Abstract
Dystroglycan is a laminin binding protein, which provides a structural link between the subsarcolemmal cytoskeleton and the extracellular matrix. It is also involved in the organization of basement membranes. So far the genomic organization of the dystroglycan gene DAG1 has not been completely investigated. Here we report the cloning and sequencing of 162 kb of dog genomic DNA containing the complete ∼71-kb canine DAG1 gene, which consists of three exons, with the translation start codon located in exon 2. Its 2679-nucleotide ORF encodes a polypeptide of 892 amino acids, which is highly similar to human, rabbit, and bovine orthologs. To further characterize the dog DAG1 gene we determined the transcription start site and several naturally occurring polymorphisms, which partially result in amino acid substitutions of the dystroglycan protein. The dog DAG1 gene was assigned to chromosome 20q15.1–q15.2 by FISH analysis. The analysis of the entire reported sequence revealed that the genes for aminomethyltransferase (AMT), bassoon (BSN), TCTA (T-cell leukemia translocation-associated) gene, and an as yet uncharacterized protein are located very close to the DAG1 gene. Therefore, this study defines a novel syntenic region among dog chromosome 20q15, human chromosome 3p21, and murine chromosome 9F.
[The sequence data described in this paper have been submitted to the EMBL nucleotide database under accession no. AJ012166.]
Dystroglycan is an important part of the skeletal muscle dystrophin-associated glycoprotein complex (DGC) (Durbeej et al. 1998; Hemler 1999). The DGC is thought to be essential for mediating mechanical forces between the actin cytoskeleton and the extracellular matrix of a contracting muscle. In the DGC dystroglycan forms a membrane-spanning link between the intracellular dystrophin and the extracellular laminin α2 chain. Besides its structural function in skeletal muscle, dystroglycan has also signaling functions and is found in many other tissues, especially the central and peripheral nervous systems, the retina, and epithelial cells. Absence of dystroglycan in gene-targeted mice is lethal during embryonic development because Reichert's membrane, an early basement membrane, is not properly formed (Williamson et al. 1997). Subsequent studies confirmed that dystroglycan is essential for the correct formation of basement membranes (Henry and Campbell 1998). The protein received additional attention as it serves as a receptor for viral and bacterial pathogens (Cao et al. 1998; Rambukkana et al. 1998).
Dystroglycan consists of two subunits, termed α- and β-dystroglycan. The two subunits are encoded by a single gene and arise through post-translational cleavage of a precursor polypeptide. The predicted molecular mass of the α-subunit is 72 kD; however due to tissue-specific glycosylation, isolated α-dystroglycan has a molecular mass between 120 and 156 kD. α-Dystroglycan is secreted and binds to laminin. β-Dystroglycan has a molecular mass of 43 kD and contains a single membrane-spanning domain. Its intracellular part binds to the carboxyl terminus of dystrophin, whereas its extracellular domain interacts with α-dystroglycan.
The dystroglycan gene DAG1 has not yet been characterized completely. Highly homologous cDNA sequences have been reported from man (Ibraghimov-Beskrovnaya et al. 1993), mouse (Brancaccio et al. 1994; Gorecki et al. 1994; Yotsomoto et al. 1996), rabbit (Ibraghimov-Beskrovnaya et al. 1992), and cattle. The human DAG1 gene also has been partially cloned; it was found that the coding portion of the gene is located on two exons, which reside on chromosome 3p21 (Ibraghimov-Beskrovnaya et al. 1993).
To elucidate the complete structure of the DAG1 gene, we cloned and sequenced the dog DAG1 gene with extensive flanking regions. The analysis of the reported sequence revealed that several other genes are in close vicinity to the DAG1 gene in the dog genome. In addition to the genomic sequence of the DAG1 gene, we determined its chromosomal localization, its transcriptional start site, and variations within the DAG1 coding sequence in different individuals.
