Abstract
Intercellular signaling by fibroblast growth factors plays vital roles during embryogenesis. Mice deficient for fibroblast growth factor receptors (FgfRs) show abnormalities in early gastrulation and implantation, disruptions in epithelial–mesenchymal interactions, as well as profound defects in membranous and endochondrial bone formation. Activating FGFR mutations are the underlying cause of several craniosynostoses and dwarfism syndromes in humans. Here we show that a heterozygotic abrogation of FgfR2-exon 9 (IIIc) in mice causes a splicing switch, resulting in a gain-of-function mutation. The consequences are neonatal growth retardation and death, coronal synostosis, ocular proptosis, precocious sternal fusion, and abnormalities in secondary branching in several organs that undergo branching morphogenesis. This phenotype has strong parallels to some Apert's and Pfeiffer's syndrome patients.
Keywords: craniosynostosis ‖ sternum ‖ kidney ‖ lung ‖ lacrimal gland
Fibroblast growth factors (Fgfs) constitute a 22-member-strong family of intercellular-signaling molecules that transduce their signal by activating specific cell surface tyrosine kinase receptors (reviewed in refs. 1–4). Fgf-receptor (FgfR) signaling relies on dimerization of two receptor molecules brought about by ligand binding in cooperation with heparan sulfate proteoglycans. In mammals, each of four FgfR genes encode an extracellular domain composed of two or three Ig-like loops, a transmembrane (TM) segment, and an intracellular tyrosine kinase. Alternative splicing of the exons that encode the third Ig loop in FgfR-1, FgfR-2, and FgfR-3 results in receptor isoforms termed IIIb or IIIc, each with distinct ligand binding specificity and tissue distributions (see Fig. 1 A and B; refs. 1 and 5). For FgfR2, IIIb and IIIc isoforms are expressed predominantly by epithelial and mesenchymal cells, respectively (6, 7). In vitro, Fgfs can promote cell proliferation, survival, differentiation, and motility (1, 8). In vivo, signaling by Fgfs governs the normal development of diverse tissues and organs in vertebrates (9, 10) as exemplified by deletion of genes encoding particular Fgfs or FgfRs (11–22). For example, targeted deletion of FgfR-2 causes early embryonic lethality, whereas mice with partial FgfR2 function show severe lung and early limb defects (16, 23, 24). A specific deletion and abrogation of FgfR2-IIIb function in mice results in agenesis of limbs, lung, and the anterior pituitary and in dysgenesis of several visceral organs (25, 26).
Figure 1.
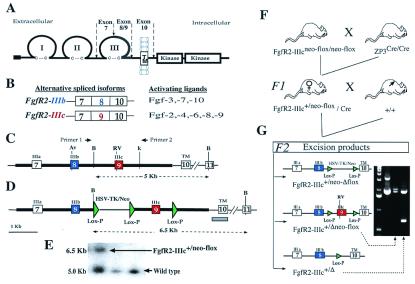
Fgf receptor structure and strategy for deletion of FgfR2 exon 9 (IIIc). (A) Schematic structure of FgfR2 highlighting the third Ig loop, a region in which alternative usage of exons 8 or 9 in FgfR2 leads to the generation of the IIIb or IIIc isoforms, respectively. TM, transmembrane domain. (B) The main ligands activating each of these isoforms. (C) Schematic depiction of mouse genomic DNA encompassing FgfR2 exons (boxes) 7, 8, 9, and 10, drawn to scale and showing main restriction enzyme sites (Av, AvaI; RV, EcoRV; B, BamHI; K, KpnI). (D) Targeting construct showing loxP sequences placed downstream of exon 9, and flanking the selectable marker gene neo driven by HSV-TK promoter located upstream of exon 9. Thick lines indicate the extent of the targeting construct. (E) Homologous recombinant 129 embryonic stem cells were identified by Southern blotting of BamHI-digested DNA by using a 450-bp genomic probe located 3′ of target vector sequences (gray bar in D). Homologous recombinant cells (FgfR2-IIIc+/neo-flox) yielded a 6.5-kb fragment and wild type yielded a 5.0-kb fragment. Both 5′ and 3′ joins were checked by PCR analysis. (F) Hemizygous (FgfR2-IIIc+/Δ) mutant mice were generated by crossing FgfR2-IIIcneo-flox/neo-flox with ZP3-Cre females (34), and then crossing F1 females carrying one copy of the targeting construct as well as the Cre transgene with wild-type males. Approximately 50% of such females gave Cre-mediated excision. (G) Of these excisions, 90% were complete (FgfR2-IIIc+/Δ); the remainder excised only the selectable marker (FgfR2-IIIc+/Δneo-flox), as determined by PCR using the pair of primers shown in C. The potential excision product, FgfR2-IIIc+/neo-Δflox, was not observed.
