Abstract
Chronic hepatitis C virus (HCV) infection patients exhibit different sustained virological responses (SVRs) following the treatment with pegylated interferon-α (IFN-α) and ribavirin. Genome-wide association studies consistently linked SVR of IFN-α-based therapy to the IL28B single-nucleotide polymorphisms (SNPs) on chromosome 19q.13 in various populations. This study was undertaken to investigate the association of IL28B SNPs with SVR in a cohort of Taiwanese chronic HCV patients. Ten SNPs of IL28B were genotyped in 728 chronic HCV patients and 960 healthy controls. Genotype distributions, allele frequencies and haplotypes were tested for SVR and susceptibility in Taiwanese chronic HCV patients. Non-genotype 1 infection (adjusted P=3.3 × 10−12, odds ratio (OR) 0.179; 95% confidence interval (CI): 0.110–0.290) and low HCV viral load (<400 000 IU ml–1) (adjusted P=3.5 × 10−9, OR 0.299; 95% CI: 0.200–0.446) were two major factors identified for high SVR. Notably, eight IL28B SNPs including previously described disease-associated SNPs (Trend test P=0.005) were significantly associated with SVR. Our data indicate that IL28B polymorphisms are the essential contributing factors for high SVR in Taiwanese chronic HCV patients. Combination of virus genotyping and host genetic data may be used to select the optimal treatment regimes in IFN-based therapy.
Keywords: hepatitis C, sustained virological response, IL28B, single-nucleotide polymorphisms
Introduction
Hepatitis C virus (HCV) infection, the leading cause of chronic liver disease, is emerging as a global concern of public health inflicting up to 3% human populations.1 HCV infection usually appears as an asymptomatic disease, which accompanies the development of chronic persistent viremia in majority individuals despite substantial virus and host cellular immune responses.2, 3, 4 HCV chronic infection associates with an intra-hepatic inflammatory infiltration, which is frequently followed by progressive fibrosis that leads to liver cirrhosis and ultimately to the development of hepatocellular carcinoma.5, 6, 7
HCV infection induces the production of interferons (IFNs), especially IFN α (IFN-α), in part through viral nucleic acid interactions with distinct pathogen-associated molecular pattern recognition receptors, such as Toll-like receptors on cell surface and/or intracellular sensors, such as retinoic acid-inducible gene I like helicases in the infected host cells.8, 9, 10, 11 The IFN signaling pathways activate IFN-stimulated genes (ISGs), which play critical roles in the host innate defense against HCV infection through repressing viral replication and enhancing cellular immune responses.9, 11, 12 Pegylated IFN-α plus ribavirin therapy became the standard therapy for HCV viral infection recently. However, the therapy failed to cure all patients who underwent the treatment. In addition, patients may encounter specific adverse effects that require therapy withdrawal.13 The interplay of the virus, environmental factors and the host immunity is implicated as the key process for the natural heterogeneity of viral clearance and liver injury.14, 15, 16, 17, 18, 19 It remains a critical issue with regard to the viral persistence and the response to anti-viral therapy in individual patient.
Interferon λ (IFN-λ) is classified as the type 3 IFN that includes three subtypes (IFN-λ1, 2 and 3). IFN-λs have pleiotropic cellular functions. The ligand receptor interaction triggers signaling pathways similar to those by IFN-α and β.20, 21 IFN-λs are produced and expressed in antigen-presenting cells. IFN-λs have lower antiviral activities than IFN-α. However, IFN-λs have critical roles in fighting against viral infection.22, 23, 24 Recently, four genome-wide association studies concurrently provided the overwhelming evidence that single-nucleotide polymorphisms (SNPs) of IFN-λ3 (also known as IL28B) on chromosome 19q13 contribute to IFN treatment response and spontaneous HCV clearance in HCV infection.25, 26, 27, 28, 29 Meanwhile, IL28B genetic variations influence the IL28B mRNA expressions, which may have a role in the regulation of intra-hepatic ISG expression.26, 27 IL28B SNPs seem to impact the treatment response to pegylated IFN-α/ribavirin for chronic HCV infection among several ethnic populations.25, 26, 27, 28, 29 In this study, we investigated whether the IL28B polymorphisms are associated with susceptibility to HCV infection and with response to the therapy of IFN and ribavirin in Taiwanese chronic HCV infection patients.
Results
Clinical characteristics of chronic HCV infection patients
This study recruited 728 Taiwanese patients with chronic HCV infection including 444 men and 284 women who received IFN-α plus ribavirin therapy. The mean ages on therapy were 51.2±10.4 (ranged 19.2–76.5) years old. Among them, 126 patients (112 patients were treated for a period of 24 weeks and 14 patients for 48 weeks) received treatment of INTRON 5MU three times a week in combination of 1200 mg ribavirin daily. The remaining 602 patients received ribavirin (1200 mg) daily and pegylated IFN-α every week (499 patients were treated for a period of 24 weeks and 103 patients for 48 weeks). Of the total 728 chronic HCV patients, 559 (76.8%) revealed sustained virological response (SVR) to IFN (INTRON or pegylated IFN-α) plus ribavirin therapy. In addition, 424 (58.2%) were identified as HCV genotype 1 viral infection and 318 (43.7%) were detected with viral load over 400 000 IU ml–1 (Table 1). The liver biopsies showed that 246 (33.8%) were in cirrhosis stage, 114 (15.7%) in severe fibrosis stage and 368 (50.5%) in mild fibrosis stage.
