Abstract
Cullin-RING ubiquitin ligase (CRL), with its founding member of SKP1-Cullins-F-box proteins (SCF) E3 ubiquitin ligase, is the largest family of E3 ligases, which requires cullin neddylation for its activation. Recently, an inhibitor of NEDD8 activating enzyme (NAE), MLN4924, was reported to block cullin neddylation and inactivate CRL/SCF E3, resulting in apoptosis induction and tumor suppression both in vitro and in vivo. We report here that apoptosis is not the sole mechanism by which MLN4924 suppresses tumor cell growth because apoptosis is moderately induced by the drug in some cancer cell lines and drug-induced growth suppression is only partially blocked by a pan-caspase inhibitor, z-VAD. MLN4924 treatment induces the characteristics of senescence phenotypes as evidenced by enlarged and flattened cellular morphology and positive staining of senescence-associated β-Gal. MLN4924-induced senescence is associated with cellular response to DNA damage, triggered by accumulation of DNA-licensing proteins CDT1 and ORC1, as a result of inactivation of CRL/SCF E3s. The senescence occurs in the manner independent of pRB/p16 and p53, but dependent on p21, a known substrate of CRL/SCF E3s and a mediator of senescence, which accumulates on CRL/SCF inactivation by MLN4924. Furthermore, MLN4924-induced senescence is irreversible and coupled with persistent accumulation of p21 and sustained activation of DNA damage response. Our study reveals a novel mechanism of MLN4924 action and showed that MLN4924 could be further developed as an effective anticancer agent by inducing apoptosis and irreversible senescence.
Introduction
The CRL (Cullin-RING ligases), with its founding member of SKP1-Cullin-F-box proteins (SCF) E3 ubiquitin ligases, is the largest cellular multiunit ubiquitin ligase [1,2] that promotes the degradation of up to 20% of ubiquitinated cellular proteins [3], thus regulating numerous biologic processes [4]. Intensive studies demonstrate that CRL/SCF ubiquitin ligases serve as promising anticancer targets [5,6] owing to the following characteristics: (a) CRL/SCF E3 ubiquitin ligase is abnormally activated in many human cancers, which contributes to uncontrolled proliferation and genomic instability [2]; (b) several subunits of CRL/SCF E3 ubiquitin ligases, such as SKP2, β-TRCP, CUL4A, and RING-finger protein ROC1 or ROC2, function as oncoproteins and are specifically overexpressed in human cancers [7–10]; (c) inhibition of CRL/SCF activity or down-regulation of these oncogenic CRL/SCF components suppresses the growth of cancer cells in vitro and in vivo [7–9]; and (d) it is a “druggable” enzyme. However, owing to its multiunit composition, it is technically challenging to set up a high-throughput screening for small-molecule inhibitors of CRL/SCF E3 ligases [11].
The core of CRL/SCF ubiquitin ligases is a cullin-RING finger protein complex [12]. In human and mouse, there are seven cullin (CUL 1–3, 4A, 4B, 5, and 7) and two RING (RBX1/RBX2) family members [1,2,6]. Activation of CRL/SCF requires cullin neddylation [13–17], a modification by adding ubiquitin-like protein NEDD8 to cullins, catalyzed by NEDD8-activating enzyme E1 (NAE), NEDD8-conjugating enzyme E2 (Ubc12), and NEDD8-E3 ligase [18]. Most recently, an NAE inhibitor, MLN4924, was identified through a highthroughput screening [3]. MLN4924 binds to NAE to create a covalent NEDD8-MLN4924 adduct that blocks NAE enzymatic activity [19]. By doing so, MLN4924 efficiently inhibits cullin neddylation, thus inactivating CRL/SCF E3 ligase to cause substrate accumulation. As a result, DNA damage response was triggered and apoptosis was induced, leading to a remarkable anticancer effect both in vitro and in vivo [3,20,21]. The findings further validate CRL/SCF E3 as a promising cancer targets [5,6] and demonstrate MLN4924 as a novel class of anticancer agent. Indeed, MLN4924 has advanced to several phase 1 clinical trials for solid tumors and hematological malignancies [22].
