Abstract
The conformational and aggregation behavior of PEG conjugates of an alanine-rich polypeptide (PEG-c17H6) were investigated and compared to that of the polypeptide equipped with a deca-histidine tag (17H6). These polypeptides serve as simple and stimuli-responsive models for the aggregation behavior of helix-rich proteins, as our previous studies have shown that the helical 17H6 self-associates at acidic pH and converts to β-sheet structures at elevated temperature under acidic conditions. In the work here, we show that PEG-c17H6 also adopts a helical structure at ambient/sub-ambient temperatures, at both neutral and acidic pH. The thermal denaturation behavior of 17H6 and PEG-c17H6 is similar at neutral pH, where the alanine-rich domain has no self-association tendency. At acidic pH and elevated temperature, however, PEGylation slows β-sheet formation of c17H6, and reduces the apparent cooperativity of thermally induced unfolding. Transmission electron microscopy of PEG-c17H6 conjugates incubated at elevated temperatures showed fibrils with widths of ~20-30 nm, wider than those observed for fibrils of 17H6. These results suggest that PEGylation reduces β-sheet aggregation in these polypeptides by interfering, only after unfolding of the native helical structure, with inter-protein conformational changes needed to form β-sheet aggregates.
Keywords: PEGylation, polypeptide, thermal denaturation, aggregation, fibril
INTRODUCTION
Advances in molecular biology and protein expression have expanded the use of protein therapeutics in the pharmaceutical industry, as well as provided numerous opportunities for the use of engineered polypeptides for a range of materials science applications. Protein therapeutics often exhibit unique, highly specific actions and fewer side effects compared to their small molecular-weight counterparts, particularly for treatment of metabolic and immunological disorders.1-4 However, a ubiquitous challenge to practical use of foldable proteins and polypeptides is preserving their functions by avoiding irreversible conformational changes during their production, processing, storage, and ultimate application.5-12 Proteins ideally have the most stable conformations in their native or folded state due to the balance between configurational entropy, solvation free energy, and energetic contributions such as hydrogen bonding, van der Waals attractions, and repulsive or attractive electrostatic interactions.6, 11, 13 In practice, aggregates composed of unfolded or misfolded proteins and polypeptides are often thermodynamically favored at finite protein concentrations; however their formation is kinetically limited, due to a need to significantly populate partially or fully unfolded monomeric intermediates.14 Exposure of proteins to harsh chemical environments, or to elevated temperature, pressure, or protein concentrations, as well as the creation of mutations, can trigger the formation of such aggregation-prone intermediate species, as observed both in vitro and in vivo.14-17 Given sufficient time, aggregates often form even under mild storage conditions.18 Thus, stabilization of the native state, and/or destabilization of the interactions between the partially unfolded intermediates, are usual options to reduce protein aggregation.19 Multiple approaches, including post-translational modifications such as glycosylation,20, 21 as well as the use of chaperones,22-24 stabilizing agents,25-30 and stabilizing fusions31-34 have been proposed to reduce protein aggregation.
In many cases, irreversible aggregates are stabilized by inter-protein β-sheets,8, 18 including proteins that are devoid of β-sheet structures, such as helical proteins, in their native state.7, 35 In addition to their use here as models for helical proteins, helical polypeptides also are of interest for use as scaffolds to present ligands with desired spatial arrangements for biotechnology and materials applications.36-41 Fundamentally, the observation that proteins without native β-sheet propensity form non-native, β-sheet rich aggregates raises questions of how primarily helical polypeptides “find” their way to β-sheet aggregates, as well as how to design them to resist aggregation without jeopardizing the helical fold.
We have previously synthesized multiple families of recombinant, glutamic acid-containing, alanine-rich helical polypeptides as scaffolds to study multivalent interactions.36-41 Given the potential role of these alanine-rich sequences as simpler models of aggregation-prone proteins, we have previously studied in detail the association behavior of one of these polypeptides (17H6, with the sequence given in Table 1). We have shown in these previous studies that the polypeptide exhibits polymorphological behavior, and self-associates to form folded, afibrillar aggregates with an average hydrodynamic radius of ~10-20 nm at acidic pH, ambient temperature, and isotonic conditions. Furthermore, 17H6 forms β-sheet fibrillar aggregates upon an increase in temperature under those solution conditions;42 qualitatively similar fibril-formation behavior has been observed in disease-related and therapeutic proteins such as α-synuclein,16 and insulin.43 Interestingly, the folded, afibrillar aggregate appears to be an on-pathway intermediate for fibril formation. Detailed characterization of the self-assembly of the fibrils indicated that they have an average width of ~7-8 nm and likely a cross-β structure; a particularly useful feature of the assembly is the periodic presentation of positively-charged patches along the fibril at sub-ambient pH (owing to the N-terminal histidine fusion tag), which has in turn permitted the assembly of 1D nanoparticle arrays.44
Table 1.
Amino acid sequences and molecular masses of the polypeptides and the conjugates.
| Symbol | Amino acid sequence | Mtheo (kDa) |
Mexp* (kDa) |
|---|---|---|---|
| 17H6 | MGH10SSGHIHM(AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGMG | 14.8 | 14.8 |
| c17H6 | (AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGS/S-lactone | 12.4 | 12.3 |
| PEG5K-c17H6 | (mPEG5K)-(AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGS/S-lactone | 17.4 | 17.5 |
| PEG10K-c17H6 | (mPEG10K)-(AAAQEAAAAQAAAQAEAAQAAQ)6AGGYGGS/S-lactone | 22.4 | 22.0 |
Peak maximum value in MALDI-TOF spectroscopy
Interestingly, cleavage of the deca-histidine tag (by cyanogen bromide (CNBr) treatment) renders the polypeptide insoluble at pH conditions below the pKa for Glu, suggesting the role of the deca-histidine tag as a solubilizing agent. Recent studies have shown that disordered flanks in some proteins,45 the acidic tail in α-synuclein (ATS) derived peptides,31-33 and late embryogenesis abundant (LEA) protein fusion34 can prevent aggregation of peptides and proteins. Similarly, conjugation of poly(ethylene glycol) to proteins has been shown to increase solution stability of hydrophobic proteins,46-48 and improve thermal stability of enzymes.49-56 Moreover, superior to the fusion tags of biological origin, PEG conjugation has also been demonstrated to increase serum half-life and reduce immunogenicity of administered proteins.57-59 PEGylation, on the other hand, may result in considerable loss of function if it disrupts the protein fold or hinders the active site. The reduction of HER-2 binding affinity of monoPEGylated scFv has been reported and attributed to both intramolecular and intermolecular blocking by PEG chains (20 kDa).60 Similarly, steric exclusion by PEG molecules of an antigen-antibody interaction has been demonstrated with increasing PEG chain length (>10 kDa).61 In the case of PEG-polyalanine conjugates (with polyalanine molecular weight ~ 0.7 kDa), self assembled fibrillar nanostructures with β-sheet conformation have been observed to switch to α-helical micellar structures with increasing PEG chain length from 1 kDa to 5 kDa.62 Conformational changes have also been reported for the other PEG containing amphiphilic63 or coiled coil64, 65 peptide hybrid systems due to the steric interference of PEG chains. Thus, it is possible that steric interference by PEG molecules could disrupt the ability of a polypeptide such as 17H6 to reversibly associate, which could disrupt fibril formation. It is also possible that PEG attachment could destabilize the native structure of the polypeptide, triggering non-native aggregation, given that the folding free energy of these polypeptides is not as high, nor the folding as cooperative, compared to larger proteins. Despite these possibilities, relatively few systematic, mechanistic studies focused on the impact of PEGylation on the self-association, unfolding, and aggregation behavior of proteins have been reported.66-69
Given the polyconformational and polymorphological behavior of 17H6, in this study we evaluated the effect of PEGylation on the conformational, aggregation and fibril formation behavior of this aggregation-prone helical alanine-rich domain, as a model system for the aggregation behavior of helix-rich proteins. The deca-histidine tag was removed and the cleaved polypeptide (alanine-rich domain) was modified with PEGs of two different molecular masses (5 kDa and 10 kDa with respective Rg values70 of ~2.1 nm and ~3 nm when untethered). The conformational stability of the conjugates was probed via circular dichroic spectroscopy, and their thermal transitions were monitored via differential scanning calorimetry. Their tendency toward fiber formation was characterized via transmission electron microscopy. While PEGylation does not abolish the folded self-association of the helical domain of the polypeptide, our data suggest it does improve resistance of the polypeptide toward subsequent, non-native β-sheet and fibril formation.