In late fetal and early neonatal life, a complex set of interactions coordinate the growth and expansion of the skull vault (calvarium) with that of the brain. Calvarial bones form by ossification of plates of neural crest-derived mesenchyme overlying the brain, and grow by appositional deposition of new bone along the periphery of each plate. Where two plates meet a suture forms, and an imbalance of bone formation at the sutures results in premature calvarial fusion (craniosynostosis) leading to serious alterations of skull shape. Genetic analysis has revealed dominant–positive mutations affecting three FGFR genes as the basis of several craniosynostosis syndromes (27–30). The most severe of these syndromes are Apert's and type 2 and type 3 Pfeiffer's syndromes, which arise through mutations in FGFR2 and are characterized by fusion of the coronal sutures and postaxial limb abnormalities (31). Moreover, a proportion of Apert's and Pfeiffer's syndrome patients show, in addition, varied abnormalities of the visceral organs which lead to genito-urinary, gastro-intestinal, and respiratory anomalies [refs. 32 and 33; Online Mendelian Inheritance in Man (OMIM) database at http://www3.ncbi.nlm.nih.gov/Omim/]. We show here that excision of a single copy of FgfR2-IIIc in mice results in a gain-of-function mutation associated with exon switching within the FgfR2 gene. The phenotype of FgfR2-IIIc hemizygotes is characterized by precocious ossification of the coronal sutures, zygomatic arch joints, the sternebrae, as well as major defects in the kidney, lung, and lacrimal glands.
Materials and Methods
Generation of Targeting Vector and Mutant Mice.
A 7.5-kb targeting construct was generated by PCR amplification (Expand PCR, Roche Molecular Biochemicals) and subsequent cloning of two fragments from a mouse FgfR2 λ clone and one fragment from plasmid pL2-Neo into a psP73 plasmid. These fragments were (i) a 3.6-kb XhoI/NotI fragment containing FgfR2 exons 7 and 8 and their flanking introns, (ii) a 2.7-kb NotI/SmaI fragment containing exon 9 and its flanking introns, and (iii) a NotI/NotI-tagged 1.2-kb fragment composed of herpes simplex virus thymidine kinase (HSV-TK)-Neo flanked by two loxP sites. The third loxP fragment was generated by the annealing of KpnI-tagged complementary loxP primers and cloned in at a KpnI site downstream of exon 9. The integrity of splice acceptor and donor sites in each exon of the targeting construct was verified by DNA sequencing. Correctly targeted ES cells were injected into blastocysts of C57BL mice to generate chimeras. Male mice containing the targeted floxed allele were crossed with ZP3-Cre transgenic female mice that express Cre recombinase in the germ cells (34). F1 female mice containing the ZP3 transgene and a floxed targeted allele were bred with C57BL mice to generate the F2 generation. Forty percent of the F2 mice showed excision of the floxed targeted allele (Fig. 1). Of these excisions, 90% were complete excisions (FgfR2-IIIc+/Δ), whereas the remainder showed partial excision (FgfR2-IIIc+/Δneo-flox) (Fig. 1G).
Bone and Cartilage Staining of Whole Skeletons.
Skeletons of whole embryos or pups were stained as described by McLeod (35).
Histochemical Detection of Alkaline Phosphatase in Coronal Sutures.
Alkaline phosphatase was detected by using the method of Drury (36). Briefly, 12-μm transverse cryostat sections through fresh-frozen 18-day-old embryos were fixed with 4% paraformaldehyde (PF) solution (pH 7.4), washed with cold 0.1 M PBS, and incubated for 30 min at 37°C with a 5:3 ratio mix of the following solutions, respectively: solution A, 300 mM sodium barbitone/5 mM CaCl2/2 mM MgSO4; solution B, 1% β-glycerophosphate. Sections were then washed in distilled water, treated for 5 min with a 2% cobalt nitrate solution, then washed and treated with 1% (NH4)2S before rinsing and clearing in alcohol series and mounting in dibutyl pthalate xylene.
Carmine (Carmalum) Staining of Lacrimal Glands.
Dissected lacrimal glands were fixed in 4% PF, washed in 0.1 M PBS, and dehydrated with 70% ethanol before staining briefly in a solution of 0.2% carmine (Sigma)/0.5% AlK(SO4)2 containing trace amounts of thymol. Excess stain was cleared in 70% ethanol.
Measurements of Blood Glucose Levels.
Blood glucose levels were measured by using 30 μl of tail blood and a Glucotrend (Roche Diagnostics) monitor according to the manufacturer's instructions.
Whole Mount in Situ Hybridization on Calvaria and Sternum.
Embryos were delivered from time-mated ZP3-Cre/FgfR2-IIIc+/neo-flox mothers, and calvarial bones and sternum were dissected to be free of overlying skin, as well as the brain and thoracic organs, in Hepes-buffered medium. Tissues were fixed overnight in 4% PF, dehydrated through an ascending methanol series in PBS/0.1% Tween-20 (Tw-20, Sigma), and stored in absolute methanol at −20°C until use. Yolk sacs from individual embryos were used for genotyping. Sense and antisense digoxigenin-labeled probes were synthesized by in vitro transcription (Roche Molecular Biochemicals kit) from appropriately linearized plasmids carrying one of the following fragments: a 450-bp FgfR2-Tk, 150-bp FgfR2-IIIb (exon 8), 148-bp FgfR2-IIIc (exon 9), 500-bp Fgf-3, 620-bp Fgf-7, or a 670-bp Fgf-10 sequence, and purified by passing through Chroma Spin −30 columns (CLONTECH) before quantifying on agarose gels.
