Table 2.
Flexibility and function of segments of PV RdRp structure
| Segment | Residues | Motif or region | Relative flexibilitya | Key residuesb | Function |
|---|---|---|---|---|---|
| S1 | 18–26 | +++ | Asn18, Pro20, Lys22, Lys24 | Template binding | |
| 32–39 | ++ | Opening/closing of NTP channel | |||
| 41–53 | +++ | Opening/closing of NTP channel | |||
| S3 | 96–117 | I and G | +++ | Leu107, Glu108, Thr114 | Template binding and opening/closing of nascent RNA channel |
| 120–149 | +++ | Lys127 | |||
| 157–179 | F | +++ | Tyr157, Lys159, Asp160, Arg163, Lys167, Arg174, Ile176, Ser179 | Triphosphate of NTP and template binding | |
| S4 | 181–200 | II | + | Ser184, Arg188, His199 | Template binding |
| 208–217 | + | Asp213 | Template binding | ||
| 226–227 | ++ | ||||
| 233–240 | A | +++ | Asp233, Asp238 | 2′-OH of NTP and metal B binding | |
| S6 | 288–292 | + | Ser288, Gly292 | Base of NTP and template binding | |
| 293–312 | B | + | Ser294, Asn297 | Base of NTP and template binding | |
| 313–317 | ++ | No known function | |||
| 323–335 | C | + | Tyr326, Gly327, Asp328, Asp329 | Primer and metal A binding | |
| 336–345 | D | +++ | |||
| 352–362 | D | +++ | Lys359 | Catalytic general acid | |
| 363–380 | E | +++ | Lys375, Arg376 | Primer binding | |
| S7 | 381–386 | +++ | Opening/closing of nascent RNA channel | ||
| 391–392 | ++ | ||||
| 405–423 | III | +++ | Lys405, Arg408, Asn409, Arg412, His413, Ser416, Leu417, Leu419, Leu420 | Template and primer binding and opening/closing of nascent RNA channel | |
| 426–461 | +++ |
The peaks in the PCA plot were identified with respect to the immediate boundary regions. Relative flexibility is assigned as follows: +++, relative displacement > 4-fold; ++, 4-fold > relative flexibility > 3-fold; and +, 3-fold > relative flexibility > 2-fold.
Residues that are important for the functions defined in the last column. However, there are no known functions for residues 313–317, which showed conserved dynamics in the studied RdRps.
The fingers and thumb subdomains contributed most to the observed motions. Motifs F and G of the fingers were highly dynamic. An interesting dichotomy was observed again when conserved structural motifs in the palm subdomain were evaluated. Motifs A, D and E were quite dynamic; motifs B and C were not. In general, all regions of PV RdRp known to interact with RNA, nucleotide or metal contained elements exhibiting both “high” and “low” flexibilities. For example, residues 18–26, 96–112, 120–149 (region I in Fig. 4a) and 405–423 (region III in Fig. 4a) interact with RNA and exhibit high mobility (Fig. 4a). However, residues 208–217 and 288–292, which also interact with RNA, exhibit low mobility (Fig. 4a). The functional significance of this observation, if any, is not clear.
Regions of PV RdRp (32–39, 41–53, 226–227 and 426–461) with no known function in nucleotidyl transfer also contributed to the motions observed during the simulation (Table 2). Interestingly, essentially all of these regions are important for optimal viral fitness at 39.5 °C38 and for the recruitment of the polymerase to the initiation complex. 39
We considered the direction of motion for PC1, which accounts for 42% of the total motions (Fig. 4c). This analysis revealed expansion–contraction and/or hinge-twisting motions of the three channels of the enzyme. First, the template–nascent RNA duplex channel (denoted as nascent RNA channel) opened and closed more than once during the simulation. Each cycle of opening and closing comprised residues of the fingers (1–16, 96–149 and 269–283) and the thumb moving in opposite directions that resembled a hinge-twisting motion. When this channel opened, the fingers subdomain moved away from the palm subdomain, and the thumb moved toward the palm with a vectorial bias in the direction of the NTP channel. Second, the NTP channel opened and closed more than once during the simulation. As observed for the nascent RNA channel, each cycle of closing included residues lining the channel (157–176 and 353–365) moving in a correlated manner with a vectorial bias in the direction of the nascent RNA channel. Opening of this channel was driven by the movement of the fingers subdomain (157–176) relative to the palm subdomain (353–365) in opposite directions. Interestingly, when the nascent RNA channel was open, the NTP channel was closed and vice versa. These motions appeared in PC1 for all picornavirus polymerases studied here (data not shown). Finally, the template channel formed by residues (17–27 and 108–116) also expanded and contracted. The opening and closing of this channel was not synchronized with either the nascent RNA or the NTP channels.
In an effort to compare the pattern of dynamics observed on the nanosecond timescale to conformational flexibility observed in numerous complex structures of PV RdRp,40 we examined the positional fluctuations of Cα atoms among these different structures that were obtained by soaking in different NTP solutions. Motifs B and C of the palm were used to superimpose these structures. We plotted the RMSD for each residue (Fig. 4d). The pattern observed for segments S1 to S6 of the RMSD plot in Fig. 4d was qualitatively similar to the pattern observed for these segments of the relative displacement plot in Fig. 4a. The dynamics of segment S7 are suppressed in the crystal structure by the crystal lattice as discussed above. These data make a very compelling link between the dynamics of PV RdRp on the nanosecond timescale and the conformational space sampled by this enzyme as shown by crystal structures.
Correlated motions exist between conserved structural/sequence motifs of PV RdRp
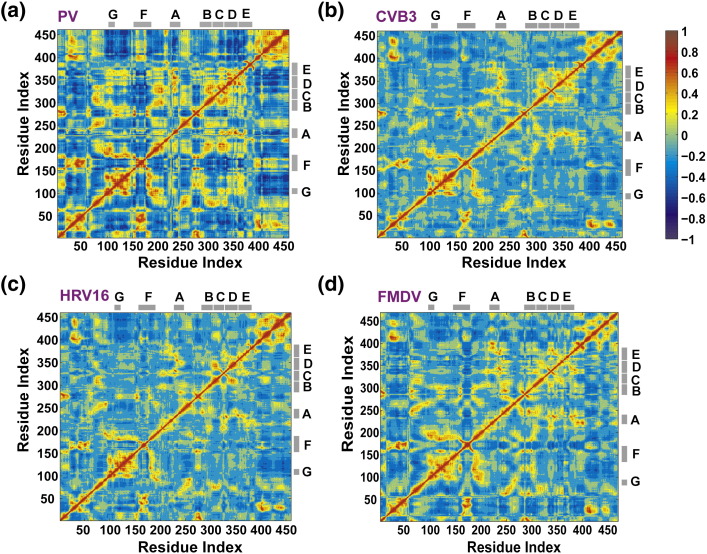
Conserved structural/sequence motifs are implicated in specific steps along the reaction coordinate. In some enzyme systems, it is believed that one approach for channeling an enzyme and its substrate down a preferred reaction pathway is to engineer into the enzyme the motions required for progression through the reaction coordinate.28, 29 Often, this outcome manifests in correlated motions of functionally important residues, motifs and/or subdomains. Therefore, we evaluated the correlation between the different motions sampled during the production run of the simulation and produced a dynamic cross-correlation map (DCCM)41 for PV RdRp (Fig. 5a).
Fig. 5.
Correlated motions in RdRps. The DCCM, which measures the correlation between the displacements of Cα atoms, calculated from simulations of RdRps from PV (a), CVB3 (b), HRV16 (c) and FMDV (d) is shown. For each enzyme, the calculated matrix whose elements are the pairwise correlation scores between its residues is visualized as a colored map. The correlation scores are encoded with a color gradient from − 1 (blue, completely anti-correlated) to + 1 (red, completely correlated). The conserved structural motifs are marked with gray bars. Residues that appeared flexible in PCA of PV RdRp (see Fig. 4a) are shown to be strongly correlated. Both positive and negative correlations have implications regarding polymerase functions as discussed in the text. The figure was prepared using MATLAB 7.6.
