Abstract
Streptococcus pneumoniae type 2 pili are recently identified fimbrial structures extending from the bacterial surface and formed by polymers of the structural protein PitB. Intramolecular isopeptide bonds are a characteristic of the related pilus backbone protein Spy0128 of group A streptococci. Based on the identification of conserved residues in PitB, we predicted two intramolecular isopeptide bonds in PitB. Using a combination of tandem mass spectrometry and Edman sequencing, we show that these bonds were formed between Lys63-Asn214 and Lys243-Asn372 in PitB. Mutant proteins lacking the intramolecular isopeptide bonds retained the proteolytic stability observed with the wild type protein. However, absence of these bonds substantially decreased the melting temperature of the PitB-derivatives, indicating a stabilizing function of these bonds in PitB of the pneumococcal type 2 pilus.
Keywords: fimbrial protein, intramolecular cross-link, proteolytic stability, thermal stability, mass spectrometry
Introduction
The human pathogen Streptococcus pneumoniae (pneumococcus) is a common cause of infections including otitis media, pneumonia, meningitis, and bacteremia [1]. As is common to many pathogenic bacteria, S. pneumoniae produces pili, fimbrious extensions on the surface of the bacteria, which can be used for interaction with host tissue and biofilm formation [2,3]. Thus, pili are recognized virulence factors, and due to their exposure on the bacterial surface they are regarded as targets for vaccine development.
Two unrelated pilus gene clusters have been identified in S. pneumoniae strains, encoding two antigenically different pili: pilus islet 1 (PI-1) [2], and pilus islet 2 (PI-2) [3]. Both pili have the typical features of Gram-positive pili that are composed of covalently polymerized subunits of a pilus backbone protein and may have one or two additional (minor pilus) proteins attached to the backbone structure [4]. The pilus proteins have the typical features of Gram-positive surface proteins that include an N-terminal signal sequence and a C-terminal cell wall sorting signal (CWSS) that contains a LPXTG motif or a variant thereof that was first found in proteins subsequently anchored to the peptidoglycan of the cell wall [5]. The covalent-linkage of pilus subunits, formed by an amide bond between the side chain of a lysine residue and the threonine in the CWSS, requires a pilus-specific sortase [6,7]. The S. pneumoniae PI-1 pili are composed of the RrgB pilus backbone protein and the two accessory pilus proteins RrgA and RrgC [2], whereas PI-2 pili are formed solely by the pilus backbone protein PitB [3]. The absence of accessory pilus proteins is a unique feature for PI-2 pili among the so far described pili in Grampositive bacteria, as is the presence of a single PI-2 pilus structure per bacterial cell [3].
In 2007, Kang et al. [6] identified intramolecular isopeptide bonds (IPBs) in the pilus backbone protein Spy0128 of the group A streptococcal (GAS) M1 pilus. These IPBs form between the ε-amino group in a lysine residue and an asparagine residue, facilitated by a glutamate [6]. The formation of these IPBs occurs spontaneously [8]. Meanwhile, IPBs have been found in several other pilus backbone proteins in Bacillus cereus [9], Corynebacterium diphtheriae [10], RrgB of S. pneumoniae [11], GBS80 of Streptococcus agalactiae [12], and the minor pilus proteins RrgA and RrgC of S. pneumoniae [11,13] and GBS52 of S. agalactiae [14].
Structural studies of pilus proteins revealed that IPBs are commonly present in protein domains with an IgG-like fold, which has first been described for repetitive CNA B and CNA A domains in the Staphylococcus aureus collagen-binding surface protein Cna [15], and that is conserved in many Gram-positive pilus proteins [6,16]. In CNA A domains, as in a CNA B domain in RrgA[13], the formation of the IPB is facilitated by an aspartic acid residue instead of a glutamate residue. However, other surface proteins that are not associated with pili, like the CNA A-similar ACE protein from Enterococcus faecalis [17] and the SspB protein from Streptococcus gordonii [18] contain IPBs, suggesting a ubiquitous role for this fold and IPBs in Gram-positive surface proteins.
Studies on the function of IPBs in pilus proteins showed that they conferred proteolytic, in particular trypsin stability to pilus proteins [9,11,19]. More recent studies identified an increased thermal stability and mechanical resilience of IPB-containing pilus proteins [19,20] and results obtained with IPB-mutant derivatives in BcpA of B. cereus and RrgB of S. pneumoniae suggested that IPBs may play a role in pilus biogenesis [9,11].