For the in situ hybridization reaction, rehydrated tissues were treated with 7 μg/ml proteinase K and refixed with 4% PF/0.2% glutaraldehyde before a 5-h prehybridization at 50°C in a solution of 50% formamide/5 mM EDTA/1.3× SSC/0.5% CHAPS (Sigma)/0.1% Tw-20/1% solution of Boehringer blocking powder (BBR)/100 μg/ml yeast tRNA/50 μg/ml heparin. Hybridization in the presence of 1 μg/ml probes was carried out overnight at 50°C. Post-hybridization washes at 50°C were as follows: hybridization solution alone, wash buffer (50% formamide/2× SSC) first without and then with 0.1% Tw-20, and wash buffer diluted 1:1 with MABT (1 M maleic acid/0.15 M NaCl/0.1% Tw-20). Tissues were then washed at room temperature with MABT before blocking for 4 h with a solution of 20% heat-inactivated goat serum/2% BBR in MABT/0.15 M NaCl/0.1% Tw-20, followed by overnight incubation at 4°C with alkaline phosphatase-coupled anti-digoxigenin antibody (Roche Molecular Biochemicals). After washing extensively in MABT, tissues were treated with NTMT (0.1 M Tris⋅HCl/0.1 M NaCl/50 mM MgCl2/0.1% Tw-20) and 2 mM levamisole. Color was developed for 1–16 h in fresh NTMT containing nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate but no levamisole. Reaction was stopped by washing tissues in PBS/0.1% Tw-20 before fixation with 4% PF and storage in glycerol.
Reverse Transcription–PCR.
Total RNA isolated by the Trizol method (GIBCO) from snap-frozen cerebral cortices (dissected free of meninges) and livers of new-born wild-type and hemizygote mice was first subjected to one round of the PCR cycle of 42°C for 15 min, followed by 95°C for 5 min, by using a reverse transcription–PCR kit (Ready-to-Go, Amersham Pharmacia) and pd(T)12–18 primers to amplify DNA from mRNA species. The products were then subjected to 32 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, using a primer from exon 7 (IIIa; CCCATCCTCCAAGCTGGACTGCCT) in conjunction with one from exon 8 (IIIb; CTGTTTGGGCAGGACAGTGAGCCA), or exon 9 (IIIc; CAGAACTGTCAACAATGCAGAGTG), or exon 10 (TM; GCTTGGTCAGCTTGTGCACAGCTGG) to amplify the IIIb, IIIc, or both isoforms of FgfR2, respectively. Reaction products were resolved alongside a 50-bp ladder on 1.5% agarose gel (see Fig. 5A).
Figure 5.
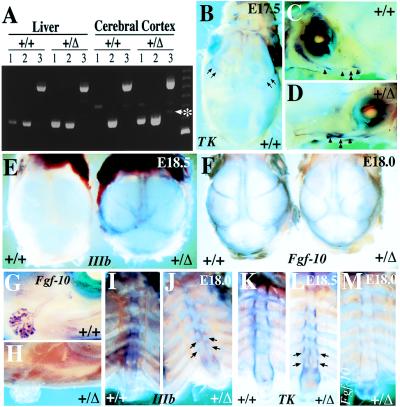
Analysis of FgfR2 and Fgf-10 expression in brain, calvaria, lacrimal glands, and sternum. (A) Reverse transcription–PCR assessment of FgfR2 expression in the liver and cerebral cortices (dissected free of meninges) of wild-type and hemizygous mice by using primers from within exons 7 and 8 [IIIb specific (320 bp), lane 1], or exons 7 and 9 [IIIc specific (310 bp), lane 2], or exons 7 and 10 [detects both isoforms (495 and 489 bp), lane 3]. Identity of bands was confirmed by DNA sequencing. Note that the IIIb-specific product, as well as a faint 345-bp band (asterisks), derived from IIIa-TM splice, is detected in the cerebral cortex and liver of hemizygous but not wild-type mice. (B–D) Expression pattern of FgfR2 in E17.5 calvaria of wild type (+/+) and hemizygote (+/Δ) as revealed by whole mount in situ hybridization (WMISH) using digoxigenin-labeled TK probe which is isoform nonspecific. Arrows in B point to staining of coronal sutures of wild type. Staining was less intense in hemizygotes (data not shown). (C and D) Bones making up the lower rim of the eye socket, the zygomatic arch (arrowheads), as well as their intervening joints are also labeled; these joints (double arrowheads) are more closely apposed in hemizygotes than in wild types. (E) WMISH with an FgfR2-IIIb-specific probe on E18.5 calvaria shows elevated expression of this isoform in calvarial sutures of hemizygotes but not in wild-type mice. Hemizygotes also present a shortened snout, wider skull, and delayed closure of the anterior fontanelle. (F) WMISH with an Fgf-10 probe shows expression of this ligand in the suture regions of both wild types and hemizygotes at E18. (G and H) In the same specimens shown in F, Fgf-10 probe labels the tips of branches within the exorbital lacrimal glands of wild type (G), whereas there is a lack of expression in hemizygote gland rudiment (H). (I–M) WMISH analysis on dissected sternums from E18 mice with an FgfR2-IIIb-specific probe (I and J), E18.5 mice using a TK probe (K and L), and E18 using an Fgf-10 probe (M). In both wild types and hemizygotes, TK and IIIb strongly label the periosteal regions of each sternebrae. However, delayed midline fusion results in two hemi sternebrae in hemizygotes (arrows in J and L), the medial rims of which also express TK and IIIb. At E18.5, when medial fusion is more advanced, TK additionally labels the periphery of the xiphoid processes, which appears bifurcated in hemizygotes. (M) Fgf-10 is expressed by interrib periosteal regions as well as the body of xiphoid.
Results
Cre-Mediated Excision of FgfR-2 Exon 9 (IIIc).
