Abstract
Recombination between chromosome-specific low-copy repeats (duplicons) is an underlying mechanism for several genetic disorders. Recently, a chromosome 15 duplicon was discovered in the common breakpoint regions of Prader–Willi and Angelman syndrome deletions. We identified previously the large HERC2 transcript as an ancestral gene in this duplicon, with ∼11 HERC2-containing duplicons, and demonstrated that recessive mutations in mouse Herc2 lead to a developmental syndrome, juvenile development and fertility 2 (jdf2). We have now constructed and sequenced a genomic contig of HERC2, revealing a total of 93 exons spanning ∼250 kb and a CpG island promoter. A processed ribosomal protein L41 pseudogene occurs in intron 2 of HERC2, and putative VNTRs occur in intron 70 (28 copies, ∼76-bp repeat) and 3′ exon 40 through intron 40 (6 copies, ∼62-bp repeat). Sequence comparisons show that HERC2-containing duplicons have undergone several deletion, inversion, and dispersion events to form complex duplicons in 15q11, 15q13, and 16p11. To further understand the developmental role of HERC2, a highly conserved Drosophila ortholog was characterized, with 70% amino acid sequence identity to human HERC2 over the carboxy-terminal 743 residues. Combined, these studies provide significant insights into the structure of complex duplicons and into the evolutionary pathways of formation, dispersal, and genomic instability of duplicons. Our results establish that some genes not only have a protein coding function but can also play a structural role in the genome.
[The sequence data described in this paper have been submitted to GenBank under accession nos. AF189221 (Drosophila HERC2 partial cDNA), AC004583 (human HERC2 exons 1–52, genomic); AF224242–AF224257 (human HERC2 exons 54–70, partial genomic sequences); AF225400–AF225409 (human HERC2 exons 71–93, partial genomic sequences). The exon-intron boundaries for exons 53–93 are derived from BACs R-142A11 and 263O22. Additional information is available as a supplementary table at www.genome.org.]
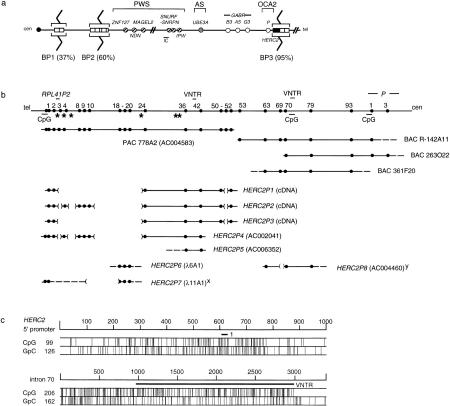
Although the underlying genetic defects in Prader–Willi syndrome (PWS) and Angelman syndrome (AS) are aberrations in imprinted gene expression, the majority of patients carry a cytogenetic deletion of chromosome 15q11–q13 (Nicholls et al. 1998). Low-copy repeats, or duplicons, have been mapped to the common deletion breakpoint regions (Buiting et al. 1998; Amos-Landgraf et al. 1999; Christian et al. 1999; Y. Ji, E. Eichler, S. Schwartz, and R.D. Nicholls, in prep.), including a large gene (HERC2) and partially duplicated paralogs, many copies of which are actively transcribed (Ji et al. 1999). The ancestral HERC2 gene gives rise to a 15.3-kb mRNA, and the duplicated paralogs to a family of 6- to 7-kb transcripts (Amos-Landgraf et al. 1999; Ji et al. 1999). The functional HERC2 gene was mapped just distal of P, suggesting the origin of the duplications in 15q13 (Fig. 1a; Ji et al. 1999). Several other ESTs also occur either within some HERC2-containing or independent duplicons but have not been characterized in detail (Christian et al. 1999). The duplicons containing HERC2 have undergone multiple genomic duplication events, resulting in at least 12 copies of partially duplicated segments, with 7 copies located at or close to the two proximal (15q11) deletion breakpoints, 3 copies at the distal (15q13) breakpoint, and 2 additional copies at the pericentromeric region of chromosome 16p (Buiting et al. 1998; Amos-Landgraf et al. 1999). The 15q11 and 15q13 duplicons (Fig. 1a) have been termed the END repeats because they flank the ends of the PWS/AS region (Amos-Landgraf et al. 1999).
Figure 1.
Genomic characterization of HERC2 and partially duplicated paralogs. (a) Model showing the position of HERC2-containing duplicons (□; HERC2, █) in chromosomes 15q11 and 15q13. These map close to or within breakpoint 2 (BP2) and BP3, whereas those shown hypothetically at BP1 are only known to map centromeric of BP2 (Amos-Landgraf et al. 1999). The frequency (%) of breakpoints (zigzag lines) at each location is shown. PWS, AS, and OCA2 are the three known disease loci in 15q11–q13 (Nicholls et al. 1998). Other symbols: (cen) centromere; (circles) genes [(hatched) paternal only; (open) nonimprinted; (shaded) maternal only]; (IC) imprinting center; (tel) telomere. (b) Genomic map with PAC/BAC clone contig of the HERC2 locus and comparison to eight of the duplicated loci. Numbers at top correspond to exon numbers of HERC2 (several exons of the flanking P gene, two VNTRs, and the RPL41P2 gene are also shown). (CpG) CpG islands; (*) simple repeats of five or more copies in 5′ HERC2 (see Table 1). Broken lines represent an unknown extent for that particular end of a clone; parentheses [( )] represent genomic deletions of HERC2 in each duplicon. (x) Intron 9 and exon 18 are joined together in this clone; (y) sequence homologous to intron 55 was also identified 7 kb 5′ of exon 63 homologous sequence in this clone. (c) CpG content of the HERC2 5′ promoter region (nucleotide 4101–5100 of PAC 778A2 sequence) and CpG island upstream and spanning the intron 70 VNTR. Exon 1 and the intron 70 putative VNTR are shown as bars.
The large 528-kD HERC2 protein contains several motifs, including three RCC1-like domains, a carboxy-terminal HECT domain, and a ZZ-type zinc finger. HERC2 is also distantly related to HERC1 (p532) and HERC3, both of which also contain a carboxy-terminal HECT domain and at least one RCC1-like domain (Ji et al. 1999). HERC1 has been suggested to function in vesicular trafficking pathways on the basis of localization to both the cytosol and the Golgi apparatus, and interaction with clathrin heavy chain (Rosa et al. 1996; Rosa and Barbacid 1997), whereas no functional studies have been performed on HERC3 (Nomura et al. 1994). Based on the conserved motifs, mouse mutation studies, and by analogy to the related HERC1, it has been suggested that human HERC2 may act as a guanine nucleotide exchange factor and E3 ubiquitin ligase, with function in protein trafficking and degradation pathways in the cell (Ji et al. 1999). Recently, the mouse Herc2 gene has been cloned, mutations of which were shown to cause severe developmental delay, jerky gait, sterility, and juvenile lethality in a recessively inherited manner (jdf2, or rjs; Lehman et al. 1998; Ji et al. 1999). Mutations of HERC2 have so far not been linked to any human genetic disorder. Although PWS and AS deletion patients are hemizygous for 3′ HERC2 (Ji et al. 1999), there is no evidence that this gene contributes to any of the PWS or AS phenotypes, consistent with a recessive gene.
To provide a framework for characterizing the role of HERC2 in human disease and the structure and evolution of chromosome 15q11–q13 duplicons, we constructed a genomic contig spanning the HERC2 locus and determined the exon–intron structure of HERC2. A highly conserved Drosophila HERC2 ortholog was also characterized. A comparison at the genomic level of the HERC2 structure with eight partially duplicated copies allows an understanding of the structure and evolutionary formation of several HERC2-containing duplicons.
