Abstract
A common allele at the TAGAP gene locus demonstrates a suggestive, but not conclusive association with risk of rheumatoid arthritis (RA). To fine map the locus, we conducted comprehensive imputation of CEU HapMap single-nucleotide polymorphisms (SNPs) in a genome-wide association study (GWAS) of 5500 RA cases and 22 621 controls (all of European ancestry). After controlling for population stratification with principal components analysis, the strongest signal of association was to an imputed SNP, rs212389 (P=3.9 × 10−8, odds ratio=0.87). This SNP remained highly significant upon conditioning on the previous RA risk variant (rs394581, P=2.2 × 10−5) or on a SNP previously associated with celiac disease and type I diabetes (rs1738074, P=1.7 × 10−4). Our study has refined the TAGAP signal of association to a single haplotype in RA, and in doing so provides conclusive statistical evidence that the TAGAP locus is associated with RA risk. Our study also underscores the utility of comprehensive imputation in large GWAS data sets to fine map disease risk alleles.
Keywords: TAGAP, genetics, rheumatoid arthritis
Introduction
Contemporary genome-wide association studies (GWASs) test common single-nucleotide polymorphisms (SNPs) for association with disease risk. The underlying biological model is that the causal mutation is in linkage disequilibrium (LD) with tagging SNPs on the genotyping array.1 If the causal mutation is also common, then it likely resides in close proximity to the disease-associated SNP. To fine map the strongest signal of association, it is necessary to perform dense genotyping in large patient collections, followed by stepwise conditional regression and haplotype analysis.2 Genome-wide imputation from a reference panel, such as HapMap, facilitates this process by providing high-density coverage of common alleles across a locus of interest.3
Rheumatoid arthritis (RA) is a common autoimmune disease of unclear etiology. Family studies estimate that >50% of the variance in disease risk is genetic.4 To date, >20 risk loci have been identified conclusively at P<5 × 10−8.5, 6 In an initial RA GWAS meta-analysis of 15 855 case–control samples, with replication in an additional 19 915 independent samples, we found strong but not conclusive evidence that a common SNP near TAGAP is associated with risk of RA (rs394581, PGWAS=5.6 × 10−4, Poverall=3.8 × 10−7 in all 35 770 case–control samples combined).7 This GWAS meta-analysis did not include complete imputation of all CEU HapMap SNPs, but rather included only 336 721 SNPs genotyped on the Affymetrix 500K platform (Affymetrix, Santa Clara, CA, USA) passing genotype quality control filters. In a follow-up GWAS meta-analysis of 5539 autoantibody-positive RA cases and 20 269 controls, we imputed 2.56 million SNPs from CEU HapMap and found continued evidence of association at the previous TAGAP RA risk SNP (rs394581, PGWAS=7.7 × 10−4).6 However, we also found suggestive evidence for additional risk alleles at the TAGAP locus, including an allele associated with risk of celiac disease and type I diabetes (TID).8, 9
The purpose of the current study is to refine the signal of association at the TAGAP locus using our second GWAS meta-analysis with genome-wide imputation. Our results show that an imputed SNP at the TAGAP locus provides a much stronger signal of association than the previously published SNP in RA.
Results and discussion
The initial goals of this study were to use genome-wide imputation and conditional analysis to determine if any of the known RA risk alleles had either (1) a better signal of association or (2) an independent, second signal of association in the associated risk locus. We included those RA risk loci with conclusive (P<5 × 10−8) and highly suggestive (P<10−6 with independent replication at P<0.01) association with risk of RA.
Our GWAS meta-analysis was conducted using the six case–control collections shown in Table 1. Within each collection, we first filtered SNPs and individuals, and then ran Eigenstrat10 on genotyped SNPs to calculate principal components, as previously described.6 We used filtered SNPs to impute 2.56 million SNPs present at >1% allele frequency in CEU HapMap.3 GWAS association analyses were performed using logistic regression (SNPTEST and R), incorporating the top five principal components as covariates to control for population stratification. We combined the results of the GWAS for each data set by weighting the logistic regression estimates (beta values) by the inverse variance of each data set.11 In our final analysis of 5 500 RA cases and 22 621 controls (all of European ancestry) with genotype data at 2.56 million SNPs, we found no evidence of systemic bias (λGC=1.01). In comparison with the published GWAS by Stahl et al.,6 here we added 2352 additional shared controls because we incorporated PC's into the GWAS to correct for stratification. In doing so, we removed 39 RA cases because they were determined to be genetic outliers by PC's in the expanded data set.