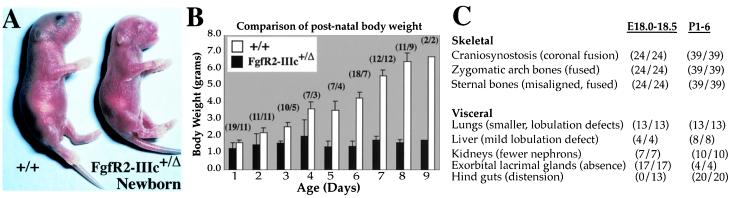
To investigate FgfR2-IIIc function, we used a Cre/loxP recombination strategy to remove from the mouse germ line exon 9 of FgfR2, which is specific for this receptor isoform (see Materials and Methods and Fig. 1 C–G). Mice containing the correctly targeted allele (composed of a floxed neo and IIIc construct) were viable and fertile. To obtain excision, male FgfR2-IIIcneo-flox/neo-flox mice were crossed with ZP3 transgenic females (34) which express Cre recombinase in the germ cells. Because excision occurs in the germ cells, F1 females containing both the ZP3-Cre transgene and a copy of the targeted floxed allele were bred with wild-type C57BL males to generate the F2 generation (Fig. 1F). All offspring containing the targeted floxed allele showed excision. Mice hemizygous or homozygous for the floxed-IIIc (FgfR2-IIIc+/Δneo-flox; Fig. 1G) gave no discernable phenotype, but mice hemizygous for FgfR2-IIIc (FgfR2-IIIc+/Δ) showed a range of bone and visceral defects (see below). FgfR2-IIIc+/Δ mice were born at the expected frequency of 50% (n = 245); at birth 90% were slightly smaller than their wild-type littermates, whereas the remainder were significantly smaller (birth weight 0.7–0.9 g; data not shown). After birth, hemizygotes did not gain weight and died within 9 days (Fig. 2). These findings are surprising because mice hemizygous for a null allele of FgfR2 gene give no detectable phenotype (16, 24) and, therefore, strongly suggest a gain-of-function mutation.
Figure 2.
Growth retardation in FgfR2-IIIc+/Δ mice. (A) Hemizygotes were identifiable at birth by their smaller sizes, characteristic dome-shaped heads, and truncated faces; the genotype was confirmed by PCR using tail genomic DNA (Fig. 1G). (B) All hemizygotes showed growth retardation after birth as reflected in a comparison of their body weights with those of wild-type littermates. Brackets show the number of hemizygotes and wild types analyzed. (C) Table summarizing the major abnormalities and their penetrance (number showing specified defects/total number of hemizygotes analyzed) at E18 and postnatally.
Bone and Visceral Defects in FgfR2-IIIc+/Δ Mice.
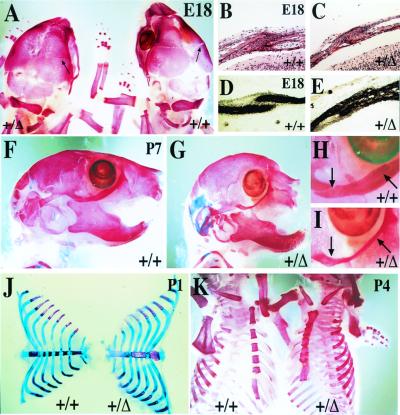
Newborn FgfR2-IIIc+/Δ mice had shortened snouts, markedly rounded skulls (Fig. 2A), and bulgy eyes (proptosis). To reveal skeletal abnormalities, bones of embryos and pups were stained with alizarin red. The coronal sutures, which separate the frontal and parietal calvarial bones, showed precocious ossification by embryonic day 18 (E18; Fig. 3 A–C). Fusion of this suture was further confirmed by histochemical staining of sections with alkaline phosphatase, a marker of mature and progenitor osteoblasts (Fig. 3 D and E). Zygomatic arch bones were also found to be fused together at their joints, a phenomenon that would result in the formation of shallow eye orbits and, thus, produce ocular proptosis (Fig. 3 F–I). Similarly, premature ossification of the coronal sutures may account for a noticeable truncation of the maxilla as well as the observed compensatory dorso ventral extension of the cranium (Fig. 3 A, F, and G).
Figure 3.
Precocious ossification of coronal sutures and the sternum in FgfR2-IIIc+/Δ mice. Skeletons stained with alizarin red to identify ossified tissue (A and K), or stained with alizarin red in combination with Alcian blue stain to additionally reveal cartilage (F–J). (A) Dorsal view of calvarial bones at E18 showing closer apposition of frontal and parietal bones at the coronal suture (arrows) in a hemizygote compared with its wild-type litter mate. (B–E) Transverse sections through the frontal and parietal bones showing fusion of coronal sutures in hemizygote in contrast to wild-type mice. Sections stained with hematoxylin–eosin (B and C) or alkaline phosphatase (D and E). (F–I) Lateral views of skulls from 7-day-old mice showing in hemizygotes rounded heads, truncated maxilla, and fusion of joints separating the zygomatic arch bones (part of the maxilla, zygomatic, and temporal bones) which make up the lower rim of the eye socket. (H and I) Detail of zygomatic arch joints shown in F and G, respectively. (J and K) Dissected and whole rib cages from 1- and 4-day-old mice, respectively, showing precocious and progressive sternal fusion in hemizygotes. Note that in these mice, individual sternebrae are thicker and less congruent, and the manubrium and xiphoid processes are bifurcated.