MATERIALS AND METHODS
Materials
The monodisperse polypeptide, 17H6, (Table 1) was expressed using an E. coli expression host, BL21(DE3)pLysS, and purified using nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography by employing pH and/or imidazole gradients as given elsewhere.38, 39 Propionaldehyde-functionalized mPEGs (polydispersity indices < 1.05) with molecular weights of 5 kDa and 10 kDa were purchased from JenKem Technology USA Inc. (Allen, TX) and Nanocs Inc. (New York, NY) respectively. Reagent grade cyanogen bromide (CNBr) and sodium cyanoborohydride (NaBH3CN) were obtained from Sigma-Aldrich. Citrate buffer (50 mM) at pH 6.0 was prepared using sodium citrate (Sigma) and citric acid (Sigma). Phosphate buffers at pH 2.3 were prepared using 10 mM o-phosphoric acid (Fisher) and salt (140 mM sodium chloride (NaCl) and 10 mM potassium chloride (KCl) (Fisher)). Phosphate buffered saline (PBS) at pH 7.4 was purchased from Thermo Scientific Inc. (Rockford, IL).
Synthesis of the conjugates
The histidine tag of 17H6 was cleaved via the cyanogen bromide (CNBr) reaction. 100 mg (~6.8 μmol) polypeptide was dissolved in 20 ml of ~62% (~16.1 M) formic acid or ~70% (~9.1 M) trifluoroacetic acid, and 500 mg (~4.7 mmol) CNBr (was added and allowed to react at room temperature for ~1 ½ days. CNBr and the cleaved histidine tag were removed using a concentrator (Pierce MWCO 9000) or a dialysis tube (ThermoFisher Snake Skin MWCO 10000) after diluting the reaction mixture with deionized water. The dialyzed/concentrated solution was freeze-dried to yield the cleaved polypeptide, c17H6 (Table 1). The yield of the purified c17H6 was obtained to be ~70 ± 10% depending on the purification protocol.
Propionaldehyde-functionalized PEG (5 kDa or 10 kDa) was conjugated to the N-terminus of c17H6 via Schiff base formation and subsequent reduction (Figure S1 (in the Supporting Information)). 150 mg (~30 μmol) mPEG5K-propionaldehyde or 300 mg (~30 μmol) mPEG10K-propionaldehyde was added to a solution of 50 mg (~4 μmol) of c17H6 (in 12.5 ml of citrate buffer containing 20 mM NaBH3CN at pH 6.0). The reaction mixture was stirred at room temperature for ~1 day. Unreacted PEG was removed via anion exchange chromatography with a HiTrap diethylaminoethane (DEAE) sepharose fast flow column (GE Healthcare Inc., Piscataway, NJ). 20 mM Tris at pH 8.0 and 20 mM Tris and 1 N NaCl at pH 8.0 were used as starting and elution buffers respectively for the DEAE column separations. MonoPEGylated conjugates, denoted as PEG5K-c17H6 and PEG10K-c17H6 (Table 1), were obtained using size exclusion chromatography with a HiPrep 16/60 Sephacryl S-100 HR model size exclusion column (GE Healthcare Inc., Piscataway, NJ). 50 mM sodium phosphate and 150 mM NaCl buffer at pH 7.2 was used as the mobile phase in SEC separations. Both anion exchange and size exclusion chromatography separations were performed on a fast protein liquid chromatography (FPLC) system (AKTA Explorer 10 FPLC, Amersham, Piscataway, NJ). The fractions obtained in SEC separations were dialysed against deionized water and the conjugate solutions in deionized water were freeze dried. The yield of the conjugates was obtained to be ~45 ± 10%. The production of monoPEGylated conjugates was confirmed by gel permeation chromatography (GPC) (Figure S2) and matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectroscopy (Table 1). Sequences and molar masses of the polypeptides and the conjugates (PEG5K- and PEG10K-17H6, indicating the molecular weight of the PEG) are given in Table 1.
Characterization methods
Circular dichroism (CD) experiments were conducted using a Jasco (Easton, MD) J-810 spectropolarimeter with a Jasco PTC-424S Peltier temperature controller. Temperature induced spectral changes were monitored from 5°C to 80°C in increments of 5°C with a temperature increment rate of 1°C/min. Prior to refolding experiments, each sample was further incubated at 80°C for 3 h. Refolding spectra were collected during cooling from 80°C to 5°C with the same increments as unfolding experiments and 1°C/min cooling rate. CD kinetic experiments were performed at 80°C for 18h followed by the refolding experiments as desribed above. All the spectra were taken between 250 nm and 190 nm at a rate of 50 nm/min and were corrected by subtraction of the corresponding buffer spectrum. Mean residue ellipticity values, [Θ]MRE (deg cm2 dmol−1) were obtained using concentration, and number of amino acids of the polypeptide and the cell pathlength.71 Helical content was estimated by the equation given in Kennedy et al. (2002).72 Thermal melting curves were constructed by recording ellipticity at 222 nm, Θ222, every 1°C during steady heating at a rate of 1°C/min. Consecutive melting curves were obtained using the same temperature range, incubation time, heating, and cooling rate as those employed in differential scanning calorimetry (DSC) experiments.
DSC experiments were carried out using a VP-DSC (Microcal, Northhampton, MA). DSC curves were taken between 5°C and 85°C at a heating rate of 1°C/min. For the consecutive DSC scans, the sample was cooled back to 5°C after reaching to 85°C (without incubation at 85°C) with a cooling rate of 1°C/min. Then, the sample was incubated at 5°C for 15 min and heated again using the same parameters of the previous heating scan.
Transmission electron microscopy (TEM) of the samples was performed using a Tecnai G2 12 TEM (FEI Company, Hillsboro, OR) at an acceleration voltage of 120 keV. The samples were stained using uranyl acetate as described in a protocol given elsewhere.42
RESULTS AND DISCUSSION
Conformation and thermal denaturation behavior at neutral pH
In the first part of this study, the effect of PEGylation on the conformational behavior of the alanine-rich polypeptide (c17H6, indicating cleavage of the deca-histidine tag from the 17H6) was investigated for comparison to our previous studies of the uncleaved polypeptide. The uncleaved 17H6 has an α-helical conformation at low and ambient temperatures, independent of pH. It was also shown that the polypeptide forms monomeric species at neutral pH, while it spontaneously forms aggregates of folded monomers at acidic pH, triggered by the protonation of glutamic acid residues. We also showed that 17H6 exhibited pH-dependent thermal denaturation behavior,42 suggesting the impact of intermolecular association on the denaturation of the polypeptide. Therefore, we investigated the conformational and thermal denaturation behavior of the PEGylated samples both at neutral and acidic pH.