This communication describes the identification of two IPBs, between Lys63 and Asn214, and between Lys243 and Asn372, in the PI-2 pilus backbone protein PitB by a combination of tandem mass spectrometry and Edman sequencing. We show that in contrast to IPBs in other pilus protein, the IPBs in PitB were dispensable for proteolytic stability of the pilus protein. However, both IPBs contributed significantly to the thermal stability of PitB, suggesting that they play a stabilizing role for PitB and thereby confer resistance of PitB to physical environmental stresses.
Material and Methods
Construction and purification of IPB deficient derivatives of PitB
The IPB deletion proteins were generated by the exchange of Lys63 and Lys243, respectively, to Ala in plasmid-encoded pitB [21] by site-specific mutagenesis using the QuikChange II XL site-specific mutagenesis Kit (Stratagene) and the proteins were expressed and purified as previously described for PitB [21]. In addition, the N-terminal 6-His-thioredoxin-tag was removed after cleavage with recombinant enterokinase (Novagen) from the wild type and mutant proteins using His-Pur Cobald-Spin columns (Pierce).
Mass spectrometry and Edman degradation of crosslinked PitB peptides
Recombinant PitB protein was trypsin-digested and chromatographed by reversed-phase high pressure liquid chromatography (RP-HPLC). The masses of peptide fragments were determined by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF/ MS), and parent ions were further analyzed by tandem mass spectrometry (MS/MS). N-terminal sequences of the recombinant protein and tryptic fragments were determined by Edman degradation (for experimental details see Supplemental Materials).
Exact mass determination of recombinant PitB and PitB-derivatives
Protein samples were analyzed on a linear trap quadrupole Fourier transform mass spectrometer (LTQ-FTMS, Thermo Scientific). Electrospray ionization mass spectrometry (ESI-MS) spectra were acquired in the ion trap and deconvoluted using ProMass (Novatia).
Proteolytic digestion of PitB and PitBK63A K243A and SDS–polyacrylamide gel electrophoresis (SDS–PAGE)
3 µg recombinant PitB and PitBK63A K243A, respectively, were incubated either with trypsin (Promega; trypsin to protein ratio 1:100 w/w) or added to a culture (at OD600nm = 0.6) of S. pneumoniae strain GA47901[21] for 16 hours at room temperature, prior to separation by SDS PAGE (12% Ready gel, BioRad) and staining with Coomassie blue. Heat-denatured protein samples were prepared by boiling (5 min) prior to proteolytic digestion.
Thermal Shift Assays
Thermal shift assays were performed basically as described by El Mortaji et al. [11]. In brief, 1 µg of protein and 2 µl of a 1 to 100 dilution of 5000×Sypro Orange (Invitrogen) were added to 50 mM Tris-HCl; 1mM CaCl2 (pH 7.8) buffer to a final volume of 25 µl. Thermal unfolding of the proteins was monitored between 20 °C and 95 °C in 1 °C increments per minutes in a iCycler IQ Real Time PCR machine (BioRad). The melting points were obtained by plotting the negative first derivative of fluorescence against temperature and the minima were referred to as the melting points.
Results
PitB contains the lysine-asparagine-glutamate triads involved in intramolecular peptide bond formation in Spy0128 of S. pyogenes
The pilus islet PI-2 of S. pneumoniae shows striking similarity to several pilus gene clusters of group A streptococci [22] in terms of the gene order and the presence of SipA, a signal-peptidase like protein, that was essential for pilus biosynthesis of these two pili, but that was absent from most other Gram-positive pilus islets [3,23]. The crystal structure of the GAS M1 pilus backbone protein Spy0128 was solved and revealed two domains, an N-terminal and a C-terminal CNA B domain that both contained an IPB [6]. However, the sequence similarity between the Spy0128 and PitB protein was very limited, with only 27% identity between the two proteins. Nevertheless, the alignment between Spy0128 and PitB revealed that the Lys-Asp-Glu residues involved in the IPBs in Spy0128 were all conserved in PitB, i.e., K63-N214-E164 and K243-N372-E325, respectively. Also conserved was the described hydrophobic and aromatic amino acid rich environment surrounding the Glu residues that provide the carbanion in the IPB formation in Spy0128 [6]. In addition, PitB encoded a Lys-residue (K195) in the position corresponding to the Lys161 in Spy0128 that has been shown to form the isopeptide bond between Spy0128 subunits [6]. Both proteins lack the YPKN-sequence of the described pilin motif that contains the Lys-residue involved in the intermolecular bond formation in most other Gram-positive pilus proteins [24]. In addition, both proteins have non-canonical LP(X)TG-motifs, with VTPTG and EVPTG, respectively, in their C-terminal CWSS (Fig. 1), as has been described earlier [3,25].