The dynamics of residues that contribute to RNA binding are negatively correlated, suggesting expansion and contraction of the nascent RNA channel (Fig. 5a). Strong negative correlations were observed between residues 96–117 and 120–149 of the fingers and residues 400–461 of the thumb, consistent with expansion and contraction of the nascent RNA channel. Residues 17–27 and 108–116, which surround the RNA template channel, were negatively correlated, suggesting expansion and contraction of the template channel. Similarly, the dynamics of residues that contribute to NTP binding are negatively correlated, also suggesting expansion and contraction of the NTP channel. Motif F is negatively correlated to motifs B–D, and both motifs D and E are negatively correlated to the thumb (Fig. 5a), influencing motions of the NTP channel. Also, the thumb residues are negatively correlated with residues 229–235 of motif A and residues 324–333 of motif C.
The motion of every conserved structural motif of the palm subdomain, regardless of the magnitude of the displacement, was positively correlated. The motions of motifs F and G are also positively correlated. In contrast to the negative correlation of the dynamics of residues of the fingers and thumb that contribute to the nascent RNA channel, we observed positive correlation between the dynamics of the fingertips and thumb.
Expansion and contraction of the nascent RNA duplex channel of PV RdRp
From the initial analysis of the complete structure of PV RdRp,17 it was clear that the narrowest portion of the nascent RNA channel was too small to accommodate duplex RNA observed in complex crystal structures.20 A representative view of this “closed” conformation is shown in Fig. 6a. It was therefore quite intriguing to observe some indication of channel opening and closing in the analyses described above.
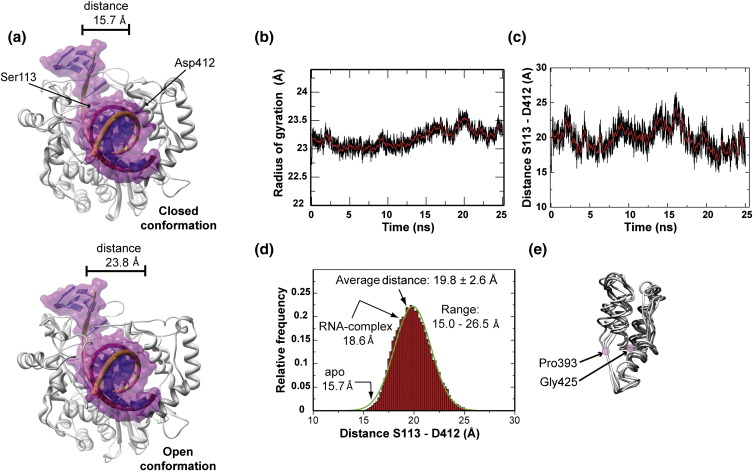
Fig. 6.
Opening and closing of the RNA-binding channel of PV RdRp. (a) The closed (top) and open (bottom) conformations of PV RdRp with modeled RNA. The RNA modeled in the apoenzyme representing the closed conformation was taken from the PV RdRp elongation complex structure (PDB code 3OL6). The open conformation derives from a snapshot from the simulation. (b) Oscillation of the radius of gyration (Rg). The time evolution of Rg for the simulated structure is shown by the black line; the running average (200-ps window) is shown by the red line. (c) Oscillation of the Cα–Cα distance of Ser113 (fingers) and Asp412 (thumb). The time evolution of distance for the simulated structure is shown by the black line; the running average (200-ps window) is shown by the red line. (d) The frequency distribution of the distances observed in (c). The green line represents the fit to a Gaussian model using the program Grace-5.1.22. (e) Hinge-twist motion of the thumb observed by superimposition of snapshots across the simulation trajectory. Residues P393 and G425 are the pivot points for the motion.
We next focused on a more direct investigation of the changes in the nascent RNA channel. First, we monitored the radius of gyration (R g) for the backbone atoms during the entire simulation (Fig. 6b). On average, R g increased by 2–4% relative to the starting structure. Conformations with R g values comparable to those of the reference structure were occasionally sampled during the first half of the trajectory. These data were consistent with expansion and contraction of the entire molecule. When two residues, Ser113 from the fingers and Asp412 from the thumb, located at opposite sides of the nascent RNA channel, were chosen as probes for the size of the channel, expansion and contraction of the channel were readily apparent (Fig. 6c). The distance between these two residues fluctuated over the entire duration of the simulation. The frequency of each distance observed during the production run of the simulation was evaluated (Fig. 6d). From this analysis, it is clear that the channel can open by as much as 10 Å from the most closed to the most open conformations, permitting the open conformation to accommodate duplex RNA (Fig. 6a). On average, the channel width was 19.8 ± 2.6 Å, which is 4.1 Å larger than observed in the crystal structure of the apoenzyme. Note that the channel width increased from 15.7 Å in the apo structure of PV RdRp to 18.6 Å upon formation of the elongation complex,20 consistent with what we observed in our simulation. The residues affected most by the closed-to-open transition are 19–24, 108–113, 120–132 and 165–170 in the fingers; 359–368 in the palm; and 407–412 and 440–450 in the thumb. The dynamics of the channel are also influenced by a hinge-twist motion of the fingers and thumb using residues P393 and G425 as pivots for the thumb (Fig. 6e).
As mentioned above, cluster analysis35 of the snapshots extracted at 10-ps intervals across the trajectory obtained from the MD production revealed four clusters (C1–C4) (Fig. S3). The most populated cluster, C1 (58%), represented the closed conformation (Fig. S3), perhaps providing an explanation for the conformation observed in the crystal. Importantly, the next most populated states represented open conformations (Fig. S3). Together, these analyses have uncovered the dynamic nature of the nascent RNA channel.
Conserved dynamics in the RdRps from CVB3, HRV16 and FMDV
The MD analysis of PV RdRp revealed putative dynamic signatures for conserved structural/sequence motifs, most of which have been assigned critical functions in the nucleotidyl transfer reaction. For PV RdRp, we extended the simulation to 52 ns. This analysis did not change conclusions reached by using the 25-ns simulation (Fig. S5). Given the availability of high-resolution structural information for a variety of related picornaviral polymerases, we were in a unique position to determine the extent to which the observations made for PV RdRp extended to others.
We used CVB3,21 HRV1622 and FMDV19 RdRps as our models. The structures of these enzymes can be superimposed with an RMSD of 0.7–1.6 Å for 439 Cα atoms. The sequences, however, vary to a much greater extent. The sequence identities are ∼ 74%, 56% and 30% for CVB3, HRV16 and FMDV, respectively, relative to PV; the sequence alignment is shown in Fig. S1.
We employed the same experimental paradigm for analysis of these enzymes as that we had used for PV RdRp. The run-times were 14 ns for CVB3 RdRp, 14 ns for HRV16 RdRp and 20 ns for FMDV RdRp. The time frame used for analysis of the simulations of these RdRps ranged from 10 to 16 ns (Fig. S2). The B-factors calculated from the simulation were in good agreement with those from X-ray, with the same caveats discussed above for PV, including restricted motions caused by crystal packing (Fig. S6). Per-residue RMSD analysis showed that the flexibility of CVB3 RdRp was on par with that of PV RdRp (Fig. S7). The overall flexibility of HRV16 RdRp was greater than observed for PV RdRp, while that of FMDV RdRp was lower (Fig. S7). Snapshots from the beginning, middle and end of the simulation (Fig. S4) revealed conformational changes for the fingers and thumb subdomains relative to the palm subdomain, suggesting opening and closing of the RNA and NTP channels.