Staining with alizarin red and Alcian blue combined also revealed abnormal ossification of the intersternebral cartilage (Fig. 3 J and K). The sternum forms by ventro medial migration of two lateral plate mesoderm-derived masses that fuse to form a single cartilaginous mass in front of the ribs (37). Individual sternebrae then form by endochondrial ossification of the interrib regions, as signals from the cartilaginous rib heads are thought to inhibit ossification where they abut the sternum (37–39). Sternal abnormalities in hemizygous mice were first seen at E16.5 as a delay in medial fusion of sternal bands (data not shown). By E18.5, the sternebrae were found to be thicker and misaligned, whereas the posterior margins of the intersternebral cartilage showed ectopic ossification. Abnormal sternebral thickening and cartilage ossification continued after birth in a posterior–anterior and caudo rostral direction (Fig. 3 J and K) and was often complete by 2–4 days after birth. Furthermore, the bone and cartilage structures that cap the ends of the sternum (the manubrium and xiphoid) showed abnormal bifurcation, probably reflecting the earlier delay in sternal band fusion.
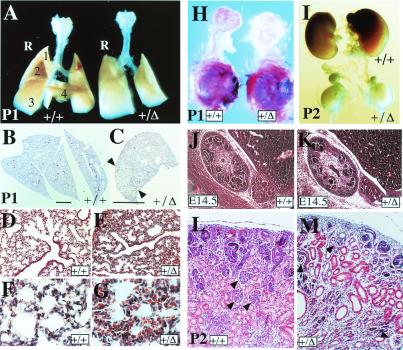
An examination of the internal organs in FgfR2-IIIc hemizygotes revealed major abnormalities in the lungs, kidneys, and lacrimal glands, organs that develop through extensive branching morphogenesis involving reciprocal loops of Fgf, Sonic-hedgehog, Bmp/Tgf-β, and Wnt signaling between mesenchymal and epithelial cells (40–44). The lungs of neonatal hemizygotes were slightly smaller than those of their wild-type littermates, but more strikingly, the right lung showed defects in lobulation—it was composed of only three partially separated pulmonary lobes compared with four distinct lobes in the wild type (Fig. 4A). Partial lobulation was further confirmed in sections of E18 lungs, which also revealed the presence of fewer bronchioles and a more densely packed mesenchyme, suggesting incomplete alveolarization of the lungs (Fig. 4 B–G). A mild lobe-separation defect was also noted in the livers of E18 and postnatal hemizygotes (data not shown). The kidneys did not grow beyond their birth size and, thus, were considerably smaller in the mutant mice at P2, although the adrenal glands were of normal size (Fig. 4I). Histological comparison at E14.5 showed that, although hemizygote kidneys were of a similar size compared to wild-type, there was a marked reduction in the number of nephrogenic units (Fig. 4 J and K). After birth, kidney architecture was found to be disorganized, and contained fewer glomeruli, dilated proximal and distal tubules, and a greater expanse of undifferentiated mesenchyme (Fig. 4 L and M). In hemizygotes, the exorbital lacrimal glands failed to form despite the presence of a rudimentary mesenchymal sac (Fig. 4H). These observations highlight a defect in secondary branching, but a more extensive study of the aforementioned signaling loops at different stages of development would help better define the underlying cause. Hemizygotes also showed postnatal distension of the hind gut, although there was no obvious defect in the stomach and gut architecture or innervation (data not shown). Moreover, despite ingesting milk, hemizygotes had a significantly lower concentration of blood glucose at P3 [3.0 ± 0.6 μM (n = 4)] compared with their wild-type littermates [5.8 ± 0.6 (n = 6)], suggesting that an undefined gastro intestinal defect resulting in malnutrition contributes to their growth retardation and early death.
Figure 4.
Visceral defects in FgfR2-IIIc+/Δ mice. (A) Comparison of whole lungs at postnatal day 1 (P1) showing the development of a single right lobe in hemizygotes compared with four distinct lobes in the wild type. (B–G) Transverse sections through the right lobes stained with hematoxylin–eosin showing partial lobe separation in the mutant right lung (indicated by arrowheads in C), as well as the development of fewer bronchioles lined with ciliated cells and fewer branched alveolar structures (D and E). (B and C, bars = 4 mm.) In mutants, lung mesenchyme is more compact and often congested with red blood cells (F and G). (H) Dissected exorbital lacrimal glands with their surrounding mesenchyme still attached to the eye and stained with the dye carmalum to reveal branching, show a clear lack of gland development in hemizygotes. (I) Comparison of kidneys at P2 reveals severe growth retardation in hemizygotes which does not seem to affect the adrenal glands lying above each kidney. (J and K) Saggital hematoxylin–eosin (H&E)-stained sections through E14.5 embryonic kidneys show the presence of fewer developing nephrons in the cortical region of hemizygotes. (L and M) H&E-stained transverse section of P2 kidneys, showing fewer and degenerating glomeruli (arrowheads), dilated proximal and distal tubules, and more undifferentiated mesenchyme in hemizygotes.
Altered FgfR2 Expression in the Brain, Cranial Sutures, and Sternal Cartilage.