Fig. 1. Alignment of pilus backbone proteins PitB of S. pneumoniae and Spy0128 of S. pyogenes M1.
Residues involved in the formation of isopeptide bonds in Spy0128 are indicated by an asterisk followed by the number for the intramolecular isopeptide bond 1 and 2, and an arrow head for the residues engaged in the intermolecular bond connecting pilus subunits. The region of the mature protein present in the recombinant PitB protein is indicated by square brackets. Signal peptide (SP) and cell wall sorting signal (CWSS) are underlined and the LPXTG-like motif is shown in bold. Accession numbers for PitB and Spy0128 are ADD91687 and AAK33238, respectively. Alignment was performed with ClustalW software.
PitB contains two intramolecular isopeptide bonds
To determine whether the putative Lys-residues in PitB indeed were involved in IPBs we analyzed the recombinant PitB protein [21] by mass spectrometry. The protein was trypsin digested and the fragments were analyzed for the two predicted non-consecutive peptides that contained IPB1 (Mr 4366.19 Da) and IPB2 (Mr 1216.74 Da), respectively. As shown in Fig. 2, evidence was obtained for the presence of both cross-links in the digests, consistent with the two predicted IPBs K63-N214 (IPB1) and K243-N372 (IPB2). The identity of the parent ions with respect to the template sequence of the IPB peptides was confirmed as follows. First, as shown for the parent ion of 1092.56+4 m/z (calculated Mr of 4366.19), its mass spectrometry fragmentation pattern was consistent with the predicted cross-linked peptide IPB1 (Fig. 2A). Similarly, fragmentation of a parent ion of 609.38+2 m/z (calculated Mr of 1216.75) afforded fragments that could be assigned to their sequence in agreement with cross-linked peptide IPB2 (Fig. 2B). Second, the two tryptic cross-linked fragments originally observed by LC-MS/MS and MALDI-TOF MS in the digests were isolated by off-line capillary reversed-phase high pressure liquid chromatography (RP-HPLC) as distinct entities (Fig. S1 in Supplemental Material), which yielded upon Edman degradation the expected PTH-amino acids in each cycle corresponding to the two peptide chains forming the respective heterodimers (see Supplemental Material for details).
Fig. 2. Identification of intramolecular isopeptide bonds (IPBs) between Lys63-Asn214 (IPB1) and Lys243-Asn273 (IPB2) of S. pneumoniae PitB.
MS/MS spectra of IPB1-containing parent ion 1092.64+ m/z (mass to charge ratio) (A) and IPB2-containing parent ion 609.42+ m/z (B). The peptides corresponding to the y-ions are indicated. (C) Exact mass determination of recombinant PitB and IPB-derivatives.
To determine whether PitB may contain additional IPBs or other modifications we analyzed the exact molecular mass of purified recombinant PitB, two single mutants, which lacked one of the IPBs (PitBK63A or PitBK243A), and an IPB double mutant (PitBK63A K243A) (Fig. 2C and Fig. S2 in Supplemental Material). The determined Mr of 37,128.9 Da for the wild type PitB was 34.2 Da lower than the calculated Mr of the linear PitB protein (Fig. 2C). This was consistent with the presence of two IPBs, due to the elimination of two ammonia-groups (17 Da) during the formation of the IPBs. The Mr of both single mutants were 18.0 Da and 15.1 Da lower respectively than calculated for the linear proteins, as expected for the loss of one ammonia group during IPB formation. The Mr of the double mutant PitBK63A K243A deviated 1.6 Da from the calculated Mr for the linear peptide, indicating that no other IPBs in an alternative position were formed. Taken together these results demonstrated that only two IPBs were present in PitB and confirmed that these involved Lys63 and Lys243, the positions homologous to the IPBs in the pilus backbone protein Spy0128 of S. pyogenes M1.