PCA analysis showed that the top six PCs accounted for 51% of the total variance for CVB3 RdRp, 56% for HRV16 RdRp and 59% for FMDV RdRp. A plot of the relative displacement as a function of residue showed remarkable similarity in the dynamics of all four RdRps (Fig. 7a). With a few exceptions, the dynamics of the conserved structural/sequence motifs A–G and functional regions I–III were conserved. The specific boundaries for the subdomains and functional motifs/regions of each RdRp are provided in Table S1. For all RdRps, motifs A, D, E, F and G and regions I and III were the most mobile; motifs B and C were the least mobile (Fig. 7a).
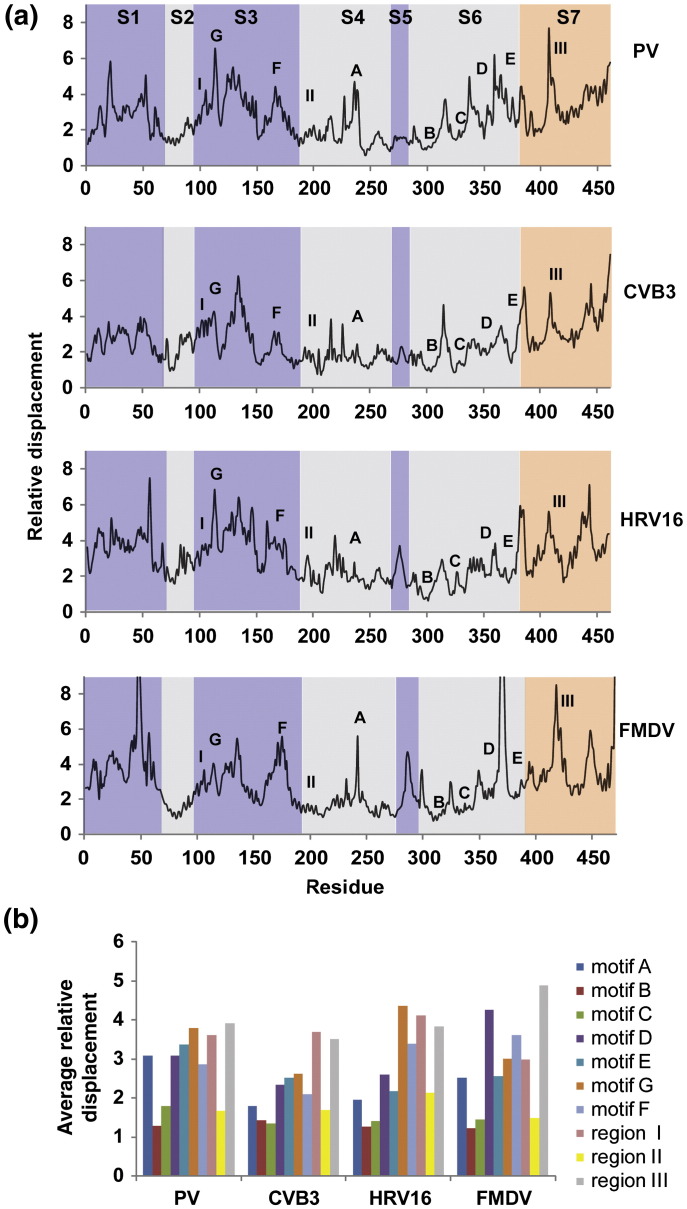
Fig. 7.
Conserved dynamics of the RdRps from PV, CVB3, HRV16 and FMDV. (a) The relative displacements of Cα atoms observed in the sum of the first six PCs (modes) are plotted for each residue. Peaks correspond to regions contributing to the major motions observed during the MD simulations. The structure has been divided into seven segments (S1–S7). The fingers subdomain is shaded purple, the palm subdomain is shaded light gray and the thumb subdomain is shaded orange. The conserved motifs A–G are indicated. Other regions of PV RdRp known to have functional significance have been labeled I–III. Dynamic regions under the peaks across the different polymerases show a striking similarity; the observed dynamic patterns are largely conserved. The major peaks in the different segments (S1–S7) observed in PV are also observed at equivalent positions for the other three RdRps. Nonetheless, differences in relative displacements of motifs A and E as well as of the region spanning residues 43–55 in the fingers are noted. Local sequence and structural variations might account for the differences in the observed dynamics. The plot of PV RdRp, shown previously in Fig. 4, is included for comparison with the other RdRps. (b) The average relative displacement calculated for the conserved motifs A–G and functional regions I–III of each RdRp is shown. The average was calculated by using the relative displacement of all residues constituting the motif as defined in Table S1.
We compared the PCA of each RdRp to that of PV, scrutinizing the conserved functional motifs/regions. In order to avoid differences due to the statistical nature of the MD simulation, we used the average values for the relative displacement for all residues of a given motif/region of each RdRp (Fig. 7b). A 3- to 4-fold difference was observed between the most and the least flexible motifs/regions of each RdRp. With this value in mind, we reasoned that a 1.5-fold difference between RdRps for any motif/region would be a reasonable cutoff for a difference that may have some functional significance. When each motif/region of the RdRps from CVB3, HRV16 and FMDV was compared to that of PV using this criterion, motif A of CVB3 and HRV16 RdRps and motif E of HRV16 RdRp showed the only differences. Outside of the conserved motifs/regions, two additional differences were noted. PV RdRp residues 43–55 were less mobile than the corresponding FMDV RdRp residues. PV RdRp residues 272–281 were less mobile than the corresponding HRV16 RdRp and FMDV RdRp residues. We did not perform all pairwise comparisons because PV RdRp is the only enzyme for which sufficient functional information exists to permit any speculation on functional consequences of altered dynamics.
We also compared the correlated nature of the motions. Again, the patterns were well conserved (Fig. 5b–d). As discussed above, some of the correlated motions govern open-to-closed transitions of the RNA template and nascent channels as well as the NTP channel. The changes in the nascent RNA channel were also observed for the other RdRps by monitoring the radius of gyration (Fig. S8) or the distance between two residues located on opposite sides of the channel (Fig. S9). For the nascent RNA channel, in all cases, a distance capable of accommodating duplex RNA (> 20 Å) was sampled most often (Fig. S9b). However, the mean distance, the frequency of distances sampled and the magnitude of the open-to-closed transition varied for all four RdRps (Fig. S9b). At the extreme, the magnitude of the transition differed by as much as 5 Å (compare CVB3 RdRp to HRV16 RdRp in Fig. S9b).
Global changes in the dynamics of PV RdRp caused by a single-amino-acid substitution conferring a high-fidelity phenotype
The analysis presented thus far has made a very compelling case for the evolution of dynamics with structure in the picornaviral RdRps. We next wanted to determine if the MD simulations could inform the molecular basis for phenotypes that were eluded by structural studies. The G64S derivative of PV RdRp exhibits an increase in incorporation fidelity relative to the WT enzyme.7, 8 This phenotype appears to arise from a decrease in the equilibrium constant for the conformational change step that leads to production of the catalytically competent RdRp–RNA–NTP complex.7 Gly64 is located some 20 Å from the catalytic center. The backbone dihedral angles of Gly64 assume a conformation that is disallowed for side chains of all other amino acids. The Gly64 backbone permits stabilization of the amino terminus in the fingers subdomain by mutual hydrogen bonds between backbone atoms and stabilizes motif A by formation of hydrogen bonds with the backbone atoms of Ala239 and Leu241. Thus, it was thought that, by changing Gly64 to Ser, these interactions would be altered, leading to misalignment of motif A, a motif known to contribute to incorporation fidelity.42 However, the X-ray crystal structure of G64S PV RdRp24 superimposed with WT PV RdRp with < 0.3-Å RMSD. Thus, the altered function is not explained by altered structure.
Again, we employed the same paradigm to evaluate the G64S derivative of PV RdRp. The run-time was 25 ns; the time frame from 3 to 25 ns was used for analysis. In this case, we restricted our focus to PCA, analysis of the average structure over the entire trajectory, correlated motions and the opening and closing of the nascent RNA channel (Fig. 8 ).