RESULTS AND DISCUSSION
Cloning of the Dog DAG1 Gene
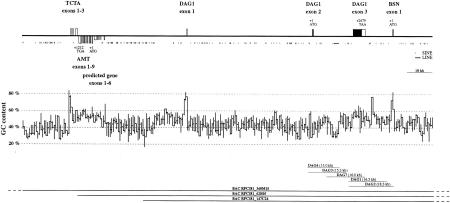
To isolate the DAG1 gene from dog we screened a recombinant phage library with radioactively labeled cDNA probes derived from the two previously reported exons of the DAG1 gene. This resulted in the isolation of five overlapping clones spanning ∼35 kb of genomic DNA. Sequence analysis of this 35-kb contig revealed the presence of two DAG1 exons with the entire ORF of the gene. However, comparison with the human cDNA sequence revealed that the exon farthest upstream on this contig lacked the 5′ end of the cDNA and started with a splice acceptor sequence. To clone the presumably missing authentic first exon of the DAG1 gene we obtained a human cDNA clone containing the missing 5′ end and screened a dog BAC library. This led to the isolation of three overlapping BAC clones spanning ∼200 kb of dog genomic sequence. A 162,073-bp fragment from the longest of these BAC clones, designated BAC RPCI81_340M15 was completely sequenced. This sequence contained the complete dog DAG1 gene, as well as several flanking genes on both sides of the DAG1 gene. The positions of the isolated clones and genes are illustrated in Figure 1.
Figure 1.
Genomic organization of the dog DAG1 locus. Exons of the genes in the analyzed region are shown as boxes. Boxes above the solid line represent exons in sense orientation; boxes below the solid line indicate exons in antisense orientation. ORFs, which are translated into proteins of known function, are indicated by numbering and the respective exons are shown as solid boxes. Untranslated exons, or exons where the translation product is not characterized, are shown as open boxes. The positions and orientations of SINE and LINE repetitive elements are indicated by arrows. (Middle) GC content. A 300-bp window size was used for the calculation of the GC content. (Bottom) The position of the isolated phage and BAC clones.
Analysis of the Genomic Structure of the Dog DAG1 Gene
Exon/intron boundaries of the canine DAG1 gene were determined by comparison of the dog genomic DNA sequence with the human cDNA sequence. In addition, a dog cDNA fragment containing the junction between the first two exons was generated by RT–PCR, sequenced, and used for comparison with the genomic sequence. From these analyses it became clear that the dog DAG1 gene consists of three exons separated by two introns. The gene spans ∼71 kb, of which 48.8 kb is contained in the first intron. All splice donor/splice acceptor sites conform to the GT/AG rule. The exon/intron junctions in the protein coding region of the DAG1 gene are well conserved between human and dog. However, at the end of the first exon the dog cDNA lacks ∼120 bp compared to the human cDNA sequence. The additional human cDNA sequence shows high homology to the beginning of the first intron of the dog genomic sequence. This is probably due to the use of a different splice donor site in humans than in dogs. The above-mentioned RT–PCR did not show any additional bands representing alternatively spliced transcripts, which could also have explained the discrepancy between human and canine cDNA sequence. The DNA sequences at the exon/intron boundaries are summarized in Table 1.
Table 1.