Temperature-induced conformational changes were monitored using CD spectroscopy. The CD spectra of the cleaved polypeptide and the conjugates at pH 7.4 are given in Figures S3, S4 and S5 for c17H6, PEG5K-c17H6, and PEG10K-c17H6, respectively. At low temperatures, the CD spectra of c17H6 and both conjugates exhibited minima at 208 and 222 nm, characteristic of the α-helical conformation; the spectra of the conjugates were essentially identical to that of the parent molecule, 17H6. The helicity values of c17H6 and the conjugates at 5°C, estimated using mean residue ellipticity values at 222 nm,72 were approximately 62-65%. There were no significant differences in the conformation and helicity of the parent polypeptide and the conjugates at neutral pH and 5°C (Table 2). These observations are consistent with previous studies describing non-specific PEG attachment to the surface amines of insulin,67 antibody fragments,73 and human serum albumin,74 in which PEGylation has been reported to have no measurable impact on the conformation of the parent peptides/proteins. On the contrary, in some cases PEG conjugation is deleterious to the secondary structure as observed in the conjugation of 5 kDa PEG to multiple lysines in cytochrome c, as suggested by changes in the CD spectrum of the protein. However, the reduction of α-helical content in that case did not result in any reported activity loss.53
Table 2.
[Θ]MRE,222 and % helicity values before unfolding and after refolding.
| Sample | pH | unfolding [Θ]MRE,222 ×10−3 (mdeg cm2 dmol−1) |
average % helicity prior to unfolding |
refolding [Θ]MRE,222 ×10−3 (mdeg cm2 dmol−1) |
average % helicity after refolding |
|---|---|---|---|---|---|
| 17H6 | 7.4 | −36.0 ± 0.1 | 60 ± 1 | −37.2 ± 1.7 | 62 ± 3 |
| c17H6 | 7.4 | −37.0 ± 0.5 | 62 ± 1 | −37.7 ± 0.5 | 63 ± 1 |
| PEG5K-c17H6 | 7.4 | −39.0 ± 2.6 | 65 ± 4 | −39.7 ± 2.7 | 66 ± 5 |
| PEG10K-c17H6 | 7.4 | −38.8 ± 0.3 | 65 ± 1 | −39.9 ± 0.2 | 67 ± 1 |
| 17H6 | 2.3 | −39.3 ± 0.7 | 65 ± 1 | −29.7 ± 0.3 | 50 ± 1 |
| PEG5K-c17H6 | 2.3 | −39.1 ± 2.5 | 65 ± 4 | −38.7 ± 2.4 | 65 ± 4 |
| PEG10K-c17H6 | 2.3 | −38.4 ± 2.5 | 64 ± 4 | −38.1 ± 2.5 | 64 ± 4 |
In order to assess the relative stabilities of the polypeptide and its modified forms, we carried out thermal denaturation experiments using CD spectroscopy and DSC. As shown in the CD spectra in Figures S3, S4 and S5, the polypeptide and the conjugates all exhibited a temperature-induced transition between the α-helical and non-helical conformation, with recovery of the α-helical structure upon cooling. The essentially complete recovery of the [Θ]MRE,222 by all of the samples upon cooling indicates complete recovery of conformational changes at neutral pH (Table 2). CD melting curves (recorded at 222 nm) and DSC thermograms of the polypeptides and the conjugates are given in Figure S6 and S7, respectively. No significant changes in the melting curves, and in the apparent melting temperature (Tm) were observed, with the peptide and both conjugates showing an apparent melting temperature of 36 ± 1°C. Tm was taken as the temperature at which the normalized ellipticity was 0.5. In the DSC curves, all the samples presented a similarly broad endotherm with a peak maximum of approximately 37 ± 1°C without establishment of clear pre- or post-transition baselines; the latter feature precludes determination of valid calorimetric or van’t Hoff enthalpies from the DSC data, and is not unexpected for unfolding that is multi-state such as for isolated helices.75 The reversibility of the transition was monitored by comparison of consecutive DSC scans (Figures S8, S9, S10). The cleaved polypeptide and the conjugates showed a quantitatively reversible transition as that observed for 17H6, corroborating the CD data. The similarities of the peak maximum values in the DSC curves, the overlay of the melting curves, and the reversibility of the transitions confirm that N-terminal PEGylation had no discernable impact on the conformational behavior, at ambient pH, of the polypeptide domain.
Despite the lack of any conformational impact of PEGylation in the case of 17H6, there are some reports indicating a modest increase in the thermal stability of globular proteins such as trypsin,49, 52, 56 laccase,54 and α-chymotrypsin55, 69 upon modifications of the surface lysines with PEG groups. The increased thermal stability observed in these previous studies was attributed to the reduced structural flexibility of the protein-PEG conjugates, indicated by H/D exchange kinetic studies.55 Therefore, in the relatively simple case of 17H6 and the conjugates, it is likely that the N-terminal conjugation of PEG moieties, as investigated in the current study, does not significantly affect the structural flexibility of the polypeptide.
Conformational and thermal denaturation behavior at acidic pH
Under acidic conditions, the potential role of the positively charged histidine tag or of the PEG is to solubilize the alanine-rich block, which has a tendency to associate via hydrophobic interactions owing to the protonation of glutamic acid residues and the inherent hydrophobicity of Ala side chains. Evaluation of the conformational and thermal denaturation behavior of these macromolecules under acidic conditions may thus give some information about the stability of the associated hydrophobic domains when a relatively hydrophilic or non-associating domain is attached. Temperature-induced conformational changes of PEG5K-c17H6 and PEG10K-c17H6 in pH 2.3 buffer, were monitored via CD spectroscopy; results are presented in Figure 1 and Figure S11, respectively. It was not possible to acquire a spectrum for the c17H6 at pH 2.3, owing to its poor solubility under these conditions. In contrast, the parent polypeptide and its conjugates were soluble at acidic pH, and an α-helical conformation was conserved in all cases. Helicity values were ~65% for all samples (Table 2) and were similar to those presented above for the samples at neutral pH, indicating that there was no significant effect of either the pH shift or the N-terminal tag on the conformation of the polypeptide at low temperature. These results are consistent with those reported for a self-associating coiled-coil peptide76, and a silk-like polypeptide,77 and tetrapeptides78 in which no significant change in conformation was observed upon PEGylation. On the contrary, introduction of PEG chains has resulted in considerable or mild/moderate conformational changes for some amphiphilic62, 63 or coiled coil64, 65 peptides respectively suggesting that the impact of PEG chain on the conformation of the self associating peptide/polypeptide domain is strongly dependent on the nature of the peptide/polypeptide block and PEG chain length.
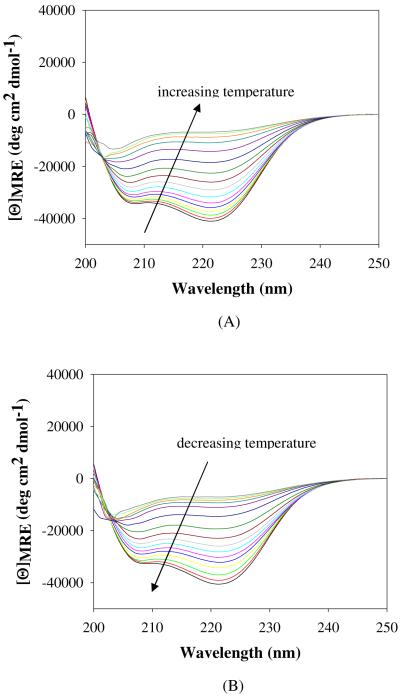
Figure 1.