RESULTS
Construction of a Genomic PAC and BAC Contig of HERC2
Of 30 positive PAC clones identified using a 5′ HERC2 probe, only 1 (778A2) contained an exonic sequence tagged site (STS) identical to HERC2. By Southern hybridization using various 5′ HERC2 probes, this PAC contained >7.9 kb of HERC2 coding sequence (data not shown). In addition, it is positive for a CpG island probe homologous to a duplicated copy of HERC2 (Amos-Landgraf et al. 1999), suggesting 778A2 contains the putative HERC2 promoter (Fig. 1b). Three BAC clones were isolated spanning 3′ HERC2 (see Methods), and all were positive by STS PCR for the 3′ UTR of HERC2, which is unique (Ji et al. 1999). Typing of other HERC2 STSs (see Methods) and those from the P gene (Lee et al. 1995) showed the extent of each genomic clone (Fig. 1b). Combined with results from PAC and BAC clone sequencing (see below), the data indicate that the contig covers all HERC2 exons, with 778A2 covering HERC2 exons 1–52, R-142A11 exons 53 to the 3′ UTR (exon 93), 263O22 exons 70–93, and 361F20 starting 5′ of exon 63 and extending beyond exon 93 (Fig. 1b).
Genomic Organization of the HERC2 Locus
We have sequenced all the exon–intron boundaries of HERC2, which demonstrates that the 15.3-kb cDNA is encoded by 93 exons (Table 1). All exon–intron boundaries conform to the consensus splice sequences (Maquat 1996). The transcription initiation of HERC2 is putatively under the control of a CpG-island promoter (Fig. 1c). Exon 1 is the smallest exon (30 bp), and exon 93 is the largest exon (997 bp), with an average exon size of 164 bp. The translational initiation codon ATG is in exon 2, and the stop codon (TAA) is in exon 93. Although the size of some 3′ introns has not been determined, intron size varies from 77 bp (intron 32) to 21,847 bp (intron 2). The first 52 exons, within PAC 778A2, occupy a genomic distance of 132 kb. Because the last 41 exons and introns span a minimum of ∼66 kb and based on the size of 3′-HERC2 BACs, we estimate that the HERC2 genomic locus spans 200–250 kb.
Table 1.
Exon–Intron Organization of the Human HERC2 Gene
| Exon no. | cDNA positiona | Exon first baseb | Intron–exon boundary | Exon size | Exon–intron boundary | Intron sizec |
|---|---|---|---|---|---|---|
| 1 | 1 | 4708 | GAGGCG | 30 | CGTCAGgtcctggcct | 610 |
| 2 | 31 | 5348 | ttacttgcagGCCTGA | 103 | ATACAGgtggggtttg | 21847d |
| 3 | 134 | 27298 | tcattgccagCTTGCA | 115 | GAAAAGgtaagggcct | 6377e |
| 4 | 249 | 33790 | aattctgcagATGATA | 135 | AACCAGgtaatcttgt | 12596 |
| 5 | 384 | 46521 | tattttctagATGTGA | 220 | AAAAAGgtaacaataa | 5062f |
| 6 | 604 | 51803 | tcgtctgcagTTCCCG | 101 | GATCAGgtacggtccc | 457 |
| 7 | 705 | 52361 | gtgcgcccagGCGAGG | 157 | GACGGGgtgagttctt | 1286 |
| 8 | 862 | 53804 | ttcatttcagGGATGT | 111 | GCTGAGgtgagggctg | 507 |
| 9 | 973 | 54422 | tctcctttagCCAAAT | 172 | ATGCACgtgagtgtca | 1346 |
| 10 | 1145 | 55940 | tcctttctagCTTTTG | 174 | CATAAGgtgtgtgtgc | 1258 |
| 11 | 1319 | 57372 | tatcttacagGGATCA | 189 | ACGCTGgtgagtgttc | 631 |
| 12 | 1508 | 58192 | ccttctttagGCCCCA | 152 | CACTGTgtatgtatcg | 2490 |
| 13 | 1660 | 60834 | tctccccaagGCCTTT | 158 | GCCATGgtacgtctgc | 85 |
| 14 | 1818 | 61077 | ctgtttgaagGCTCCA | 114 | AGAACGgtacgtagag | 2448 |
| 15 | 1932 | 63639 | tattttttagGGCAAG | 252 | TGCAAGgtgagtgcaa | 1947 |
| 16 | 2184 | 65838 | tcccttgtagGGAAGA | 194 | GCCCAGgtactgaata | 3515 |
| 17 | 2378 | 69547 | ttcctcttagAGCTTT | 201 | CTTCAGgtattcatga | 743 |
| 18 | 2579 | 70491 | ctcctttcagTTGCAT | 229 | GCGCAGgtgggcctgg | 92 |
| 19 | 2808 | 70812 | tctatttcagTTTCAG | 125 | ATCCAGgtatggcttt | 1353 |
| 20 | 2933 | 72290 | tgaattacagGATATT | 179 | TCTTAGgtaaatcgta | 5603 |
| 21 | 3112 | 78072 | tcctgtatagAAACAT | 185 | TTTCTAgtaagttgct | 1653 |
| 22 | 3297 | 79910 | cttcttcaagGTCCAG | 156 | TTACTGgtaccttttg | 675 |
| 23 | 3453 | 80741 | ttaaatgtagGTGTTC | 186 | TAATGGgtacggcgtc | 7108g |
| 24 | 3639 | 88035 | gtatatacagAGTCAT | 171 | TTCTTGgtaagattac | 384 |
| 25 | 3810 | 88590 | tttgtttcagCTCAGT | 104 | TTGGAGgtgaggctgt | 1000 |
| 26 | 3914 | 89694 | atttttccagCCTGAC | 151 | GTGCCAgtaagaaaat | 2676 |
| 27 | 4065 | 92521 | cattttccagAATGGC | 215 | GTGAAGgtgagctagg | 273 |
| 28 | 4280 | 93009 | tctgttgcagGACTTT | 133 | ATTTAGgtaaggagct | 102 |
| 29 | 4413 | 93244 | ctttttacagGTCATG | 128 | ATTAAGgtgatagatt | 92 |
| 30 | 4541 | 93464 | aactttgcagACTCAT | 196 | AGAGAGgtaagaatgt | 2645 |
| 31 | 4737 | 96305 | taaaacacagTTCCTA | 134 | CCCAAGgtgcagtatt | 519 |
| 32 | 4871 | 96958 | cctttgttagGACAAA | 174 | AAACAGgtaacatttg | 77 |
| 33 | 5045 | 97209 | tattttgcagTTGGAG | 137 | TATAGGgtaaaacgtt | 113 |
| 34 | 5182 | 97459 | tatttgaaagGGAACC | 152 | AGCTTGgtgagtcaat | 785 |