Although the biochemical mechanisms of MLN4924 action are well defined [19], induction of apoptosis is the only known biologic mechanism by which MLN4924 suppresses cancer cell growth [3,20,21]. In the present study, we report that MLN4924-mediated apoptosis induction only partially contributes to the growth suppression and that the other death mechanism involves MLN4924-triggered senescence, which is an irreversible and p21-dependent process, likely as a consequence of prolonged DNA damage response. Our study demonstrates that by inducing both apoptosis and senescence through inactivation of CRL/SCF E3 ligases, MLN4924 acts as an effective anticancer agent for targeted cancer therapy.
Materials and Methods
Cell Culture and Drug Treatment
Human cancer lines, HCT116 colon (p53-wt), H1299 lung (p53-null), and U87 glioblastoma (inactive wt p53) were purchased from ATCC (Manassas, VA). HCT116-p21+/+ versus HCT116-p21-/- and MEF-p21+/+ versus MEF-p21-/- cells were kindly provided by Drs Vogelstein and Roberts, respectively, and were authenticated by immunoblot analysis with demonstration of expected expression of p21 in p21+/+ cells but not in p21-/- cells. Other lines were authenticated by expected p53 expression using immunoblot analysis. Cells were grown at 37°C in 5% CO2 in McCoy's medium (HCT116 cells) or Dulbecco modified Eagle medium (H1299, U87, and MEF cells) supplemented with 10% fetal bovine serum. Cells were treated with MLN4924 (a gift from Millennium Pharmaceutical, Inc, Cambridge, MA), followed by various growth assays and immunoblot analysis.
ATPlite Cell Proliferation Assay
Cells were seeded in 96-well plate and treated with MLN4924 for 72 to 96 hours, followed by ATPlite assay [8,9,23].
Clonogenic Survival Assay
Single-cell suspension was seeded in 60-mm dishes and treated with MLN4924 for 9 days. Colonies were stained and counted [8,9].
SA-β-Galactosidase Staining
The expression of senescence-associated β-galactosidase was determined by SA-β-Galactosidase (SA-β-Gal) staining [8,9,24] after exposing cells to MLN4924 at 0.1 µM for 72 to 80 hours.
Immunoblot Analysis
Cell lysates were prepared for immunoblot analysis using antibodies against p16, total and pRB, WEE-1, and caspase-3 (Santa Cruz Biotechnology, Santa Cruz, CA); p53 (Calbiochem, Gibbstown, NJ); ORC-1, CDT1, and p21 (BD Biosciences, San Diego, CA); β-actin (Sigma, St Louis, MO); phospho-gamma H2A (Ser 139; Millipore, Billerica, MA); and poly (ADP-ribose) polymerase and phospho-CHK1 (Cell Signaling, Danvers, MA).
FACS Analysis and DNA Fragmentation Assay
Cells were treated with various doses of MLN4924 for various periods, followed by FACS analysis and DNA fragmentation assay [8,9,23].
Microscopy
Cellular morphology and SA-β-Gal staining were captured by Olympus 1X71 using Olympus LCP LAN F1 lens and Olympus DP70 cameras (Olympus Optical Co. Ltd, Center Valley, PA). The acquisition software used is the Olympus DP Controller 2002 (Olympus Optical Co, Ltd).