CD spectra of 50 μM PEG5K-c17H6 in pH 2.3 buffer during (A) unfolding upon increasing temperature from 5°C to 80°C and (B) refolding upon decreasing temperature from 80°C to 5°C subsequent to the incubation at 80°C for 3 h.
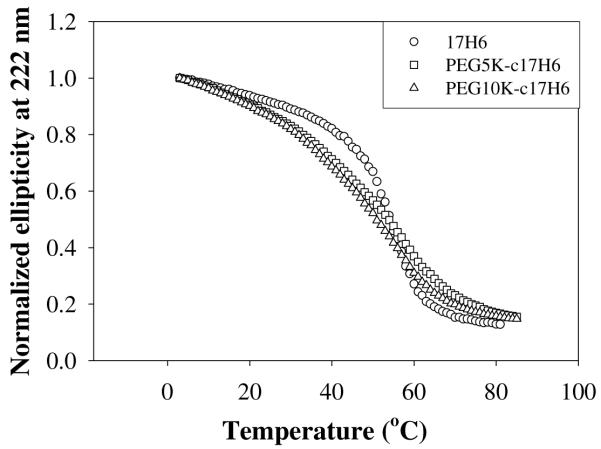
However, a significant difference between the thermal denaturation behavior of 17H6 and the conjugates was revealed by comparison of the extent of refolding after extended heating at high temperatures. The differences in the [Θ]MRE,222 and % helicity values prior to unfolding and after refolding are presented in Table 2. Unlike the polypeptide equipped with the deca-histidine tag, the PEGylated polypeptides showed essentially complete refolding upon cooling after incubation at 80°C for 3 h, with no significant loss compared to their initial α-helical contents. To assess whether PEGylation thermodynamically stabilizes native α-helical structures relative to unfolded monomers, CD melting curves (Figure 2) were compared. Apparent melting temperatures from the CD melting curves were estimated as ~54°C for 17H6 and PEG5K-c17H6 and ~51°C for PEG10K-c17H6 at a concentration of 50 μM. These similar or even reduced apparent melting temperatures of the conjugates, when compared to 17H6, clearly indicate that the high temperature stability of the conjugates is not due to conformational stabilization of the native α-helical conformation upon PEGylation.
Figure 2.
Comparison of CD melting curves of 17H6 (circles), PEG5K-c17H6 (squares), and PEG10K-c17H6 (triangles) at pH 2.3 (C = 50 μM).
Although there is no significant difference in the apparent melting temperatures of the polypeptide and the conjugates, the shapes of the melting curves were observed to be quite different. Initially, as shown in Figure 2, 17H6 unfolds to a lesser extent with increasing temperature than do the conjugates, but then at temperatures above ~45°C, unfolds with a greater temperature sensitivity (i.e., much more “cooperatively”) than do the conjugates. Together, this leads to net mid-point unfolding temperatures that are similar between 17H6 and the conjugates. Given the improved thermal stability of PEGylated enzymes on the basis of hydrogen-bonding contacts in the conjugates,49, 51 as well as the decreased temperature sensitivity of the conformation of amphipilic and coiled coil peptides upon PEGylation,63, 64 it was expected that the conjugates would be more resistant to unfolding, as opposed to the experimental observations. However, as previously shown, the native self-association of 17H6 at low pH provides some of the conformational stability, with higher Tm values for self-associated helix multimers than for the isolated monomer.42 Small-angle neutron scattering experiments (SANS, unpublished data) suggest that 17H6 has a higher aggregation number than the conjugates; the greater number of molecules in the aggregates would provide additional interactions within the aggregate structure to resist unfolding. This presumably is similar in spirit to the behavior of multimeric proteins such as β-glucosidase,79 or transthyretin,80 in which native multimer formation provides significant stabilization of the individual folded chains. Thus, it is likely that the difference in initial aggregation state/compactness of the polypeptide and the conjugates has significant effect on their relative unfolding free energies.
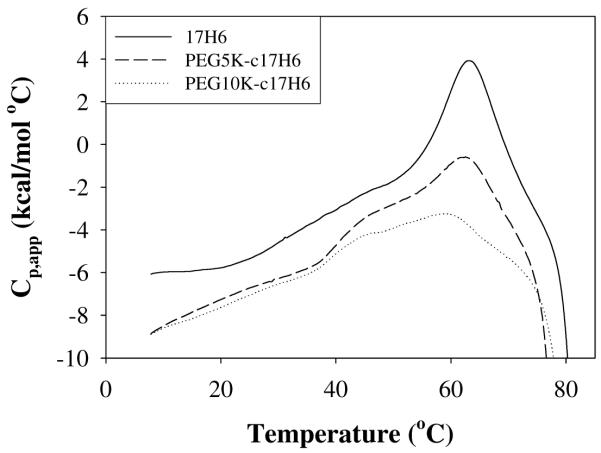
Detailed analysis of the thermal transitions of 17H6 and the conjugates, at acidic pH and concentrations of 100 μM (monomer basis), was conducted via DSC (Figure 3). For 17H6 and the conjugates, the DSC curves show one major endotherm with a low-temperature shoulder, and a high-temperature exotherm consistent with irreversible aggregate formation, confirming that the thermal transition has multiple components, in contrast to the neutral pH case. One major difference in the DSC curves of 17H6 and the conjugates is the relative magnitudes of the endotherms at ~60°C, which may represent a difference in packing/aggregation of the folded aggregate (i.e., low temperature state) and unfolding of the helix, suggesting a correlation of the peak area with the aggregation number determined by SANS. Any interpretation of slight differences in the position of exotherms is complicated by the difficulties in the baseline determination, making conclusions about variations in the onset of aggregation with PEGylation difficult. However, it is clear from these DSC curves that PEGylation does not completely prevent aggregation of the 17H6 at high temperatures. Previous reports of PEG conjugation to aggregation-prone proteins have yielded variable results with respect to aggregation. In most cases, the aggregation propensity of the conjugates was observed to be lowered compared to that of the parent proteins, at least putatively due to steric hindrance by PEG molecules.66-68, 77 However, in a specific case in which PEG is attached to a buried amino acid of granulocyte colony stimulating factor (G-CSF), PEGylated G-CSF exhibited faster aggregation compared to G-CSF, which is likely a result of exposure of some hydrophobic residues.81 In the case of 17H6, N-terminal conjugation with PEG at least slowed the conversion from helical aggregates to β-sheet aggregates (see also the discussion of Figure 5 below).
Figure 3.
Comparison of the first DSC scans of 17H6 (solid line), PEG5K-c17H6 (dashed line), and PEG10K-c17H6 (dotted line) in pH 2.3 buffer (C = 100 μM).
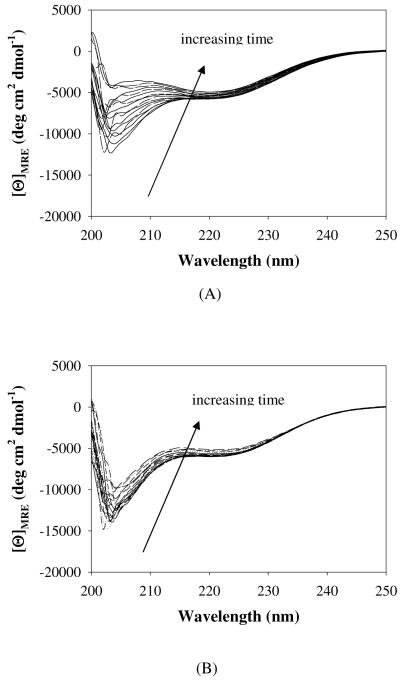
Figure 5.