| 35 | 5334 | 98396 | tgctcattagGTATCC | 192 | TGATTGgtaggtctgc | 6010hi |
| 36 | 5526 | 104598 | gctccatcagGCCCCA | 188 | GATCAGgtactcagag | 1383 |
| 37 | 5714 | 106169 | gctttctcagGATGGG | 193 | ACTCTGgtgggtgact | 1780 |
| 38 | 5907 | 108142 | ttttttttagAAGCCG | 183 | AGACATgtatggaatg | 2686 |
| 39 | 6090 | 111011 | cctcctgcagCTTCTC | 182 | AGGCAGgtaatgtgct | 818 |
| 40 | 6272 | 112011 | cttttatcagATCTTA | 147 | TCAGAGgtgggtggcc | 383 |
| 41 | 6420 | 112541 | tcctcctcagAGTCCA | 197 | GAAGGGgtgggtttgt | 103 |
| 42 | 6617 | 112841 | tctgcactagGCCCAG | 231 | AAACCAgtaggtgaac | 1158 |
| 43 | 6848 | 114230 | tcctctccagCTCCCT | 139 | TTGCAGgtcagtacat | 1299 |
| 44 | 6987 | 115668 | ccttccgcagGACAAG | 144 | ACACAGgtgtcttttt | 4619 |
| 45 | 7131 | 120431 | ttaaattcagATGATG | 143 | CTTGAGgtacagccat | 3652 |
| 46 | 7274 | 124226 | gtctgcccagGCTGCT | 268 | TGCCTGgtacttcgtt | 97 |
| 47 | 7542 | 124591 | cctctcttagGTGTGG | 137 | TCCATGgtcagtgcct | 558 |
| 48 | 7679 | 125286 | tttcttgcagTCTACT | 99 | ATTCAGgtgagtaatt | 2686 |
| 49 | 7778 | 128071 | ttaattctagGTGGGA | 169 | TTATAGgtgagcacat | 97 |
| 50 | 7947 | 128337 | acatctgtagGCTATC | 126 | TGAAAGgtaatattat | 1808 |
| 51 | 8073 | 130271 | tctgtcctagCTTTCA | 109 | GGTTACgtgagttatt | 106 |
| 52 | 8182 | 130486 | ttacaaatagGTGTGA | 140 | AACCAGgtatggcaga | >2015 |
| 53 | 8322 | 348 | cacgaaatagGTCAGT | 191 | GGAAAGgtagcatcta | >800 |
| 54 | 8513 | ttttctaaagCACTGG | 106 | TGTCAGgtaattcagt | 82 | |
| 55 | 8619 | tttttcctagGTGGAA | 92 | ACAGAGgtaagtagct | >1200 | |
| 56 | 8711 | ttgtgtctagTATCAC | 176 | CGGAAGgtgagaacca | >1200 | |
| 57 | 8887 | gtttggttagCCTCAT | 112 | TCCAAGgtacggcctg | >1150 | |
| 58 | 8999 | tctgttgtagATAAAG | 82 | TTGCAGgtatgattat | 110 | |
| 59 | 9081 | ttccttgcagTGACTG | 144 | ACTCAGgtacaagcca | >1100 | |
| 60 | 9225 | aatgttgcagGTGGCC | 91 | CAGAATgtaagggtac | 291 | |
| 61 | 9316 | cttgttgtagGAACTG | 178 | AAAATGgtgattatac | 181 | |
| 62 | 9494 | aattgagtagGTGAAA | 82 | ATGAAGgtgagtggct | 86 | |
| 63 | 9576 | ttaaacctagGTTTGG | 172 | GACATGgtacgtaaac | >1000 | |
| 64 | 9748 | cttctctcagGGGAAA | 145 | GGGCAGgtaaggctgc | 891 | |
| 65 | 9893 | tcgtaaacagGTGTAT | 226 | ACTTAGgtaacacaga | >1280 | |
| 66 | 10119 | cctcttatagGCGTGC | 172 | TGCCAGgtaggcttct | 893 | |
| 67 | 10291 | gtcctgacagAGATGC | 184 | AGAGAGgtaaaagcga | 578 | |
| 68 | 10475 | cttggactagATTGTT | 141 | AAAGAGgtgaagaggc | >1260 | |
| 69 | 10616 | tttgacttagGATGTT | 192 | CCACAGgtgagtctca | >1070 | |
| 70 | 10808 | tcctgaccagGTGGCA | 154 | TACCAGgtacaggggc | >6385 | |
| 71 | 10962 | tgtccttcagGTGCAG | 108 | GGTCAGgtaaggacag | 1433 | |
| 72 | 11070 | cctcccgcagGCCGAG | 132 | CTGCTGgtaaggaagg | 436 | |
| 73 | 11202 | tcattcacagGCCCTA | 159 | CCCTAGgtaaatgcca | 85 | |
| 74 | 11361 | ctcatttcagCTGCCA | 119 | GCTTTGgtatggagca | 904 | |
| 75 | 11480 | gtctccttagAGTTCT | 126 | TTTAAGgtaatgtttc | 434 | |
| 76 | 11606 | gtgcctgcagGTACTG | 156 | GATGAGgtattgcatg | 392 | |
| 77 | 11762 | ttttgtttagGTGGCT | 116 | GAACAGgttattatat | 101 | |
| 78 | 11878 | tgtattgtagGCGACC | 199 | GGGAAGgtaagagctc | >12000 | |
| 79 | 12077 | cttttcttagCTGTAT | 215 | CAGAAGgtatgatgtg | >4000 | |
| 80 | 12292 | ttttttgcagTCCGTG | 178 | AAGCTGgtgaggaggc | 391 | |
| 81 | 12470 | ttctctgaagGTGGAG | 162 | ATGAAGgtatttccca | 1507 | |
| 82 | 12632 | ccccctctagATTGAT | 92 | TACCTGgtacgcaaga | 200 | |
| 83 | 12724 | ttgtgtgcagGGGCAA | 140 | AGGATGgtaggtaggg | 4971 | |
| 84 | 12864 | cttcattcagGTGAGG | 188 | GCACAGgtgagtccgg | 770 | |
| 85 | 13052 | tgtgttctagGTCCCC | 198 | GGAAAGgtattcaagt | 2606 | |
| 86 | 13250 | tgtgttttagGAGGCG | 84 | ATCCAGgtagcacatg | >5000 | |
| 87 | 13334 | tcctctccagGTCAAA | 142 | TTGTGGgtgagaactt | >200 | |
| 88 | 13476 | tcccctgcagGTGAAT | 195 | TCCTGGgtgagctact | >2700 | |
| 89 | 13671 | gaatgatcagGTGTGT | 113 | AGTGAGgtaactccct | 627 | |
| 90 | 13784 | tccttaaaagGTTGAT | 191 | CTATAGgttggtggtc | 935 | |
| 91 | 13975 | tttcttccagACTCCA | 106 | ACGATGgtatgccgac | 290 | |
| 92 | 14081 | cctcccgcagGTGTGT | 213 | ATCCAGgtaggctcct | 1035 | |
| 93 | 14294 | tgctcaacagGTGTTG | 997 | TGACATaaaagtgtagj |
GenBank accession no. AF071172 (Ji et al. 1999).
Exons 1–52, GenBank accession no. AC004583 (PAC 778A2; exon 53, GenBank accession no. AQ388928 (BAC R-142A11 end).
(>) Minimum intron size based on contiguous flanking sequence.
(TA)22 begins at position 25031b.
(CA)15 begins at position 28775b.
(TG)18 begins at position 47153b.
(PuT)66 begins at position 81120b. Pu = purine, with (GT)20 the longest pure repeat.
(TCTG)5 begins at position 102604b.
(TG)20 begins at position 102638b.
Polyadenylation takes place within or just 5′ of the four underlined adenine residues.