Results
Apoptosis Is Not the Only Mechanism by Which MLN4924 Induced Growth Suppression
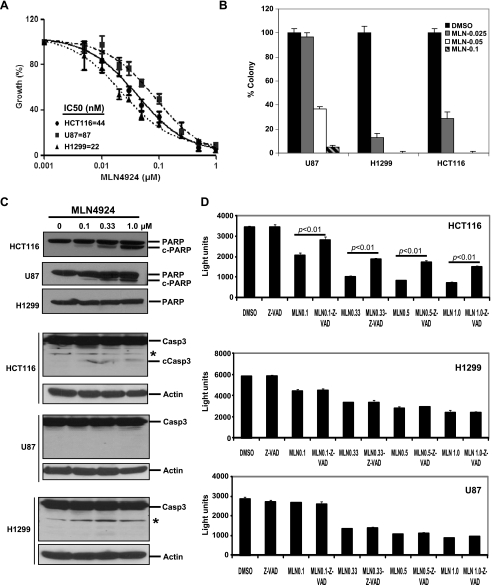
MLN4924, a potent NAE inhibitor, was recently reported to block cullin neddylation, thus inactivating CRL/SCF E3 ligase to cause accumulation of a number of its substrates, leading to suppression of tumor cell growth both in vitro and in vivo by inducing abnormal cell cycle progression and apoptosis [3,20,21]. We determined the efficacy of MLN4924 against the growth of several human cancer cell lines, including HCT116, H1299, and U87 cells. MLN4924 effectively inhibited cancer cell proliferation with the half-maximal inhibitory concentration (IC50) value ranging from 22 to 87 nM (Figure 1A). MLN4924 was also a potential inhibitor of cancer cell clonogenic survival with an IC50 less than 50 nM (Figure 1B). To explore the apoptosis mechanism of MLN4924 action, we measured formation of cleaved PARP and caspase-3, two hallmarks of apoptosis. As shown in Figure 1C, MLN4924 caused a limited cleavage of PARP and caspase-3, particularly at a low drug concentration in HCT116 cells, but not in H1299 cells even at high drug concentrations. A minor cleavage of PARP, but not of caspase-3, was observed in U87 cells. We further determined the involvement of apoptosis in MLN4924-induced growth suppression by a rescuing experiment using z-VAD, a pan-caspase inhibitor and expected a full rescue if apoptosis is the sole mechanism. Consistent with our PARP and caspase-3 cleavage results, z-VAD only partially rescued MLN4924-induced growth suppression in HCT116 cells, but not in U87 and H1299 cells (Figure 1D). Taken together, the findings indicated that MLN4924-induced apoptosis only partially contributes to observed growth suppression and suggested the involvement of other cell killing mechanism.
Figure 1.
Effect of MLN4924 on cell growth and apoptosis. (A) Sensitivity of cancer cells to MLN4924: Cells were seeded in 96-well plates in triplicate and treated with various MLN4924 doses for 72 hours, followed by ATPlite assay. Shown is mean ± SEM (n = 2). (B) Effect of MLN4924 on clonal survival of human cancer cells: Cells were seeded in 60-mm dishes in duplicate and treated with MLN4924 at indicated concentration (µM) for 9 days, followed by colony staining and counting. A representative experiment is shown (n=3). (C) Induction of cleavage of PARP and caspase-3: Cells were treated with MLN4924 for 24 hours, followed by immunoblot analysis. cPARP indicates cleaved PARP; cCasp3, cleaved caspase-3. *Nonspecific band. (D) Partial rescue of MLN4924-induced growth suppression by z-VAD: Cells were seeded in 96-well plates in triplicate and treated with 40 µM of z-VAD for 4 hours before MLN4924 (µM) treatment for 96 hours, followed by ATPlite assay with light units reflecting cell growth. Shown is a representative experiment (n = 3). Paired Student's t test was performed.
Senescence Is Induced by Low Dose of MLN4924
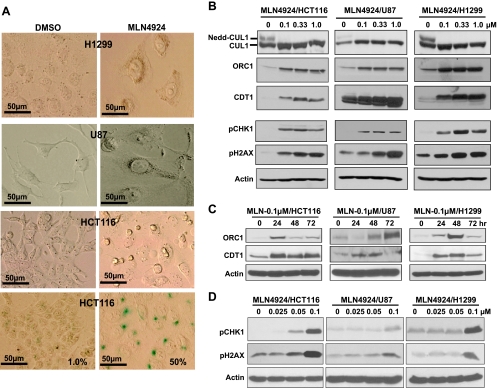
We recently showed that small interfering RNA silencing of ROC1/RBX1, a RING component of CRL/SCF E3 ligase required for its activity, induced, in addition to apoptosis, senescence, which was associated with DNA damage response [8]. We therefore determined whether pharmacological inactivation of CRL/SCF E3 ligase by MLN4924 would also induce senescence. Indeed, all three lines of cancer cells after MLN4924 treatment at 0.1 µM concentration demonstrated an enlarged and flattened shape, a reminiscence of senescence phenotype (Figure 2A, top three panels). SA-β-Gal staining showed that ∼50% of HCT116 cells were stained positively compared with only ∼1% cells with positive staining in the DMSO control group (Figure 2A, bottom panel). MLN4924-induced SA-β-Gal staining was also seen in H1299 and U87 cells after prolonged staining (data not shown). Thus, MLN4924 could also induce senescence in addition to apoptosis.