Table 1. Clinical characteristics and SVR analysis of Taiwanese patients with chronic HCV infection.
| Clinical characteristic | Total (N=728) | SVR positive (N=559) | SVR negative (N=169) | P-value |
|---|---|---|---|---|
| Age on IFN therapy | 51.2±10.4 | 50.1±10.5 | 54.8±9.3 | <0.001* |
| Male | 444 (61.0%) | 361 (64.6%) | 83 (49.1%) | <0.001* |
| Liver cirrhosis | 246 (33.8%) | 155 (27.7%) | 91 (53.8%) | <0.001* |
| HCV genotype: | ||||
| Genotype 1 | 424 (58.2%) | 281 (50.3%) | 143 (84.6%) | <0.001* |
| Non-genotype 1 | 304 (41.8%) | 278 (49.7%) | 26 (15.4%) | |
| HCV RNA ≥0.4 miu | 318 (43.7%) | 204 (36.5%) | 114 (67.5%) | <0.001* |
| BMI ≥27 kg m–2 | 146 (20.1%) | 106 (19.0%) | 40 (23.7%) | 0.181 |
| Adherence | 483 (66.3%) | 386 (69.1%) | 39 (23.1%) | 0.005* |
| Regimen | ||||
| INTRON+RBV | 126 (17.3%) | 98 (17.5%) | 28 (16.6%) | 0.772 |
| PEG-IFN+RBV | 602 (82.7%) | 461 (82.5%) | 141 (83.4%) | |
Abbreviations: BMI, body mass index; HCV, hepatitis C virus; PEG IFN, pegylated interferon; RBV, ribavirin; SVR, sustained virological response.
*P<0.05.
Clinical characteristics affect SVR during the treatment of chronic HCV infection
We examined the possible factors that may contribute to the SVR of chronic HCV infection. As shown in Table 2, age at therapy and gender were important host factors that affected the SVR. Patients with younger age at therapy were more likely to have higher SVR rate in chronic HCV infection (adjusted P=0.0004, odds ratio (OR) 0.962; 95% confidence interval (CI): 0.942–0.983). The SVR rate tended to be higher (81.3%) in men than in women (69.8%) (adjusted P=0.0093, OR 1.705; 95% CI: 1.140–2.549). Non-cirrhosis (mild and severe fibrosis) is another critical factor that demonstrated high SVR in comparison with cirrhosis (adjusted P=4.1 × 10−6, OR 0.379; 95% CI: 0.251–0.573). Body mass index below 27 kg m–2 showed modest effect on the higher SVR (adjusted P=0.0486, OR 0.616; 95% CI: 0.381–0.997). There was no significant SVR difference between the patients with INTRON plus ribavirin (76.6%) and those with pegylated IFN-α plus ribavirin (77.8%) albeit the percentage of patients with high viral RNA were lower in INTRON plus ribavirin group (19.0%) than that in pegylated IFN-α plus ribavirin (48.8%) group. Viral genotypes were significantly associated with SVR. Patients with non-HCV genotype 1 infection (91.4% 278 of 304) were more likely to reveal SVR than the patients with HCV genotype 1 infection (66.3%) (adjusted P=3.6 × 10−12, OR 0.178; 95% CI: 0.110–0.290). In addition, 86.6% of the patients with viral load <400 000 IU ml–1 demonstrated SVR as compared with 64.2% of the patients with HCV viral load over 400 000 IU ml–1 (adjusted P=2.3 × 10−9, OR 0.292; 95% CI: 0.195–0.437). Therefore, viral load appeared to contribute significantly to the SVR in the therapy.
Table 2. Clinical characteristics significantly affect SVR by multiple varieties analysis in 728 Taiwanese chronic HCV infection patients.
| Clinical characteristics | N |
Stepwise logistic regression |
||
|---|---|---|---|---|
| OR | 95% CI | P-value | ||
| Age on IFN therapy | 728 | 0.962 | 0.942–0.983 | 0.0004 |
| Gender: female | 284 | 1.705 | 1.140–2.549 | 0.0093 |
| BMI: below 27 kg m–2 | 582 | 0.616 | 0.381–0.997 | 0.0486 |
| HCV RNA viral load ≤ 400 000 IU ml–1 | 410 | 0.292 | 0.195–0.437 | 2.3 × 10−9 |
| HCV genotype: Non-genotype 1 | 304 | 0.178 | 0.110–0.290 | 3.6 × 10−12 |
| Liver biopsy: non-cirrhosis | 482 | 0.379 | 0.251–0.573 | 4.1 × 10−6 |
Abbreviations: BMI, body mass index; CI, confidence interval; HCV, hepatitis C virus; OR, odds ratio; SVR, sustained virological response.
IL28B polymorphisms are associated with chronic HCV infection
Ten SNPs of IL28B genetic variations were genotyped in 728 chronic HCV infection patients and 960 healthy controls. As shown in Table 3, genotype distributions and allele frequencies of two IL28B SNPs were significantly different between patients with chronic HCV infection and the normal healthy donors. The C allele of SNP rs28416813 appeared to be a risk allele for HCV infection (Trend test P=1.0 × 10−6 with 100 000 permutations; G vs C adjusted P=6.4 × 10−7, OR 0.026, 95% CI: 0.006–0.110; genotypes CG+GG vs CC adjusted P=1.5 × 10−5, OR 0.012, 95% CI: 0.002–0.089). In addition, the SNP rs8099917 genotypes appear to be a predisposition factor for HCV infection (genotype GG vs GT+TT adjusted P=0.001, OR 0.034; 95% CI: 0.004–0.254). Therefore, the SNP rs8099917 GG could be defined as the protective genotype. Notably, the SNP rs8099917 GG genotype frequency of the normal control males tested negative for HCV antibody was 21.5 times of that in the male patients with HCV infection (Table 4). Significant deviations from Hardy–Weinberg equilibrium in genotype and allele distributions were observed for SNP rs28416813 and rs8099917 in both HCV patients and normal controls (P<0.05).
Table 3. Allele and genotype distributions of IL28B SNPs in 728 Taiwanese chronic HCV infection patients and 960 normal controls.
| SNP marker | Risk allele | RAF (normal) | RAF (HCV) | Ptrend | Genotypes/alleles |
Age, sex adjusted |
|
|---|---|---|---|---|---|---|---|
| P-value | OR (95% CI) | ||||||
| rs28416813 | C | 0.975 | 0.999 | 1.0 × 10−6 | C/G+G/G vs C/C | 1.5 × 10−5 | 0.012 (0.002–0.089) |
| G vs C | 6.4 × 10−7 | 0.026 (0.006–0.110) | |||||
| rs8099917 | T | 0.936 | 0.947 | 0.2006 | G/G vs G/T+T/T | 0.0010 | 0.034 (0.004–0.254) |
| G vs T | 0.0425 | 0.713 (0.515–0.989) | |||||
Abbreviations: CI, confidence interval; HCV, hepatitis C virus; OR, odds ratio; RAF (normal) and RAF (HCV), risk allele frequency in normal controls and HCV patients; SNP, single-nucleotide polymorphism.