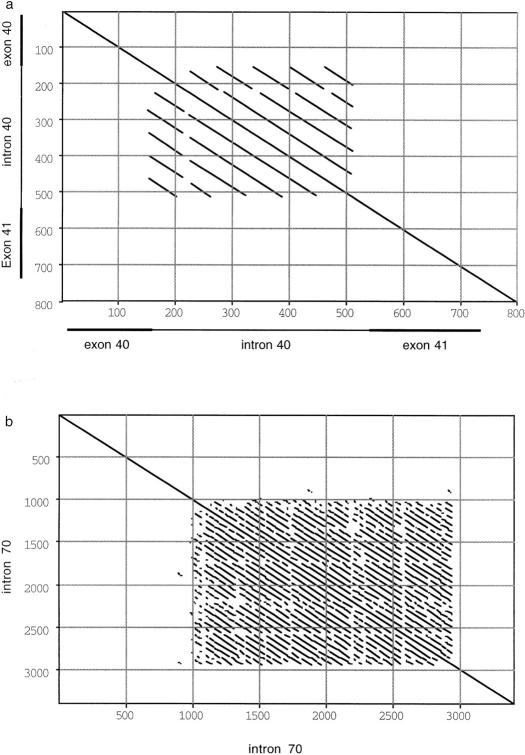
The genomic sequence of 5′ HERC2 (exons 1–52) has a moderate level of genome-wide repetitive elements that comprise 46% of the total sequence (SINE 21.20%, LINE 18.22%, LTR 3.48%, and other elements, 2.08%), with a GC content of 43.5%. Several simple repeats were identified, including five dinucleotide repeats with copy numbers >15, and a tetranucleotide repeat (Table 1; Fig. 1b). In addition, 43 copies of clustered TGAG were identified in intron 38, in a TG-rich region (109,548–110,202 nucleotides of PAC 778A2). We also identified two putative variable number of tandem repeat (VNTR) sequences within HERC2. One is located in intron 40, with six copies of an ∼62-bp sequence (range 60–63), each of >90% identity (Fig. 2a). Interestingly, the first copy of this repeat starts within exon 40, and the other five copies span virtually all of intron 40. Analysis of BAC 263O22 sequence identified, in intron 70, 28 copies of an ∼76-bp repeat monomer (range 64–79), each with >84% identity (Fig. 2b). This VNTR is adjacent to an intronic CpG island (Fig. 1c) that is of unknown function. With four to six CpG dinucleotides per repeat, a 3-kb CpG island can be defined that is larger than the average size for a CpG island (Bird 1986).
Figure 2.
Analysis of putative VNTRs in HERC2 intron 40 (a) and intron 70 (b). (a) Eight hundred base pairs of sequence (PAC 778A2, 112001–112800) is aligned against itself. Position 11–157 corresponds to exon 40, and 541–737 corresponds to exon 41. (b) The first 3400 bp of sequence from contig 27 (BAC 263O22) is aligned against itself. The parameters used for both analyses were a window of 30 nucleotides containing no less than 65% identity.
In comparing the 5′ (exons 1–52) genomic sequence with HERC2 cDNA, 13 sequence changes were identified. Two were cDNA sequence errors, at positions 328 (A replaces T) and 1228 (G replaces C), neither of which changes the translated protein sequence. Eleven changes represent putative single nucleotide polymorphisms (SNPs) (Supplemental table available at www.genome.org), five of which are silent and do not affect corresponding amino acid sequences. Six would cause amino acid changes, although three occur at amino acids that differ between human and mouse and may not be significant. Of the others, one is conservative (Leu → Phe), one may be conservative (His → Arg) if a positively charged residue is sufficient here, and one is nonconservative (Ser → Arg). These putative SNPs will aid assessment of the role of HERC2 in human disease.
A BLAST search of repeat-masked HERC2 genomic sequence identified multiple ESTs, but most show only 84%–99% homology to the genomic sequence, suggesting that they are transcribed from HERC2-related duplicated segments. These ESTs can be assigned to four classes. (1) These include ESTs that contain HERC2 exon-related sequence representing HERC2-related pseudogenes (Buiting et al. 1998; Ji et al. 1999), and (2) rare ESTs that contain homologous sequence to Alu or L1 elements, which may represent nonfunctional transcripts. A 99% homologous sequence to IMAGE clone 120151 (AF129928), identified in HERC2 intron 4, is actually mostly part of an ancient L1 element (69.2% identity) and hence most likely does not represent a unique gene in the chromosome 15 duplicon, contrary to a previous report (Christian et al. 1999). (3) A small number of ESTs contain only unique sequence, but as these ESTs do not cluster, they may not represent new genes and, rather, reflect background transcription in the genome. (4) A processed pseudogene for ribosomal protein L41 occurs within intron 2 of HERC2 (Fig. 1b). This L41 pseudogene (RPL41P2) was inserted into the 3′ end of an Alu repetitive element, with a 14-bp direct repeat (AAAAAATTATCTGG) flanking both ends of the pseudogene, whereas the functional L41 gene maps to chromosome 12 (Kenmochi et al. 1998).
Comparison of HERC2 to Partially Duplicated Paralogs
Our characterization of the HERC2 genomic locus now allows direct sequence comparisons to the HERC2-related portions of chromosome 15q11, 15q13, and 16p11.2 duplicons (see introductory section). For the HERC2P1 and P3 cDNA (Ji et al. 1999), HERC2 exons 3–23 and exon 51 are deleted, whereas exons 1, 2, 24–50, and 52 are present, and exons 53–93 are absent (Fig. 1b). For HERC2P2 (Ji et al. 1999), exons 4 and 8–10 are present as well (Fig. 1b). Most cDNA clones derived from HERC2P2–3 have retained intron 40, with six copies of the VNTR, whereas HERC2P1 has five VNTR copies. We noticed previously that the 3′ ends of these cDNAs do not match HERC2 cDNA sequence (Ji et al. 1999), nor do they contain intron 52 sequence, which suggests that the 3′ ends originate from sequence homologous to a 3′-HERC2 intron not covered by contiguous sequence (see below for origin of this sequence).
The sequence (AC002041) of a 234-kb BAC clone that maps to chromosome 16p11.2, and contains HERC2P4, has a contiguous 36.6-kb segment (from 50.8 kb to 87.4 kb) homologous to HERC2, including exons 24–42 (Fig. 1b). There are several deletions of 5′ HERC2 that include exons 3, 5–7, and 11–23 and flanking intron sequences. The genomic sequence of HERC2P4 also contains sequence homologous to the HERC2 CpG island, with exon 1 and 2, as well as exons 4 and 8–10 present, which is the same pattern as the cDNA representing HERC2P2 (Fig. 1b). This suggests that missing exons in the HERC2P1–3 cDNAs do result from genomic deletions, as we suggested previously (Ji et al. 1999), and not from alternative splicing (Christian et al. 1999). The last 8891 bp of sequence (AC006352) of a newly identified 126-kb BAC is homologous to HERC2 between exons 36 and 42 (Fig. 1b), in reverse orientation (an overlapping clone is needed to characterize the 5′ end of this paralog). This clone represents the putative second chromosome 16 locus (HERC2P5) based on the presence of identical diagnostic nucleotide sequences (Buiting et al. 1998; Ji et al. 1999). Both chromosome 16 loci have only three copies of the intron 40 VNTR. The homology of both chromosome 16 loci to HERC2 stops in intron 42 at an identical nucleotide. An L1 element is present in both chromosome 16 loci, but not in HERC2, immediately after the homologous sequence. The HERC2-related sequences between the two chromosome 16 loci are 99.64% identical, which is consistent with allelism; however, the flanking 60.5 kb of sequence shared between the two chromosome 16 BAC clones have a significantly lower (98.46%) sequence identity (E.E. Eichler, pers. comm.), suggesting that the two loci are paralogous and that they diverged from an ancestral sequence near the time of divergence of chimpanzee and human (∼6 mya; Goodman 1999). Therefore, either the HERC2-related sequence in one of the two chromosome 16 paralogous loci is of recent evolutionary origin, or gene conversion within HERC2-related sequence has led to homogenization of these sequences.