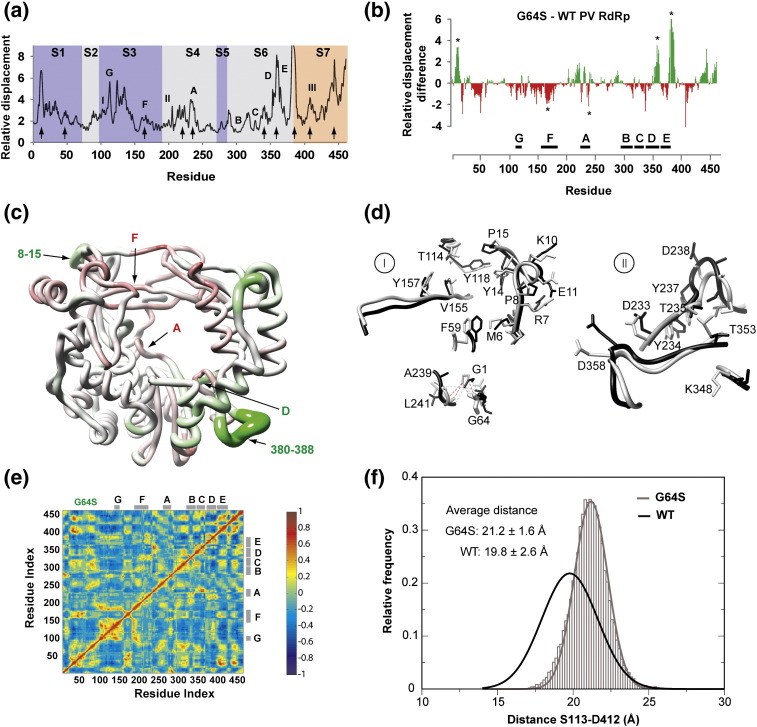
Fig. 8.
Global changes in the dynamics of PV RdRp anti-mutator (G64S 3Dpol). (a) The relative displacements of Cα atoms observed in the sum of the first six PCs (modes) are plotted for each residue. Regions showing visually observable differences of the relative displacement between G64S and WT are indicated by arrows. (b) The differences between relative displacements of WT and G64S are shown. Positive differences (green bars) indicate regions in G64S that are more flexible than WT, negative differences (red bars) indicate less flexible regions. (c) Mapping of the differences between G64S and WT onto the structure of the WT PV RdRp. The differences are shown as color gradients of green (positive) and red (negative); the structure is rendered as a tube whose radius indicates the magnitude of the difference (large radius corresponds to regions that are more flexible in G64S, and small radius corresponds to regions that are less flexible compared to WT). (d) Structural perturbations caused by the G64S substitution. The average structures for WT (black) and G64S (gray) are shown. (d-I) shows changes from position 64 to the N-terminal residues (8–15) and motif F (162–172). (d-II) shows changes in motif D (353–362). The hydrogen-bonding network involving residues Gly1, Gly64, A239 and Leu241 remains in both enzymes. (e) The DCCM calculated for G64S PV RdRp as done for the WT enzymes in Fig. 5. (f) Opening and closing of the template–nascent RNA duplex channel. The frequency distribution for the Cα–Cα distance of Ser113 (fingers) and Asp412 (thumb) in G64S (gray curve) is shown. The black curve corresponds to the WT and is shown for comparison. A shift to the more open conformations is observed in G64S relative to the WT.
For PCA, the top six modes accounted for 62% of the total variance sampled during the simulation. The plot of the relative displacement as a function of residue (Fig. 8a) was visually different from that of the WT enzyme (Fig. 4a). The single-amino-acid substitution produced changes in the magnitudes of the dynamics observed across the entire structure (Fig. 8a). Several areas stand out (indicated by arrows in Fig. 8a). Residues 8–15, 219–224, 231–233 (motif A), 353–362 (motif D), 380–388 and 440–446 were more flexible in G64S mutant, whereas residues 19–22, 41–53, 161–172 (motif F), 238–239 (motif A), 337–338 and 407–411 were less flexible. Strikingly, the altered dynamics caused by this one substitution were more substantial in several functional regions than observed when the sequence differed by 70% as is the case for the FMDV RdRp. Together, the observations reveal a physical difference between G64S and WT PV RdRps that may account for the observed biochemical and biological phenotypes.7, 8 In addition, these observations strengthen further the argument that structure, dynamics and function coevolve in this system.
In order to highlight the regions of PV RdRp most affected by G64S substitution, we subtracted the relative displacement observed at each position of WT PV RdRp from the corresponding relative displacement observed for G64S PV RdRp. This difference was plotted as a function of residue (Fig. 8b). Values greater than zero indicated greater flexibility of the mutant; values less than zero indicate lower flexibility. Three regions stand out that are more flexible in G64S: residues 8–15, 354–362 (motif D) and 380–388. On the other hand, regions of motifs A and F appeared more flexible in the WT. Mapping the differences plotted in Fig. 8b onto the structure of PV RdRp indicated that, generally, regions surrounding the catalytic center were more flexible in the WT, whereas regions that were more flexible in G64S are located on the exterior of the molecule (Fig. 8c).
The change of Gly64 to Ser led to visible changes in the average structure that provide a plausible explanation for the global alterations observed in the PCA (Fig. 8a and b). The averaged structures of G64S and WT PV RdRps superimpose with an RMSD value of 1.8 Å. A closer look at position 64 in the averaged structure showed different phi–psi conformations for Gly64 (phi = 169.7°, psi = − 170.7°) in the WT and Ser64 (phi = − 159.5°, psi = 155°) in the mutant enzymes. The changes in conformations of Ser64 altered the conformations of adjacent residues (59–65) even more dramatically; consequently, the side chain of Phe59 moved out of position, which, in turn, induced Met6 to move in order to maintain the hydrophobic interaction with Phe59 (Fig. 8d-I). The movement of Met6 and the adjacent residues influenced the conformations of the Pro8–Pro15 turn. Interestingly, as a result of these conformational changes, the side chain of Glu11 moved away from Arg7, breaking the hydrogen-bonding interaction observed in the WT and forming a new interaction with Lys10 in G64S. This rearrangement of interactions removed restrictions on the movement of the Pro8–Pro15 turn, which was shown to be more flexible in PCA (Fig. 8b and c).
Moreover, the conformational changes in residues 8–15 caused the side chain of Tyr14 to move out of position, which, in turn, influenced the nearby residues: Thr114 and Tyr118 (Fig. 8d-I). The side chains of Thr114 and Tyr118 moved closer to Val115 and Tyr157 of motif F, enhancing their hydrophobic interactions and adding more restrictions on the motions of motif F, in agreement with the PCA (Fig. 8b and c).
Overall, the G64S substitution did not affect the hydrogen-bonding network that mediates the interactions between the backbone atoms of residues Gly1 at the N-terminus and Gly64/Ser64, Ala239 and Leu241 of motif A (Fig. 8d-I). However, a new transient interaction between the backbone oxygen atom of Gly1 and the backbone nitrogen atom of Asn63 was noted for the G64S enzyme. The restricted motions of Ser64 relative to Gly64 reduced the flexibility of residues in motif A, as shown in PCA (Fig. 8b and c).
Additionally, the altered conformations of residues of motif A in the vicinity of Ser64 propagated to residues Tyr234, Thr235 and Tyr237 (Fig. 8d-II). As a result, the positioning of the nearby residue Thr353 in motif D was influenced, leading to further changes in conformations of residues 353–362. Interestingly, Thr353 in G64S moved away from its position in the WT enzyme, resulting in a loss of a potential interaction between the side chains of Thr353 and Lys348 (a hydrogen bond between Thr353 and Lys348 was transiently observed in the trajectory of the WT). Consequently, residues 353–362 become less restricted in the mutant enzyme relative to the WT, in agreement with what we observed in PCA (Fig. 8b and c). Finally, the conformational changes in residues of motif D influenced the positioning of the nearby residues Phe377 and Arg379, leading to the conformational changes in residues 380–388. In the mutant enzyme, residues 380–388 moved closer to the thumb with a dramatic rearrangement of the side chain of Tyr384.