Figure 2.
Induction of senescence (A) and CRL/SCF E3 substrates and DNA damage response (B–D). Cells were treated with MLN4924 at 0.1 µM for 80 hours, followed by morphological observation (A, top three panels) and SA-β-Gal staining (A, bottom). Photographs were taken using Olympus DP70 camera (x400) with size bar shown. Cells were treated with MLN4924 for 24 hours at various concentrations (B, D) or for indicated time periods at 0.1 µM (C), followed by immunoblot analysis.
It has been recently shown that DNA rereplication triggers a DNA damage response to mediate oncogene-induced senescence [25,26]. Because both CDT1 and ORC1 are critically involved in DNA replication [27–29] and are substrates of CRL/SCF E3 ligases [30–33], we determined whether CDT1 and ORC1 were accumulated on MLN4924 treatment. We first confirmed that MLN4924 indeed inhibited cullin-1 neddylation even at 0.1 µM (Figure 2B, top). We then found that MLN4924 induced a dose- (Figure 2B, panels 2 and 3) and time-dependent (Figure 2C) accumulation of CDT1 and ORC1 in all three lines. Consistent with a recent report showing that CDT1 overexpression caused double-strand breaks and triggered a DNA damage response, followed by senescence induction [34], we found that MLN4924 treatment also triggered DNA damage response in all three cancer lines, as evidenced by induction of phospho-CHK1 and phospho-H2AX, which was again in a dose-dependent manner (Figure 2, B, panels 4 and 5, and D). These results suggest that senescence induction is a universal response of cancer cells to MLN4924, which is consistent with the notion that, by inhibiting cullin neddylation, MLN4924 inactivates CRL/SCF E3 ligases and causes accumulations of DNA-licensing proteins, CDT1 and ORC1, to trigger DNA damage response, leading to senescence.
MLN4924-Induced Senescence Is Independent of p16/pRB/p53, But Dependent on p21, a Substrate of CRL/SCF E3 Ubiquitin Ligases
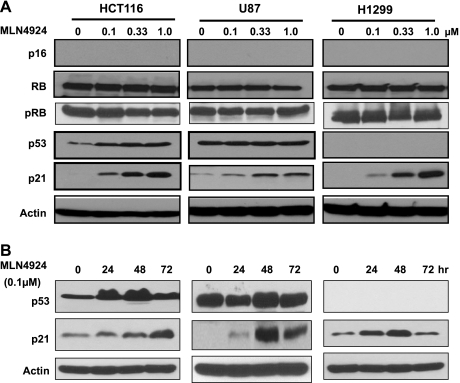
The p16/pRB and p53/p21 axes are two major senescence-triggering pathways in response to stresses [35]. To elucidate the mediators of MLN4924-induced senescence, we determined the activation status of these pathways in MLN4924-treated cells. Although p16 is undetectable, consistent with the fact that these cell lines are p16-null [36,37], both total and phosphorylated levels of RB, a reported SCF substrate only when E7 oncoprotein or EB virus laten antigen 3C was present [38,39], did not change on MLN4924 treatment (Figure 3A), excluding the involvement of the p16/pRB axis. We then investigated the role of the p53/p21 axis in MLN4924-mediated senescence. As shown in Figure 3 (A and B), p53 was accumulated in HCT116 cells after drug treatment in dose- and time-dependent manners. However, p53, expressed at a high basal level, was not further induced on drug treatment in U87 cells harboring a nonfunctional wild-type p53 [8,40]. As expected, no p53 was detectable in p53-null H1299 cells [41]. The fact that senescence induction in all three cancer lines, regardless of their p53 status, suggests that MLN4924-induced senescence may not require a functional p53.