Two other related loci have been partially sequenced previously (HERC2P6 and P7; Amos-Landgraf et al. 1999). HERC2P6 (AF140516, AF140517) contains homologs of exons 18–20 with flanking intronic sequences, but this locus has not been further characterized. HERC2P7 (Fig. 1b) contains homologous sequences to the HERC2 promoter and exon 1 (AF140519), as well as intron 9 and exons 18–20 (AF140518). This locus has intron 9 joined to the 3′ half of exon 18, and the HERC2P7 EST (AF071178) was also found to contain this fusion, with intron 18 properly spliced. Two other ESTs (AA535902, AI688214) contain this fusion. They are 98% identical to AF071178 and 99% identical to each other. This suggests that there are additional loci with such a fusion event and that the retention of intron 9 in mRNA is not an isolated event.
The sequence (AC004460) of another 114-kb genomic clone contains a duplicated segment of HERC2 exons 63–79, excluding exon 69 (Fig. 1b). The 3′ boundary of this duplicon (denoted HERC2P8) may be 500 bp into intron 79, although as this is only 5 kb from the BAC end and as HERC2 genomic sequence is not complete for this region, HERC2P8 may contain additional 3′ sequence homologous to HERC2. The putative VNTR in intron 70 is present but has only 11 repeat copies. Within HERC2P8, 7 kb 5′ of exon 63, there is a small fragment (160 bp) homologous to HERC2 intron 55, in the same orientation as exons 63–79, suggesting that the duplicated segment is at least 36 kb. Interestingly, we also identified homologous sequence (97%) to the 3′ end of the HERC2P1–3 cDNAs in this clone, as two exons flank either side of the exon 63 homologous sequence but lie in the reverse orientation to HERC2. Each of these two sequences is flanked by consensus exon–intron boundaries, and the second of these two exons represents the last exon for the HERC2P1–3 cDNAs, including the polyadenylation signal. These observations suggest that there has been an inversion event during the formation of the current HERC2P1–3 duplicons, probably involving intron 52 (see above) to intron 63.
Characterization of the Drosophila HERC2 Ortholog
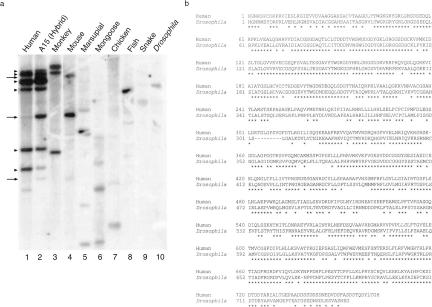
Our previous studies demonstrated that the human and mouse HERC2 cDNAs show an extraordinary 96% amino acid sequence identity (Ji et al. 1999). We extended this observation by Southern blot analysis of genomic DNA from several animal species (Fig. 3a). Under moderate hybridization conditions, HERC2-homologous sequences are detected in all mammals and other vertebrate species tested, as well as in the fruit fly. Nevertheless, no HERC2 homolog is present in the sequenced genomes of Caenorhabditis elegans (The C. elegans Sequencing Consortium 1998) and Saccharomyces cerevisiae (Mewes et al. 1997). In contrast, database search identified a Drosophila EST (AA567486) highly homologous to part of the HECT domain of HERC2. Using nested primers from the 5′ end of the Drosophila EST and a conserved primer from the RLD3 encoded domain of human and mouse HERC2, we isolated additional Drosophila HERC2 cDNA sequences (see Methods). Combined, we have obtained 2945 bp of Drosophila HERC2 cDNA, encoding a partial open reading frame (ORF) of 743 amino acids. The overall amino acid sequence identity between HERC2 and the carboxy-terminal Drosophila ORF is 70%, with long stretches of sequence identity between the two species (Fig. 3b).
Figure 3.
Identification of an evolutionary highly conserved Drosophila HERC2 gene. (a) Zooblot analysis of HERC2. A15 is a mouse–human somatic cell hybrid of which the only retained human chromosome is chromosome 15. (Monkey) African green monkey; (marsupial) Sminthopsis macroura. Arrows or arrowheads indicate bands in A15 that are of mouse or human origin, respectively. (Lanes 1–3) A 3-day autoradiograph exposure; (lanes 4–10) a 7-day exposure. (b) Amino acid sequence comparison of the carboxy-terminal 762 residues of human HERC2 with the partial Drosophila HERC2 (743 residues). Asterisks (*) indicate identical residues.
A BLAST search using the partial Drosophila cDNA identified a working draft sequence from Drosophila BAC R30J04 (GenBank accession no. AC008338) as containing HERC2. HERC2-homologous sequences in the Drosophila BAC span the entire human gene, with only minor gaps, starting from residue 14 of HERC2 amino acid sequence. Sequence identity is highest in regions with functionally important motifs, including the three RCC1-like domains and the HECT domain (Ji et al. 1999). The ZZ-type zinc finger in human and mouse HERC2 (Ji et al. 1999) has degenerated in Drosophila, whereas the DOC domain (Grossberger et al. 1999) immediately following the zinc finger is conserved. Six introns have been identified in Drosophila HERC2, none of whose positions are conserved in the human ortholog. Nevertheless, the complete gene structure will not be known until the BAC sequence is completed. Based on several lines of evidence, Drosophila HERC2 maps to band 19C–E of the X chromosome. This location for R30J04 was mapped by the Berkeley Drosophila Genome Project. Sequence identical to AC008338 is also present in BACs R41N19 (GenBank accession no. AC009217; 19A–C) and 48H01 (BAC end sequence; 19C1–C2). Furthermore, two STSs mapped to the same band as R30J04, Dm25C7 and Dm0500, are within the Drosophila HERC2 gene. Finally, R30J04 also contains two known genes mapped previously to the 19C–D region, pp4 (19C1–2; Helps et al. 1998) and PBPRP-2 (19D; Pikielny et al. 1994).
DISCUSSION
Sequence analysis has revealed that the HERC2 genomic locus, encoding a putative giant protein of 528 kD (Ji et al. 1999), comprises 93 exons. The only other characterized genes with >90 exons are the type VII collagen gene (COL7A1), with 118 exons (Christiano et al. 1994; Kivirikko et al. 1996), and the perlecan gene (HSPG2), with 94 exons (Cohen et al. 1993). The largest gene identified in chromosome 22 sequence has only 54 exons (Dunham et al. 1999). Therefore, with the possible exception of the 3-megadalton titin protein that is uncharacterized at the genomic level, the number of exons in COL7A1, HSPG2, and HERC2 may represent the upper limit that a gene can have. This maximum size may result from the accuracy and time with which splicing can occur for such highly fragmented genes. Interestingly, ENU mutagenesis studies suggest that Herc2 is the most mutable mouse locus studied so far (Walkowicz et al. 1999; E.M. Rinchik, pers. comm.). This may be due to the large number of exon–intron boundaries, because the mouse gene is likely to have the same structure, or to the large target size of the 15.3-kb Herc2 exons and encoded ORF. We have identified three splice site mutations in ENU-generated jdf2 animals (Ji et al. 1999). Alternatively, the exceptionally high mouse–human identity suggests that even minor changes of HERC2 amino acid sequence could lead to dysfunction of the protein, consistent with the high rate of ENU mutation.