We also analyzed the DCCM calculated for G64S (Fig. 8e) and compared it to that of WT PV RdRp (Fig. 5a). Generally, correlations in the case of G64S were weaker than those of WT. In many places in the DCCM, changes in correlations can be observed in the mutant enzyme compared to the WT. For example, motifs D and E revealed positive correlations to the thumb in G64S simulation; in contrast, the same residues were shown to be negatively correlated in the WT. Also, the negative correlations between motif F and motifs B–D seemed less pronounced in G64S compared to WT. Motifs B and C showed stronger positive correlations with the thumb in G64S relative to WT. Similar to WT, residues surrounding the NTP and nascent RNA channels were negatively correlated, suggesting that opening and closing of these channels occurred normally and in synchrony (data not shown).
Analysis of the expansion and contraction of the nascent RNA channel by probing the distance between Cα atoms of Ser113 and Asp412 showed a shift of ∼ 1 Å in the average distance toward a more open conformation of G64S relative to WT (Fig. 8d). Moreover, the distribution of the sampled distance is narrower than that of the WT, suggesting that G64S is more rigid than WT.
Discussion
This study was motivated by our desire to determine if dynamics could provide insight into the physical basis for biochemical and biological phenotypes of PV RdRp not explained by structures.25 It has been suggested by some that rate-limiting conformational changes in enzymes that occur on the millisecond timescale may be determined by atomic fluctuations and correlated motions on the picosecond-to-nanosecond timescale.30 Atomistic MD simulations provide a tractable approach to assess the picosecond-to-nanosecond dynamics of a protein for which X-ray crystal structure data are available.
We performed a 25-ns simulation of the dynamics of PV RdRp. In order to scrutinize the trajectory, we calculated B-factors for each residue and compared these to those obtained from the X-ray crystal structure data (Fig. 2). Overall, the calculated B-factors were in good agreement with experimental values except for residues participating in crystal contacts. Until recently, there was no empirical information available on the dynamics of PV RdRp or any RdRp for that matter.26, 43 So, how does one really convince the bench biologist and biochemist that these computational data are functionally relevant? In the case of the picornaviruses, X-ray crystal structures are available for many RdRps in the family including CVB3, HRV16 and FMDV.19, 21, 22 The biochemical parameters are nearly identical for the PV and FMDV RdRps, which are at the extremes of this group [30% identity on the amino acid level (Fig. S1)].10 The relevance of the algorithm as applied to PV RdRp could be put to test by applying it to the other picornaviral RdRps. We reasoned that any dynamic features of functional significance should be conserved across the family. To our delight, we observed that the overall pattern of flexibility as a function of residues was conserved for all four RdRps evaluated (Fig. 7). The conservation in dynamics even extended to correlated motions observed between sequence/structural motifs of functional importance (Fig. 7). Together, our results engender confidence in the utility of MD simulations and suggest conservation in the dynamics of the picornaviral RdRps. As discussed in more detail below, the possibility exists that the dynamics are conserved for function, thus providing additional evidence for a role of dynamics in enzyme catalysis27, 44 and contributing another enzyme system to use to interrogate this property of enzymes that is likely essential.
One of the most striking observations made during the course of this study is that the magnitude of the intersubdomain conformational changes on the nanosecond timescale was large (Figs. 3 and Fig. S7). For all four polymerases analyzed, both the fingers and the thumb subdomains moved relative to each other. The changes in intersubdomain distances sampled during the course of the nanosecond trajectories were in the 4- to 10-Å range. Conformational changes of this magnitude are often observed on the microsecond–millisecond timescale in enzymes and appear as rate-limiting events in the kinetic mechanism.30, 45 In the case of the RdRp, it is clear that the fingers and thumb subdomains undergo conformational changes of this magnitude during the catalytic cycle. Therefore, it is possible that conformational states of the RdRp required for function are sampled on the nanosecond timescale to enable conformational changes that occur on longer timescales.
The Ghose laboratory has used NMR of the RdRp from φ6 to study the dynamics of this RdRp.43 φ6 RdRp is a member of a lineage in the RdRp superfamily different from the picornavirus RdRps.46 The φ6 RdRp was labeled with [methyl-13C]Ile. Ile residues are present in all three subdomains of this RdRp. In the palm subdomain, Ile could be used to probe the dynamics of motifs B, C, D and E. Order (s 2) parameters were used to probe dynamics on the picosecond-to-nanosecond timescale. As observed here, the φ6 palm subdomain exhibited the least overall flexibility of the three subdomains. In addition, φ6 motifs D and E were most flexible, and motif C was the least mobile on this timescale in the apoenzyme. These same residues exhibiting dynamics on the picosecond-to-nanosecond timescale also exhibited dynamics on the timescale of catalysis. Our MD data on the picornaviral RdRps, when considered in the context of the NMR data for the φ6 RdRp, suggest conservation of the picosecond-to-nanosecond timescale dynamics of functional elements across the RdRp superfamily and a potential linkage of these fast dynamics to catalysis.
Assembly of picornaviral RdRps with duplex RNA is slow.47 It is known that the rate-limiting step for assembly is a conformational change in the enzyme.47 The simulation revealed that the interaction of the fingertips with the thumb that leads to a completely encircled active site is stable throughout the entire trajectory for all enzymes. This observation is consistent with the proposal that release of the fingertips from the thumb limits association with and/or dissociation from primed templates in vitro.48 For PV RdRp, residues of the fingertips (28–42 and 162–173), palm (214–219 and 376–380) and thumb subdomains move as a unit (Fig. 3). However, these same residues of the fingertips and palm are negatively correlated (Fig. 5). The thumb appears to be serving as a latch to constrain the complete opening of the channel for the nascent RNA duplex. Therefore, when or if the fingertips–thumb interactions are disrupted, one might expect a concerted opening of the channels for the nascent RNA duplex to enter. This observation applies to all of the picornaviral RdRps studied here (Figs. 3 and Fig. S7). The laboratory of Kao has shown that the RdRp from hepatitis C virus, an enzyme that also contains a completely encircled active site, is capable of using circular templates.48 This observation has been interpreted to mean that the fingertips–thumb interaction is broken during the course of assembly.
The negative correlations of the PV RdRp fingertips (28–42 and 167–173) and palm (214–219 and 376–380) extend to the fingers to include residues (18–24, 43–57 and 157–176) (Fig. 5) and contribute to expansion and contraction of the channel for the nascent RNA duplex, even in the context of the completely encircled active site. The expansion and contraction of the nascent RNA channel are most influenced by residues (96–117 and 120–149) of the fingers and residues (400–461) of the thumb (Fig. 6). Crystal structures of PV RdRp show that the channel for the nascent RNA duplex is of insufficient diameter to accommodate duplex RNA. The expansion of the channel observed during the simulation showed that PV RdRp samples conformations sufficient for duplex RNA (Fig. 6). Similar observations were made for the other RdRps, although the average diameter and magnitude of the expansion and contraction motions varied (Fig. S9). For every RdRp, conformations of the nascent RNA channel larger than those of duplex RNA (20 Å) were also sampled (Fig. S9). Of note, differences in the extent of expansion are also observed in complexes of PV and FMDV RdRps with nucleic acid.20, 49 This “hyper-expansion” suggests that translocation can proceed by using a passive mechanism without the need to actively disengage the RdRp from its interactions with the major and minor grooves of the nascent RNA duplex. The “Brownian” nature of polymerase translocation has been suggested by others.50
Another interesting feature of the opening and closing of the nascent RNA duplex channel was that these motions were correlated with the opening and closing of the NTP channel (Fig. 4c). When the RNA channel was open, the NTP channel was closed and vice versa. Of note, motions of residues lining the NTP channel were biased in the direction of the nascent RNA channel (Fig. 4c). Given that the solvent and solutes flow through these channels, it is possible that these motions serve to pump reactants and products in and out of the active site. Closing of the NTP channel would force its contents into the nascent RNA channel and perhaps force its opening. Closing of the nascent RNA channel may also facilitate release of unincorporated NTP or inorganic pyrophosphate out of the active site through the NTP channel. The motions for the nascent RNA channel are more complex than those of the NTP channel. Therefore, a directional bias is not as obvious. Such an anti-correlated relationship between the nascent RNA duplex channel and the NTP channel has never been discussed. However, an examination of RdRp crystal structures supports this conclusion. The best example is provided by the rabbit hemorrhagic disease virus RdRp in which open and closed states of the nascent RNA duplex channel were present in the crystal.51 When the nascent RNA duplex channel is open, the NTP channel is closed and vice versa.