Exon–Intron Junctions of the Dog DAG1 Gene
| No. | Exon (bp) | Splice donor | Intron (bp) | Splice acceptor | ||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − | 2 | 2 | 8 | |||||||||||||||||||||||||||||||||||||||
| . | . | . | g | c | g | t | c | g | c | a | c | g | G | A | C | T | G | |||||||||||||||||||||||||
| − | 1 | 0 | 8 | − | 1 | 0 | 7 | |||||||||||||||||||||||||||||||||||
| 1 | 121 | G | C | C | C | G | g | t | a | c | g | t | g | c | t | c | . | . | . | 48842 | . | . | . | t | c | c | a | a | a | c | c | a | g | G | C | T | C | A | ||||
| 2 | 7 | 6 | 2 | 7 | 7 | |||||||||||||||||||||||||||||||||||||
| 2 | 383 | G | T | C | A | A | T | g | t | g | a | g | a | c | t | g | g | . | . | . | 16535 | . | . | . | t | c | t | c | t | t | t | t | a | g | G | T | G | T | C | A | ||
| V | a | l | L | y | s | V | a | l | S | e | r | |||||||||||||||||||||||||||||||
| 4 | 7 | 3 | 1 | |||||||||||||||||||||||||||||||||||||||
| 3 | >4455 | A | A | A | A | A | T | A | A | A | C | A | A | T | T | C | C | A | . | . | . | |||||||||||||||||||||
Transcribed sequences are shown in uppercase letters, and nontranscribed regions in lowercase letters. Untranslated regions are shown in italics. The conserved GT/AG exon/intron junctions are shown in boldface type. For the first exon the transcription start site is shown instead of a splice acceptor; for the last exon the first of two closely associated polyadenylation signals is shown in bold italics instead of a splice donor. Amino acid residues are indicated with respect to each boundary. Numbers refer to the corresponding positions in the DAG1 cDNA starting with +1 at the adenosine of the translation initiation codon.
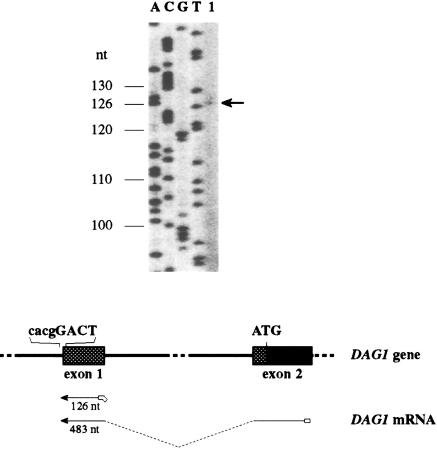
The transcription start site of the dog DAG1 gene was determined by primer extension analysis with two different primers located in the first and second exon, respectively (Fig. 2). The longest extension product of both primers corresponded to the same G residue 228 nucleotides upstream from the start codon of the dog cDNA sequence. The region immediately downstream of the transcription start site is well conserved between human and dog. The reported human cDNA sequence starts only two nucleotides downstream of the dog transcription start site, therefore supporting the results of the primer extension analyses.
Figure 2.
Determination of the transcription start site of the DAG1 gene. (Top) A detail of the sequencing gel with the DNA marker sequence (lanes A, C, G, T) and the product of the primer extension reaction of primer exon1_Ext (lane 1) is shown. Sizes are indicated on the left side of the gel. The size of the extension product is actually 5 nucleotides longer than the size of the first exon, as the primer contained a RsaI tag at its 5′ end for cloning purposes. (Bottom) The diagram outlines the experimental strategy. Primers exon1_Ext and exon2_Ext (open boxes) were annealed to total RNA and elongated as described in Methods. The thin dotted line indicates the spliced first intron. Important DNA sequence elements (transcription start site, ATG start codon) are shown. Crosshatched boxes within exons represent the 5′ UTR.
The promoter of the dog DAG1 gene is GC-rich with a GC-content of 78.7% in the 300 nucleotides preceding the transcription start site (see also Fig. 1). Like other GC-rich promoters it does not contain a TATA box or a CAAT box.
Analysis of the Dog DAG1 cDNA Sequence
The dog DAG1 cDNA sequence is ∼4.5 kb long, with an ORF of 1679 nucleotides (including the stop codon) and a 5′ UTR of 228 nucleotides. At the 3′ end two canonical polyadenylation signals, AATAAA, are found within 350 bp similar to the human DAG1 gene. Depending on which polyadenylation signal is used this would result in 3′ UTRs of 2.1 and 2.4 kb, respectively. Upstream from the main ORF is a very short ORF encoding for only seven amino acids. A similar upstream ORF is also present in the human DAG1 cDNA sequence. According to the criteria of Kozak (1996), such an upstream ORF is compatible with translation of a second ORF from the same transcript, although reduced protein yields are to be expected. The translation product starting at the second ATG is 892 amino acids long and exhibits 94%, 92%, and 91% identity to the corresponding human, rabbit, and bovine dystroglycan precursor proteins. The dog protein sequence lacks a stretch of three amino acids shortly after the signal peptide, which is present in all of the other species but otherwise contains no insertions or deletions.