Table 1. Case–control samples for GWAS meta-analysis.
| Case collection | Control collection | Geographical origin | Antibody status | Cases | Controls | Genotyping platform | |
|---|---|---|---|---|---|---|---|
| Meta-analysis | Brigham Rheumatoid Arthritis Sequential Study (BRASS) | Shared controls | Boston, USA | 100% CCP+ | 478 | 1631 | Affymetrix 6.0 |
| Canada | Canada and shared controls | Toronto, Canada | 100% CCP+ | 589 | 1554 | Illumina 370K | |
| Epidemiological Investigation of Rheumatoid Arthritis (EIRA) | EIRA | Sweden | 100% CCP+ | 1142 | 1072 | Illumina 317K | |
| 5500 cases, 22 621 controls | North American Rheumatoid Arthritis Consortium (NARAC) I | Shared controls | North America | 100% CCP+ | 868 | 1194 | Illumina 550K |
| NARAC III | Shared controls | North America | 100% CCP+ | 902 | 6613 | Illumina 317K | |
| Wellcome Trust Case Control Consortium (WTCCC) | Shared controls from WTCCC | UK | 100% RF+ or CCP+ | 1521 | 10557 | Affymetrix 500K |
Abbreviations: CCP, cyclic citrullinated peptide; GWAS, genome-wide association study; RF, rheumatoid factor.
The characteristics of the six case–control collections used in the GWAS meta-analysis are shown.
To refine the signal of association at each RA risk locus, we conditioned on the previously described SNP genotype to look for additional statistical evidence of association at other nearby SNPs. For each of the known RA risk loci, we assessed association statistics, both before and after conditional analysis, of SNPs within a 1-Megabase (Mb) region centered on the known RA risk allele. In our analysis conditional on the previously associated SNP, the TAGAP locus had a clear signal that was different than the previously reported SNP (Supplementary Table 1). The TNFAIP3, CCL21, CTLA4-CD28 and ANKRD55-IL6ST loci showed evidence of independent, second signals of association, consistent with previous reports.6, 7, 12 No other loci showed strong evidence for association after analysis conditional on the previously associated RA SNP (conditional P<10−4). Because of these findings, we focused solely on refining the signal of association at the TAGAP locus. We note, however, that our study design has limited power to detect independent rare alleles or common alleles of more modest effect.
In unconditional analysis, the new TAGAP SNP, rs212389, reached a conservative level of genome-wide significance in our GWAS meta-analysis (P=3.9 × 10−8, odds ratio=0.87). As shown in Figure 1a, the new TAGAP SNP, rs212389, was five orders of magnitude more significant than the previously reported TAGAP SNP (rs394581). These two SNPs are located only 7.3 kb apart and are in LD with one another (r2=0.59 and D′=0.92). Importantly, the new TAGAP SNP had not been genotyped directly in any of the six collections.
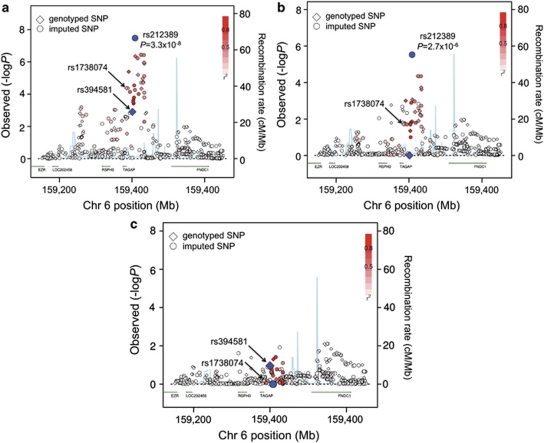
Figure 1.