Ptrend value were calculated by the Cochran–Armitage trend test with permutation=100 000.
Table 4. Genotype frequencies of IL28B SNPs in normal controls and patients with chronic HCV infection, SVR positive, SVR negative.
| Clinical |
SVR positive |
SVR negative |
HCV |
Normal |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| characteristic/ IL28B SNPs | Male (N=360) | Female (N=199) | Total (N=559) | Male (N=83) | Female (N=86) | Total (N=169) | Male (N=443) | Female (N=285) | Total (N=728) | Male (N)a | Female | Total |
| rs12980275 | ||||||||||||
| AA | 0.903 | 0.950 | 0.920 | 0.807 | 0.791 | 0.799 | 0.885 | 0.902 | 0.892 | 0.870 | 0.883 | 0.876 |
| AG | 0.097 | 0.045 | 0.079 | 0.193 | 0.209 | 0.201 | 0.115 | 0.095 | 0.107 | 0.127 | 0.112 | 0.120 |
| GG | 0.000 | 0.005 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.001 | 0.004 | 0.005 | 0.004 |
| rs8105790 | ||||||||||||
| CC | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.004 | 0.002 | 0.003 |
| CT | 0.094 | 0.045 | 0.077 | 0.205 | 0.186 | 0.195 | 0.115 | 0.088 | 0.104 | 0.101 | 0.105 | 0.103 |
| TT | 0.906 | 0.955 | 0.923 | 0.783 | 0.814 | 0.799 | 0.883 | 0.912 | 0.894 | 0.895 | 0.892 | 0.894 |
| rs11881222 | ||||||||||||
| AA | 0.911 | 0.960 | 0.928 | 0.795 | 0.791 | 0.793 | 0.889 | 0.909 | 0.897 | 0.881 | 0.892 | 0.886 |
| AG | 0.089 | 0.040 | 0.072 | 0.193 | 0.209 | 0.201 | 0.108 | 0.091 | 0.102 | 0.115 | 0.106 | 0.111 |
| GG | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.004 | 0.002 | 0.003 |
| rs8103142 | ||||||||||||
| CC | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.006 | 0.002 | 0.004 |
| CT | 1.000 | 1.000 | 1.000 | 0.988 | 1.000 | 0.994 | 0.998 | 1.000 | 0.999 | 0.994 | 0.998 | 0.996 |
| TT | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rs28416813 | ||||||||||||
| CC | 1.000 | 1.000 | 1.000 | 0.998 | 1.000 | 0.994 | 0.998 | 1.000 | 0.999 | 0.914 | 0.998 | 0.953 |
| CG | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.082 | 0.000 | 0.044 |
| GG | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.004 | 0.002 | 0.003 |
| rs4803219 | ||||||||||||
| CC | 0.917 | 0.960 | 0.932 | 0.807 | 0.791 | 0.799 | 0.896 | 0.909 | 0.901 | 0.88 | 0.881 | 0.883 |
| CT | 0.083 | 0.040 | 0.068 | 0.193 | 0.209 | 0.201 | 0.104 | 0.091 | 0.099 | 0.115 | 0.119 | 0.117 |
| TT | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rs12979860 | ||||||||||||
| CC | 0.917 | 0.960 | 0.932 | 0.795 | 0.779 | 0.787 | 0.894 | 0.905 | 0.898 | 0.885 | 0.892 | 0.888 |
| CT | 0.083 | 0.040 | 0.068 | 0.193 | 0.221 | 0.207 | 0.104 | 0.095 | 0.100 | 0.109 | 0.105 | 0.107 |
| TT | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.006 | 0.002 | 0.004 |
| rs8099917 | ||||||||||||
| GG | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.043 | 0.002 | 0.024 |
| GT | 0.092 | 0.045 | 0.075 | 0.217 | 0.174 | 0.195 | 0.115 | 0.084 | 0.103 | 0.060 | 0.103 | 0.080 |
| TT | 0.908 | 0.955 | 0.925 | 0.771 | 0.826 | 0.799 | 0.883 | 0.916 | 0.896 | 0.897 | 0.895 | 0.896 |
| rs7248668 | ||||||||||||
| AA | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.006 | 0.002 | 0.000 | 0.001 | 0.004 | 0.002 | 0.003 |
| AG | 0.092 | 0.035 | 0.072 | 0.169 | 0.221 | 0.195 | 0.106 | 0.091 | 0.100 | 0.098 | 0.097 | 0.097 |
| GG | 0.908 | 0.965 | 0.928 | 0.819 | 0.779 | 0.799 | 0.892 | 0.909 | 0.898 | 0.899 | 0.901 | 0.900 |
| rs10853728 | ||||||||||||
| CC | 0.653 | 0.714 | 0.674 | 0.554 | 0.547 | 0.550 | 0.634 | 0.663 | 0.646 | 0.678 | 0.676 | 0.678 |
| CG | 0.319 | 0.261 | 0.299 | 0.349 | 0.430 | 0.391 | 0.325 | 0.312 | 0.320 | 0.281 | 0.294 | 0.287 |
| GG | 0.028 | 0.025 | 0.027 | 0.096 | 0.023 | 0.059 | 0.041 | 0.025 | 0.034 | 0.041 | 0.029 | 0.036 |
Abbreviations: HCV, hepatitis C virus; SNP, single-nucleotide polymorphism; SVR, sustained virological response.
Normal male (N=514, 514, 514, 514, 514, 514, 514, 513, 514, 513, 513); normal female (N=445, 446, 444, 446, 446, 446, 446, 446, 422, 445); normal total (N=959, 960, 958, 960, 960, 960, 959, 960,935, 958).