Analysis of Polymorphisms Within the DAG1 Coding Sequence
To search for different DAG1 alleles within the dog population we isolated genomic DNA from four different dogs and amplified the three DAG1 exons with their flanking sequences. To amplify the complete 4.5-kb third exon, five overlapping PCR products from this exon were generated. The PCR-amplified exons from the four dogs were directly sequenced and the sequences compared to each other. A total of eight polymorphic sites within the dog DAG1 cDNA sequence were identified and are summarized in Table 2. Three of these eight polymorphisms led to amino acid exchanges. All of these amino acid exchanges are clustered in a relatively short region near the amino terminus of the protein. To verify that the protein-changing mutations are not caused by PCR artifacts, the amplification and sequencing of these sites was repeated with identical results.
Table 2.
Polymorphisms in the Dog DAG1 Sequence
| Location of polymorphic site (exon) | Nucleotide positiona | cDNA positionb | Nucleotide polymorphism | Amino acid substitution |
|---|---|---|---|---|
| 2 | 113708 | +99 | A → G | silent |
| 3 | 130489 | +345 | G → C | E115 → D115 |
| 3 | 130760 | +616 | G → A | D206 → N206 |
| 3 | 130817 | +673 | G → A | V225 → M225 |
| 3 | 131869 | +1725 | T → C | silent |
| 3 | 132229 | +2085 | G → A | silent |
| 3 | 132562 | +2418 | G → C | silent |
| 3 (3′ UTR) | 134574 | +4430 | G → T |
Numbering refers to GenBank accession no. AJ012166.
+1 corresponds to the adenosine of the start codon ATG of the DAG1 cDNA.
Analysis of the Genomic Organization of DAG1 and Flanking Genes
Comparisons of the entire 162,073 bp of the reported sequence with the nucleotide database using the BLAST program revealed the presence of four other putative genes flanking the DAG1 gene (Fig. 1). Upstream from DAG1 on the same DNA strand as DAG1, the dog homolog of the human T-cell leukemia translocation-associated (TCTA) gene is located. This gene was first identified at the site of a t(1;3) translocation commonly occurring in T-cell acute lymphoblastic leukemia (Aplan et al. 1995). In humans it contains a short ORF that encodes a protein of 103 amino acids without homology to any other known protein. In the canine TCTA gene the corresponding ORF cannot be translated because an upstream ATG in a different reading frame is present, which is followed by a stop codon after only a few codons. Therefore, the dog TCTA seems to be a pseudogene.
Immediately downstream of the TCTA gene but on the complementary DNA strand, the gene for aminomethyltransferase AMT is located. AMT, or glycine cleavage system T-protein, is a mitochondrial enzyme involved in glycine metabolism (Nanao et al. 1994). The gene consists of nine exons and encodes a protein with 91% identity to the human AMT.
Adjacent to the AMT gene, the program GeneMark of the EBI online tools predicted another protein coding region. Sequence comparisons revealed homologies to several ESTs but to no known genes. Based on the homology to human ESTs with GenBank/EMBL/DDBJ accessions numbers H86023 and AA701640, a putative gene with six exons could be localized.
Downstream of the DAG1 gene, again on the same strand as DAG1, the gene for the brain-specific synapse-associated protein bassoon (BSN) follows. Bassoon has been identified as a component of the synaptic cytoskeleton (tom Dieck et al. 1998; Winter et al. 1999). The human gene is >100 kb in size with a large first intron. In the reported dog sequence there is only the first exon of BSN present, which is to be expected when one assumes that the first intron of the dog BSN is similar in size to the first intron of the human gene.