TAGAP results from GWAS meta-analysis. (a) Results conditional only on the top five principal components (PCs). (b) Results conditional on previous RA SNP, rs394581 (in addition to five PCs). (c) Results conditional on new RA SNP, rs212389 (in addition to five PCs). In each plot, genotyped SNPs (diamonds) and imputed SNPs (circles) are shown across a 500-kb window, where the color of the symbol indicates LD (as measured by r2) to the new RA SNP (rs212389). Two genes, TAGAP and RSPH3, map within the recombination hotspots (shown in blue); three other genes map to the region (EZR, OSTCL/LOC202459 and FNDC1).
Conditional analysis revealed that the new TAGAP SNP, rs212389, better explains the association at this locus than the previously associated RA TAGAP SNP. After conditioning on the previously reported TAGAP SNP (rs394581), the new TAGAP SNP (rs212389) remained highly significant (P=2.2 × 10−6; Figure 1b). In contrast, conditioning on the new TAGAP SNP abrogated any remaining signal across the entire 1-Mb region, including any signal at the previously reported TAGAP SNP (rs394581; P=0.07; Figure 1c).
The same TAGAP locus is associated with risk of celiac disease and TID.8, 9 The celiac/TID risk allele appears different than the RA risk allele, as the new RA SNP demonstrates evidence of association even after analysis conditioning on the celiac/TID SNP, rs1738074 (P=1.7 × 10−4). These results are consistent with the patterns of LD between the pairs of SNPs: the celiac/TID SNP, rs1738074, has r2=0.32 with the new RA risk SNP, rs212389, and r2=0.35 with the previously reported RA risk SNP, rs394581.
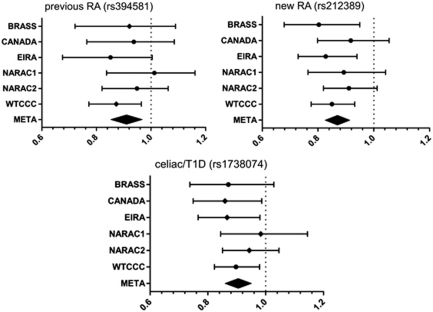
As the new TAGAP SNP was imputed in all six GWAS collections, we wanted to ensure that the signal of association was not driven by any one GWAS collection, and that the association was not an artifact of imputation. As shown in Figure 2, the new TAGAP SNP (rs212389) demonstrated a signal of association in all six GWAS collections. For the new TAGAP SNP, the imputation scores were higher for those samples genotyped using the Illumina platform (Illumina, San Diego, CA, USA; Epidemiological Investigation of Rheumatoid Arthritis (EIRA), North American Rheumatoid Arthritis Consortium (NARAC) 1 & 2, and Canada; Supplementary Table 2).
Figure 2.
TAGAP results for previous RA SNP (rs394581), new RA SNP (rs212389) and celiac/TID SNP (rs1738074), within each GWAS collection. In each plot, the point estimate of the odds ratio (OR) is shown, with 95% confidence intervals (CI). Genotyped SNPs are indicated by diamonds and imputed SNPs by circles. The meta-analysis of all six collections is indicated by the filled diamond at the bottom of the graph, where the edges of the diamond indicate the 95% CI. For the new RA SNP (rs212389), the OR is 0.87 (95% CI 0.83–0.91).
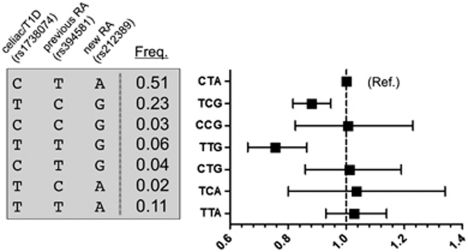
To further fine map the signal of association, we used our GWAS meta-analysis to construct haplotypes generated by these three SNPs (new RA SNP, previous RA SNP and celiac/TID SNP). As the imputation scores were higher for those samples genotyped using the Illumina platform, we analyzed only on those 3501 RA cases and 10 433 controls genotyped on Illumina (see Table 1). As shown in Figure 3, our haplotype analysis indicated that the best genetic model is one in which the haplotype tagged by the G allele of rs212389 (new RA) and the T allele of rs1738074 (celiac/TID SNP) confers protection from RA (P=1.1 × 10−6). A similar but more significant result is obtained when we use all six GWAS collections in our analysis (P=1.8 × 10−8).