Figure 3.
MLN4924 induces p21. Cells were treated with MLN4924 for 24 hours at various concentrations (A) or with 0.1 µM of MLN4924 for various time points (B), followed by immunoblot analysis with indicated antibodies.
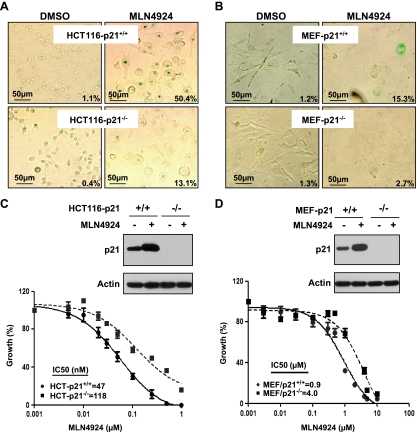
In contrast, p21, a known substrate of CRL/SCF [2,42–44] and an effective senescence mediator [35,45], was accumulated in a dose- and time-dependent manner in all three lines of cancer cells treated with MLN4924 (Figure 3, A and B), suggesting that p21 may play an essential role in MLN4924-induced senescence. To test this hypothesis, we used two pairs of cell lines: HCT116-p21+/+ versus HCT116-p21-/- and mouse embryonic fibroblasts (MEFs) with p21+/+ versus p21-/- background. As shown in Figure 4A, MLN4924 treatment induced senescence in 50% of HCT116-p21+/+ cells, but only 13% of HCT-116-p21-/- cells, demonstrating that p21 confers an approximate four-fold increase in sensitivity. Likewise, p21+/+ MEFs were approximately five-fold more sensitive than p21-/- MEFs to MLN4924-induced senescence with 15% versus 2.7% of population undergoing senescence, respectively (Figure 4B). Thus, MLN4924-induced senescence seems to be largely p21-dependent.
Figure 4.
MLN4924-induced senescence is p21 dependent. Cells were treated with MLN4924 at 0.1 µM (HCT116; A) or 1.0 µM (MEFs; B) for 80 hours, followed by SA-β-Gal staining. The percentage of cell population undergoing senescence were quantified by counting blue cells out of total cells in four to five independent areas. Shown is percent β-Gal-positive cells averaged from two independent experiments. Photographs were taken using Olympus DP70 camera (x200) with size bar shown. P < 0.01 by Student's t test between paired p21+/+ and p21-/- cells in both HCT116 and MEFs on MLN4924 treatment. Paired HCT116 cells (p21+/+ vs p21-/-) (C) or MEF cells (p21+/+ vs p21-/-) (D) were treated with MLN4924 at 0.1 or 1.0 µM for 80 hours, respectively, followed by immunoblot analysis (top), or with MLN4924 at various concentrations for 72 hours, followed by ATPlite assay. Shown is mean ± SEM from two independent experiments (bottom).
p21-Mediated Senescence Contributes to MLN4924-Induced Growth Suppression
To determine whether senescence induction directly contributes to MLN4924-mediated growth suppression, we compared the drug sensitivity of paired HCT116 cells with or without p21 and found that HCT116-p21+/+ cells were ∼2.5-fold more sensitive to MLN4924 than HCT116-p21-/- cells with an IC50 value of 47 versus 118 nM (Figure 4C). Similarly, p21+/+ MEF cells were approximately four-fold more sensitive to the drug than p21-/- MEF cells with an IC50 value of 0.9 versus 4.0 µM (Figure 4D). Detection of basal and induced levels of p21 was shown in p21+/+ but not in p21-/- cells (Figure 4, C and D). Taken together, p21-mediated senescence directly contributed to MLN4924-induced growth suppression. However, the fact that MLN4924 still caused growth suppression of p21-/- cells suggests that induction of senescence is not the only mechanism by which MLN4924 suppresses cell growth.