The GC content of the reported sequence correlates extremely well with the presence of genes. Three CpG islands with very high scores were detected using the program CPG of the EBI at the 5′ ends of the genes TCTA, DAG1, and BSN. Two more CpG islands with moderately high scores were detected at the 5′ end of the novel predicted gene and at the 3′ end of the DAG1 gene. The GC content of the reported sequence is 46%. This relatively low value is due to the presence of the polyadenosine tracts of 153 copies of a dog tRNA–SINE element (Das et al. 1998; Bentolila et al. 1999). In the second DAG1 intron there is also an incomplete LINE-1 element with a relatively low GC content. The distribution of the GC content and repetitive elements along the sequence are illustrated in Figure 1.
Chromosomal Localization of the Sequenced Contig
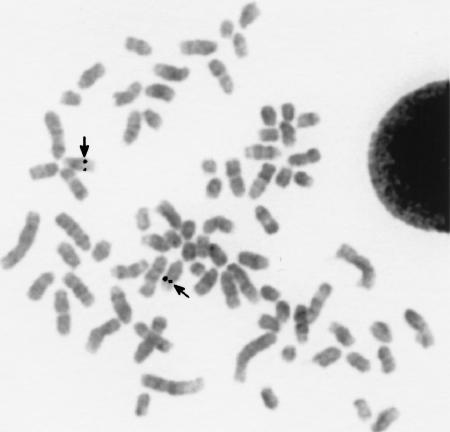
To determine the chromosomal location of the dog DAG1 gene as well as the other genes in the reported sequence, we used the genomic phage clone DAG2 as probe in FISH analysis. As shown in Figure 3, a hybridization signal was detected on CFA 20q15.1–q15.2. All of the genes described (AMT, BSN, DAG1, TCTA) have been localized in human to HSA 3p21, which is a region within the evolutionarily conserved chromosome segment shared with CFA 20q11–15.2 (Breen et al. 1999a). Therefore, these four genes seem to form a syntenic region between human and dog, and the information about the relative position and orientation of these genes in the dog genome might promote human gene mapping efforts as well. In mice, Bsn and Dag1 have been mapped to chromosome 9F. Assuming synteny between dog and mouse in this region, one would also expect to find the murine Amt and Tcta genes on chromosome 9F.
Figure 3.
Chromosomal assignment of the dog DAG1 gene by FISH analysis. The biotinylated genomic phage clone DAG2 containing the 3′ end of the dog DAG1 gene was hybridized to metaphase chromosomes of a normal dog. Double signals are visible on both CFA 20q15.1–q15.2, indicated by arrows. The chromosomes are counterstained with propidium iodide. They were subsequently identified by DAPI staining.
In conclusion, for the first time, our results provide the complete genomic structure of the DAG1 gene containing three exons. The reported 162 kb of DNA sequence adds significantly to our knowledge of the dog genome, as there are currently only four other database entries with >10 kb of dog DNA sequence available, of which the 16-kb mitochondrial sequence is the longest. As predicted, the localization of a total of five genes on the sequenced contig defines a syntenic region between dog chromosome 20q15.1–q15.2, human chromosome 3p21, and murine chromosome 9F.
METHODS
DNA Library Screening
A dog genomic DNA library constructed in λFIX II (Stratagene, Heidelberg, Germany) was screened according to standard protocols (Ausubel et al. 1995). Two 32P-labeled DNA fragments were used simultaneously as probes: the insert of the IMAGE clone 267199 containing 1.2 kb of exon 3 of the human DAG1 gene, and a 270-bp human DNA fragment of exon 2 that was PCR-amplified on human genomic DNA. The IMAGE clone was provided by the Resource Center of the German Human Genome Project at the Max-Planck-Institute for Molecular Genetics (RZPD, IMAGp998C08586). To isolate the missing 5′ end of the dog DAG1 gene, the dog BAC library RPCI81 (Osoegawa et al. 1998) was also screened. High-density BAC filters were probed according to the RPCI protocols (http://bacpac.med.buffalo.edu/) with a 32P-labeled 670-bp EcoRI–EcoRV fragment of the human cDNA clone HD-4 (Ibraghimov-Beskrovnaya et al. 1993) corresponding to exons 1 and 2 of the DAG1 gene.