Recently, Shiraishi et al. (1999) isolated DNA fragments representing methylated CpG islands in human adenocarcinomas of the lung, one (AB077148) that is 99% identical to the HERC2 CpG island promoter. A PCR-based investigation of methylation status of AB077148 in genomic DNA showed differential methylation in both cancerous and noncancerous lung tissue of the same patients, suggesting it may be imprinted (Shiraishi et al. 1999). However, HERC2 is expressed from both maternal and paternal alleles and hence is not imprinted in either human or mouse (Gabriel et al. 1998, 1999; Ji et al. 1999). Consistent with this, Herc2 mutations in the jdf2 syndrome are recessive (Lehman et al. 1998; Ji et al. 1999). The differential methylation observed by Shiraishi et al. (1999) could result from an inability of the PCR-based assay to distinguish the HERC2 promoter and the estimated nine additional duplicated copies (Amos-Landgraf et al. 1999; this paper). Because at least four copies of HERC2-related CpG islands are pericentromeric in 15q11.1 and 16p11, these may be methylated. Alternatively, it is possible that the “noncancerous tissue” (Shiraishi et al. 1999) is actually precancerous and that HERC2 and/or related sequences may be targets of silencing by methylation during tumorigenesis.
The HERC2 gene is evolutionary highly conserved, with human and Drosophila HERC2 showing 70% identity over the carboxy-terminal 743 amino acids. Further analysis of Drosophila HERC2 genomic sequence suggests that much of the protein is functionally important, particularly the three RCC1-like domains and the HECT domain. In contrast, the ZZ zinc finger (Ji et al. 1999) is not present in the fly HERC2, suggesting some differences in protein–protein interactions compared with human HERC2. The high degree of homology across mammalian and invertebrate species indicates that HERC2 plays a conserved role in the cell. Identification of mutations in Drosophila HERC2, based on chromosomal location, may allow comparison to the jdf2 mice to better understand the developmental function of HERC2, which may help predict more accurately the potential human phenotype expected for HERC2 mutations.
In the finished genomic sequence, the only gene identified other than HERC2 was a processed pseudogene (RPL41P2), inserted into an Alu element in HERC2 intron 2. Although it is unknown whether the two putative VNTRs identified within HERC2 are polymorphic in human populations, this appears likely as there is variation between the copy number of both VNTRs in HERC2 and that for HERC2-containing duplicons. Most copies of the intron 40 VNTR contain exon 40 coding sequence and the exon–intron boundary, although the effect on intron 40 splicing is unknown. Interestingly, this intron is not spliced in most transcripts from HERC2P1–3, despite retention of five to six VNTR copies. The large VNTR in intron 70 is preceded by a CpG-rich region. The same genomic region in mouse has been suggested to contain a regulatory element for p gene expression (Walkowicz et al. 1999). It is possible that this VNTR, together with the CpG island, has this function in mouse and human. However, further studies in the mouse and analysis of whether transcripts are produced from the intronic CpG island will be necessary to determine its function.
Previous evidence suggests that there are seven HERC2-containing duplicons in 15q11, including five with a copy of the HERC2 5′ CpG island and two copies that do not have the CpG island but contain other 5′ sequences (Amos-Landgraf et al. 1999). Similarly, three duplicons in 15q13 each carry the 5′ CpG island (Amos-Landgraf et al. 1999), the most proximal of which is the ancestral HERC2 gene (Ji et al. 1999). Our previous (Buiting et al. 1998; Amos-Landgraf et al. 1999) and current studies also define two HERC2-containing duplicons in chromosome 16p11.2, for a total of 12 loci containing sequence from the 5′ half of HERC2. Although exon 93 of HERC2 is unique in the human genome (Ji et al. 1999), we demonstrated here that HERC2P8 contains sequences paralogous to intron 55 to exon 79, in the absence of further 5′ sequences, suggesting that there are even more than 12 HERC2 duplicons in the genome. However, many of these duplicons may be highly fragmented and represent distinct subfamilies. Christian et al. (1999) independently identified duplicons at or near the proximal and distal PWS/AS breakpoints, by STS content mapping in YACs and by interphase FISH. Proximal breakpoint 2 (15q11) was suggested to be a single duplicon of ∼400 kb in size, in inverted orientation to two duplicon copies in 15q13 (Christian et al. 1999). However, these studies cannot discriminate between closely related and closely spaced repeats, nor divergent copies; hence, we suggest that these studies have underestimated the number of duplicons in the breakpoint regions. Our results indicate that one end of the END repeat duplicons is within 3′ HERC2 (between exons 79 and 93). The HERC2-related content is 36.6 kb in the most rearranged/deleted duplicon in 16p11.2, but HERC2 sequences from exon 1 to at least exon 79 (spanning 150–200 kb) are included in some chromosome 15 duplicons. However, the endpoint of the duplicon 5′ of HERC2 is unknown at this time. Christian et al. (1999) identified five ESTs and two PAC ends homologous to genes within the duplicated regions, which suggests that additional genes or pseudogenes may be present in the duplicons or that several classes of unrelated duplicons may be interspersed in these regions. However, not all these ESTs represent new genes. One EST (A006B10) is from a duplicated HERC2 locus, and a second (A008B26) is homologous to HERC2 intron 4 (and corresponds to an L1 element). According to their putative map positions (Christian et al. 1999), the two PAC-end gene sequences should be within the 3′ end of the HERC2 locus and may lie in unfinished intronic regions, perhaps adjacent to the CpG island in intron 70. This leaves three potential genes within the END repeats outside and telomeric of HERC2. One is MYLE, a 1-kb transcript encoding a putative 68-amino-acid protein, whereas the other two ESTs (SHGC17218 and SGC32610) have not been characterized. Taken together, duplicons in the PWS/AS deletion breakpoint regions are clearly complex in structure and arrangement, with HERC2 a major component. It will, however, be necessary to build and sequence complete clone contigs of these duplicons to gain a full understanding of the complexity of these sequences.
We have shown previously that a stable putative fusion HERC2 to HERC2-related transcript could be detected by Northern analysis in one of five PWS/AS deletion patients (Amos-Landgraf et al. 1999). Given that distal breakpoints could also occur in HERC2-related sequences telomeric of HERC2 and that some fusion genes may not produce stable transcripts, many PWS/AS deletion breakpoints may occur within the HERC2-related portions of the END repeats. The newly identified microsatellite and VNTR sequences within HERC2 should help identify the positions of PWS/AS breakpoints within or distal to HERC2. Similar studies will also determine the potential role of HERC2-containing duplicons in other chromosome 15q11–q13 rearrangements, including duplications (Clayton-Smith et al. 1993a; Browne et al. 1997; Repetto et al. 1998), triplications (Schinzel et al. 1994; Cassidy et al. 1996), inversions (Clayton-Smith et al. 1993b), and inverted duplications [inv dup(15)] (Robinson et al. 1993; Huang et al. 1997; Wandstrat et al. 1998). Other chromosome-specific duplicons are implicated in many additional chromosomal rearrangements (Lupski 1998; Y. Ji, E. Eichler, S. Schwartz, and R.D. Nicholls, in prep). Although some involve a simple, low-copy repeat, others show a complexity comparable to the duplicons we have described in 15q11, 15q13, and 16p11 (Y. Ji, E. Eichler, S. Schwartz, and R.D. Nicholls, in prep). For example, four genes occur in the large (>200-kb) duplicons mediating the deletion in Smith–Magenis syndrome (Chen et al. 1997). Duplicons in chromosome 22q11 (LCR22s) are also very complex, with eight copies of the LCR22s ranging from ∼20 kb to >200 kb (Dunham et al. 1999; Edelmann et al. 1999a,b). Multiple genes/pseudogenes map within each of the LCR22 duplicons (Collins et al. 1997; Dunham et al. 1999; Edelmann et al. 1999a,b), with duplications, deletions, and inverted duplications predominantly mediated by three of these duplicons (Edelmann et al. 1999b). The mechanism in homologous chromosome rearrangements involving simple duplicons (Y. Ji, E. Eichler, S. Schwartz, and R.D. Nicholls, in prep.) is now thought to involve double-strand break repair (Lupski 1998; Lopes et al. 1999), but the mechanisms in cases involving complex duplicons are not known as breakpoints have not been characterized. Further studies of complex duplicons will determine how and why these sequences are genetically unstable in both evolutionary terms of expansion and dispersal, and their role in mediating chromosome rearrangements in genetic diseases.