p21 Is a Survival Factor That Protects HCT116 Cells from Apoptosis Induced by MLN4924
We next determined relative contribution of MLN4924-induced apoptosis to growth suppression in paired HCT116 cells with or without p21 expression. It has been well established that p21 plays a survival role in preventing p53-dependent apoptosis on DNA damage, particularly in HCT116 cells [46]. We, therefore, hypothesized that in contrast to MLN4924-induced senescence, HCT116-p21-/- cells would be more sensitive than p21+/+ cells to MLN4924-induced apoptosis. Indeed, using two independent measures of apoptosis, namely, FACS analysis for sub-G1 apoptotic population (Figure 5A) and DNA fragmentation (Figure 5B), we found that MLN4924 induced doseand time-dependent apoptosis in both lines, with HCT116-p21-/- cells being about two-fold more sensitive than p21+/+ cells. Thus, p21 also plays a survival role against apoptosis induced by MLN4924, an enzyme inhibitor that does not directly cause DNA damage. Given the fact that HCT116-p21+/+ cells were more sensitive to senescence but more resistant to apoptosis with an overall higher sensitivity to drug-induced growth suppression (2.5-fold lower IC50 than that in HCT116-p21-/- cells), we concluded that senescence contributes more than apoptosis to MLN4924-induced growth suppression.
Figure 5.
p21-null HCT116 cells are more sensitive to apoptosis induced by MLN4924. (A) Paired HCT116 cells were treated with various doses of MLN4924 for 24 hours, followed by FACS analysis for sub-G1 apoptotic population. Shown is mean ± SEM from two independent experiments. *P < 0.05 by Student's t test. (B) Paired HCT116 cells were treated with various doses of MLN4924 for 24 or 48 hours, respectively, followed by DNA fragmentation assay, run in 1.8% agarose gel. Lanes 1 and 6, DMSO controls; lanes 2/3 and 7/8, 0.3 µM for 24/48 hours; lanes 4/5 and 9/10, 1.0 µM for 24/48 hours, respectively. M indicates molecular marker.
MLN4924-Induced Senescence Is Irreversible, Which Is Associated with Prolonged p21 Accumulation and DNA Damage Response
Having established that senescence induction is a critical anticancer mechanism by MLN4924, we next determined whether MLN4924-induced senescence is reversible. Cells were treated with MLN4924 at 0.1 µM for 72 hours to induce senescence-like morphologic changes, followed by either drug removal and cell culture in drug-free medium for additional 72 hours or continued incubation in drug-containing medium for an additional 72 hours. As shown in Figure 6A for both HCT116 and H1299 cells as well as U87 cells (data not shown), the senescence-like morphologic changes were similar between two groups of cells regardless of MLN4924 removal, suggesting that removal of MLN4924 failed to revert the senescence phenotype. The clonogenic survival assay also showed that MLN4924-induced suppression of clonal survival was not reversible either after the removal of the drug (Figure 6B, and data not shown for U87 cells). Thus, MLN4924-induced senescence is irreversible.
Figure 6.
MLN4924-induced senescence is irreversible. Cells were treated with MLN4924 at 0.1 µM for 72 hours to induce senescence. Cells were washed and either cultured in drug-free medium for additional 72 hours (MLN-72h-washed) or were continuously cultured in drug-containing medium for additional 72 hours (MLN-144h), followed by photography using Olympus DP70 camera (x400) with size bar shown (A), colony staining (B), and immunoblot analysis (C).