A gain-of-function mutation suggested that in the absence of exon 9 (IIIc), there was a splicing switch to include exon 8 (IIIb) (see Fig. 1). To examine this possibility, we performed semiquantitative reverse transcription–PCR on RNA isolated from cerebral cortex, a tissue that predominantly expresses the IIIc isoforms of FgfRs 1–3 (45), only to discover significant up-regulation of FgfR2-IIIb in hemizygote brain (Fig. 5A). DNA sequence analysis of the PCR products also revealed an additional faint band (Fig. 5A, asterisk) that derives from RNA with an exon 7 (IIIa) to 10 (TM) splice. We would not expect this RNA to make a significant contribution to the phenotype because it would encode a product which, because of a translational termination in TM caused by a change in the reading frame, would lack TM and kinase domains as well as part of the ligand binding domain. Ectopic expression of FgfR2-IIIb at sites of FgfR2-IIIc expression would, however, make FgfR2-IIIc-expressing cells responsive to a much greater number of Fgfs. We were particularly interested to determine whether abnormal FgfR2-IIIb expression is implicated in the premature ossification of the coronal sutures and zygomatic arch, as well as the abnormal fusion of sternebrae. For this analysis, dissected calvaria and rib cages of E17.5 to E18.5 fetuses were subjected to whole mount in situ hybridization using FgfR2 TK and probes specific for FgfR2 exons IIIb or IIIc, although the IIIc probe did not work well in our hands. At E17.5, wild-type mice showed FgfR2 staining in the osteogenic fronts of the coronal sutures, the anterior rims of the frontal bones, the presumptive metopic suture, and the rims of the nasal bones (Fig. 5B). Staining was also seen at the periphery of the zygomatic arch bones (Fig. 5 C and D). The IIIb-specific probe gave a similar but very weak signal (data not shown), suggesting that TK expression also represents the normal pattern of IIIc expression. Surprisingly, at E18.5 heterozygous mice showed a much stronger staining for FgfR2-IIIb at the calvarial sutures, a stain that was still barely detectable in sutures of wild-type mice (Fig. 5E). Because FgfR2-IIIb signaling relies predominantly on Fgf-3, Fgf-7, and more importantly, Fgf-10, we used antisense probes to examine expression of each of these ligands in dissected calvaria. Fgf-10, but not Fgf-3 or Fgf-7, was detected in the suture region, providing strong evidence for a potential source of activating ligand for cells expressing elevated FgfR2-IIIb (Fig. 5F). Interestingly, Fgf-10 expression was also detected in lacrimal glands of wild types but not hemizygotes (Fig. 5 G and H).
A similar set of experiments on dissected sternums also showed a correlation between the abnormalities observed and the pattern of FgfR2-IIIb expression (Fig. 5 I and J). FgfR2-IIIb was found to be normally expressed at the periphery of the sternebrae, but with the incomplete medial fusion which appeared to create two independent ossification centers, FgfR2-IIIb was also expressed at the midline in hemizygotes. The TK probe gave a similar pattern of expression, and Fgf-10 was also found to be expressed in the periosteal regions of both wild types and hemizygotes (Fig. 5M). These findings indicate that FgfR2-IIIb signaling is involved in normal endochondrial bone formation in the sternum, and that in the mutant mice a more extensive expression pattern causes excessive ossification in the intersternebral cartilage.
Discussion
A number of syndromic craniosynostoses have been associated with dominantly acting mutations of FGFR1, FGFR2, and FGFR3 (reviewed in refs. 27–30). Most frequently, these mutations occur in FGFR2, either in exon IIIc or the region linking the second and third Ig loops (see Fig. 1). Biochemical analysis shows that these mutations can result in ligand-independent receptor activation, increased ligand receptor binding times, or a broadening of receptor–ligand binding specificity (29, 46, 47). Expression pattern analysis for FgfRs 1–3 in rodent calvaria suggests a possible role for FgfR2-IIIc in maintaining sutural cell proliferation, and paradoxically, the potential to promote osteogenic differentiation (48–52). In favor of the latter property, experimental application of Fgf2 or Fgf4, both ligands for FgfR2-IIIc, causes premature differentiation of such cells and suture fusion (49, 52). Similarly, ectopic expression of Fgf3 and Fgf4 in a mutant mouse that develops bulgy eyes also results in craniosynostosis (53). Collectively, these observations suggest that elevated FgfR2 signaling is the underlying cause of precocious ossification of sutural cells.
Here we confirm FgfR2 expression in the calvarial sutures. We show that coronal sutures are the first to express this receptor, and that FgfR2 is additionally expressed in zygomatic arch joints. More importantly, we demonstrate a mechanism for elevated FgfR2 signaling as the underlying cause of synostosis. Thus, through a splice-switch mechanism, FgfR2-IIIb expression is substantially elevated in calvarial sutures and zygomatic joints of FgfR2-IIIc+/Δ hemizygotes, rendering cells that would predominantly express FgfR2-IIIc responsive to a broader set of Fgf ligands. We showed that Fgf-10, a ligand for FgfR2-IIIb, is normally expressed in the suture region, strongly suggesting that such cells become aberrantly activated, which leads to abnormal levels of FgfR2 signaling and their premature differentiation.
In addition to synostosis of coronal and zygomatic sutures, FgfR2-IIIc+/Δ mice also show abnormal endochondrial ossification of the sternum. It is not clear whether this abnormality is caused by inappropriate FgfR2 expression in the sternebrae, or is secondary to the delayed medial fusion and rib misalignment, causing additional centers of ossification, perhaps because of a perturbation of the proposed negative signaling from the ribs. Nonetheless, our observations suggest that FgfR2 signaling is involved in normal sternum development, and, therefore, may play a role in both endochondrial and membranous bone formation.