METHODS
Isolation of PAC and BAC Clones
We screened a human genomic PAC library (RPCI-4) with a 1.1-kb HERC2 cDNA probe (probe C, cDNA coordinates 2612–3714; Ji et al. 1999) using standard hybridization and washing conditions (Church and Gilbert 1984) and isolated a total of 30 positive clones. These may represent six or more different loci because the library has a fivefold coverage of the human genome and probe C contains a 5′ region of HERC2 that is duplicated. PCR primers RN304 (Ji et al. 1999) and RN305 (5′-ACCAGCCACTCTGCAGCACG-3′) were used to amplify a 107-bp STS (which corresponds to part of exon 18 of HERC2) from seven of the PAC clones, and the products were cloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA) and sequenced. Five contained sequence identical to HERC2P6 (λ6A1; Ji et al. 1999), one identical to HERC2P7 (λ11A1; Ji et al. 1999), and PAC 778A2 has the same sequence as HERC2 cDNA. An EagI sequence variant present in HERC2 but not HERC2P6 nor HERC2P7 was identified in this STS. Based on STS PCR and EagI digestion, none of the remaining 23 PACs contain sequence identical to HERC2.
Two STSs were used to screen by PCR the RPCI-11 human BAC library (Research Genetics, Huntsville, AL). A single positive clone (263O22) was isolated using a 171-bp STS [primers RN638 (5′-TCGTGAGTCGTCTTGATTGTAT-3′, starts from nucleotide 14940 of HERC2 cDNA) and RN637 (5′-CTTCTGGTTTTTCATTTTGGTT-3′, ends in nucleotide 15110)] from the unique 3′ UTR of HERC2 (Ji et al. 1999). Multiple positive genomic clones were isolated using primers RN911 (5′-GTTTGGTATTTTCCTGGGGTGATG-3′) and RN912 (5′-ACCCCCTGTCCATTTAGTCTCTCA-3′), which corresponds to a duplicated portion of HERC2 cDNA sequence (9576–9681) (see Results). Only one (361F20) was also positive for the HERC2 3′ UTR and hence is derived from the HERC2 locus. By screening the TIGR BAC-end database (http://www.TIGR.ORG) with HERC2 cDNA sequence, we identified R-142A11 as positive for HERC2 exon 53 (see Results), and further characterization of this BAC showed that it is also positive for the HERC2 3′ UTR.
Other STS primers used for typing PACs and BACs in this study include RN599 (5′-ACTGGACTGGGTTGCTATCAGAAAT-3′) and RN600 (5′-CACAAAAATCAAAGTCATCA CAGTTT C-3′) for HERC2 exons 51–52 and RN687 (5′-AGTGATGGGTCTGTGAATGG-3′) and RN690 (5′-TTCCCCATCATTTTCTCCCAGCAG-3′) for HERC2 exons 72–74, as well as primers for exons of the P gene (Lee et al. 1995).
Sequence Analysis of HERC2 Genomic Clones and Related Sequences
PAC 778A2 and BAC 263O22 were shotgun subcloned into an M13 phage vector and sequenced. PAC 778A2 sequence was finished, whereas BAC 263O22 was only partially sequenced and assembled into 22 contigs of at least two overlapping sequence reads. From the latter sequence, 22 exons were identified, including HERC2 exons 70–93 with the exception of exons 80 and 88. P exon 3 is present in one 263O22 sequence contig. Both ends of BAC R-142A11 were sequenced (Cleveland Genomics, Cleveland, OH). To identify exon–intron boundaries for exons 54–69, as well as exons 80 and 88, primers were designed from HERC2 cDNA sequence (primer sequences available from the corresponding author) and used to directly sequence from BAC clones R-142A11 and 263O22 (Cleveland Genomics).
HERC2 exons and polymorphisms were identified by pairwise sequence alignment (http://dot.imgen.bcm.tmc.edu:9331) of genomic sequence with HERC2 cDNA sequence (GenBank accession no. AF071172; Ji et al. 1999). RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) was used to analyze genome-wide repetitive elements and simple repeats and to calculate repeat and GC contents. The MacVector software package was used to characterize the VNTR sequences. EST and gene homologs were identified using BLAST (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast). BLAST and pairwise sequence alignments were used to analyze the duplicated HERC2 loci.
Analysis of Drosophila HERC2
Southern hybridization of zooblots was performed by standard methods (Sambrook et al. 1989), with probe C (Ji et al. 1999), using 30% formamide prehybridization and hybridization solutions, and a final wash at 45°C with 2× SSC and 0.1% SDS. A Drosophila EST (GenBank accession no. AA567486) homologous to 3′ HERC2 was identified in the dbEST database by BLAST. Sequence analysis revealed a 1372-bp cDNA fragment, with a 661-bp ORF and a 3′ UTR of 711 bp. Two nested primers were designed from the 5′ end of this clone and used, together with a human HERC2 primer (RN667; Ji et al. 1999), to PCR amplify an additional 5′ cDNA from an oocyte SMART cDNA library (Clontech, Palo Alto, CA). Primers RN808 (5′-GAGAGCAAAGGCACTGGAATCACC-3′) and RN667 were used for first-round PCR (annealing at 55°C for 30 sec and extension at 68°C for 2 min), then RN807 (5′-TTCCTGGGCGAGATGTGCGTGTAG-3′) and RN667 were used for nested PCR (annealing at 60°C for 30 sec, and extension at 72°C for 2 min). PCR was performed with the Advantage cDNA PCR Kit (Clontech); the 1.6-kb nested PCR product was cloned into the pCR2.1 vector (Invitrogen) and sequenced. A BLAST search of the High Throughput Genomic Sequences database, using compiled Drosophila HERC2 cDNA sequence, identified BAC R30J04 (GenBank accession no. AC008338) as containing Drosophila HERC2. BLAST searches and pairwise sequence alignments, using the human HERC2 protein sequence and AC008338 translated in all six frames, identified additional amino-terminal Drosophila HERC2 sequences in this BAC.
Acknowledgments
We thank Drs. Todd A. Gray and Evan E. Eichler for helpful discussions, James M. Amos-Landgraf and Tao Pan for technical contributions, and Drs. Evan E. Eichler and Peter J. Harte for critical reading of the manuscript. This work was funded by a Neuromuscular Disease Research Program Grant from the Muscular Dystrophy Association (R.D.N.) and the National Institutes of Health (R.A.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rxn19@po.cwru.edu; FAX (216) 368-3432.