IL28B polymorphisms are associated with SVR in the treatment of chronic HCV infection
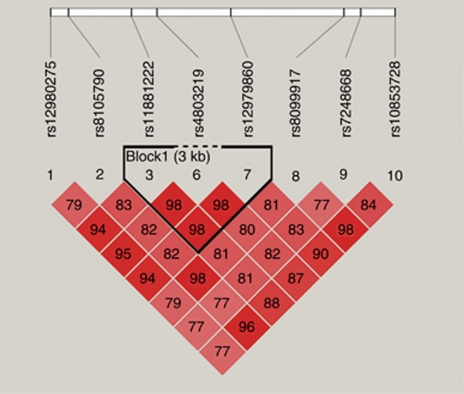
Patients with chronic HCV infection may manifest heterogeneous clinical characteristics and exhibit different SVRs to IFN therapy. Hence, we stratified patients into two groups in accordance with SVR to the treatment. As shown in Table 3, genotype distributions of the eight SNPs (rs12980275, rs8105790, rs11881222, rs4803219, rs12979860, rs8099917, rs7248668 and rs10853728) were significantly different between SVR positive and negative patients (Trend test P=0.005 with 100 000 permutations). The advantageous alleles of IL28 polymorphisms had similarly high frequencies in SVR positive groups (Table 5). Consequently, we examined whether IL28B SNP haplotypes were associated with SVR based on linkage disequilibrium blocks (Figure 1). As shown in Table 6, IL28B haplotypes ACC (rs11881222, rs4803219 and rs12979860), AT (rs12980275 and rs8105790), ATA (rs12980275, rs8105790 and rs11881222) and TA (rs8105790 and rs11881222) revealed as low responder alleles. Our data suggest that genetics of IL28B may impact the SVR in the treatment of HCV infection in Taiwanese.
Table 5. Association of IL28B SNPs with SVR in Taiwanese patients with chronic HCV infection.
| SNP marker | Risk allele | RAF (NR) | RAF (SVR) | Ptrend | Genotypes/alleles |
Age, sex adjusted |
Stepwise logistic regression |
||
|---|---|---|---|---|---|---|---|---|---|
| P-value | OR (95% CI) | P-value | OR (95% CI) | ||||||
| rs4803219 | C | 0.899 | 0.966 | 2.0 × 10−6 | C/T vs C/C | 3.7 × 10−7 | 0.258 (0.153–0.435) | 1.4 × 10−7 | 0.191 (0.103–0.354) |
| T vs C | 7.4 × 10−7 | 0.284 (0.172–0.467) | 2.1 × 10−7 | 0.213 (0.119–0.382) | |||||
| rs12979860 | C | 0.891 | 0.966 | 1.0 × 10−6 | C/T+T/T vs C/C | 4.4 × 10−8 | 0.236 (0.140–0.395) | 3.1 × 10−8 | 0.181 (0.099–0.331) |
| T vs C | 3.5 × 10−8 | 0.252 (0.155–0.411) | 2.9 × 10−8 | 0.200 (0.113–0.353) | |||||
| rs11881222 | A | 0.894 | 0.964 | 1.0 × 10−6 | A/G+G/G vs A/A | 2.1 × 10−7 | 0.256 (0.153–0.428) | 1.1 × 10−7 | 0.195 (0.107–0.356) |
| G vs A | 1.6 × 10−7 | 0.272 (0.167–0.443) | 9.0 × 10−8 | 0.214 (0.121–0.376) | |||||
| rs12980275 | A | 0.899 | 0.959 | 4.9 × 10−5 | A/G+G/G vs A/A | 3.4 × 10−6 | 0.302 (0.182–0.500) | 2.0 × 10−6 | 0.241 (0.134–0.433) |
| G vs A | 1.0 × 10−5 | 0.340 (0.211–0.549) | 4.3 × 10−6 | 0.275 (0.158–0.476) | |||||
| rs8105790 | T | 0.896 | 0.962 | 9.0 × 10−6 | C/C+C/T vs T/T | 1.3 × 10−6 | 0.282 (0.169–0.471) | 5.5 × 10−7 | 0.222 (0.123–0.400) |
| C vs T | 9.5 × 10−7 | 0.298 (0.184–0.484) | 7.5 × 10−7 | 0.248 (0.143–0.431) | |||||
| rs8099917 | T | 0.896 | 0.962 | 5.0 × 10−6 | G/G+G/T vs T/T | 9.6 × 10−7 | 0.277 (0.165–0.462) | 4.3 × 10−7 | 0.217 (0.120–0.392) |
| G vs T | 7.1 × 10−7 | 0.293 (0.180–0.476) | 6.2 × 10−7 | 0.245 (0.141–0.426) | |||||
| rs7248668 | G | 0.896 | 0.964 | 4.0 × 10−6 | A/A+A/G vs G/G | 6.1 × 10−7 | 0.268 (0.160–0.450) | 3.6 × 10−6 | 0.246 (0.136–0.445) |
| A vs G | 4.5 × 10−7 | 0.283 (0.173–0.462) | 4.5 × 10−6 | 0.270 (0.154–0.472) | |||||
| rs10853728 | C | 0.746 | 0.924 | 0.0015 | G/G+C/G vs C/C | 0.0007 | 0.533 (0.371–0.767) | 0.0006 | 0.485 (0.322–0.731) |
| G/G vs C/G+C/C | 0.0109 | 0.330 (0.141–0.775) | 0.0117 | 0.291 (0.111–0.759) | |||||
| G vs C | 0.0002 | 0.564 (0.418–0.761) | 0.0002 | 0.527 (0.375–0.740) | |||||
Abbreviations: CI, confidence interval; HCV, hepatitis C virus; NR, no response; OR, odds ratio; RAF, risk allele frequency; SNP, single-nucleotide polymorphism; SVR, sustained virological response.
Risk allele, the allele with higher frequency in cases compared with controls.
Ptrend value was calculated by the Cochran–Armitage trend test with 100 000 permutations.
Figure 1.
Pairwise linkage disequilibrium (LD) patterns for eight polymorphisms through IL28B regions.