To address the molecular basis for the irreversibility of MLN4924-induced senescence, we determined the dynastic changes of a subset of the CRL/SCF substrates (WEE1, ORC1, and p21) and of DNA damage response proteins (pCHK1 and pH2AX) at 72 hours after MLN4924 treatment and thereafter at the various time points after the drug removal. As shown in Figure 6C, the CRL/SCF substrates WEE1 [47] and ORC1 [31] were expectedly accumulated at 72 hours after MLN4924 treatment and subsequently decreased on drug removal, suggesting that drug removal resulted in reactivation of CRL/SCF E3 ubiquitin ligase. Interestingly, p21, on accumulation by MLN4924, remained elevated in HCT116 cells or even continued to accumulate in H1299 cells (Figure 6C) after drug removal. Moreover, two phosphorylated proteins, pCHK1 and pH2AX, reflecting the cellular DNA damage response, which is a potential trigger of cell senescence [25,26], also remained at the elevated levels on drug removal (Figure 6C). The results suggest that persistent p21 accumulation and sustained activation of DNA damage response after drug removal contributed to the irreversibility of MLN4924-induced senescence.
Discussion
Induction of apoptosis was previously defined as the mechanism of MLN4924 action for growth suppression of solid and hematopoietic tumor cells [3,20,21]. While this article was at the late stage of preparation, Lin et al. [48] reported that MLN4924 could also induce senescence in PC3 prostate cancer cells with both p53-null and PTEN-null background. Here we showed in three cancer lines that, in addition to apoptosis induction, MLN4924 also induces cellular senescence after a prolonged treatment at low drug concentrations, which contributes to growth suppression. Our mechanistic study revealed that MLN4924-induced senescence is associated with DNA damage response, likely triggered by DNA rereplication due to accumulation of DNA-licensing proteins (e.g., CDT1 and ORC1), resulting from inactivation of CRL/SCF E3 ligases. Furthermore, the senescence occurs in a manner independent of p16/pRB/p53, but dependent of p21, and is irreversible. Irreversibility of MLN4924-induced senescence is associated with sustained elevation of p21 and prolonged activation of DNA damage response.
While this article was under the review, Lin et al. [49] reported similar findings that MLN4924 triggered checkpoint activation and induced apoptosis and senescence, with main focus in HCT116 cells. Comparison of our study with the study by Lin et al. revealed several similarities and discrepancies. The similarities include the following: 1) MLN4924 triggers DNA damage response by inducing CDT1 accumulation, 2) MLN4924 induces both apoptosis and senescence, and 3) senescence is p21 dependent and irreversible. The discrepancies from our study include the following: 1) senescence is p53 dependent as well, although to a lesser extent; and 2) p53/p21-null cells were more sensitive to the drug [49]. These discrepancies can be partly explained by the fact that our senescence study was conducted under low drug concentration (0.1 µM) with prolonged period of exposure (72–80 hours) versus their high drug concentration (1 µM) with short exposure (8 hours) [49]. In addition, based on their study mainly conducted in a single HCT116 line, Lin et al. [49] concluded that apoptosis, not senescence, might be more important for the antiproliferative effect of MLN4924. Although our study was not geared to determine which mechanism is the major contributor to MLN4924-induced growth suppression rather than focus on senescence as a novel mechanism, we did observe that 1) apoptosis contributed less than senescence to MLN4924-induced growth suppression in HCT116 cells (Figures 4 and 5) and 2) apoptosis contributed little, if any, to drug-induced growth suppression in H1299 and U87 cells (Figure 1). Nevertheless, a drug concentration-dependent switch between apoptosis and senescence in HCT116 cells with or without p53 or p21 was previously reported after treatment with anticancer drug camptothecin [50]. Taken together, it seems that MLN4924 induces both apoptosis and senescence, and the contribution of each mechanism to its growth suppression activity would likely be dependent on cellular context, drug concentrations used, and duration of drug treatment.
In summary, our study elucidated a new mechanism of MLN4924 action by inducing senescence to suppress cancer cell growth. In addition, it provides three useful aspects with regards to future development of MLN4924 as a novel class of anticancer agent [22]. First, MLN4924 induces senescence largely independent of p53. Virtually all human cancers regardless of p53 status can be treated by the drug. Second, MLN4924 induces senescence irreversibly at low drug concentrations, which makes it possible to use low doses of the drug to achieve a greater therapeutic index, given the fact that normal fibroblasts are much more resistant to the drug. Third, prolonged p21 accumulation and DNA damage response could serve as useful biomarkers for target modulation by MLN4924.