The template channel is formed by residues 17–27 and 108–116 of the fingers subdomain. Residues 17–27 exhibited substantial flexibility (Fig. 3a and b). In this study, flexibility has correlated strongly with function. However, detailed information on the functions/interactions of this channel was not available until recently.20 The new structural data have revealed that residues 17–27 line the channel and interact with RNA near the single-strand/double-strand junction. Of particular importance is Pro20. This residue exhibited a significant change in the psi dihedral angle during simulation where psi changed from 140° in the average structure to − 5.8° in the crystal structure, consistent with the flexible nature of this residue. Pro20 functions to unstack template bases at the + 2 and + 3 positions. In addition to facilitating these specific interactions of the template channel with RNA, the dynamic nature of residues forming this channel should permit entry/exit of the RNA into the RdRp catalytic site.
When known functional sequence/structural motifs were overlaid on the dynamics (relative displacement) of residues, most of these motifs exhibited the greatest flexibility (Fig. 4a). Of course, some were exceptionally stable on this timescale (e.g., motifs B and C in Fig. 4a). The observed flexibility may facilitate interactions that employ an induced-fit mechanism for binding. The more static motifs may employ a lock-and-key mechanism for binding. It is well known that motifs A, D and F contribute to interactions with incoming nucleotide that dictate both ground-state binding and isomerization events that lead to a catalytically competent complex.42, 52 In contrast, motif B plays an indirect role in selection of rNTP by forming a hydrogen bond with Asp238; motif C and the portion of motif A containing Asp233 serve as ligands for the Mg2+ required for catalysis, and substantial changes in the positions of these motifs have never been noted when crystal structures of different complexes have been evaluated. Interestingly, regardless of the magnitude of flexibility, all sequence/structural motifs participating in the same aspect of the catalytic cycle exhibited correlated motions (Fig. 5). Such a high level of coordination between structural elements might be required to facilitate function.
Flexible regions of no known functional relevance to nucleotidyl transfer were also noted (Fig. 7a and Table 2). Interestingly, essentially all of these regions are important for optimal viral fitness at 39.5 °C. Diamond and Kirkegaard have shown that alanine substitutions at Lys38, Glu39, Lys51, Asp53, Glu226, Glu227, Glu426, 428, Arg455 and 456 produce viruses incapable of multiplication, having diminished fecundity or exhibiting small-plaque phenotypes.38 Our studies of the complex used for initiation of genome replication have implicated the carboxy-terminal helix of the thumb of PV RdRp in recruitment of the enzyme to and retention in the initiation complex.39 That study specifically identified Asp406 and Arg455; this study identified these residues as being highly dynamic (Table 2). Additional studies are warranted to understand the mechanistic basis for the diminished viral fitness associated with mutagenesis of the most dynamic regions of PV RdRp. It is intriguing to speculate that functional domains of proteins of known structure but unknown function could be identified by virtue of the magnitude of their flexibility and a correlated nature of the motions observed for flexible regions.
The conserved nature of the dynamics observed for all of the WT picornaviral RdRps engendered sufficient confidence in the algorithm to put the overall approach to the ultimate test by evaluating a PV RdRp derivative that exhibited an anti-mutator phenotype7, 8 that could not be explained by structure.24 The derivative changes Gly64, a residue remote from the active site, to Ser and is referred to as G64S PV RdRp. The impact of this one change on the nanosecond timescale dynamics of PV RdRp was more substantial than differences observed between evolutionarily distinct WT enzymes (compare Fig. 7, Fig. 8). The G64S substitution is in the fingers and affected the dynamics observed in every subdomain. The core of the polymerase became less flexible, while loops on the surface became more flexible (Fig. 8b and c). The changes in dynamics led to differences in the average structure of the mutant relative to the WT enzyme (Fig. 8d). In addition, the correlated motions of the mutant were weaker than those of the WT enzyme; reversed and mixed correlations were also noted (Fig. 8e).
Kinetic analysis of G64S PV RdRp revealed that the equilibrium constant for the conformational change step that occurs after nucleotide binding that is a prerequisite to catalytic competence is shifted to the left for a correct nucleotide.7 Therefore, transit through this step would be even more difficult for an incorrect nucleotide or nucleotide analogue. Kinetic and/or structural studies have implicated motifs A, D and F in this conformational change step.20, 23, 43, 49, 52 The flexibility of these motifs was altered in the mutant (Fig. 8a–c). In addition, the correlated motions involving motif D were perturbed (Fig. 8e). Notwithstanding, numerous changes in flexibility and correlated motions in other regions were present. The extents to which these changes impact functions remain unclear. Finally, the average size of the catalytic site was larger for the mutant relative to WT (Fig. 8f).
Together, the altered dynamics observed for G64S PV RdRp could explain the higher barrier to formation of the catalytically competent state. The reduced flexibility of the catalytic core will limit conformational sampling of the core that may be necessary to achieve catalytic competence. Perhaps, this reduced flexibility would affect the incorrect nucleotides more than the correct nucleotides, hence increasing fidelity. The observations made here suggest two interesting lines of experimental investigation. Clearly, rigidification of the catalytic core is caused by propagation of the restricted motion of the G64S backbone. Dissemination of the reduced flexibility is mediated by an interacting (allosteric) network of amino acids, including, but not limited to, those shown in Fig. 8d. It may be possible to suppress the effects of G64S by introducing glycine residues in the vicinity of the site of interaction between Gly64 and motif A. Alternatively, it may be possible to prevent dissemination of the effects of the G64S substitution to motifs F and D by introducing “functionally compensatory” substitutions at various positions on the allosteric pathway. In addition, if a specific distance is required for interactions in the active site that lead to catalytic competence, this state will be sampled less often, given the more expanded nature of the catalytic site. The effect of the reduced sampling could be more severe for incorrect nucleotide incorporation than for correct incorporation, leading to the higher-fidelity phenotype.
Importantly, the properties of G64S PV RdRp observed by using the MD simulations relate to the properties of the enzyme in solution, as has been shown recently by using NMR.26 The Boehr laboratory has compared the [methyl-13C]Met resonances of WT 3Dpol and G64S RdRp. Consistent with our studies, the G64S substitution caused global changes in the structure/dynamics of the enzyme, including the fingers (Met6, Met95), palm (Met354) and thumb (Met392) subdomains.
Collectively, our studies support the hypothesis that dynamics, even on the picosecond-to-nanosecond timescale, may be the missing link to our understanding of RdRp fidelity and likely that of the other classes of nucleic acid polymerases.25 We conclude that dynamics on the picosecond-to-nanosecond timescale as reported by all-atom MD simulations represents an important, underutilized tool for elucidation of the dynamics–function relationships of polymerases.