PitB intramolecular isopeptide bonds are not required for proteolytic stability
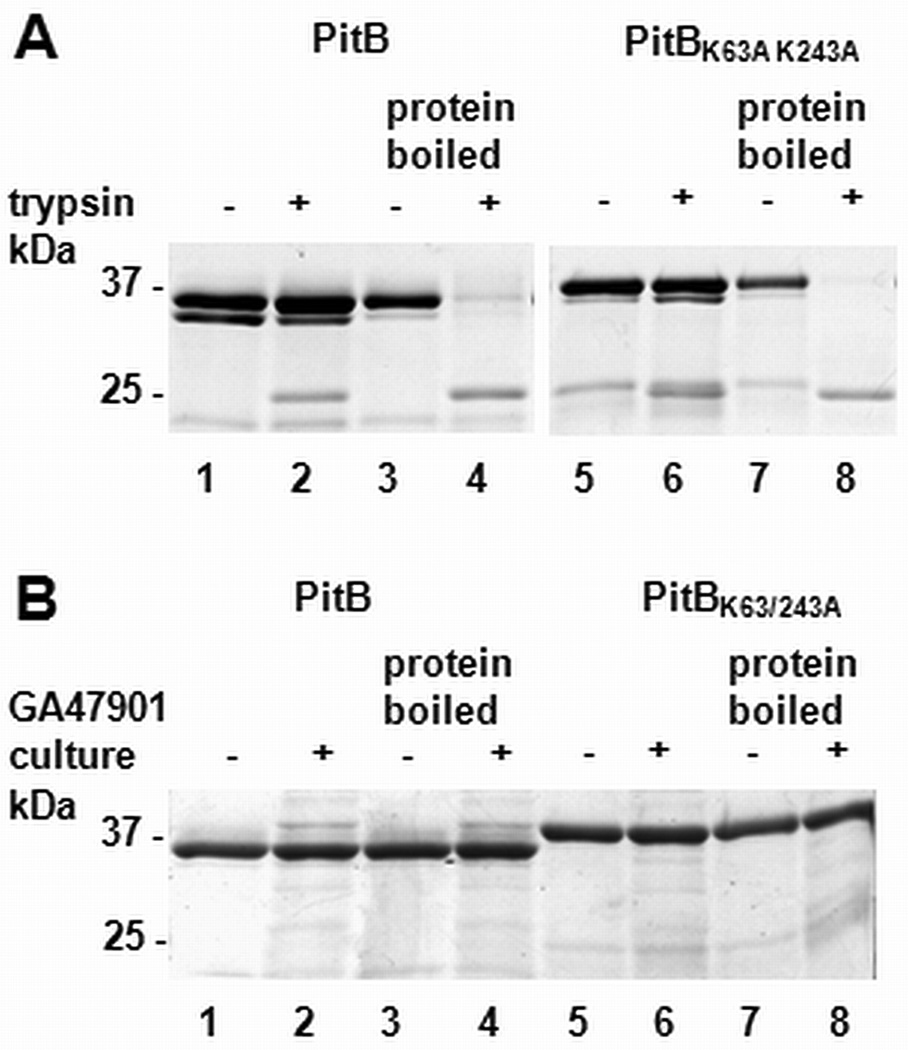
The first described function of IPBs in a pilus backbone proteins was to confer proteolytic stability [6]. To determine whether the identified IPBs in PitB were conferring trypsin resistance to PitB, the wild type PitB and the IPB double mutant derivative were treated with excess amounts of trypsin and subsequently analyzed by SDS-PAGE (Fig. 3A). Incubation with trypsin did not result in substantial proteolytic digestion of wild type PitB. A characteristic degradation product of about 24 kDa was observed, consistent with a trypsin susceptibility located between the predicted N-terminal and the C-terminal domains. Surprisingly, the mutant protein appeared to be as trypsin resistant as the wild type protein. The mutant protein produced a characteristic degradation product of 25 kDa, however the overall amount of protein and the amount of degradation product did not differ between the wild type and the mutant PitB, indicating that both proteins were trypsin resistant. Applying longer incubation times did not increase the amount of the degradation products in the wild type sample nor the mutant protein (data not shown). In a control experiment, PitB wild type and mutant protein was heat-denatured prior to trypsin digestion. This treatment resulted in an almost complete degradation of the proteins, with only one band of defined degradation product present, confirming the activity of trypsin under the assay conditions. These results indicated that IPBs were dispensable for trypsin resistance of PitB. Maintenance of trypsin resistance in the absence of IPBs has been observed previously in several IPB single mutants of the PI-1 pilus proteins RrgB (2 out of 3 IPBs) and RrgC (1 out of 2 IPBs) of S. pneumoniae [11].