In Apert's and Pfeiffer's syndromes, craniosynostosis is often accompanied by syndactyly of fore and hind limbs or broader thumbs and great toes, respectively (30, 31, 54, 55). Limb and sternum are both derived from lateral plate mesoderm, and it is interesting that although FgfR2-IIIc+/Δ mice do not show limb defects, they do manifest precocious sternal thickening and fusion. Species differences may explain this phenomenon because a similar relationship was reported in glypican-3 knockouts. Here, a mutation in the human gene causes limb polydactyly, whereas glypican-3-deficient mice develop sternal defects (56).
A significant number of patients with Apert's and Pfeiffer's syndromes also show visceral and growth abnormalities (32, 54), further strengthening the phenotypic resemblance of these human syndromes to FgfR2-IIIc+/Δ mice. Although missense mutations are most frequently associated with Apert and Pfeiffer patients, a point mutation and Alu insertions close to or within exon 9 (IIIc) have been shown to affect FgfR2 splicing. These latter mutations cause inappropriate expression of FgfR2-IIIb in cultured fibroblasts of the patients, and this has been suggested as an underlying cause of limb defects (55, 57). It is possible that ectopic expression of FgfR2-IIIb in visceral mesenchyme disrupts the normal mesenchymal–epithelial interactions by, for example, sequestering Fgf7 and Fgf10, normally produced by mesenchymal cells to act instructively on the adjacent epithelial cells. Alternatively, FgfR2-IIIb up-regulation could perturb the normal program of mesenchymal cell differentiation. The observation that, in hemizygotes, kidney and lung mesenchyme are disorganized, and mesenchymal cells in the rudimentary lacrimal glands seem not to express Fgf-10, would be consistent with this possibility. However, the observed visceral defects do not obviously match those reported for mice deficient for Fgf-10, Fgf-7 (19, 21, 58), or FgfR2-IIIb (25), suggesting that the visceral phenotypes arise through a more complex set of interactions governing mostly secondary branching. Nevertheless, it is interesting that FgfR2-IIIb is up-regulated in cultured fibroblasts of some Apert's and Pfeiffer's syndrome patients (54, 55, 57). Hence, the mice described in this report could serve as an attractive model for further investigations of the molecular mechanisms governing normal bone growth, as well as mechanisms underlying craniosynostosis and the associated visceral defects in some Apert's and Pfeiffer's syndromes.
Acknowledgments
We thank Dr. Anton Burns (Netherland Cancer Institute) for donation of the ZP3-Cre colony and the staff at the Imperial Cancer Research Fund's transgenic unit for their assistance with the generation and breeding of the described transgenic mice. We also thank Profs. Giorgio Gabella, Gordon Stamp, and Adrian Woolf, and Dr. Paul Winyard for help and advice. We thank Dr. David Ish-Horowitz (Imperial Cancer Research Fund, London) and Prof. Gillian Morriss-Kay (Oxford University) for critically reading the manuscript. M.K.H. is supported by an Imperial Cancer Research Fund fellowship. This work was also supported by a grant from the Human Frontier Science Program (to C.D.).
Abbreviations
- Fgf
fibroblast growth factor
- FgfR
Fgf receptor
- TM
transmembrane
- PF
paraformaldehyde
- En
embryonic day n
- Pn
postnatal day n
- TK
thymidine kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 3641.
References
- 1.McKeehan W L, Wang F, Kan M. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 2.Ornitz D M. BioEssays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura T, Nakatake Y, Konishi M, Itoh N. Biochem Biophys Acta. 2000;1492:203–206. doi: 10.1016/s0167-4781(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita T, Yoshioka M, Itoh N. Biochem Biophys Res Commun. 2000;277:494–498. doi: 10.1006/bbrc.2000.3696. [DOI] [PubMed] [Google Scholar]
- 5.Ornitz D M, Xu J S, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G X, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 6.Peters K, Werner S, Chen G, Williams L. Development (Cambridge, UK) 1992;114:233–243. doi: 10.1242/dev.114.1.233. [DOI] [PubMed] [Google Scholar]
- 7.Orr-Urtreger A, Bedford M, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 8.Basilico C, Moscatelli D. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi T P, Rossant J. Curr Opin Genet Dev. 1995;5:485–491. doi: 10.1016/0959-437x(95)90053-j. [DOI] [PubMed] [Google Scholar]
- 10.Martin G. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 11.Deng C X, Wynshawboris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi T P, Harpal K, Henkemeyer M, Rossant J. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 13.Feldman B, Poueymirou W, Papaioannou V E, Dechiara T M, Goldfarb M. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 14.Colvin J S, Bohne B A, Harding G W, McEwen D G, Ornitz D M. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 15.Deng C, Wynshaw B A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 16.Arman E, Haffnerkrausz R, Chen Y, Heath J K, Lonai P. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dono R, Texido G, Dussel R, Ehmke H, Zeller R. EMBO J. 1998;17:4213–4225. doi: 10.1093/emboj/17.15.