Table 6. Association of IL28B haplotypes with SVR in patients with chronic HCV infection.
| Haplotype |
Frequency |
Associated haplotype |
EM algorithm* |
Logistic regressiona |
Stepwise logistic regression |
||||
|---|---|---|---|---|---|---|---|---|---|
| SVR−(N=169) | SVR+(N=559) | P-value | 95% CI | P-value | 95% CI | P-value | 95% CI | ||
| rs11881222+rs4803219+rs12979860 | 0.891 | 0.964 | ACC | <0.0001 | 3.31 (2.08–5.27) | 6.7 × 10−8 | 3.80 (2.34–6.16) | 4.1 × 10−8 | 4.82 (2.75–8.44) |
| 0.098 | 0.033 | GTT | <0.0001 | 0.32 (0.20–0.51) | 1.3 × 10−6 | 0.29 (0.17–0.48) | 3.8 × 10−7 | 0.22 (0.12–0.39) | |
| rs12980275+rs8105790 | 0.887 | 0.949 | AT | 0.0001 | 2.36 (1.53–3.62) | 1.1 × 10−5 | 2.73 (1.74–4.27) | 5.0 × 10−6 | 3.32 (1.99–5.53) |
| rs12980275+rs8105790+rs11881222 | 0.885 | 0.949 | ATA | 0.0001 | 2.43 (1.58–3.72) | 6.0 × 10−6 | 2.80 (1.79–4.36) | 4.0 × 10−6 | 3.34 (2.00–5.56) |
| rs8105790+rs11881222 | 0.884 | 0.954 | TA | <0.0001 | 2.73 (1.76–4.22) | 1.0 × 10−6 | 3.15 (2.00–4.96) | 4.0 × 10−7 | 3.88 (2.29–6.55) |
Abbreviations: CI, confidence interval; EM, expectation maximization; HCV, hepatitis C virus; OR, odds ratio; SVR, sustained virological response.
Adjusted for sex and age.
*P-value with 10 000 permutations.
IL28B polymorphisms are associated with HCV genotypes of chronic HCV infection
We also investigated possible interactions of IL28B polymorphisms with other clinical characteristics of chronic HCV infection in Taiwanese. We observed significant association (P<0.05) between HCV genotypes and IL28B SNP rs8099917 (Trend test P=0.03 with 100 000 permutations; G vs T adjusted P=0.0318, OR 1.729, 95% CI: 1.049–2.851) and between HCV genotypes and rs7248668 (Trend test P=0.028 with 100 000 permutations; A vs G adjusted P=0.0267, OR 1.777, 95% CI: 1.069–2.956). Our data suggest that IL28B gene products may associate with certain strains of HCV infection. Next, we performed multivariate analysis to comparatively evaluate the independent contribution of the significant IL-28B SNPs and the non-genetic clinical characteristics. Notably, rs12979860 is the only IL28B SNP showed significant association in multivariate analysis (P=2.3 × 10−8, OR 0.177, 95% CI: 0.096–0.324).
Discussion
Chronic HCV infection involves in a complex interaction of virus with host innate and adaptive immunity. Although the acute infections are highly amenable to therapy, approximately 80% HCV infections progress to viral persistence.2, 5 Several host factors including alcohol intake, age at the time of infection, gender and co-infection with the hepatitis B virus or human immunodeficiency virus are known to affect disease courses in HCV infection.1, 5, 13, 14 Accumulating evidence supports a critical role of host genetics in immune responses, which may predict the treatment response and outcome of viral clearance.18, 30, 31 Consistent with previous reports, this study established a predictive role of IL28B polymorphisms in the treatment of chronic HCV infection with IFN-α plus ribavirin in Taiwanese patients.
HCV evades the host immune surveillance by triggering production of viral inhibitory enzymes and negative signal regulatory proteins that block signal transduction pathways of human innate immune system.19, 32 In addition, HCV employs multiple escaping strategies including viral genetic variability (genotype switches), baseline viral load, quasi-species entity and high viral turnover to protect HCV from host humoral and cellular immunity.16, 19, 32, 33 Our data provided direct evidence that HCV genotype 1 and high basal viral load are two key factors for SVR in the treatment of Taiwanese HCV patients.
IFN-λ1 (IL29), IFN-λ2 (IL28A) and IFN-λ3 (IL28B) belong to type II cytokine family induced by virus infections. Besides their antivirus activities, IFN-λs are capable of modulating innate immune response effects.21 IFN-λs bind to the heterodimeric receptors of specific IFN-λR1 and common IL10R2, which form a trimeric ISG factor complex that triggers a cascade of signal transductions.34 The binding of ISG factor to IFN response elements results in steady upregulations of several ISGs with antiviral effects.35, 36, 37, 38 In animal models, the antiviral activity of Toll-like receptor signaling pathways induced by Toll-like receptor-3 and 9 agonists was significantly decreased in IL28R1 knockout mice.39 In gene expression comparison studies, peripheral blood and liver tissue cells in chronic HCV infection with low SVR in response to IFN/ribavirin therapy display a defect in ISG production, which may be due to pre-activated states in blunted IFN cell signaling.40, 41, 42 These findings point to the critical roles of host genetic variations in IFN-α-mediated innate immune signal pathways that may affect chronic HCV infection course and treatment response. Indeed, certain host genetic polymorphisms involving in IFN signal pathways and ISGs have shown modest effects on IFN-α treatment response in patients infected with HCV.43, 44, 45
HCV persistent infection induces a cascade of innate and adaptive immune cellular dysfunctional changes.4, 17 IFN-λs produced by monocyte-derived dendritic cells and plasmacytoid dendritic cells facilitate viral clearance by influencing innate and adaptive immunities.22, 46, 47 IL29 and IFN-α modulate the co-stimulatory molecules expressions on plasmacytoid dendritic cells and lead to signature cytokines production.48 In addition, IL29 modulates the Th1/Th2 differentiation that favors the pro-inflammatory (Th1) response, which enhance adaptive cellular immunity to eradicate virus infection.49, 50, 51, 52 During vaccination, IL28B increases the cytolytic CD8T cell differentiations and suppresses the of T regulatory cell productions, which protect animals from mortality after a lethal influenza challenge.52 These immunological responses collectively suggest IFN-λs are critical in suppressing viral replication during HCV chronic infection. However, the mechanisms that IL28B SNPs are associated with the response to IFN-α-based therapy remain to be elucidated in humans.