CD spectra, collected at 1 h intervals upon incubation at 80°C for 18 h, of 50 μM solutions of (A) PEG5K-c17H6, (B) PEG10K-c17H6, in pH 2.3 buffer.
Consecutive DSC scans of the conjugates under acidic conditions were also taken to provide complementary information about the reversibility of the transition obtained from CD data, although at a higher concentration owing to signal-to-noise limitations in the DSC experiment (Figures 4, S12). Consecutive DSC scans of 17H6 indicated a gradual decrease in the peak intensity in subsequent heating-cooling cycles, suggesting irreversible aggregation.42 However, those of the PEGylated samples showed that after the drop of intensity observed between the first and second scans, no changes in the following scans were observed. To check whether the changes in the DSC data are associated with conformational changes in the polypeptide, CD melting experiments at 222 nm were performed under the same conditions (concentrations of 100 μM and same heating/cooling rates). Although it was not possible to collect a full-wavelength spectrum at the higher concentrations, reliable ellipticity values at 222 nm were possible to obtain. The resulting melting curves are presented in Figure S13, showing behavior that is semi-quantitatively similar to that observed in the DSC experiments; 17H6 is conformationally reversible in the first two heating/cooling cycles, but loses reversibility in the third and subsequent cycles (Figure S13A), exhibiting β-sheet conformation at the end of the fifth scan (data not shown). PEGylated samples, on the other hand, experienced no loss of reversibility of the conformational transition (Figure S13B-C), nor any measurable change in the reliable portion of the spectra for these high peptide concentrations (~210-250 nm) after heating (i.e., no loss of the helical conformation after heating). These findings, along with those below, suggest that some portion of the aggregation is reversible in the case of the PEGylated conjugates and irreversible conformational changes are significantly less than those observed for 17H6 upon thermal denaturation and refolding.
Figure 4.
Consecutive DSC scans of PEG5K-c17H6 in pH 2.3 buffer. The first scan is given as dotted line; the other scans are represented as solid line.
β-Sheet tendency and fibril formation behavior at acidic pH
It was previously shown that 17H6 undergoes β-sheet formation and fibrillization upon incubation at low pH and 80°C for prolonged time periods.42 Similar experiments were carried out with PEGylated samples in order to assess the tendency of the conjugates to form β-sheet structures relative to the unmodified polypeptide. CD spectra for samples (50 μM) in pH 2.3 buffer at 80°C were collected at various intervals over a period of 18 h (Figure 5), and then full-wavelength spectra of the samples during cooling were collected at various temperatures from 80°C to 5°C as shown in Figure 6. The PEG5K-c17H6 conjugate ultimately adopted β-sheet structure (CD minimum at ca. 218 nm) upon incubation at 80°C (Figure 5A), with only a small fraction of the initial α-helical structure recovered upon cooling (Figure 6A). PEG10K-c17H6, on the other hand, showed mainly a signal at 205 nm consistent with a non-helical structure (Figure 5B), with this signal only decreasing in intensity over time. In the refolding experiments for PEG10K-c17H6 shown in Figure 6B, it is apparent that the PEG10K-c17H6 conjugate regains a greater amount of helicity than the PEG5K-c17H6, and although the high-temperature spectrum did not show a significant contribution from β-sheet, that some fraction of the original α-helical structure (~30%) was lost for the PEG10K-c17H6 conjugate.
Figure 6.
CD spectra obtained upon cooling of solutions of (A) PEG5K-c17H6, (B) PEG10K-c17H6 from 80°C to 5°C, at 5°C temperature intervals, subsequent to the incubation at 80°C for 18 h.
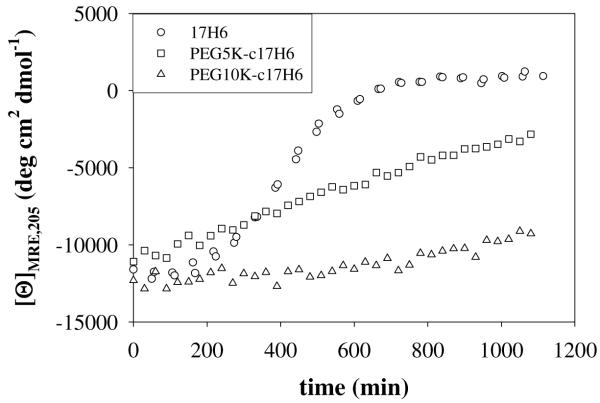
The rate at which the random-coil structure was transformed to β-sheet structure for 17H6 and the conjugates was monitored by evaluating the change in [θ]MRE,205 as a function of time (Figure 7). As shown in Figure 7, 17H6 showed a sigmoidal time course for β-sheet formation whereas PEGylated samples showed mainly linear trends with a slowed rate of β-sheet formation; this effect was more prominent for the 10K PEG. It is apparent from these data that the conjugation of the PEG chain reduces the rate of β-sheet formation of the polypeptide domain. It seems plausible that this is due to predominantly steric hindrance, perhaps via the relatively large PEG end-groups limiting the number of amino acids per 17H6 chain that can be proximal to form hydrogen bonds within the β-sheets, although variations in the hydrophobic-hydrophilic balance of these conjugates may also play a role. A similar effect has been previously observed for proteins modified with PEGs of similar length as those employed here; specifically, longer PEG chains (ca. 2000 or 5000 Da) have been reported to reduce the aggregation rate more efficiently than shorter PEG chains (ca. 750 or 1000 Da) for trypsin,56 and insulin.66, 67 These findings support that the improvement in the apparent thermal stability of the alanine-rich block imparted by the N-terminal PEG segments (compared to stabilization imparted by the deca-histidine tag) is likely due to the stabilization not of the native helical structure (see above), but of the non-helical intermediate structures that form prior to β-sheet aggregation; the PEG likely reduces intermolecular interactions between random coil or early-stage β-sheet structures, but still allows native self-association when Glu side chains are protonated and the molecule is folded.
Figure 7.
Comparison of the change in [θ]MRE,205 over time, with incubation at 80°C, of 17H6 (circles),42 PEG5K-c17H6 (squares), and PEG10K-c17H6 (triangles).