DNA Sequence Analysis
Isolated phage or BAC DNA was restricted with appropriate restriction enzymes, and the resulting fragments were subcloned into the polylinker of pGEM-4Z (Promega, Mannheim, Germany). Recombinant plasmid DNA was sequenced with the thermosequenase kit (Amersham–Pharmacia, Freiburg, Germany) and a LICOR 4000L automated sequencer. Within extremely GC-rich regions, some fragments were sequenced manually with the T7 sequencing kit and α-35S-labeled thio–dATP as label. The plasmid clones were subcloned further with different restriction enzymes. After sequencing a collection of plasmid subclones, existing gaps were closed by a primer walking strategy until both strands were completely sequenced. Sequence data were analyzed with Sequencher 3.1.1 (GeneCodes, Ann Arbor, MI) and MacDNASIS 3.6 (Hitachi, Hess, Oldendorf, Germany). Further analyses were performed with the online tools of the European Bioinformatics Institute (http://www.ebi.ac.uk/).
PCR Amplification and Sequencing of DAG1 Exons
PCR was performed in 100-μl reactions containing 100 ng of genomic dog DNA, 200 μm dNTPs, 1 μm each primer, and 2.5 units of Taq polymerase in the reaction buffer supplied by the manufacturer (Qiagen, Hilden, Germany). After a 5-min initial denaturation at 94°C, 32 cycles of 1 min at 94°C, 1 min at the annealing temperature of the specific primer pair, and 1.5 min at 72°C were performed in a Hybaid Omnigene thermocycler (MWG AG Biotech, Ebersberg, Germany). Primers were designed to amplify the complete coding sequence, all exon/intron junctions, and 5′- and 3′ UTRs. Because the last exon could not be amplified in a single PCR, it was divided into four overlapping fragments (a,b,d,e). Fragment b was difficult to sequence; therefore, a smaller subfragment of b, termed c, was also amplified for sequence analysis. Primer sequences and annealing temperatures are given in Table 3. The amplified products were purified by electrophoresis through 1% agarose gels and sequenced directly using IRD800-labeled internal primers.
Table 3.
Primer Sequences
| Primer (exon)a | Sequence (5′-3′) | Length | Tm (°C) | Product (bp) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1, F | A | G | C | C | T | G | G | G | T | C | G | G | T | T | G | T | A | C | G | 19 | |||||
| 1, R | A | G | C | A | G | G | T | G | T | T | C | C | T | C | G | G | G | A | G | 19 | 60 | 684 | |||
| 2, F | A | T | T | G | T | G | T | C | T | T | C | A | T | G | A | C | T | T | C | 19 | |||||
| 2, R | G | A | A | T | T | C | T | T | C | C | T | C | A | T | T | C | A | C | 18 | 50 | 953 | ||||
| 3a, F | A | G | C | T | G | T | T | G | T | G | A | T | C | A | T | T | G | T | C | 19 | |||||
| 3a, R | G | A | G | T | T | G | T | G | G | T | G | G | T | T | G | A | A | G | 18 | 54 | 1122 | ||||
| 3b, F | T | C | C | C | A | C | G | A | C | T | A | C | C | A | C | C | A | A | G | 19 | |||||
| 3b, R | A | C | C | G | G | T | C | T | C | C | A | C | A | G | A | C | A | A | C | 19 | 55 | 1507 | |||
| 3c, F | G | C | C | G | G | C | A | C | C | T | C | C | A | G | T | T | T | A | T | C | C | 21 | |||
| 3c, R | G | T | C | A | T | G | T | T | C | T | T | G | G | G | A | C | G | G | G | A | G | C | 22 | 68 | 514 |
| 3d, F | C | C | T | T | T | A | C | G | G | C | C | C | C | C | A | T | G | G | A | G | 20 | ||||
| 3d, R | C | G | C | C | A | C | C | A | G | A | C | C | T | T | T | C | C | C | A | G | 20 | 65 | 1296 | ||
| 3e, F | T | C | C | C | A | A | C | A | G | T | C | C | A | C | A | G | C | C | C | 19 | |||||
| 3e, R | T | C | C | C | C | T | C | T | T | C | C | C | T | C | T | G | T | C | A | C | A | 21 | 61 | 1402 | |
(F) Forward; (R) reverse.