Conclusion
We have performed the first comparative analysis of the dynamics of members of the RdRp superfamily of enzymes by using all-atom MD simulations on the nanosecond timescale. At the extremes were the RdRps from PV and FMDV whose primary structures are only 30% identical. The pattern of dynamics observed for all RdRps was conserved and was in good agreement with empirical data on the dynamics of a phage RdRp. The congruence of so much computational and empirical data lends credence to the use of MD simulation for evaluation of RdRp dynamics and leads us to conclude that RdRp dynamics coevolve with the RdRp tertiary structure. The major contributors to the motions on the nanosecond timescale were conserved structural regions required for function. Motions involved in identical function were correlated regardless of location in the structure. Changing a single amino acid residue of PV RdRp remote from the catalytic site that increases polymerase fidelity causes a global change in the observed dynamics of this derivative that extends to the active site. We conclude that RdRp dynamics are governed by long-range networks of interacting residues that have evolved for function, including polymerase fidelity. This groundwork sets the stage for elucidation of the dynamics–function relationships of this important class of enzymes and exploration of RdRp dynamics as a tractable target for development of antivirals and attenuated viruses.
Methods
MD simulation
The starting coordinates of the polymerases were obtained from the following deposited X-ray structures in the Research Collaboratory for Structural Bioinformatics PDB:53 PV (PDB code 1RA6 17), G64S (2IJF 24), CVB3 (3CDU 21), HRV16 (1XR7 22) and FMDV (1U09 19). The X-ray structures were edited in Coot54 to match the amino acid composition of the WT proteins. The structural waters observed in the crystal structures were kept during the simulations; any other nonprotein atoms were removed from the starting structures.
MD simulations were carried out using SANDER/PMEMD in AMBER10 package55 with the AMBER99 force field. Prepared structures were immersed in a truncated octahedron solvent box filled with TIP3P water molecules56 and neutralized by adding the appropriate number of sodium ions. A minimal distance of 20 Å between any protein atom and the edge of the solvent box was imposed. All simulations were carried out with an integration time step of 1 fs and applying periodic boundary conditions using a cutoff radius of 9 Å for the nonbonded interactions. The neighbor pair list was updated every 10 steps. Electrostatic interactions were calculated with the Particle Mesh Ewald method.57 The SHAKE algorithm58 was used to constrain all bond lengths involving hydrogen atoms.
The following protocol was used to carry out simulations. First, the coordinates of the simulated system were relaxed to remove any steric clashes between the atoms of proteins, waters and ions. This was carried out in multiple steps using SANDER: (1) Short energy minimization of waters and ions only. (2) Short constraint dynamics under conditions of constant pressure and temperature (NPT ensemble) for 100 ps. Only the atoms of waters and ions were allowed to move while constraining the protein atoms using a force constant of 500 kcal/mol Å2. (3) Energy minimization of the protein side chains in addition to the waters and ions. (4) Short constraint dynamics run similar to step 2 but allowing the side-chain atoms in addition to the waters and ions to move. (5) All atoms of the entire system were energy minimized, applying convergence criteria for the energy gradient distance root-mean-square (the root-mean-square of the cartesian elements of the gradient) of 0.1 kcal/mol Å. Subsequently, the system was slowly heated to 300 K over a period of 150 ps under conditions of constant volume and temperature (NVT ensemble) and applying a Berndsen thermostat.59 This was followed by a 200-ps NVT dynamics before switching to NPT dynamics for another 150 ps, applying Berendsen's method59 with temperature and pressure coupling constants of 1 ps (weak coupling). Finally, the NPT dynamics were continued for the remainder of the MD simulation utilizing the parallel version of PMEMD in AMBER10. The total simulation times for the different systems were 25 ns (PV WT and G64S derivative), 20 ns (FMDV) and 14 ns (CVB3 and HRV16). Snapshots of the MD trajectories were retained every 1 ps for analysis. The first 2 ns (PV), 3 ns (G64S) or 4 ns (CVB3, HRV16 and FMDV) were considered as equilibration times and discarded from analysis. All MD simulations were carried out on 128 Cray XT3 processors, TeraGrid resources managed by the Pittsburgh Supercomputing Center, and 32 Intel Xeon E5450 Quad-Core processors managed by the High Performance Computing group at The Pennsylvania State University.
Simulation analyses
All analyses of MD trajectories were performed using the Ptraj module of AMBER 10. All structure figures were generated using Chimera.60
Root-mean-square deviation
To determine the equilibration time of the MD simulation, we calculated the RMSD by aligning snapshots from the MD trajectory to a reference structure using the definition:
where the coordinate rti ,min corresponds to the optimal superimposition of the configuration at time t and the reference structure and α is the number of atoms used for the structural alignment. From the plot of RMSD as a function of time, the equilibration time was determined as the time point at which abrupt changes in the RMSD were no longer observed.
Per-residue RMSD
To get information on the extent of structural changes during simulations, we calculated the per-residue RMSD over the last 10 ns of MD trajectories. Structures extracted at intervals of 10 ps were aligned to the minimized starting structure as a reference using the backbone atoms of either the palm or the thumb residue. The per-residue RMSD for a certain residue was calculated by averaging the RMSDi ,n between residue i in the structure of frame n along the MD trajectory and the equivalent residue in the reference structure using the following definition:
where the RMSD between equivalent residues is averaged for all atoms, and N is the total number of frames used for the calculation. High values of per-residue RMSD were considered as an indicator for the existence of conformational changes sampled during simulations.
B-factors
B-factors were obtained from the calculated root-mean-square fluctuations (RMSFs) using the relation:
where r i are the coordinates for atom i, and the averaging is calculated over the last 5 ns of the MD trajectory. Calculated B-factors can be compared directly to experimental B-factors from crystal structures.
Clustering
To identify distinct conformations generated over the course of MD simulation, we performed the cluster analysis. Snapshot structures extracted from the equilibrated trajectory at 10-ps intervals along the production run were grouped into clusters using the refinement (means) algorithm35 in Ptraj. Initially, the number of clusters was determined by carrying out the cluster analysis using the average-linkage algorithm35 in Ptraj. In addition, the two-dimensional (2D)-RMSD plot, generated from the corresponding 2D-RMSD matrix, helped to determine the number of clusters before applying the means algorithm. The 2D-RMSD matrix was obtained by performing the RMSD comparison of the MD trajectory structures sampled at 10-ps intervals along the production run against all other structures in the same portion of the trajectory. Visual inspection of the 2D-RMSD plot provided an estimate for the number of distinct conformations sampled during the simulation.
Radius of gyration
The spread of a simulated structure from its center was monitored by calculating the radius of gyration (R g) over the length of the entire MD trajectory. The R g was calculated using the following relation:
where r i are the coordinates for atom i, w i is the mass of atom i and R c is the center of mass of the N atoms.
Dynamic cross-correlation map
To yield information about correlated motions sampled during MD simulations, we carried out the DCCM analysis41 in which the cross-correlation of the atomic displacements of Cα atoms was examined across the MD production run. For the displacement vectors Δr i and Δr j of atoms i and j, the cross-correlation is given by:
matrix C whose elements c ij are given by the equation above was calculated for Cα atoms in Ptraj. The calculated matrix was visualized and analyzed using MATLAB 7.6 (The MathWorks, Natick, MA). For completely correlated motions c(i,j) = 1, and for completely anti-correlated motions c(i,j) = − 1. Complete correlation means that the motions have the same phase as well as the same period. Deviations from 1 (or − 1) imply either that the motions of i and j are less correlated (or anti-correlated) or that they deviate from motion along a straight line.
Principal component analysis
To separate major motions from irrelevant noise sampled during the simulation, we carried out PCA36, 37 over the entire MD production run by diagonalizing the variance/covariance matrix (S) of the positional deviations of Cα atoms with respect to the average structure. The elements of this matrix are given by:
where s ij is an element of the covariance matrix S, ri is the coordinate x, y, z of Cα i atom of the trajectory, r j is the coordinate x, y, z of Cα j atom of the trajectory, 〈r i〉 is the coordinate x, y, z of the average structure, 〈r j〉 is the coordinate x, y, z of the average structure and 〈 〉t denotes the time average over the MD trajectory.