REFERENCES
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstradt A, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Browne CE, Dennis NR, Maher E, Long FL, Nicholson JC, Sillibourne J, Barber JCK. Inherited interstitial duplications of proximal 15q: Genotype-phenotype correlations. Am J Hum Genet. 1997;61:1342–1352. doi: 10.1086/301624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Gross S, Ji Y, Senger G, Nicholls RD, Horsthemke B. Expressed copies of the MN7 (D15F37) gene family map close to the common deletion breakpoints in the Prader-Willi/Angelman syndromes. Cytogenet Cell Genet. 1998;81:247–253. doi: 10.1159/000015039. [DOI] [PubMed] [Google Scholar]
- Cassidy SB, Conroy J, Becker L, Schwartz S. Paternal triplication of 15q11-q13 in a hypotonic, developmentally delayed child without Prader-Willi or Angelman syndrome. Am J Med Genet. 1996;62:206–207. [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology [published erratum appears in Science 1998 283 35] Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Christiano AM, Hoffman GG, Chung-Honet LC, Lee S, Cheng W, Uitto J, Greenspan DS. Structural organization of the human type VII collagen gene (COL7A1), composed of more exons than any previously characterized gene. Genomics. 1994;21:69–79. doi: 10.1006/geno.1994.1239. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, Webb T, Cheng XJ, Pembrey ME, Malcolm S. Duplication of chromosome 15 in the region 15q11-13 in a patient with developmental delay and ataxia with similarities to Angelman syndrome. J Med Genet. 1993a;30:529–531. doi: 10.1136/jmg.30.6.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, Driscoll DJ, Waters MF, Webb T, Andrews T, Malcolm S, Pembrey ME, Nicholls RD. Difference in methylation patterns within the D15S9 region of chromosome 15q11-q13 in first cousins with Angelman syndrome and Prader-Willi syndrome. Am J Med Genet. 1993b;47:683–686. doi: 10.1002/ajmg.1320470519. [DOI] [PubMed] [Google Scholar]
- Cohen IR, Grassel S, Murdoch AD, Iozzo RY. Structural characterization of the complete human perlecan gene and its promoter. Proc Natl Acad Sci. 1993;90:10404–10408. doi: 10.1073/pnas.90.21.10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, Mungall AJ, Badcock KL, Fay JM, Dunham I. The organization of the γ-glutamyl transferase genes and other low copy repeats in human chromosome 22q11. Genome Res. 1997;7:522–531. doi: 10.1101/gr.7.5.522. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet. 1999a;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999b;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Gabriel JM, Higgins MJ, Gebuhr TC, Shows T, Saitoh S, Nicholls RD. A model system to study genomic imprinting of human genes. Proc Natl Acad Sci. 1998;95:14857–14862. doi: 10.1073/pnas.95.25.14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel JM, Merchant M, Ohta T, Ji Y, Caldwell RG, Ramsey MJ, Tucker JD, Longnecker R, Nicholls RD. A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and Angelman syndrome. Proc Natl Acad Sci. 1999;96:9258–9263. doi: 10.1073/pnas.96.16.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. The genomic record of Humankind's evolutionary roots. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossberger R, Gieffers C, Zachariae W, Podtelejnikov AV, Schleiffer A, Nasmyth K, Mann M, Peters JM. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J Biol Chem. 1999;274:14500–14507. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- Helps NR, Brewis ND, Lineruth K, Davis T, Kaiser K, Cohen PT. Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J Cell Sci. 1998;111:1331–1340. doi: 10.1242/jcs.111.10.1331. [DOI] [PubMed] [Google Scholar]
- Huang B, Crolla JA, Christian SL, Wolf-Ledbetter ME, Macha ME, Papenhausen PN, Ledbetter DH. Refined molecular characterization of the breakpoints in small inv dup(15) chromosomes. Hum Genet. 1997;99:11–17. doi: 10.1007/s004390050301. [DOI] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, Tanaka T, Page DC. A map of 75 human ribosomal protein genes. Genome Res. 1998;8:509–523. doi: 10.1101/gr.8.5.509. [DOI] [PubMed] [Google Scholar]
- Kivirikko S, Li K, Christiano AM, Uitto J. Structure of mouse type VII collagen reveals evolutionary conservation of functional protein domains and genomic organization. J Invest Dermatol. 1996;106:1300–1306. doi: 10.1111/1523-1747.ep12349019. [DOI] [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Jong MT, Fukai K, Spritz RA. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics. 1995;26:354–363. doi: 10.1016/0888-7543(95)80220-g. [DOI] [PubMed] [Google Scholar]
- Lehman AL, Nakatsu Y, Ching A, Bronson RT, Oakey RJ, Keipo-Hrynko N, Finger JN, Durham-Pierre D, Horton DB, Newton JM, et al. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc Natl Acad Sci. 1998;95:9436–9441. doi: 10.1073/pnas.95.16.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Tardieu S, Silander K, Blair I, Vandenberghe A, Palau F, Ruberg M, Brice A, LeGuern E. Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: A study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum Mol Genet. 1999;8:2285–2292. doi: 10.1093/hmg/8.12.2285. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- Mewes HW, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver SG, et al. Overview of the yeast genome [published erratum appears in Nature 1997, 387: 737] Nature (Suppl.) 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Repetto GM, White LM, Bader PJ, Johnson D, Knoll JHM. Interstitial duplications of chromosome region 15q11q13: Clinical and molecular characterization. Am J Med Genet. 1998;9:82–89. doi: 10.1002/(sici)1096-8628(19980901)79:2<82::aid-ajmg2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Binkert F, Gine R, Vazques C, Miller W, Rosenkranz W, Schinzel A. Clinical and molecular analysis of five inv dup(15) patients. Eur J Hum Genet. 1993;1:37–50. doi: 10.1159/000472386. [DOI] [PubMed] [Google Scholar]
- Rosa JL, Barbacid M. A giant protein that stimulates guanine nucleotide exchange on ARF1 and Rab proteins forms a cytosolic ternary complex with clathrin and Hsp70. Oncogene. 1997;15:1–6. doi: 10.1038/sj.onc.1201170. [DOI] [PubMed] [Google Scholar]
- Rosa JL, Casaroli-Marano RP, Buckler AJ, Vilaro S, Barbacid M. p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins [published erratum appears in EMBO J. 1996, 15: 5738] EMBO J. 1996;15:4262–4273. [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schinzel AA, Brecevic L, Bernasconi F, Binkert F, Berthet F, Wuilloud A, Robinson WP. Intrachromosomal triplication of 15q11-q13. J Med Genet. 1994;31:798–803. doi: 10.1136/jmg.31.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M, Chuu YH, Sekiya T. Isolation of DNA fragments associated with methylated CpG islands in human adenocarcinomas of the lung using a methylated DNA binding column and denaturing gradient gel electrophoresis. Proc Natl Acad Sci. 1999;96:2913–2918. doi: 10.1073/pnas.96.6.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkowicz M, Ji Y, Ren X, Horsthemke B, Russell LB, Johnson DK, Rinchik EM, Nicholls RD, Stubbs L. Molecular characterization of radiation- and chemically-induced mutations associated with neuromuscular tremors, runting, juvenile lethality, and sperm defects in jdf2 mice. Mamm Genome. 1999;10:870–878. doi: 10.1007/s003359901106. [DOI] [PubMed] [Google Scholar]
- Wandstrat AE, Leana-Cox J, Jenkins L, Schwartz S. Molecular cytogenetic evidence for a common breakpoint in the largest inverted duplications of chromosome 15. Am J Hum Genet. 1998;62:925–936. doi: 10.1086/301777. [DOI] [PMC free article] [PubMed] [Google Scholar]