Fig. 3. Proteolytic stability of recombinant PitB and PitBK63A K243A.
Recombinant PitB (lanes 1–4) and PitBK63A K243A (lane 5–8) were treated either with trypsin (A) or added to a culture of S. pneumoniae strain GA47901 (B) prior to separation by SDS PAGE. Whether trypsin or culture was present in the reaction and whether the samples were boiled prior to incubation is indicated on top of the gels.
Since PitB forms pilus structures on the surface of S. pneumoniae, we asked whether they contribute to proteolytic stability of PitB against pneumococcal proteases, which might be present on the pneumococcal surface or secreted into the medium. PitB and the IPB-double mutant were added to exponentially growing cultures (OD600nm= 0.6) of S. pneumoniae strain GA47901 [21]. Stability of PitB and the IPB double mutant appeared not to be affected in the presence of actively growing pneumococci (Fig. 3B). Heat-denatured protein samples of the wild type and the mutant proteins showed no degradation either, suggesting that no pneumococcal proteases with broad substrate specificity were active under our experimental conditions. Thus, our results demonstrate that IPBs were dispensable under natural environmental conditions.
The IPB double mutant (Fig. 3A and B) and the single mutants (data not shown) migrated slightly slower on an SDS-PAGE gel than the wild type protein, with the effect more pronounced for the double mutant. This suggested that the IPBs support a more compact protein conformation. Similar changes in the migration of IPB-containing pilus proteins on SDS-PAGE gels have been observed previously [11,26].
Taken together, we were unable to find evidence for a stabilizing function of IPBs in PitB against proteolysis. Both the wild type and the mutant protein were markedly stable towards trypsin and in their natural environment. It cannot be ruled out that IPBs may protect PitB against other environmentally encountered proteases not present under our experimental conditions, like proteases of bacteria in the same environmental niche or of the host.
Intramolecular peptide bonds confer thermal stability to PitB
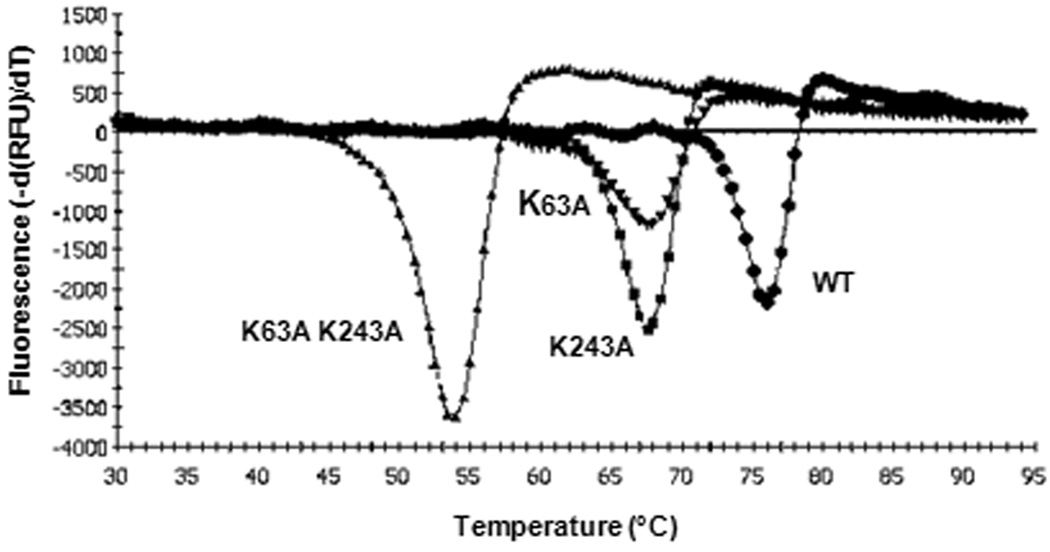
To determine whether the IPBs in PitB may contribute to resistance against physical stresses, we analyzed the thermal stability of the wild type protein and the single and double IPB mutant proteins by thermal shift assays [11]. The melting temperature of the wild type protein of 75.9 °C was far higher than for the two single mutants PitBK63A (67.6 °C) and PitBK243A (67.5 °C), respectively (Fig. 4). However, loss of both IPBs in the double mutant PitBK63A K243A further decreased the melting temperature to 54.0 °C, demonstrating that the IPBs conferred substantial thermal stability to PitB. Increased thermal stability due to IPBs was observed previously in Spy0128 of S. pyogenes and RrgB and RrgC of S. pneumoniae, and has been discussed as an indication for a stabilizing function of IPBs for the IgG-like fold [10,11].