An orthogonal set of eigenvectors (or modes) is obtained by diagnolization of matrix S; the identified modes describe the directions of maximum variation in the observed conformational space spanned in the simulation trajectory. The variance around an average structure can be characterized directly as atomic displacements or motion. In this analysis, our focus was to characterize regions of RdRp structures that contribute most to the major motions described by the top principal modes. To do so, for each Cα atom, we summed and normalized contributions to the total variance along the top six modes to the average of the least contributing residues (5% of total residues). The relative displacements were plotted as a function of residue (Cα atoms).
Acknowledgements
This study was funded by a grant from National Institutes of Health (AI045818 to C.E.C.) and a seed grant from the Penn State Huck Institute of the Life Sciences and Material Research Institute (to C.M.C. and C.E.C.). This research was also supported in part by the National Science Foundation through TeraGrid resources provided by Pittsburgh Supercomputing Center (grant number MCB080068N to C.M.C.). We acknowledge the Information Technology Services-High Performance Computing group and the Materials Simulation Center at Penn State University for their computer resources and Dr. Ping Lin for the valuable time and help. We thank Jamie Arnold, Philip Bevilacqua, David Boehr, Janna Maranas, Scott Showalter and Eric Smidansky for comments on the manuscript.
Edited by D. Case
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2011.04.078
Contributor Information
Coray M. Colina, Email: colina@matse.psu.edu.
Craig E. Cameron, Email: cec9@psu.edu.
Appendix A. Supplementary data
References
- 1.World Health Organization. (2008). Laboratory surveillance for wild and vaccine-derived polioviruses, January 2007–June 2008. Wkly. Epidemiol. Rec.83, 321–328. [PubMed]
- 2.Jegouic S., Joffret M.L., Blanchard C., Riquet F.B., Perret C., Pelletier I. Recombination between polioviruses and co-circulating Coxsackie A viruses: role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathog. 2009;5:e1000412. doi: 10.1371/journal.ppat.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Clercq E. The design of drugs for HIV and HCV. Nat. Rev., Drug. Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 4.Graci J.D., Cameron C.E. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol. 2008;3:553–566. doi: 10.2217/17460794.3.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graci J.D., Cameron C.E. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antiviral Chem. Chemother. 2004;15:1–13. doi: 10.1177/095632020401500101. [DOI] [PubMed] [Google Scholar]
- 6.Sidwell R.W., Huffman J.H., Khare G.P., Allen L.B., Witkowski J.T., Robins R.K. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 7.Arnold J.J., Vignuzzi M., Stone J.K., Andino R., Cameron C.E. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer J.K., Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl Acad. Sci. USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vignuzzi M., Stone J.K., Andino R. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Arias A., Arnold J.J., Sierra M., Smidansky E.D., Domingo E., Cameron C.E. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J. Virol. 2008;82:12346–12355. doi: 10.1128/JVI.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu C.J., Zheng C.Y., Zhang Q., Shi L.L., Li Y., Qu S.F. An antiviral mechanism investigated with ribavirin as an RNA virus mutagen for foot-and-mouth disease virus. J. Biochem. Mol. Biol. 2006;39:9–15. doi: 10.5483/bmbrep.2006.39.1.009. [DOI] [PubMed] [Google Scholar]
- 12.Sierra M., Airaksinen A., Gonzalez-Lopez C., Agudo R., Arias A., Domingo E. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 2007;81:2012–2024. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vignuzzi M., Stone J.K., Arnold J.J., Cameron C.E., Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vignuzzi M., Wendt E., Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 15.Castro C., Arnold J.J., Cameron C.E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 2005;107:141–149. doi: 10.1016/j.virusres.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrer-Orta C., Agudo R., Domingo E., Verdaguer N. Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase. Curr. Opin. Struct. Biol. 2009;19:752–758. doi: 10.1016/j.sbi.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Thompson A.A., Peersen O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrer-Orta C., Arias A., Perez-Luque R., Escarmis C., Domingo E., Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template–primer RNA. J. Biol. Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 20.Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruez A., Selisko B., Roberts M., Bricogne G., Bussetta C., Jabafi I. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J. Virol. 2008;82:9577–9590. doi: 10.1128/JVI.00631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love R.A., Maegley K.A., Yu X., Ferre R.A., Lingardo L.K., Diehl W. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure. 2004;12:1533–1544. doi: 10.1016/j.str.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 23.Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcotte L.L., Wass A.B., Gohara D.W., Pathak H.B., Arnold J.J., Filman D.J. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J. Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron C.E., Moustafa I.M., Arnold J.J. Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase? Curr. Opin. Struct. Biol. 2009;19:768–774. doi: 10.1016/j.sbi.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., Welch J.L., Arnold J.J., Boehr D.D. Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry. 2010;49:9361–9371. doi: 10.1021/bi100833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammes-Schiffer S., Benkovic S.J. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 28.Henzler-Wildman K., Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 29.Henzler-Wildman K.A., Thai V., Lei M., Ott M., Wolf-Watz M., Fenn T. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 30.Henzler-Wildman K.A., Lei M., Thai V., Kerns S.J., Karplus M., Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 31.McCammon J.A., Gelin B.R., Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 32.Dodson G.G., Lane D.P., Verma C.S. Molecular simulations of protein dynamics: new windows on mechanisms in biology. EMBO Rep. 2008;9:144–150. doi: 10.1038/sj.embor.7401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karplus M. Molecular dynamics simulations of biomolecules. Acc. Chem. Res. 2002;35:321–323. doi: 10.1021/ar020082r. [DOI] [PubMed] [Google Scholar]
- 34.O'Reilly E.K., Kao C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 35.Shao J., Tanner S.W., Thompson N., Cheatham T.E. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 2007;3:2312–2334. doi: 10.1021/ct700119m. [DOI] [PubMed] [Google Scholar]
- 36.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 37.Garcia A.E. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 38.Diamond S.E., Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J. Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen M., Reitman Z.J., Zhao Y., Moustafa I., Wang Q., Arnold J.J. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2–3Dpol complex. J. Biol. Chem. 2008;283:875–888. doi: 10.1074/jbc.M707907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson A.A., Albertini R.A., Peersen O.B. Stabilization of poliovirus polymerase by NTP binding and fingers–thumb interactions. J. Mol. Biol. 2007;366:1459–1474. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichiye T., Karplus M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 42.Gohara D.W., Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry. 2004;43:5149–5158. doi: 10.1021/bi035429s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren Z., Wang H., Ghose R. Dynamics on multiple timescales in the RNA-directed RNA polymerase from the cystovirus phi6. Nucleic Acids Res. 2010;38:5105–5118. doi: 10.1093/nar/gkq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karplus M., Kuriyan J. Molecular dynamics and protein function. Proc. Natl Acad. Sci. USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benkovic S.J., Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 46.Bruenn J.A. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991;19:217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer–template substrate (sym/sub) J. Biol. Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 48.Ranjith-Kumar C.T., Kao C.C. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA. 2006;12:303–312. doi: 10.1261/rna.2163106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrer-Orta C., Arias A., Perez-Luque R., Escarmis C., Domingo E., Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl Acad. Sci. USA. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 51.Ng K.K., Cherney M.M., Vazquez A.L., Machin A., Alonso J.M., Parra F., James M.N. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 2002;277:1381–1387. doi: 10.1074/jbc.M109261200. [DOI] [PubMed] [Google Scholar]
- 52.Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westbrook J., Feng Z., Jain S., Bhat T.N., Thanki N., Ravichandran V. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen W.L., Chandrasekhar J., Madura J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 57.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 58.Ryckaert J.P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 59.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 60.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.