The morphologies of the species that form at high temperature and prolonged incubation were characterized via TEM and are shown in Figure 8 and 9 for PEG5K-c17H6 and PEG10K-c17H6, respectively. The images were acquired after first cooling the samples to room temperature, thus the images represent what remained aggregated after cooling; the DSC data (Figure 4) suggests that much of the sample remains aggregated after cooling for the samples at low pH. Non-uniform and wider fibrils (~20-30 nm width) were observed for both the PEG5K-c17H6 and PEG10K-c17H6 conjugates (Figure 8 and 9) compared to the previously reported widths of the 17H6 fibrils (~7.5 nm width42, 44), but there was no clear dependence of fibril width on PEG chain length. In the case of PEG10K-c17H6, fibrils like those in Figure 8 were present in small amounts, but the samples showed primarily short fibrils and bead-like structures (Figure 9). Similarly, shorter fibrils have been observed for other PEGylated, β-sheet-forming polypeptides; in the case of the PEG-polypeptide-PEG conjugates (for silk-like polypeptides) shorter fibrils were observed with increasing PEG molecular weight, indicating significant interference of the longer PEG chains with the stacking of the β-sheet structures.77 Pronounced morphogenic effects of the PEG chain length (350, 1200, 1800 Da) have also been observed for PEG-hydrophobic tetrapeptide (F4 or V4) conjugates. For PEG-F4 conjugates,78 nanotubes, fibrils, and worm-like micelles have been obtained as the PEG chain increases in length, similar to the behavior of PEG-polyalanine conjugates.62 On the other hand, an increase in PEG chain length increases the population of uniform structures (fibrils) over ill-defined aggregates (plank-like structures) in the case of PEG-V4 hybrids. Hence, the differences in the structures obtained from the self-assembly of PEG-F4 and PEG-V4 conjugates have clearly indicated that the nature of the peptide block is also important in the self-assembly process.78
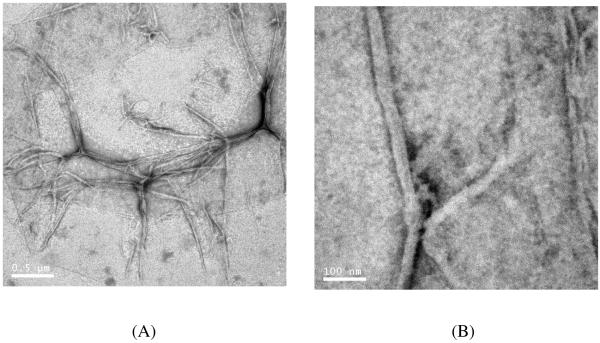
Figure 8.
TEM images of PEG5K-c17H6 at pH 2.3, after incubation at 80°C for 18 h. Scale bars: (A) 500 nm, (B) 100 nm.
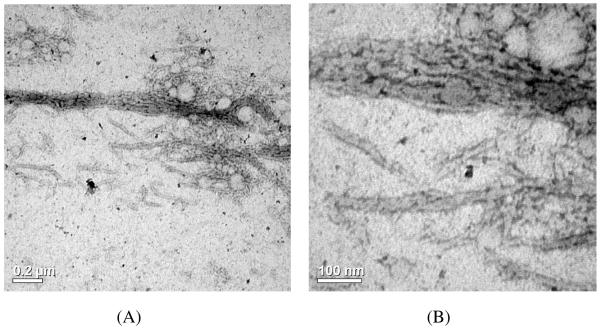
Figure 9.
TEM images of PEG10K-c17H6 at pH 2.3, after incubation at 80°C for 18 h. Scale bars: (A) 200 nm, (B) 100 nm.
Most previous studies on the morphogenic effect of PEG conjugation, on the other hand, have focused on PEG-amyloid peptide conjugates. The major reported impact of PEG on fibril formation has been to inhibit the lateral fibril interactions.63, 82 Previous SANS studies and negative-stained TEM images of PEG-amyloid peptide conjugates have shown that PEG coats the surface of the fibrils.82-84 In cryo-TEM images of an amyloid β-peptide fragment attached to different molecular weights of PEG, no significant difference in the width of the fibrils was observed, in agreement with the core-shell nature of the fibrils.85
In our study, we also observed the lack of impact of PEG chain length on the stained portion of the fibrils, consistent with these previous findings. However, the width of the fibrils formed by the conjuguates is significantly greater than those of the 17H6 fibrils.42, 44 The greater width of the PEGylated polypeptides is not a result of visualization of the PEG fragments, as these polymers are not stained by the negative staining method. In our proposed model of the fibril self-assembly of 17H6, the ~7.5 nm fibril width is ascribed to an anti-parallel, folded β-sheet structure.44 Thus, it is possible that the conjugation of PEG chains of even moderate molecular weight and size (Rg values of ~2-3 nm) interferes with folding of the polypeptide chains during or prior to the fibril formation, allowing longer sections of the chain to adopt intra- (and/or inter-) molecular β-sheet structures during fibrillization, which results in wider fibrils; alternatively, the N-terminal regions of the PEGylated peptides could simply be more difficult to pack tightly within the “spine” of the fibrils, and so could be forced to be more flexible and expanded – thus giving effectively wider fibrils due to the steric constraints of packing the PEG chains along the corona or periphery of the fibrils. More detailed investigations of the fibrillization behavior of conjugates with various length PEGs could provide insight into the contacts made between amino acids during aggregation and fibril formation. Although determination of detailed reasons for the differences between the morphology of the polypeptide 17H6 and its conjugates is beyond the scope of this report, the results clearly show that N-terminal PEGylation does not completely prevent the non-native aggregation product, fibrils, but that as PEG chain length increases, fibril formation is impeded.
CONCLUSIONS
In this study, the effect of PEG conjugation on the folded conformation, thermal denaturation, and fibrillization behavior of an alanine-rich helical polypeptide was investigated. It was observed that N-terminal PEGylation did not change the helicity of the polypeptide at low temperature, independent of solution pH. The thermal stability and denaturation behavior at neutral pH was also unaffected. However, the impact of PEGylation was pronounced at acidic conditions, under which the polypeptide has been demonstrated to self-associate. PEGylation retarded β-sheet formation and fibrillization, and this was attributed to net reduction of attractive interactions between aggregation-prone intermediate unfolded structures, as well as to steric interference with the chain mis- or re-folding that is necessary for conversion to β-sheet fibrils. The conjugation of PEG of higher molecular weight more significantly retarded formation of β-sheet fibrils, consistent with the increased steric interference expected for the larger macromolecule conjugate. It was also observed that PEGylation changed the fibril morphology to some degree, possibly by interfering with the folding of β-sheet structures in the alanine-rich domain. In conclusion, N-terminal PEGylation did not significantly change the native conformational behavior but improved resistance of the pre-fibril, unfolded structures to formation of non-native, β-sheet inter-protein contacts. Our results suggest the utility for these PEGylated polypeptides as useful model systems for the investigation of the aggregation properties of helix-rich proteins with additional application in the control of assembled nanostructures.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by grants from the National Institutes of Health (NIH) and the National Center for Research Resources (NCRR), a component of the NIH (1-P20-RR017716, 1-RO1-EB006006, and 5-P20-015588 (instrument facilities)). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIH. This work was also supported by the Center for Neutron Science at UD under award (U.S. Dept. of Commerce) #70NANB7H6178. TEM resources provided by the W. M. Keck College of Engineering Electron Microscopy Laboratory are acknowledged for the microscopy aspects of this work.