Fig. 4. Thermal shift assays with recombinant PitB and PitB-derivatives.
Melting points were determined for PitB (WT, 75.9 °C; ●), PitBK63A (67.6 °C; ▼), PitBK243A (67.5 °C; ■), and PitBK63A K243A (54.0 °C; ▲).
Discussion
PitB is similar to M1 pili protein Spy0128 of GAS, with high sequence similarity limited to the conserved residues involved in intra- and intermolecular bonds and their immediate environment (Fig. 1). However, PitB and several GAS pilus backbone proteins [25,27] share additional features that separate these pilus proteins from other Gram-positive pilus proteins, including a modified CWSS with (Q/E/V)(V/T)PTG instead of an LPXTG-motif. In addition, motifs that have been described tentatively to be involved in the formation of IPBs are absent from PitB and these GAS pilus protein: The KVD-like motif [11], which includes the IPB-involved Lys-residue and the E-box motif EXAPXGY [28]. The latter contains a Glu residue, which facilitates IPB formation in C. diphtheriae [11]. A KVD-like motif is present in PitB adjacent to K243, with K244 as the lysine of the putative KVD-like motif, but determination of the molecular mass of the mature proteins showed that in the absence of K243 in the PitBK243A mutant K244 is not used alternatively for IPB formation. Therefore, motifs involved in the IPB and intermolecular bond formation in PI-2 and GAS pili still await identification.
The identification of IPBs in PitB that are formed by amino acids residues homologous to those in IPBs of Spy0128 suggests a folding of PitB in an N-terminal and C-terminal domain similar to the two domains observed in the Spy0128 crystal structure [6]. The location of the IPBs delineates the potentially surface exposed regions of PitB. Since in PI-2 pili the backbone protein PitB mediates attachment to eukaryotic cells, in contrast to other Gram-positive pili that employ accessory pilin proteins for adhesion, these exposed regions in PitB are candidates for host interactions and therefore demand further characterization.
In addition to proteolytic and thermal stability, mechanical properties of pilus proteins have been studied. Alegre-Cebollada et al. analyzed the mechanical properties of Spy0128 of GAS and found that this pilus backbone protein was not expandable due to the two IPBs. Supposedly, this rigidity allows M1 pili to withstand forces acting on bacterial surface structures that could otherwise result in the unfolding of the protein [20].
Recently, additional protein modifications in Gram-positive pilus proteins have been described. A disulfide bond was identified in pilus protein SpaA of C. diphtheriae [10] and a very unusual intramolecular thioester bond was observed in the adhesin Spy0125 of the GAS M1 pilus, which is formed between a cysteine and a glutamine residue, and is supposedly important in host interaction [29]. However, PitB lacks cysteine residues, ruling out the presence of disulfide or thioester bonds and the determination of the molecular weight of the full length protein is inconsistent with additional protein modifications.
Taken together, we found that PitB has two IPBs, and that these bonds confer thermal stability to PitB, likely stabilizing the overall conformation of the protein. Future work will have to determine the role of these IPBs in the overall pilus structure, whether they are a feature of PI-2 relevant for host-interactions of this important human pathogen and whether they may be sites of conserved epitopes that may be a basis of broadly cross-reactive vaccines.
Supplementary Material
Fig. S1: RP-HPLC peptide map of a tryptic digest of recombinant PitB.
Fig. S2: Mass determination of PitB (A) and the derivatives PitBK63A (B), PitBK243A (C), and PitBK63 K243 (D) by ESI-MS.