Figure 3.
TAGAP results for haplotypes created by the celiac/TID SNP (rs1738074), previous RA SNP (rs394581) and new RA SNP (rs212389). Seven haplotypes are created by the three SNPs. The frequency of each haplotype from the control population is shown. The point estimate of the odds ratio (OR) and 95% confidence intervals (CI) are shown.
To test whether risk is best explained by the rs212389-G/rs1738074-T common haplotype, rather than either allele alone, we performed an analysis in which we compared three risk models: G allele of rs212389 (new RA), T allele of rs1738074 (celiac/TID) and rs212389-G/rs1738074-T common haplotype. Consistent with our other analyses, we found that the rs212389-G/rs1738074-T model was significantly better than either model, which included only a single SNP (P=0.01; Supplementary Table 3). Although this analysis suggests that the causal allele resides on the rs212389-G/rs1738074-T, additional genotyping in large multi-ethnic sample collections will be required to formally identify the best genetic model at the TAGAP locus.
Under a model in which the causal allele is common and tagged by the common G-rs212389/T-rs1738074 haplotype, then the causal allele should be catalogued within the 1000 Genomes Project, as this haplotype is common (present in ∼31% of control chromosomes). We used phased 1000 Genomes data to identify all variants in high LD (r2>0.80) with the new risk haplotype (Supplementary Table 4). At this LD threshold, we identified only 10 potential causal variants. However, none of these variants were within the protein-coding sequence of TAGAP or other neighboring genes, and none disrupted an obvious non-coding functional motif (for example, transcription factor binding site).
Although TAGAP is the most promising biological candidate gene, four other genes map to the region of LD: EZR, OSTCL/LOC202459, RSPH3 and FNDC1 (Figure 1). TAGAP gene is expressed at high levels in the immune system, including B- and T-lymphocyte cells, dendritic cells, natural killer cells and monocytes. The protein product is predicted to function as a Rho GTPase-activating protein. Otherwise, little is known about the function of TAGAP in the immune system.
There are several important limitations of our study. First, we did not genotype the new RA-associated SNP in any samples. The imputation quality score (Supplementary 2) and the consistency of the effect across the different cohorts (Figure 2) provide strong evidence that the signal of association at the imputed SNP is not because of a technical artifact. Second, no re-sequencing was done to discover variants not present in either HapMap or 1000 Genomes data. Formally speaking, our results could be consistent with a genetic model in which multiple causal rare variants reside on the common G-rs212389/T-rs1738074 haplotype, rather than a single causal allele that resides on this haplotype. Third, our study is underpowered to detect independent alleles of modest effect, especially those that are of low allele frequency (for example, 1–5%). Last, we did not perform experiments to determine whether the new RA SNP (rs212389) or any variants from the 1000 Genomes data in LD with the common G-rs212389/T-rs1738074 haplotype are functional.
In conclusion, our study provides conclusive evidence that the TAGAP locus is associated with risk of RA. In doing so, we have refined the signal of association at the TAGAP locus either to rs212389 (new RA SNP) or to a haplotype tagged by the G allele of rs212389 (new RA SNP) and the T allele of rs1738074 (celiac/TID SNP). Comprehensive imputation was essential to refine this association, as the new SNP was not genotyped directly on either the Affymetrix or Illumina platforms.
Acknowledgments
RMP is supported by grants from the NIH (R01-AR057108, R01-AR056768, U01-GM092691), and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. SR is supported by an NIH Career Development Award (1K08AR055688-01A1). FASK is supported by a Marie Curie International Outgoing Fellowship for Career Development from the European Community's FP7 (Grant Agreement number PIOF-GA-2009-237280).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary Material
References
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22:109–118. doi: 10.1097/BOR.0b013e328336474d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, et al. Shared and distinct genetic variants in Type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.