Primer Extension Analysis
Total RNA from dog skeletal muscle was prepared with Trizol (Life Technologies, Karlsruhe, Germany). Ten picomoles of IRD800-labeled primer were annealed to 2 μg of total RNA and reverse transcribed in a 20-μl reaction volume with 200 units of MMLV reverse transcriptase (Life Technologies, Karlsruhe, Germany). After a 50-min incubation at 44°C and 10 min at 75°C, 5 μg of RNase A was added and the reaction was maintained at 37°C for 30 min. The synthesized cDNA was precipitated by addition of 4 μl of 0.1 m spermine and subsequent centrifugation for 40 min at 22,000 rpm and 4°C. The cDNA pellet was dissolved in 2.5 μl of formamide loading dye and denatured at 70 °C for 5 min. One microliter was loaded on an automated sequencer together with a sequencing reaction as size standard. The primer sequences for the experiment were exon1_Ext, 5′-CGTACCGGGCGCTGAGAG-3′; exon2_Ext, 5′-GGTCCTCACCCTCTGTAC-3′.
FISH
A total of 150 ng of the dog DAG1 gene phage DAG2 was labeled with digoxigenin-11–dUTP and hybridized to dog metaphase preparations as described by Breen et al. (1999b). The chromosomes were counterstained in DAPI prior to being mounted with Vectashield (Vector Laboratories, Burlingame, CA) and sealed with a coverslip. Images were captured using an Axiophot fluorescence microscope (Zeiss, Jena, Germany), equipped with an FITC/Texas Red/DAPI filter set and a cooled KAF1400 CCD camera (Photometrics, Munich, Germany), both driven by SmartCapture software (Vysis Inc., Downers Grove, IL). Hybridization signals were scored from 30 metaphase preparations using image-enhanced DAPI banding and assigned according to the DAPI-banded ideogram of Breen et al. (1999c).
Acknowledgments
We thank Kevin P. Campbell for the generous gift of the human dystroglycan cDNA clone HD-4. We also thank Pieter de Jong and Joe Catanese for providing the canine RPCI81 BAC filters and clones, together with excellent information about these resources on the World Wide Web. M.B. (AHT) is supported by funds from the Guide Dogs for the Blind Association.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tleeb@zucht.tiho-hannover.de; FAX 49-511-9538582.
REFERENCES
- Aplan PD, Johnson BE, Russell E, Chervinsky DS, Kirsch IR. Cloning and characterization of TCTA, a gene located at the site of a t(1;3) translocation. Cancer Res. 1995;55:1917–1921. [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: Greene Publishing Associates/Wiley-Interscience; 1995. [Google Scholar]
- Bentolila S, Bach JM, Kessler JL, Bordelais I, Cruaud C, Weissenbach J, Panthier JJ. Analysis of major repetitive DNA sequences in the dog (Canis familiaris) genome. Mamm Genome. 1999;10:699–705. doi: 10.1007/s003359901074. [DOI] [PubMed] [Google Scholar]
- Brancaccio A, Ruegg MA, Engel J. Cloning and sequencing of mouse skeletal muscle alpha-dystroglycan. Matrix Biol. 1994;14:681–685. doi: 10.1016/s0945-053x(05)80032-0. [DOI] [PubMed] [Google Scholar]
- Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999a;61:145–155. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- Breen M, Langford CF, Carter NP, Holmes NG, Dickens HF, Thomas R, Suter N, Ryder EJ, Pope M, Binns MM. FISH mapping and identification of canine chromosomes. J Hered. 1999b;90:27–30. doi: 10.1093/jhered/90.1.27. [DOI] [PubMed] [Google Scholar]