In therapy with pegylated IFN-α plus ribavirin, the adverse events and failure of response sometimes occurred.13 The novel HCV treatment strategies are still under development to improve sustained response rates. IFN-λs have several important roles in controlling chronic HCV infections and may be useful in future therapy. IFN-λs can enhance the subsaturating levels of IFN-α and increase the antiviral efficacy.35 The combination of IFN-λs and IFN-αs may provide the additive therapy effect through the complementary roles of two types of cytokines.37, 38, 53 As the distribution of IFN-λ receptors are restricted on plasmacytoid dendritic cells, peripheral B cell, hepatocytes and epithelial cells, IFN-λs may be used to target specific cell responses and to avoid the adverse events of therapy of IFN-αs.53, 54
Genome-wide association studies in independent populations confirmed that IL28B genetic variations are associated with HCV IFN-based therapy response and spontaneous clearance of HCV.25, 26, 27, 28, 29 In this study, we compared genotypes and alleles frequency of IL28B SNPs between normal controls and patients with chronic HCV infection in Taiwanese. In Caucasians and Africans, IL28B SNP rs12979860 CC genotype and C allele are associated with spontaneous resolution of HCV infection, however, no differences in genotype and allele distributions were observed for IL28B SNP rs12979860 between HCV patients and normal controls in Taiwanese. Paradoxically, rs8099917 GG genotype and rs28416813 G allele that associated with low response in Australians and Japanese were significantly enriched in Taiwanese normal male population. On the other hand, eight SNPs with strong linkage disequilibrium demonstrate significant associations with SVR on single point analysis. However, haplotype analysis failed to increase the significance of association, which is different from the results of previous studies.26, 27 These findings suggest that various IL28B SNPs are associated with the treatment response of HCV in Taiwanese. Nevertheless, the exact causal roles of IL28B polymorphisms remain to be elucidated.
The combinational treatment of pegylated IFN-α and ribavirin is widely applied to chronic HCV infection. The treatment results in sustained clearance of virus and clinical improvement in various ethnic populations.13, 55, 56, 57, 58 However, the response rates showed ethnic difference with poor response in patients of African ancestry and Hispanic ancestry in comparison with patients of Caucasian ancestry.55, 56 We observed higher SVR rates in Taiwanese chronic HCV infection who received IFN-α and ribavirin combination therapy, similar to the previous findings in Asians.57, 58 The IL28B genetic variations affect mRNA expressions, indicating functional polymorphisms in regulatory regions may interfere with treatment responses.26, 27 This study confirmed that IL28B SNPs are associated with SVR of HCV treatment response across all ethnic groups. The advantageous IL28B SNPs have significantly higher frequencies in Asian populations than in populations with African and Caucasian ancestry origins, which may provide explanation for the ethnic differences in SVR in IFN-based therapy among Asians, Europeans and Africans.
Recognizing the cost-effective concern and the potentially serious adverse effects, investigators and clinicians put emphasis on the importance of predicting the response to IFN-based biologic therapy. The longer duration of treatment achieved significantly higher SVR rates than the shorter course therapy, which may indicate the specific refractory cases with genotype 1 infection.58, 59 Human genome variations explain part of the different effective responses. The combination of data including virus genotypes, viral load, immunological cellular and gene expression changes, and host genetic variations may be critical to determine individual appropriate treatment doses and duration, which is valuable to minimize the adverse effects of biologic agents. This study could provide another critical evidence for usefulness of application of genetic data in clinical settings for predicting the treatment response. In Asians, patients carrying IL28B low response alleles and genotypes could be identified for longer antiviral treatment. However, SVR of HCV infection is determined by multiple genetic loci. The practical utility of genetic data in treatment choice remains to be determined for IFN-based therapy.
In conclusion, IFN-λs appear as critical functional immune response molecules that could be therapeutic target in the treatment of persistent HCV infections. Genetic data of IL28B SNPs may provide novel guidelines in determining optimal treatment duration with IFN-based therapies.
Materials and methods
Study subjects
This study recruited 728 patients from the Hepatology clinics of Linkou Medical Center, Chang Gung Memorial Hospital. Hepatology specialists confirmed the chronic HCV infection clinically and verified the diagnosis with liver biopsies in all examined patients. Two cohorts of patients with chronic hepatitis C were analyzed. Patients in cohort A were treated with INTRON plus ribavirin (126 patients, January 1988 to October 2001) and patients in cohort B were treated with pegylated IFN plus ribavirin (604 patients, November 2001 to December 2008). All patients were positive for anti-HCV (for >6 months) and for HCV–RNA. In addition, HCV from each patient was genotyped and the infections were verified with pathological findings before the enrolment for treatment. The patients in cohort A received INTRON (IFN-α 2b, Schering Co., Kenilworth, NJ, USA) 5 MU three times a week and ribavirin 1200 mg per day. The cohort B patients received ribavirin 1200 mg per day and PEGINTRON (15 μg kg–1 week–1, Peginterferon-α 2b, Schering Co.) or PEGASYS (180 μg week–1, Peginterferon-α 2a, Roche). For the purpose of this study, a total of 960 healthy blood donors (514 men and 446 women) were recruited following a questionnaire survey to exclude any donors with autoimmune diseases including rheumatoid arthritis, systemic lupus erythematous, ankylosing spondylitis, and autoimmune thyroiditis, diabetes mellitus, viral hepatitis (HBV and HCV) infections and cardiovascular diseases. The age of healthy control donors ranges from 18 to 64 years old with a mean age of 40.3±10.7.