Footnotes
aybentop@iyte.edu.tr, cjr@udel.edu, kiick@udel.edu)
SUPPORTING INFORMATION Synthesis scheme of the conjugates, GPC curves of c17H6 and the conjugates, additional CD and DSC data and TEM images are included. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Pavlou AK, Reichert JM. Nat. Biotechnol. 2004;22:1513–1519. doi: 10.1038/nbt1204-1513. [DOI] [PubMed] [Google Scholar]
- (2).Roque ACA, Lowe CR, Taipa MA. Biotechnol. Prog. 2004;20:639–654. doi: 10.1021/bp030070k. [DOI] [PubMed] [Google Scholar]
- (3).Leader B, Baca QJ, Golan DE. Nat. Rev. Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- (4).Huang CJ, Lowe AJ, Batt CA. Appl. Microbiol. Biotechnol. 2010;87:401–410. doi: 10.1007/s00253-010-2590-7. [DOI] [PubMed] [Google Scholar]
- (5).Andya JD, Maa YF, Costantino HR, Nguyen PA, Dasovich N, Sweeney TD, Hsu CC, Shire SJ. Pharm. Res. 1999;16:350–358. doi: 10.1023/a:1018805232453. [DOI] [PubMed] [Google Scholar]
- (6).Krishnan S, Chi EY, Webb JN, Chang BS, Shan DX, Goldenberg M, Manning MC, Randolph TW, Carpenter JF. Biochemistry. 2002;41:6422–6431. doi: 10.1021/bi012006m. [DOI] [PubMed] [Google Scholar]
- (7).Chi EY, Krishnan S, Kendrick BS, Chang BS, Carpenter JF, Randolph TW. Protein Sci. 2003;12:903–913. doi: 10.1110/ps.0235703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chi EY, Krishnan S, Randolph TW, Carpenter JF. Pharm. Res. 2003;20:1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- (9).Bischof JC, He XM. Ann. N. Y. Acad. Sci. 2005;1066:12–33. doi: 10.1196/annals.1363.003. [DOI] [PubMed] [Google Scholar]
- (10).Frokjaer S, Otzen DE. Nat. Rev. Drug Discovery. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- (11).Rathore N, Rajan RS. Biotechnol. Prog. 2008;24:504–514. doi: 10.1021/bp070462h. [DOI] [PubMed] [Google Scholar]
- (12).Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS. Pharm. Res. 2010;27:544–575. doi: 10.1007/s11095-009-0045-6. [DOI] [PubMed] [Google Scholar]
- (13).Dobson CM, Sali A, Karplus M. Angew. Chem., Int. Ed. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- (14).Sanchez-Ruiz JM. Biophys. Chem. 2010;148:1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- (15).Fink AL. Folding Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- (16).Uversky VN, Li J, Fink AL. J. Biol. Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- (17).Mahler HC, Friess W, Grauschopf U, Kiese S. J. Pharm. Sci. 2009;98:2909–2934. doi: 10.1002/jps.21566. [DOI] [PubMed] [Google Scholar]
- (18).Weiss WF, Young TM, Roberts CJ. J. Pharm. Sci. 2009;98:1246–1277. doi: 10.1002/jps.21521. [DOI] [PubMed] [Google Scholar]
- (19).Vagenende V, Yap MGS, Trout BL. Biochemistry. 2009;48:11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- (20).Sola RJ, Al-Azzam W, Griebenow K. Biotechnol. Bioeng. 2006;94:1072–1079. doi: 10.1002/bit.20933. [DOI] [PubMed] [Google Scholar]
- (21).Sola RJ, Griebenow K. J. Pharm. Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Giffard RG, Xu LJ, Heng Z, Carrico W, Ouyang YB, Qiao YL, Sapolsky R, Steinberg G, Hu BR, Yenari MA. J. Exp. Biol. 2004;207:3213–3220. doi: 10.1242/jeb.01034. [DOI] [PubMed] [Google Scholar]
- (23).Goyal K, Walton LJ, Tunnacliffe A. Biochem. J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kovacs D, Kalmar E, Torok Z, Tompa P. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Samuel D, Kumar TKS, Ganesh G, Jayaraman G, Yang PW, Chang MM, Trivedi VD, Wang SL, Hwang KC, Chang DK, Yu C. Protein Sci. 2000;9:344–352. doi: 10.1110/ps.9.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Shiraki K, Kudou M, Fujiwara S, Imanaka T, Takagi M. J. Biochem. 2002;132:591–595. doi: 10.1093/oxfordjournals.jbchem.a003261. [DOI] [PubMed] [Google Scholar]
- (27).Bondos SE, Bicknell A. Anal. Biochem. 2003;316:223–231. doi: 10.1016/s0003-2697(03)00059-9. [DOI] [PubMed] [Google Scholar]
- (28).Okanojo M, Shiraki K, Kudou M, Nishikori S, Takagi M. J. Biosci. Bioeng. 2005;100:556–561. doi: 10.1263/jbb.100.556. [DOI] [PubMed] [Google Scholar]
- (29).Kar K, Kishore N. Biopolymers. 2007;87:339–351. doi: 10.1002/bip.20834. [DOI] [PubMed] [Google Scholar]
- (30).Hamada H, Arakawa T, Shiraki K. Curr. Pharm. Biotechnol. 2009;10:400–407. doi: 10.2174/138920109788488941. [DOI] [PubMed] [Google Scholar]
- (31).Park SM, Jung HY, Chung KC, Rhim H, Park JH, Kim J. Biochemistry. 2002;41:4137–4146. doi: 10.1021/bi015961k. [DOI] [PubMed] [Google Scholar]
- (32).Park SM, Ahn KJ, Jung HY, Park JH, Kim J. Protein Eng., Des. Sel. 2004;17:251–260. doi: 10.1093/protein/gzh029. [DOI] [PubMed] [Google Scholar]
- (33).Lee EN, Kim YM, Lee HJ, Park SW, Jung HY, Lee JM, Ahn YH, Kim J. Pharm. Res. 2005;22:1735–1746. doi: 10.1007/s11095-005-6489-4. [DOI] [PubMed] [Google Scholar]
- (34).Singh J, Whitwill S, Lacroix G, Douglas J, Dubuc E, Allard G, Keller W, Schernthaner JP. Protein Expression Purif. 2009;67:15–22. doi: 10.1016/j.pep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- (35).Roberts CJ, Darrington RT, Whitley MB. J. Pharm. Sci. 2003;92:1095–1111. doi: 10.1002/jps.10377. [DOI] [PubMed] [Google Scholar]
- (36).Farmer RS, Kiick KL. Biomacromolecules. 2005;6:1531–1539. doi: 10.1021/bm049216+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang Y, Kiick KL. J. Am. Chem. Soc. 2005;127:16392–16393. doi: 10.1021/ja055102+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Farmer RS, Argust LM, Sharp JD, Kiick KL. Macromolecules. 2006;39:162–170. doi: 10.1021/ma051534t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Farmer RS, Top A, Argust LM, Liu S, Kiick KL. Pharm. Res. 2008;25:700–708. doi: 10.1007/s11095-007-9344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Liu S, Kiick KL. Macromolecules. 2008;41:764–772. doi: 10.1021/ma702128a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Top A, Kiick KL. Adv. Drug Delivery Rev. 2010;62:1530–1540. doi: 10.1016/j.addr.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Top A, Kiick KL, Roberts CJ. Biomacromolecules. 2008;9:1595–1603. doi: 10.1021/bm800056r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sharma N, Top A, Kiick KL, Pochan DJ. Angew. Chem., Int. Ed. 2009;48:7078–7082. doi: 10.1002/anie.200901621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Abeln S, Frenkel D. PLoS Comput. Biol. 2008;4:e1000241. doi: 10.1371/journal.pcbi.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Basu A, Yang K, Wang ML, Liu S, Chintala R, Palm T, Zhao H, Peng P, Wu DC, Zhang ZF, Hua J, Hsieh MC, Zhou J, Petti G, Li XG, Janjua A, Mendez M, Liu J, Longley C, Zhang Z, Mehlig M, Borowski V, Viswanathan M, Filpula D. Bioconjugate Chem. 2006;17:618–630. doi: 10.1021/bc050322y. [DOI] [PubMed] [Google Scholar]