- Breen M, Bullerdiek J, Langford N. Standardisation of the GTG and DAPI banded karyotypes of the domestic dog (Canis familiaris) using chromosome specific paint probes. Chromosome Res. 1999c;7:401–406. doi: 10.1023/a:1009224232134. [DOI] [PubMed] [Google Scholar]
- Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- Das M, Chu LL, Ghahremani M, Abrams-Ogg T, Roy MS, Housman D, Pelletier J. Characterization of an abundant short interspersed nuclear element (SINE) present in Canis familiaris. Mamm Genome. 1998;9:64–69. doi: 10.1007/s003359900681. [DOI] [PubMed] [Google Scholar]
- Durbeej M, Henry MD, Campbell KP. Dystroglycan in development and disease. Curr Opin Cell Biol. 1998;10:594–601. doi: 10.1016/s0955-0674(98)80034-3. [DOI] [PubMed] [Google Scholar]
- Gorecki DC, Derry JMJ, Barnard EA. Dystroglycan: Brain localisation and chromosome mapping in the mouse. Hum Mol Genet. 1994;3:1589–1597. doi: 10.1093/hmg/3.9.1589. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Dystroglycan versatility. Cell. 1999;97:543–546. doi: 10.1016/s0092-8674(00)80764-3. [DOI] [PubMed] [Google Scholar]
- Henry MD, Campbell KP. A role for dystroglycan in basement membrane assembly. Cell. 1998;95:859–870. doi: 10.1016/s0092-8674(00)81708-0. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Milatovich A, Ozcelik T, Yang B, Koepnick K, Francke U, Campbell KP. Human dystroglycan: skeletal muscle cDNA, genomic strutcure, origin of tissue specific isoforms and chromosomal localization. Hum Mol Genet. 1993;2:1651–1657. doi: 10.1093/hmg/2.10.1651. [DOI] [PubMed] [Google Scholar]
- Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- Nanao K, Takada G, Takahashi E, Seki N, Komatsu Y, Okamura-Ikeda K, Motokawa Y, Hayasaka K. Structure and chromosomal localization of the aminomethyltransferase gene (AMT) Genomics. 1994;19:27–30. doi: 10.1006/geno.1994.1007. . [Erratum Genomics 20: 519.] [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, de Jong PJ. An improved approach for the construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Rambukkana A, Yamada H, Zanazzi G, Mathus T, Salzer JL, Yurchenco PD, Campbell KP, Fischetti VA. Role of α-dystroglycan as a Schwann cell receptor for Mycobacterium leprae. Science. 1998;282:2076–2079. doi: 10.1126/science.282.5396.2076. [DOI] [PubMed] [Google Scholar]
- Tom Dieck S, Sanmarti-Vila L, Langnaese K, Richter K, Kindler S, Soyke A, Wex H, Smalla KH, Kämpf U, Fränzer JT, et al. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J Cell Biol. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RA, Henry MD, Daniels KJ, Hrstka RF, Lee JC, Sunada Y, Ibraghimov-Beskrovnaya O, Campbell KP. Dystroglycan is essential for early embryonic development: disruption of Reichert's membrane in Dag1-null mice. Hum Mol Genet. 1997;6:831–841. doi: 10.1093/hmg/6.6.831. [DOI] [PubMed] [Google Scholar]
- Winter C, tom Dieck S, Boeckers TM, Bockmann J, Kämpf U, Sanmarti-Vila L, Langnaese K, Altrock W, Stumm M, Soyke A, et al. The presynaptic cytomatrix protein bassoon: Sequence and chromosomal localization of the human BSN gene. Genomics. 1999;57:389–397. doi: 10.1006/geno.1999.5788. [DOI] [PubMed] [Google Scholar]
- Yotsomoto S, Fujiwara H, Horton JH, Mosby TA, Wang X, Cui Y, Ko MSH. Cloning and expression analyses of mouse dystroglycan gene: Specific expression in maternal decidua at the peri-implantation stage. Hum Mol Genet. 1996;5:1259–1267. doi: 10.1093/hmg/5.9.1259. [DOI] [PubMed] [Google Scholar]