HCV infection assays
Clinical characteristics including liver biochemical test, HCV genotypes, HCV viral load, liver histology and therapeutic responses to the combination regimen (IFN-α plus ribavirin). The HCV antibody was tested by the Abbott AxSYM anti-HCV 3.0 (Abbott Laboratories, Abbott Park, IL, USA). The HCV RNA detection was carried out with Cobas Amplicor HCV v2.0 kit (Roche Diagnostics Co., Indianapolis, IN, USA; lower limit of detection of at concentrations of 50 IU ml–1) and RNA was quantitated by the VERSANT HCV RNA 3.0 Assay (bDNA) (Bayer Diagnostics, Berkeley, CA, USA; lower limit of detection of at 615 IU ml–1) or COBAS TaqMan HCV Test (TaqMan HCV; Roche Molecular Systems Inc., Branchburg, NJ, USA; lower limit of detection of at concentrations of 15 IU ml–1). The baseline levels of HCV–RNA were determined by VERSANT HCV RNA 3.0 Assay in 669 patients and confirmed by Cobas Amplicor HCV v2.0 before January, 2008. Thereafter, COBAS TaqMan HCV Test was applied in 59 patients. HCV RNA level <2 MEQ (equal to 400 000 IU ml–1) was defined as low viral load. HCV genotypes were analyzed using a genotype-specific probe-based assay in the 50 untranslated region.
Clinical SVR and liver biopsy
SVR was defined when serum HCV–RNA was undetectable at the end of therapy and at 24-week follow-up after the treatment. Non-response was designated when viral RNA reappears within 6 months after the cessation of treatment; or when viral load decreases <2 logs after 12-week treatment, or when viral RNA was detectable at the end of treatment. The liver biopsy was interpreted and graded by Ishak classification. In the analysis, we categorized fibrosis into cirrhosis (fibrosis score 5 or 6), severe fibrosis (fibrosis score 4) and mild fibrosis (fibrosis score 1 or 2 or 3).
Nucleic acid isolation
Anti-coagulated peripheral blood was obtained from healthy normal blood donors and HCV patients. Genomic DNA was isolated from EDTA anti-coagulated peripheral blood using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA) as previously described.60
Taqman-based assay for the genotyping of SNPs
The oligonucleotide sequences flanking IL28B polymorphisms were designed as primers for Taqman allelic discrimination assays. Allele-specific primers were labeled with a fluorescent dye (FAM or VIC) and used in the PCR reaction. Aliquots of the PCR products were genotyping using allele specific probe of SNPs on a real-time PCR Thermocylcler (ABI, Foster City, CA, USA).
Statistical analysis
We carried out single-locus analyses of IL28B polymorphisms for 960 normal healthy controls and 728 patients with chronic HCV infection. Three χ2 tests: the genotype test, the allele test and Cochran–Armitage trend test were performed, and associations with SNPs (P<0.05) were identified using the SAS/Genetics software package release 8.2 (SAS Institute, Cary, NC, USA). For the analysis of risk genotypes/alleles, we used logistic regression models adjusted for age and sex and calculated P-values, ORs, and their 95% CIs. Linkage disequilibrium between marker loci was assessed and haplotype blocks were constructed using Haploview 4.1 (Broad Institute of MIT and Harvard; http://www.broad.mit.edu/mpg/haploview). For the markers within the same haplotype block, we used disease status (case vs control) and five clinical characteristics: body mass index, drug, HCV genotype, HCV RNA viral load and SVR as traits and tested for the haplotype–trait association utilizing SAS HAPLOTYPE procedure. To investigate the genetic association with clinical characteristics including body mass index, drug, HCV genotype, HCV RNA viral load and SVR for the case group, we controlled for each of these five characteristics and performed stepwise logistic regression analyses. Additionally, we examined the association of SVR with age on IFN therapy, gender, body mass index, HCV RNA viral load, HCV genotype, adherence and degrees of liver fibrosis in the chronic HCV infection patients using Student's t-test and χ2 test. The 5% level of significance for P-values was used for all the analyses.
Acknowledgments
We thank Shin Chu Blood Donor Center for samples collection. This study was supported by grants from the Chang Gung Memorial Hospital (No. CMRPG 380681 and CMRPG 360113).
The authors declare no conflict interest.
References
- Shepard CW, Finelli L, Alter M. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachiadakis I, Pollara G, Chain BM, Naoumov NV. Is hepatitis C virus infection of dendritic cells a mechanism facilitating viral persistence. Lancet Infect Dis. 2005;5:296–304. doi: 10.1016/S1473-3099(05)70114-6. [DOI] [PubMed] [Google Scholar]
- Bialek SR, Terrault NA. The changing epidemiology and natural history of hepatitis C virus infection. Clin Liver Dis. 2006;10:697–715. doi: 10.1016/j.cld.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. Lancet. 1997;349:825–832. doi: 10.1016/s0140-6736(96)07642-8. [DOI] [PubMed] [Google Scholar]
- Blonski W, Reddy KR. Hepatitis C virus infection and hepatocellular carcinoma. Clin Liver Dis. 2008;12:661–674. doi: 10.1016/j.cld.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Cheng G, Zhong J, Chung J, Chisari FV. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci USA. 2007;104:9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Dolganiuc A, Mandrekar P. Pattern recognition receptors: a contemporary view on liver diseases. Hepatology. 2006;44:287–298. doi: 10.1002/hep.21308. [DOI] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanto T, Hayashi N. Innate immunity in hepatitis C virus infection: Interplay among dendritic cells, natural killer cells and natural killer T cells. Hepatol Res. 2007;37 (Suppl 3:S319–S326. doi: 10.1111/j.1872-034X.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- National Institute of Health U.S.A. NIH consensus statement on management of hepatitis C. NIH Consens State Sci Statements. 2002;19:1–46. [PubMed] [Google Scholar]
- Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- Lechner F, Wong DKH, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimme R, Lohmann V, Weber F. A target on the move: innate and adaptive immune escape strategies of hepatitis C virus. Antiviral Res. 2006;69:129–141. doi: 10.1016/j.antiviral.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Missale G, Cariani E, Ferrari C. Role of viral and host factors in HCV persistence: which lesson for therapeutic and preventive strategies. Dig Liver Dis. 2004;36:703–711. doi: 10.1016/j.dld.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Szabo G, Dolganiuc A. Hepatitis C and innate immunity: recent advances. Clin Liver Dis. 2008;12:675–692. doi: 10.1016/j.cld.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Li M, Liu X, Zhou Y, Su SB. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol. 2009;86:23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, et al. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- Ank N, West H, Bartholdy C, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;10:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure—a genome-wide association study. Gastroenteroloy. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AR, Jagger E, Post JJ, Crooks LA, Rawlinson WD, Hahn YS, et al. Host and viral factors in the immunopathogenesis of primary hepatitis C virus infection. Immunol Cell Biol. 2007;85:24–32. doi: 10.1038/sj.icb.7100010. [DOI] [PubMed] [Google Scholar]
- Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nature Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- Gale M, Jr, Foy EM. Evasion of intracellular host defense by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- Hiroishi K, Ito T, Imawari M. Immune responses in hepatitis C virus infection and mechanisms of hepatitis C virus persistence. J Gastroenterol Hepatol. 2008;23:1473–1482. doi: 10.1111/j.1440-1746.2008.05475.x. [DOI] [PubMed] [Google Scholar]
- Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem. 2009;284:20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Butera M, Nelson DR, Liu C. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J. 2005;2:80. doi: 10.1186/1743-422X-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliaccetti NE, Eduardo R, Kleinstein SH, Mu XJ, Bandi P, Robek MD. Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem. 2008;283:30079–30089. doi: 10.1074/jbc.M804296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- Taylor MW, Tsukahara T, Brodsky L, Schaley J, Sanda C, Stephens MJ, et al. Changes in gene expression during pegylated interferon and ribavirin therapy of chronic hepatitis C virus distinguish responders from nonresponders to antiviral therapy. J Virol. 2007;81:3391–3401. doi: 10.1128/JVI.02640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci USA. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel TM, Morgan TR, Bonkovsky HL, Naishadham D, Pfeiffer RM, Wright EC, et al. Variants in interferon-alpha pathway genes and response to pegylated interferon-Alpha2a plus ribavirin for treatment of chronic hepatitis C virus infection in the hepatitis C antiviral long-term treatment against cirrhosis trial. HALT-C Trial Group. Hepatology. 2009;49:1847–1858. doi: 10.1002/hep.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Yee LJ, Im K, Rhodes SL, Tang Y, Tong X, et al. Association of single nucleotide polymorphisms in interferon signaling pathway genes and interferon-stimulated genes with the response to interferon therapy for chronic hepatitis C. Virahep-C Study Group. J Hepatol. 2008;49:184–191. doi: 10.1016/j.jhep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Sun J, Meng L, Heathcote J, Edwards AM, McGilvray ID. ISG15, a ubiquitin-like interferon-stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. J Gen Virol. 2010;91:382–388. doi: 10.1099/vir.0.015388-0. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Shingai M, Matsumoto M, Wakita T, Seya T. Hepatitis C virus-infected hepatocytes extrinsically modulate dendritic cell maturation to activate T cells and natural killer cells. Hepatology. 2008;48:48–58. doi: 10.1002/hep.22337. [DOI] [PubMed] [Google Scholar]
- Decalf J, Fernandes S, Longman R, Ahloulay M, Audat F, Lefrerre F, et al. Plasmacytoid dendritic cells initiate a complex chemokine and cytokine network and are a viable drug target in chronic HCV patients. J Exp Med. 2007;204:2423–2437. doi: 10.1084/jem.20070814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megjugorac NJ, Gallagher GE, Gallagher G. Modulation of human plasmacytoid DC function by IFN-{lambda}1 (IL-29) J Leukoc Biol. 2009;86:1359–1363. doi: 10.1189/jlb.0509347. [DOI] [PubMed] [Google Scholar]
- Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, et al. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Dai J, Eskdale J, Gallagher GE, Megjugorac NJ, Gallagher G. Interferon-lambda1 (interleukin-29) preferentially down-regulates interleukin-13 over other T helper type 2 cytokine responses in vitro. Immunology. 2008;125:492–502. doi: 10.1111/j.1365-2567.2008.02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennechet FJ, Uzé G. Interferon-lambda-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood. 2006;107:4417–4423. doi: 10.1182/blood-2005-10-4129. [DOI] [PubMed] [Google Scholar]
- Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N, et al. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009;113:5868–5877. doi: 10.1182/blood-2008-11-190520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers LJ, Cassidy W, Howell CD, Hu S, Reddy KR. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology. 2004;39:1702–1708. doi: 10.1002/hep.20212. [DOI] [PubMed] [Google Scholar]
- Muir AJ, Bornstein JD, Killenberg PG. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265–2271. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- Missiha S, Heathcote J, Arenovich T, Khan K. Impact of Asian race on response to combination therapy with peginterferon alfa-2a and ribavirin in chronic hepatitis C. Am J Gastroenterol. 2007;102:2181–2188. doi: 10.1111/j.1572-0241.2007.01431.x. [DOI] [PubMed] [Google Scholar]
- Liu CH, Liu CJ, Lin CL, Liang CC, Hsu SJ, Yang SS, et al. Pegylated interferon-alpha-2a plus ribavirin for treatment-naïve Asian patients with hepatitis C virus genotype 1 infection: a multicenter, randomized controlled trial. Clin Infect Dis. 2008;47:1260–1269. doi: 10.1086/592579. [DOI] [PubMed] [Google Scholar]
- Ueda T, Chung H, Kudo M, Kitai S, Ishikawa E, Yada N, et al. Prolonged PEG-IFN and RBV is effective in patients with HCV genotype 1 and high viral load who achieved virological response later than 24 weeks. Intervirology. 2010;53:55–59. doi: 10.1159/000252785. [DOI] [PubMed] [Google Scholar]
- Chen JY, Wang CM, Ma CC, Luo SF, Edberg JC, Kimberly RP, et al. Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum. 2006;54:3908–3917. doi: 10.1002/art.22220. [DOI] [PubMed] [Google Scholar]