Acknowledgments
The authors thank Millennium Pharmaceutical, Inc, for providing MLN4924 and Drs Vogelstein and Roberts for paired p21-null cells.
Footnotes
This work is supported by the National Cancer Institute grants (CA111554 and CA118762) to Y.S. and is partially supported by Fudan University in China and Chinese National Nature Science Foundation grant (31071204) to L.J.
References
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 3.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 4.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 5.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, Palacios J, Nathanson KL, Garcia MJ, Benitez J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11:R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y. Overview of approaches for screening for ubiquitin ligase inhibitors. Methods Enzymol. 2005;399:654–663. doi: 10.1016/S0076-6879(05)99043-5. [DOI] [PubMed] [Google Scholar]
- 12.Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, Pan ZQ. The SCF(HOS/β-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc Natl Acad Sci USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 19.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 21.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-κB-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 22.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 23.Bockbrader KM, Tan M, Sun Y. A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells. Oncogene. 2005;24:7381–7388. doi: 10.1038/sj.onc.1208888. [DOI] [PubMed] [Google Scholar]
- 24.Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated β-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- 25.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 26.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 27.Rialland M, Sola F, Santocanale C. Essential role of human CDT1 in DNA replication and chromatin licensing. J Cell Sci. 2002;115:1435–1440. doi: 10.1242/jcs.115.7.1435. [DOI] [PubMed] [Google Scholar]
- 28.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- 31.Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- 32.Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753–3756. doi: 10.1074/jbc.C500464200. [DOI] [PubMed] [Google Scholar]
- 33.Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V, Kouloukoussa M, et al. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 35.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawabe S, Roth JA, Wilson DR, Meyn RE. Adenovirus-mediated p16INK4a gene expression radiosensitizes non-small cell lung cancer cells in a p53-dependent manner. Oncogene. 2000;19:5359–5366. doi: 10.1038/sj.onc.1203935. [DOI] [PubMed] [Google Scholar]
- 37.Kim SK, Wang KC, Cho BK, Lim SY, Kim YY, Oh CW, Chung YN, Kim CY, Lee CT, Kim HJ. Adenoviral p16/CDKN2 gene transfer to malignant glioma: role of p16 in growth, invasion, and senescence. Oncol Rep. 2003;10:1121–1126. [PubMed] [Google Scholar]
- 38.Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, Harper JW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci USA. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asai A, Miyagi Y, Sugiyama A, Gamanuma M, Hong SH, Takamoto S, Nomura K, Matsutani M, Takakura K, Kuchino Y. Negative effects of wild-type p53 and s-Myc on cellular growth and tumorigenicity of glioma cells. Implication of the tumor suppressor genes for gene therapy. J Neurooncol. 1994;19:259–268. doi: 10.1007/BF01053280. [DOI] [PubMed] [Google Scholar]
- 41.Robinson M, Jiang P, Cui J, Li J, Wang Y, Swaroop M, Madore S, Lawrence TS, Sun Y. Global GeneChip profiling to identify genes responsive to p53-induced growth arrest and apoptosis in human lung carcinomas. Cancer Biol Ther. 2003;2:406–415. doi: 10.4161/cbt.2.4.437. [DOI] [PubMed] [Google Scholar]
- 42.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 45.Lundberg AS, Hahn WC, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Curr Opin Cell Biol. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- 46.Yu J, Tiwari S, Steiner P, Zhang L. Differential apoptotic response to the proteasome inhibitor Bortezomib [VELCADE, PS-341] in Bax-deficient and p21-deficient colon cancer cells. Cancer Biol Ther. 2003;2:694–699. [PubMed] [Google Scholar]
- 47.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Z, Wei W, Dunaway S, Darnowski JW, Calabresi P, Sedivy J, Hendrickson EA, Balan KV, Pantazis P, Wyche JH. Role of p21 in apoptosis and senescence of human colon cancer cells treated with camptothecin. J Biol Chem. 2002;277:17154–17160. doi: 10.1074/jbc.M112401200. [DOI] [PubMed] [Google Scholar]