- (47).Stigsnaes P, Frokjaer S, Bjerregaard S, van de Weert M, Kingshott P, Moeller EH. Int. J. Pharm. 2007;330:89–98. doi: 10.1016/j.ijpharm.2006.09.002. [DOI] [PubMed] [Google Scholar]
- (48).Yu PZ, Qin D, Qin GH, Fan B, Ma GH, Su ZG. Process Biochem. 2009;44:1340–1345. [Google Scholar]
- (49).Gaertner HF, Puigserver AJ. Enzyme Microb. Technol. 1992;14:150–155. doi: 10.1016/0141-0229(92)90174-m. [DOI] [PubMed] [Google Scholar]
- (50).Longo MA, Combes DJ. Chem. Technol. Biotechnol. 1999;74:25–32. [Google Scholar]
- (51).Zhang ZD, He ZM, Guan GQ. Biotechnol. Tech. 1999;13:781–786. [Google Scholar]
- (52).He ZM, Zhang ZD, He MX. Process Biochem. 2000;35:1235–1240. [Google Scholar]
- (53).Garcia-Arellano H, Valderrama B, Saab-Rincon G, Vazquez-Duhalt R. Bioconjugate Chem. 2002;13:1336–1344. doi: 10.1021/bc025561p. [DOI] [PubMed] [Google Scholar]
- (54).Lopez-Cruz JI, Viniegra-Gonzalez G, Hernandez-Arana A. Bioconjugate Chem. 2006;17:1093–1098. doi: 10.1021/bc0503465. [DOI] [PubMed] [Google Scholar]
- (55).Rodriguez-Martinez JA, Sola RJ, Castillo B, Cintron-Colon HR, Rivera-Rivera I, Barletta G, Griebenow K. Biotechnol. Bioeng. 2008;101:1142–1149. doi: 10.1002/bit.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Treethammathurot B, Ovartlarnporn C, Wungsintaweekul J, Duncan R, Wiwattanapatapee R. Int. J. Pharm. 2008;357:252–259. doi: 10.1016/j.ijpharm.2008.01.016. [DOI] [PubMed] [Google Scholar]
- (57).Molineux G. Cancer Treat. Rev. 2002;28:13–16. doi: 10.1016/s0305-7372(02)80004-4. [DOI] [PubMed] [Google Scholar]
- (58).Harris JM, Chess RB. Nat. Rev. Drug Discovery. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- (59).Veronese FM, Pasut G. Drug Discovery Today. 2005;10:1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- (60).Kubetzko S, Sarkar CA, Pluckthun A. Mol. Pharmacol. 2005;68:1439–1454. doi: 10.1124/mol.105.014910. [DOI] [PubMed] [Google Scholar]
- (61).Das R, Baird E, Allen S, Baird B, Holowka D, Goldstein B. Biochemistry. 2008;47:1017–1030. doi: 10.1021/bi702094j. [DOI] [PubMed] [Google Scholar]
- (62).Choi YY, Jang JH, Park MH, Choi BG, Chi B, Jeong B. J. Mater. Chem. 2010;20:3416–3421. [Google Scholar]
- (63).Hamley IW, Ansari A, Castelletto V, Nuhn H, Rosler A, Klok HA. Biomacromolecules. 2005;6:1310–1315. doi: 10.1021/bm049286g. [DOI] [PubMed] [Google Scholar]
- (64).Vandermeulen GWM, Tziatzios C, Klok HA. Macromolecules. 2003;36:4107–4114. [Google Scholar]
- (65).Vandermeulen GWM, Tziatzios C, Duncan R, Klok HA. Macromolecules. 2005;38:761–769. [Google Scholar]
- (66).Uchio T, Baudys M, Liu F, Song SC, Kim SW. Adv. Drug Delivery Rev. 1999;35:289–306. doi: 10.1016/s0169-409x(98)00078-7. [DOI] [PubMed] [Google Scholar]
- (67).Hinds KD, Kim SW. Adv. Drug Delivery Rev. 2002;54:505–530. doi: 10.1016/s0169-409x(02)00025-x. [DOI] [PubMed] [Google Scholar]
- (68).Rajan RS, Li TS, Aras M, Sloey C, Sutherland W, Arai H, Briddell R, Kinstler O, Lueras AMK, Zhang Y, Yeghnazar H, Treuheit M, Brems DN. Protein Sci. 2006;15:1063–1075. doi: 10.1110/ps.052004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Rodriguez-Martinez JA, Rivera-Rivera I, Sola RJ, Griebenow K. Biotechnol. Lett. 2009;31:883–887. doi: 10.1007/s10529-009-9947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Rubinson KA, Krueger S. Polymer. 2009;50:4852–4858. [Google Scholar]
- (71).Kelly SM, Jess TJ, Price NC. Biochim. Biophys. Acta, Proteins Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- (72).Kennedy RJ, Tsang KY, Kemp DS. J. Am. Chem. Soc. 2002;124:934–944. doi: 10.1021/ja016285c. [DOI] [PubMed] [Google Scholar]
- (73).Lu YL, Harding SE, Turner A, Smith B, Athwal DS, Grossmann JG, Davis KG, Rowe AJ. J. Pharm. Sci. 2008;97:2062–2079. doi: 10.1002/jps.21170. [DOI] [PubMed] [Google Scholar]
- (74).Meng F, Manjula BN, Smith PK, Acharya SA. Bioconjugate Chem. 2008;19:1352–1360. doi: 10.1021/bc7003878. [DOI] [PubMed] [Google Scholar]
- (75).Scholtz JM, Marqusee S, Baldwin RL, York EJ, Stewart JM, Santoro M, Bolen DW. Proc. Natl. Acad. Sci. U. S. A. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Pechar M, Kopeckova P, Joss L, Kopecek J. Macromol. Biosci. 2002;2:199–206. [Google Scholar]
- (77).Smeenk JM, Schon P, Otten MBJ, Speller S, Stunnenberg HG, van Hest JCM. Macromolecules. 2006;39:2989–2997. [Google Scholar]
- (78).Tzokova N, Fernyhough CM, Butler MF, Armes SP, Ryan AJ, Topham PD, Adams DJ. Langmuir. 2009;25:11082–11089. doi: 10.1021/la901413n. [DOI] [PubMed] [Google Scholar]
- (79).Dwevedi A, Dubey VK, Jagannadham MV, Kayastha AM. Appl. Biochem. Biotechnol. 2010;162:2294–2312. doi: 10.1007/s12010-010-9003-3. [DOI] [PubMed] [Google Scholar]
- (80).Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA. Curr. Opin. Struct. Biol. 2010;20:54–62. doi: 10.1016/j.sbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Veronese FM, Mero A, Caboi F, Sergi M, Marongiu C, Pasutt G. Bioconjugate Chem. 2007;18:1824–1830. doi: 10.1021/bc070123+. [DOI] [PubMed] [Google Scholar]
- (82).Burkoth TS, Benzinger TLS, Jones DNM, Hallenga K, Meredith SC, Lynn DG. J. Am. Chem. Soc. 1998;120:7655–7656. [Google Scholar]
- (83).Burkoth TS, Benzinger TLS, Urban V, Lynn DG, Meredith SC, Thiyagarajan P. J. Am. Chem. Soc. 1999;121:7429–7430. [Google Scholar]
- (84).Thiyagarajan P, Burkoth TS, Urban V, Seifert S, Benzinger TLS, Morgan DM, Gordon D, Meredith SC, Lynn DG. J. Appl. Crystallogr. 2000;33:535–539. [Google Scholar]
- (85).Castelletto V, Newby GE, Zhu Z, Hamley IW, Noirez L. Langmuir. 2010;26:9986–9996. doi: 10.1021/la100110f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.