ACKNOWLEDGEMENTS
We thank Dr. Christian Klein (Bruker Daltonics) for his help with interpretation of ESI-MS/MS data of the crosslinks and Dr. Fred Strobel of the Department of Chemistry (Emory University) for exact mass determinations by ESI MS. This work was supported by funds from NIH grant #R01AI 070829 (DSS) and a VA Merit Award from the Dept. of Veterans Affairs (DSS). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnoli F, Moschioni M, Donati C, Dimitrovska V, Ferlenghi I, Facciotti C, Muzzi A, Giusti F, Emolo C, Sinisi A, Hilleringmann M, Pansegrau W, Censini S, Rappuoli R, Covacci A, Masignani V, Barocchi MA. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J Bacteriol. 2008;190:5480–5492. doi: 10.1128/JB.00384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 5.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 6.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 7.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in group B streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 8.Zakeri B, Howarth M. Spontaneous intermolecular amide bond formation between side chains for irreversible peptide targeting. J Am Chem Soc. 2010;132:4526–4527. doi: 10.1021/ja910795a. [DOI] [PubMed] [Google Scholar]
- 9.Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci U S A. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci U S A. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Mortaji L, Terrasse R, Dessen A, Vernet T, Di Guilmi AM. Stability and assembly of pilus subunits of Streptococcus pneumoniae. Journal of Biological Chemistry. 2010;285:12405–12415. doi: 10.1074/jbc.M109.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vengadesan K, Ma X, Dwivedi P, Ton-That H, Narayana SVL. A model for group B streptococcus pilus type 1: the structure of a 35-kDa C-terminal fragment of the major pilin GBS80. Journal of Molecular Biology. 2011;407:731–743. doi: 10.1016/j.jmb.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izoré T, Contreras-Martel C, El Mortaji L, Manzano C, Terrasse R, Vernet T, Di Guilmi AM, Dessen A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure. 2010;18:106–115. doi: 10.1016/j.str.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, Bice T, Hook M, Narayana SV. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 16.Hendrickx APA, Budzik JM, Oh S-Y, Schneewind O. Architects at the bacterial surface — sortases and the assembly of pili with isopeptide bonds. Nat Rev Micro. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Ponnuraj K, Xu Y, Ganesh VK, Sillanpaa J, Murray BE, Narayana SV, Hook M. The Enterococcus faecalis MSCRAMM ACE binds its ligand by the collagen hug model. J Biol Chem. 2007;282:19629–19637. doi: 10.1074/jbc.M611137200. [DOI] [PubMed] [Google Scholar]
- 18.Forsgren N, Lamont RJ, Persson K. Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J Mol Biol. 2010;397:740–751. doi: 10.1016/j.jmb.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem. 2009;284:20729–20737. doi: 10.1074/jbc.M109.014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alegre-Cebollada J, Badilla CL, Fernández JM. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. Journal of Biological Chemistry. 2010;285:11235–11242. doi: 10.1074/jbc.M110.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zähner D, Gudlavalleti A, Stephens DS. Increase in pilus islet 2-encoded pili among Streptococcus pneumoniae isolates, Atlanta, Georgia, USA. Emerg Infect Dis. 2010;16:955–962. doi: 10.3201/eid1606.091820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kratovac Z, Manoharan A, Luo F, Lizano S, Bessen DE. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J Bacteriol. 2007;189:1299–1310. doi: 10.1128/JB.01301-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zähner D, Scott JR. SipA is required for pilus formation in Streptococcus pyogenes serotype M3. J Bacteriol. 2008;190:527–535. doi: 10.1128/JB.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 25.Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci U S A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quigley BR, Zähner D, Hatkoff M, Thanassi DG, Scott JR. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol Microbiol. 2009;72:1379–1394. doi: 10.1111/j.1365-2958.2009.06727.x. [DOI] [PubMed] [Google Scholar]
- 27.Barnett TC, Patel AR, Scott JR. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J Bacteriol. 2004;186:5865–5875. doi: 10.1128/JB.186.17.5865-5875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 29.Pointon JA, Smith WD, Saalbach G, Crow A, Kehoe MA, Banfield MJ. A highly unusual thioester bond in a pilus adhesin is required for efficient host cell interaction. Journal of Biological Chemistry. 2010;285:33858–33866. doi: 10.1074/jbc.M110.149385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: RP-HPLC peptide map of a tryptic digest of recombinant PitB.
Fig. S2: Mass determination of PitB (A) and the derivatives PitBK63A (B), PitBK243A (C), and PitBK63 K243 (D) by ESI-MS.