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyers E N, Lewandoski M, Martin G R. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 19.Min H, Danilenko D, Scully S, Bolon B, Ring B, Tarpley J, DeRose M, Simonet W. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortega S, Ittmann M, Tsang S H, Ehrlich M, Basilico C. Proc Natl Acad Sci USA. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Meyers E N, Lewandoski M, Martin G R. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arman E, Haffnerkrausz R, Gorivodsky M, Lonai P. Proc Natl Acad Sci USA. 1999;96:11895–11899. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X L, Weinstein M, Li C L, Naski M, Cohen R I, Ornitz D M, Leder P, Deng C X. Development (Cambridge, UK) 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 25.DeMoerlooze L, SpencerDene B, Revest J M, Hajihosseini M, Rosewell I, Dickson C. Development (Cambridge, UK) 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 26.Celli G, Larochelle W J, MacKem S, Sharp R, Merlino G. EMBO J. 1998;17:1642–1655. doi: 10.1093/emboj/17.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeMoerlooze L, Dickson C. Curr Opin Genet Dev. 1997;7:378–385. doi: 10.1016/s0959-437x(97)80152-9. [DOI] [PubMed] [Google Scholar]
- 28.Jabs E. Clin Genet. 1998;53:79–86. doi: 10.1111/j.1399-0004.1998.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 29.Naski M, Ornitz D. Front Biosci. 1998;3:781–794. doi: 10.2741/a321. [DOI] [PubMed] [Google Scholar]
- 30.Wilkie A O. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- 31.Wilkie A O, Slaney S F, Oldridge M, Poole M D, Ashworth G J, Hockley A D, Hayward R D, David D J, Pulleyn L J, Rutland P. Nat Genet. 1995;9:165–172. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- 32.Cohen M J, Kreiborg S. Am J Med Genet. 1993;45:758–760. doi: 10.1002/ajmg.1320450618. [DOI] [PubMed] [Google Scholar]
- 33.Park W J, Theda C, Maestri N E, Meyers G A, Fryburg J S, Dufresne C, Cohen M M, Jabs E W. Am J Hum Genet. 1995;57:321–328. [PMC free article] [PubMed] [Google Scholar]
- 34.Lewandoski M, Wassarman K, Martin G. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 35.McLeod M. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 36.Drury R, Wellington E. Carlton's Histological Technique. New York: Oxford Univ. Press; 1967. [Google Scholar]
- 37.Chen J. J Anat Lond. 1952;86:373–386. [PMC free article] [PubMed] [Google Scholar]
- 38.Braun T, Rudnicki M, Arnold H-H, Jaenisch R. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- 39.Tsuyoshi T, Moribe H, Kondoh H, Higashi Y. Development (Cambridge, UK) 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 40.Dudley A, Godin R, Robertson E. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogan B. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 42.Markarenkova H, Ito M, Govindarajan V, Faber S, Sun L, McMahon G, Overbeek P, Lang R. Development (Cambridge, UK) 2000;127:2563–2572. doi: 10.1242/dev.127.12.2563. [DOI] [PubMed] [Google Scholar]
- 43.Motoyama J, Liu J, Ding Q, Post M, Hui C-C. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- 44.Weaver M, Dunn N, Hogan B. Development (Cambridge, UK) 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 45.Hajihosseini M K, Dickson C. Mol Cell Neurosci. 1999;14:468–485. doi: 10.1006/mcne.1999.0800. [DOI] [PubMed] [Google Scholar]
- 46.Anderson J, Burns H, Enriquez-Harris P, Wilkie A O, Heath J. Hum Mol Genet. 1998;7:1475–1483. doi: 10.1093/hmg/7.9.1475. [DOI] [PubMed] [Google Scholar]
- 47.Yu K, Herr A B, Waksman G, Ornitz D M. Proc Natl Acad Sci USA. 2000;97:14536–14541. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iseki S, Wilkie A O, Heath J K, Ishimaru T, Eto K, Morriss-Kay G M. Development (Cambridge, UK) 1997;124:3375–3384. doi: 10.1242/dev.124.17.3375. [DOI] [PubMed] [Google Scholar]
- 49.Iseki S, Wilkie A O, Morriss-Kay G M. Development (Cambridge, UK) 1999;126:5611–5620. doi: 10.1242/dev.126.24.5611. [DOI] [PubMed] [Google Scholar]
- 50.Johnson D, Iseki S, Wilkie A O, Morriss-Kay G M. Mech Dev. 2000;91:341–345. doi: 10.1016/s0925-4773(99)00278-6. [DOI] [PubMed] [Google Scholar]
- 51.Rice D, Aberg T, Chan Y-S, Tang Z, Kettunen P, Pakarinen L, Maxson R, Jr, Thesleff I. Development (Cambridge, UK) 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- 52.Kim H J, Rice D P C, Kettunen P J, Thesleff I. Development (Cambridge, UK) 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- 53.Carlton M, Colledge W, Evans M. Dev Dyn. 1998;212:242–249. doi: 10.1002/(SICI)1097-0177(199806)212:2<242::AID-AJA8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 54.Plomp A, Hamel B C, Cobben J, Verloes A, Offermans J, Lajeunie E, Fryns J, de Die-Smulders C. Am J Med Genet. 1998;75:245–251. [PubMed] [Google Scholar]
- 55.Tsukuno M, Suzuki H, Eto Y. J Craniofacial Genet Dev Biol. 1999;19:183–188. [PubMed] [Google Scholar]
- 56.Paine-Saunders S, Viviano B, Zupicich J, Skarnes W, Saunders S. Dev Biol. 2000;225:179–187. doi: 10.1006/dbio.2000.9831. [DOI] [PubMed] [Google Scholar]
- 57.Oldridge M, Zackai E H, McDonald-McGinn D M, Iseki S, Morriss-Kay G M, Twigg S R, Johnson D, Wall S A, Jiang W, Theda C, et al. Am J Hum Genet. 1999;64:446–461. doi: 10.1086/302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L, Degenstein L, Fuchs E. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]